Abstract
Background
Although many prognostic single-gene (SG) lists have been identified in cancer research, application of these features is hampered due to poor robustness and performance on independent datasets. Pathway-based approaches have thus emerged which embed biological knowledge to yield reproducible features.
Methods
Pathifier estimates pathways deregulation score (PDS) to represent the extent of pathway deregulation based on expression data, and most of its applications treat pathways as independent without addressing the effect of gene overlap between pathway pairs which we refer to as crosstalk. Here, we propose a novel procedure based on Pathifier methodology, which for the first time has been utilized with crosstalk accommodated to identify disease-specific features to predict prognosis in patients with hepatocellular carcinoma (HCC).
Findings
With the cohort (N = 355) of HCC patients from The Cancer Genome Atlas (TCGA), cross validation (CV) revealed that PDSs identified were more robust and accurate than the SG features by deep learning (DL)-based approach. When validated on external HCC datasets, these features outperformed the SGs consistently.
Interpretation
On average, we provide 10.2% improvement of prediction accuracy. Importantly, governing genes in these features provide valuable insight into the cancer hallmarks of HCC. We develop an R package PATHcrosstalk (available from GitHub https://github.com/fabotao/PATHcrosstalk) with which users can discover pathways of interest with crosstalk effect considered.
Research in context.
Evidence before this study
Deep learning-based approaches have provided inspiring prediction performance in cancer research. However, a recent study employing autoencoder, a deep learning framework, to obtain mRNA features for risk stratification in hepatocellular carcinoma, reported prediction accuracy around 0.7 in multiple datasets. This moderate performance can be largely attributed to the high heterogeneity of hepatocellular even using the competent deep learning framework. Pathifier, a pathway-based methodology, derives pathway deregulation score, a robust feature of interpretability, using mRNA expression data. This approach is promising for noisy expression data and has not been used in hepatocellular carcinoma.
Added value of this study
We found that the association between pathway deregulation and prognosis in patients with hepatocellular carcinoma could be affected by gene overlap between pathway pairs, developed and validated a risk stratification model. This model is based on the pathway deregulation score of 13 pathways/sub-pathways by Pathifier that differentiate patients into moderate or aggressive risk subtypes with significantly survival difference in both the training and validation datasets. Our study provides substantial improvement of prediction accuracy compared with the deep learning-based study, using the same training cohort (N = 355) as well as other three validation cohorts (N = 231, 221, 41). Our results suggest that pathway deregulation score-based approach is more accurate than deep leaning based method in risk classification. We also provided a nomogram to predict prognosis of patients with hepatocellular carcinoma, which was based on 13 identified pathways/sub-pathways.
Implications of all available evidence
For the first time, we identified prognosis associated pathways with removal of gene overlap between pathway pairs and validated a risk stratification model based on deregulation score of 13 pathways/sub-pathways. This model can provide more accurate and robust prediction than the counterpart by deep learning-based approach, with strong potential in future clinical application. Moreover, the dominant genes in these pathways/sub-pathways could provide more focused insights for therapeutic targets discovery of hepatocellular carcinoma. This study has obtained high prediction accuracy in hepatocellular carcinoma which is highly heterogenous and caused by intricate etiologic factors, and can thus be applied to other tumor types with promising findings.
Alt-text: Unlabelled Box
1. Introduction
High-throughput expression data in combination with machine learning is a widely adopted strategy to identify prognostic genes with which cancer patients can be stratified into risk subgroups with distinct outcomes. With the hope that the proposed gene-lists can aid the clinicians in therapy decision in cancer [1], several commercialized gene-signatures for risk prediction have been approved in clinical practice, especially for breast cancer [2,3]. In spite of relative success in prognosis prediction, it was demonstrated that the prognostic gene-lists were highly unstable in prediction performance and biomarkers identified were greatly influenced by the selection of samples in the training datasets [4]. Due to inherent noise of expression data and tumor heterogeneity, poor feature robustness and prediction performance have been observed in multiple studies [4,5]. In addition, small sample size which is not nearly comparable to the number of features (genes) for each sample can lead to a technical hurdle termed “curse of dimensionality”.
Attempts to decode cancer have thus brought about systematic approaches which borrow information from molecular mechanisms among individual genes, proteins or metabolites such as predefined pathways from biological databases e.g. Kyoto Encyclopedia of Genes and Genomes (KEGG) [6], Reactome [7] to extract more stable and interpretable features for risk prediction. Among the pathway-based approaches, most of them estimate a pathway's activity for whole samples without information on its deregulation in a particular sample [8], and some others aim to discover novel molecular mechanisms other than established pathways [9], the few exceptions are PARADIGM and Pathifier [10,11]. PARADIGM approach determines a score for each pathway and sample utilizing verified connection information and functional assembly of this pathway, which can be inappropriate to complex or incomplete pathways due to its demanding input of pathway mechanism, while Pathifier algorithm only calls for expression data of genes belong to each pathway to estimate coarse-grained scores which represent the extent of pathways' deregulation, by determining the deviation of diseased sample from normal counterpart on the basis of expression data. Several studies have applied Pathifier to tumor subtyping and prognosis prediction successfully [12,13].
However, most existing pathways used are general rather than disease-specific and disease progression can only affect them partially. For pathway pairs with many genes in common which we refer to as crosstalk, taking into consideration the impact of the overlapping genes on the PDS quantification of the two pathways can contribute to the identification of disease-associated features, which may become significant when we eliminate the effect of the common part on the significance of the pathways. Although it is intuitive that pathways can influence each other, especially when they share genes, the presence of this phenomenon has not been studied in PDS estimation, which is obtained on the basis of expression variation of genes in each pathway. For two pathways with shared genes: if expression levels of these genes govern the deviation from normal samples in both pathways, the sub-pathway consisting of these common dominant genes should be more related to disease progression; if the expression variations of shared genes provide minimal contribution to the aberration of each pathway, the two remaining sub-pathways after the removal of shared genes can be more disease relevant. In similar manner, crosstalk could have effect on the significance of PDS in associated with survival. To the best of our knowledge, PDS has never been used in complex diseases such as cancer with crosstalk accommodated among existing pathways to discover disease-specific sub-pathways for prognosis prediction.
Due to inherent heterogeneity, cancers are stratified into subtypes with distinct clinical outcome for personalized therapy. One instance is found in HCC, where molecular subtypes have been identified to show distinct differences in terms of survival [14,15].The prognosis of HCC varies across different subtypes and stages, with an average 5-year survival rate of 18% [16]. The first line treatment (Sorafenib) shows modest effect on prognosis improvement [17]. High heterogeneity in HCC, as well as intricate etiologic factors such as HBV, HCV and alcohol, poses challenges for accurate and robust prediction of prognosis in HCC patients. To fill this gap, considerable efforts have been made to identify subtypes to delineate the heterogeneity of HCC using mRNA expression data [[18], [19], [20], [21]]. A recent study based on deep learning (DL) framework has embedded survival information as part of the procedure to obtain SGs in prediction of two subtypes with significant survival differences [15], and concluded that moderate prediction accuracy by DL-based approach is caused by heterogeneous nature of HCC due to various risk factor. We have thus turn to more robust PDS rather than mRNAs from noisy expression data, hoping for a more accurate prediction for HCC prognosis.
In this study, we discovered pathway-level features for accurate subtype prediction in HCC based on Pathifier, as opposed to previous studies which derived features without crosstalk effect under consideration. We obtained PDSs from three relatively large HCC cohorts (The Cancer Genome Atlas (TCGA) [22], LIRI-JP [23] and NCI [24]), applied crosstalk correction and discovered survival associated pathways respectively. Afterwards, we identified 76 common pathways shared by these three cohorts, rather than use top features arbitrarily from one cohort of them. Unsurprisingly, survival significant sub-pathways from 8 pathways were discovered after correction, while the 8 predefined pathways were not. We merged and reduced these 76 pathways to 13 non-redundant features, which we believed to be HCC survival specific. Performance was evaluated using CV within the HCC dataset (N = 355) from TCGA cohort, and demonstrated that discovered PDS features provided higher accuracy and stability in classifying survival-risk subgroups than the SGs from the DL-based study [15]. Subsequently, we evaluated this learned model from TCGA data on three external HCC datasets and showed that these features could predict risk subgroups in HCC with elevated accuracy than the SGs by DL-based approach consistently. Notably, the prediction accuracy and robustness of these PDS features also outperformed that of the SGs from the three-omics DL-based design in TCGA cohort, which derived SG features from not only expression profile but also methylation data as well as microRNA expression data. In short, we developed the PDS-based approach for HCC which could improve the prediction accuracy substantially compared to DL-based approach; additionally, we evaluated the removal of crosstalk on PDS between pathway pairs.
In addition to discrete risk classification for HCC, the PDS features were applied to characterize gradual progression of survival-risk using an independent unsupervised sorting algorithm SPIN (Sorting Points Into Neighbourhoods; ref. [25]) for above four HCC cohorts, the ranking profiles of which were not only able to characterize risk progression continuously and provide individualized risk prediction in HCC samples, but reproduced the survival-risk distinction of subtypes classified aforementioned. Finally, we found that the dominant genes in discovered pathways consisted of multiple HCC therapeutic targets identified in previous studies and novel biological markers for further investigation.
2. Material and methods
2.1. Outline of the prediction pipeline
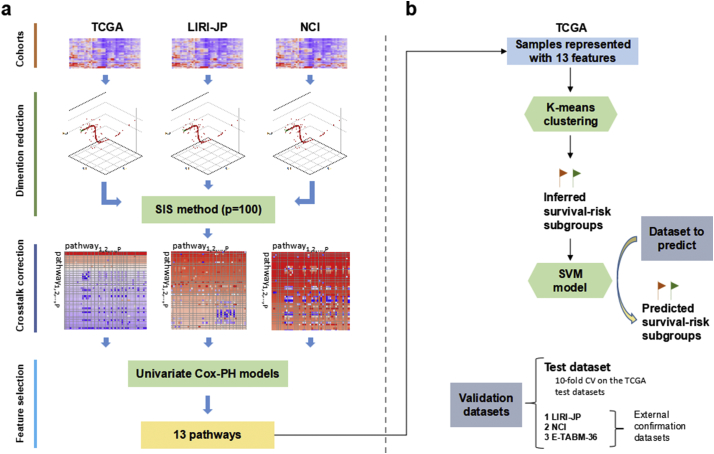
We summarized the overall workflow (Fig. 1) and the motivation behind the pipeline design in each step. Given a series of expression profiles, the beginning stage focused on extracting HCC-risk specific pathways whose PDS were used to build prediction model (Fig. 1a). To eliminate false positive features as much as possible, we only adopted the survival-associated PDSs shared by the three large HCC cohorts (TCGA, LIRI-JP, NCI) rather than use features from any one of them. We estimated PDS for each pathway and sample utilizing Pathifier algorithm, which quantified the extent of pathway deregulation by quantifying the deviation of the diseased sample from the normal in terms of expression profile of genes in this pathway. This PDS measurement ranged from 0 to 1 and was able to characterize pathway deregulation of HCC samples from normal state continuously during cancer initiation and progression. Subsequently, we kept the top survival-correlated features utilizing Sure Independence Screening (SIS) method with the cutoff threshold of 100 which was much larger than the default n/log(n) for each cohort, where n is the sample size [26].
Fig. 1.
Overall workflow. The workflow includes two steps: (a) obtaining survival-associated features; (b) building model and predicting labels for external cohorts. (a) Expression data from the 3 cohorts is transformed to Pathways Deregulation Scores (PDS) using Pathifier; then, 100 survival-correlated pathways screened with Sure Independence Screening (SIS) for each cohort as well as the screened pathways only shared by the other two cohorts are subject to crosstalk correction; then the common corrected pathways shared by the three cohorts are identified; then the redundant features are filtered out, leaving 13 pathways. (b) K-mean clustering is applied to samples represented by these features in TCGA cohort to identify survival-risk groups. Then the features as well as the survival-risk labels are used to build an SVM model(s) to predict the survival-risk labels of new datasets.
With the 100 survival-correlated pathways from each cohort, we explored the crosstalk effect on the significance of these pathways as well as the common ones between the other two cohorts. With the hypothesis that crosstalk was mainly attributed to gene-overlap between pathways, then strong correlations of PDS between pathway pairs with many common genes were expected. To demonstrate it, we calculated Jaccard similarity index and PDS correlation for each pathway pair with >3 genes in common. Furthermore, we evaluated the effect of crosstalk removal on survival-significance shift of pathways: for each pathway pair [i, j] with over 3 overlapping genes, the notation Pi\j was defined as the set of elements in pathway Pi excluding the intersection with Pj; similarly, with the notations Pj\i we denoted the genes that were in pathway Pi but not in pathway Pj; additionally, with the notation Pi∩j we defined the overlap between pathway Pi and Pj. Then we calculated the p-values of these sub-pathways in univariate Cox-PH model built with survival information and each PDS as the predictor. After crosstalk correction, significant (FDR p-value < 0.01) pathways/sub-pathways were identified for the three cohorts separately, with the common ones as candidate features for prediction. Within these features, we noticed that multiple sub-pathways from the same existing pathways were identified as significant features and showed considerable correlation of PDSs, among which we chose the representative sub-pathway with lowest mean and SD in p-values in all three cohorts. Finally, 13 pathway/sub-pathways were selected to build classifier for HCC risk.
To compare with SGs from the DL-based study, we employed the same procedure to build and validate models. First of all, K-means clustering using the features obtained was applied to get labels for HCC samples in TCGA cohort with the same cluster number of two. With these labels, we built a classifier using Support Vector Machine (SVM) framework (Fig. 1). To predict TCGA held-out test data in CV, we built an SVM classifier where the 355 TCGA samples were split into 10 folds for model training and test with a 6/4 ratio rather than 10-fold CV to keep enough test samples for survival analysis. To predict the other three HCC cohorts, we trained the classification model using whole TCGA samples. To avoid any unfair comparisons, we have used the three same metrics (C-index, Brier score and Log-rank p-value) to evaluate performance [27,28].
2.2. Clinical and gene expression data
Clinical data and normalized mRNA expression data of three large independent cohorts (TCGA cohort, LIRI-JP cohort, and NCI cohort) were directly obtained from The Cancer Genome Atlas (TCGA) portal (https://tcga-data.nci.nih.gov/tcga/) using TCGA-Assembler (v1.0.3; ref. [29]), International Cancer Genome Consortium (ICGC) portal (https://dcc.icgc.org/projects/LIRI-JP; ref. [23]), National Cancer Institute (NCI) data acquired from Gene Expression Omnibus (GEO) database GSE14520 Affymetrix high-throughput GeneChip HG-U133A microarray dataset [24], respectively. The expression data of TCGA and LIRI-JP cohorts were RNA sequencing (RNA-Seq) data, while NCI cohort expression data was from microarray platform. To compare with the DL-based study, we also prepared another small validation cohort E-TABM-36 from mRNA microarray platform [30].
After removing patients without proper survival (or censor) time information or expression data, 355, 231, 221 and 41 patients with HCC remained in each of the above four cohorts. Clinical characteristics of these cohorts were provided in Supplementary Table S1. We used the first three datasets to discover HCC survival related pathways due to their relatively large sample sizes.
2.3. Pathway Deregulation Score (PDS)
In the dP-dimensional space SP, where each dimension characterizes the expression intensity of specific gene in pathway P, and each point in this space represents a sample. To depict the variability (e.g., due to disease progression) within the samples, then a principal curve passing across the middle of the sample points can be found [31], where samples with close projections show similar behaviour of pathway P. Then we determined the distance DP(i) between the projections of sample i and the normal samples along the curve as PDS.
When extracting common features across the three large HCC cohorts, we adopted the 2/3 power transformation of the expression data from RNA-seq and microarray platform to stabilize variance instead of the aggressive log2 transformation, aiming to ensure that the curve found can explain sample variability close to reality. Log2-transformed expression datasets downloaded from microarray platform were converted to original scale before power transformation. In addition, with only 5 non-tumoral samples (3 cirrhosis and 2 non-cirrhosis) in E-TABM-36 cohort, we borrowed normal samples from NCI cohort to assist PDS estimation after removing batch effect using sva R package [32], as expression data of these two cohorts were both from the microarray platform.
Gene sets of 322 pathways were obtained from the KEGG database (http://www.kegg.jp/; [6]). Identity of genes in gene sets was decided by their Ensembl IDs. Gene sets with <3 genes varying in the data were omitted, leaving 320 KEGG pathways. PDS score was calculated for each pathway.
2.4. Variance stabilization
Some genes had a large variation in expression levels, while some genes showed a smaller variation which could also influence the functionality of a pathway. Thus, we divided each gene's expression by the standard deviation (SD) of its expression in normal tissues. To eliminate the genes of which variations were mainly due to noise, we kept 5000 genes in KEGG pathway gene sets with highest Median Absolute Deviation (MAD) over all samples for RNA-seq data in TCGA and LIRI-JP cohorts, while for NCI and E-TABM-36 cohorts, we adopted the top 7000 probes to ensure the number of genes was comparable to the above two cohorts due to redundant probes of microarray platform.
2.5. Feature prescreening
We applied prescreening procedure to remove survival irrelevant pathways to accelerate calculation in the steps afterwards. For each cohort, we utilized Sure Independence Screening (SIS) method to keep survival-correlated pathways with the limit of cutoff threshold n/log(n) or 100 if n/log(n) smaller than 100, where n was the sample size. [26].
2.6. Crosstalk correction and crosstalk matrix
For two pathways Pi and Pj with overlapping genes, Pi\j refers to the remaining genes in pathway Pi when removing the overlapping genes with Pj; similarly, Pj\i denotes the set of genes in Pj after subtracting genes in Pi, and Pi∩j represents the set of genes that are in both Pi and Pj. Then, the PDS of these sub-pathways (Pi∩j, Pi\j, and Pj\i) are calculated, which is referred to crosstalk correction.
For each pair of pathways [i, j] with no <3 common genes, we applied crosstalk correction, and computed the p-value for each pathway/sub-pathway using univariate Cox proportional hazards (Cox-PH) model built with survival information as well as its PDS [33]. The correction step yields a k × k matrix, where k is the number of pathways, the matrix of p-values can be conveniently represented with a heatmap of the negative log p-values. In this matrix, cell [i, j] denotes the significance of sub-pathway Pi\j. The rows and columns of this matrix are organized according to significance of the original pathways in descending order, each of which can be found in the diagonal cell [i, i] for Pi. This matrix is called as crosstalk matrix, which can demonstrate the crosstalk effect between pathways conveniently.
2.7. Sub-pathway selection
After crosstalk correction, we selected significant features (FDR p-value <.01) for each cohort, and the common significant sub-pathways/pathways in three cohorts can be easily obtained. To determine the optimal gene set for each sub-pathway, we chose the gene-list of this sub-pathway from one of these three cohorts, whose PDS showed most robust significance (lowest mean and SD of p-values) across three cohorts in the univariate Cox-PH model.
2.8. K-means clustering and supervised classification
Similar to the procedures in the DL-based study, we performed K-means clustering using the obtained features to get labels for TCGA cohort with the same cluster number of two. With these labels, we built a classifier utilizing SVM framework (Fig. 1). In CV with TCGA dataset, we trained a classification model where the 355 HCC samples were split into 10 folds for model training and test with a 6/4 ratio. To predict the other three cohorts, we built a classification model using whole TCGA samples.
As to data normalization, two scaling steps were employed on both the training data and validation datasets. Median scaling was applied to discovered pathways of each cohort at first, where PDS of each pathway was scaled by its median and MAD across all samples. Then, each feature was scaled and centred by the mean and Standard Deviation (SD) of its equivalent from the TCGA training dataset.
We used the e1071 package [34] available from https://CRAN.R-project.org/package=e1071 to build SVM classifiers. The optimal hyperparameters of the classifier were determined in CV using grid search algorithm.
2.9. Evaluation metrics for models
We used the same three metrics with the DL-based study which reflected the prediction accuracy.
2.9.1. Concordance index (C-index)
This metrics can quantify the proportion of patient pairs from a cohort whose risk prediction are in good agreement with survival outcome [27]. Generally, higher C-index score means more accurate in prediction performance, and a score close to 0.50 implies prediction no better than random. To calculate C-index, a Cox-PH model was built with the cluster labels and survival outcome from training data and used to predict survival using the labels of the test data. The C-index was calculated with R survcomp package [35].
2.9.2. Log-rank p-value
The log-rank test compares the survival difference of two groups at each observed event time (R survival package [36] available from http://CRAN.R-project.org/package=survival). Kaplan-Meier analysis was applied to obtain survival-curve plot of HCC subtypes.
2.9.3. Brier score
The metrics calculates the mean of the difference between the observed and the predicted survival beyond a certain time in survival analysis [28]. A smaller score implies higher accuracy. The score is obtained using R survcomp package.
2.10. The DL-based approach
We compared the prediction accuracy of the pathway-based features with SGs from recently reported DL-based approach using the same four cohorts [15]. In step 1 of the DL-based approach, the author used mRNA features in the TCGA cohort as input for the DL framework of autoencoder; then 100 nodes from the bottleneck layer were respectively used to build univariate Cox-PH model for feature selection (log-rank p-value < 0.05); then group labels of each sample were determined by K-means clustering with these features. In step 2, the mRNA features were ordered according to the correlation with the cluster labels indicated by ANOVA test F values, common features with the validation data were kept, then the top 100 of which were utilized to train classification model for survival-risk labels prediction of validation datasets.
2.11. Functional analysis
2.11.1. Clinical covariate analysis
Using Fisher exact tests, we examined the associations of inferred subgroups with other clinical covariates, including grade, stage, cirrhosis and multinodular.
2.11.2. TP53 mutation analysis
The somatic mutation frequency distributions of the TP53 gene between HCC survival subgroups were compared with Fisher exact test for TCGA and LIRI-JP cohorts, both of which had sequencing data for HCC samples.
2.12. Construction of the nomogram
To provide individualized risk prediction of HCC subtype, a nomogram was constructed using clinical characteristics and 13 identified features. As the classifier above was built with SVM model, we thus used VRPM package to generate a color-based nomogram to explain the SVM classifier [37]. To make it more concise, we set the contribution of interaction between predictors to be zero.
3. Results
3.1. Crosstalk affects pathway deregulation on survival significance
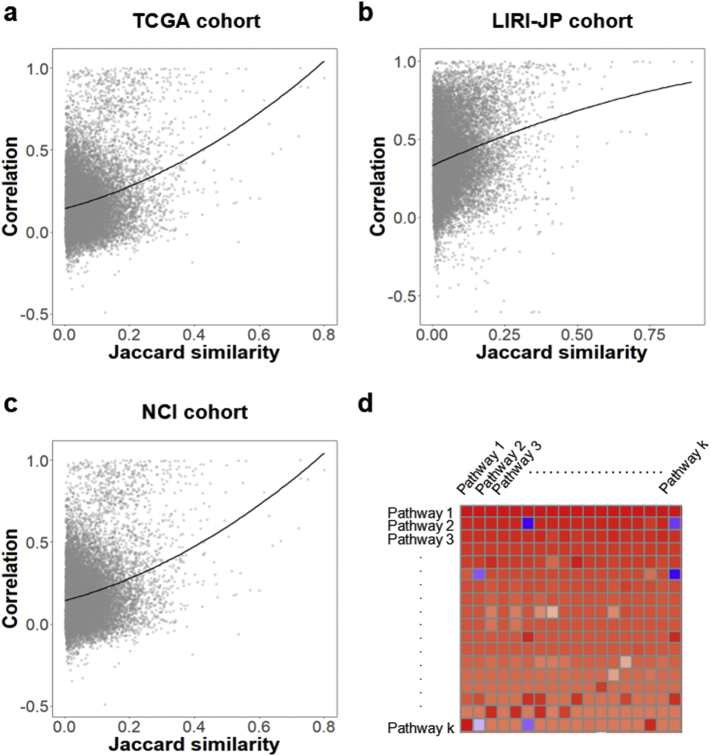
Crosstalk effect was discussed in classical over-representation studies [38], but never addressed for Pathifier methodology. We came up with the hypothesis that strong correlations of PDS between pathway pairs could be expected if the expression levels of common genes between them governed the deregulation of these two pathways. To validate it, we computed the Jaccard similarity index [39] of each pair of survival-correlated pathways with at least 3 common genes, and the Pearson correlation coefficient between their PDSs. The Jaccard similarity index JS was defined as follows:
where ∣Pi ∩ Pj∣ represented number of genes in both pathway Pi and Pj, and ∣Pi ∪ Pj∣ denoted the size of gene set from the union of pathway Pi and Pj. Pathway pairs with high Jaccard index showed considerable correlation in PDSs of all three cohorts (Fig. 2), implying the presence of crosstalk in prescreened PDS features [40].
Fig. 2.
Crosstalk effect on PDS correlation and example of crosstalk matrix. (a) TCGA cohort. (b) LIRI-JP cohort. (c) NCI cohort. Each point represents the Pearson correlation coefficient between PDSs of an overlapping pathway pair with the Jaccard similarity index. The lines represent the fitting of quadratic models. Both three cohorts show strong correlations when Jaccard similarity indices are high. (d) Example of a crosstalk matrix. On the diagonal, we find the significance in univariate Cox-PH model built with survival information and original PDS of K pathways is ordered by p-value. The p-values in the matrix have been log-transformed (base 10 log), and the sign of the result has been inverted. The color of the cell represents the p-value: bright red for p-values close to zero, bright blue for p-values close to 1. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Furthermore, to demonstrate the effect of removing common genes on the significance of the pathways, we applied crosstalk correction for those survival-correlated features. Among the pathways including the 100 pathways screened by SIS for each cohort, and the common ones only discovered from the other two cohorts, we calculated the crosstalk matrix as illustrated in the Materials and Methods section to show the significance shifts of these pathways after crosstalk correction for each cohort. The matrices illustrated the effect of the common genes removed on the p-values of pathways in different cases (Fig. S1). Fig. 2d gave an heatmap example of crosstalk matrix, where negative log transformation of original p-values of pathway i were placed in the diagonal cell [i,i] and recalculated negative log p-values after removing overlap part with pathway j were shown in cell [i,j]. After crosstalk correction, high significance (bright red in cell [2]) of pathway 2 disappeared (deep blue in cell [2,6]) when the crosstalk due to pathway 6 eliminated, while pathway k (firebrick in cell [k,k]) became more significant (bright red in cell [k,1]) when the crosstalk due to pathway 1 eliminated. The heatmaps in Supplementary Fig. S1 gave more detailed representations of cross matrices for three cohorts (TCGA, LIRI-JP and NCI). For simplicity, we represented the pathways with the KEGG map IDs in the heatmaps. These findings implied that overlapping gene sets could affect considerably to the significance of related pathways.
In order to discover survival-significant pathways/sub-pathways, we conducted overlapping analysis on significant features (FDR p-value < 0.01) after crosstalk correction among three cohorts and identified 76 common sub-pathways and pathways. Among them, survival significant subsets from 8 pathways were uncovered after correction, while the original pathways were not identified as significant, including pyrimidine metabolism, cGMP-PKG pathway, HIF-1 pathway, sphingolipid pathway, cellular senescence, toll-like receptor pathway, salmonella infection and pancreatic cancer. Notably, 70 features from 7 groups represented deregulations of sub-pathways from the corresponding 7 pathways after removing overlapping gene with others. For those in the same group, we chose the representative sub-pathway with lowest mean and SD in p-values among all three cohorts. Finally, we identified 13 non-redundant sub-pathways/pathways whose PDSs were significantly associated with survival in HCC (Table 1).
Table 1.
The list of 13 identified survival significant sub-pathway/pathways and function summary.
| Pathway identifier | Function summary |
|---|---|
| 04071\04012 | Sphingolipid signaling pathway\ErbB signaling pathway |
| 00240\00230 | Pyrimidine metabolism\Purine metabolism |
| 04218\04110 | Cellular senescence\Cell cycle |
| 05167\01521 | Kaposi sarcoma-associated herpesvirus infection\EGFR tyrosine kinase inhibitor resistance |
| 05132 ∩ 05231 | Salmonella infection ∩ Choline metabolism in cancer |
| 00520 | Amino sugar and nucleotide sugar metabolism |
| 04022 ∩ 04210 | cGMP-PKG signaling pathway ∩ Apoptosis |
| 00480 | Glutathione metabolism |
| 00030 | Pentose phosphate pathway |
| 04620\05130 | Toll-like receptor signaling pathway\Pathogenic Escherichia coli infection |
| 04540 | Gap junction |
| 05212\05206 | Pancreatic cancer\MicroRNAs in cancer |
| 04066\05211 | HIF-1 signaling pathway\Renal cell carcinoma |
i\j represents the set of genes that are in KEGG pathway i but not in KEGG pathway j.
i∩j represents the set of genes that are in both KEGG pathway i and pathway j.
3.2. Performance comparison within TCGA dataset
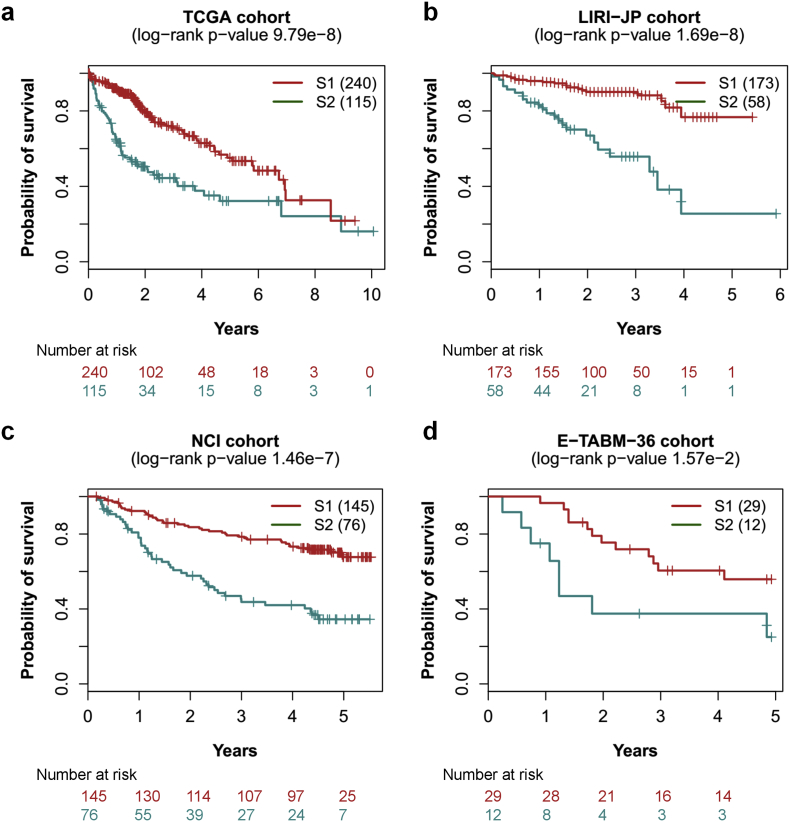
To compare the classification performance of the 13 features with the 100 SGs by the DL-based approach, we implemented the feature selection and model building of the DL-based procedure proposed by Chaudhary et al. [15] using our curated TCGA dataset. Due to the stochastic gradient descent algorithm in optimization process, we repeated the training process for 100 times using autoencoder and chose the optimal split with similar ratio of 103/252 (vs. 105/255 by Chaudhary et al.) and drastic survival difference between the split subgroups (log-rank p-value = 8.37e-7). Then group labels were utilized to build an SVM classification model using CV, where the 355 TCGA samples were split into 10 folds and used for training and test with a 6/4 ratio. We assessed the prediction accuracy with C-index as well, which measured the proportion of all patient pairs whose risk prediction were consistent with observed survival outcomes [41]. Furthermore, the error of the model fitting on survival information was evaluated with Brier score [28].
We observed that PDS features produced considerable improvement in prediction accuracy in terms of C-index and more significant log-rank p-value in survival difference between survival-risk subgroup S1 and S2 compared with the 100 SGs derived using DL-based approach (Table 2). Also, we obtained low Brier error rates in model fitting. Compared to the DL-based study in CV, on average, the test data from TCGA HCC samples produced higher C-index (0.77 ± 0.05 vs. 0.70 ± 0.08), low Brier score (0.21 ± 0.02 vs. 0.21 ± 0.02), and more significant average log-rank p-value (5.85e-4 vs. 3.89e-3) on survival difference (Table 2). Meanwhile, the lower SD of C-index (0.05 vs. 0.08) in our result indicated more robust performance of prediction in CV within TCGA dataset.
Table 2.
Performance of cross-validation based robustness of SVM classifier on test set in TCGA cohort and external validation on three confirmation cohorts using 13 features in comparison with the DL-based approach implemented by us as well as Chaudhary et al.
| Cohort | Omics data type | PDS-based method |
DL-based method |
DL-based method by Chaudhary et al. |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Samples (N) | C-index | Brier score | Log-rank P | Samples (N) | C-index | Brier score | Log-rank P | Samples (N) | C-index | Brier score | Log-rank P | ||
| TCGA | RNA-Seq | 355 | 0.77 (±0.05) | 0.21 (±0.02) | 5.85e-4 | 355 | 0.70 (±0.08) | 0.21 (±0.02) | 3.89e-3 | 360 | 0.68 (±0.07) | 0.20 (±0.02) | 0.01 |
| LIRI-JP | RNA-Seq | 231 | 0.83 | 0.15 | 1.69e-8 | 231 | 0.84 | 0.15 | 1.07e-7 | 230 | 0.75 | 0.16 | 4.4e-04 |
| NCI | mRNA microarray | 221 | 0.76 | 0.17 | 1.46e-7 | 221 | 0.62 | 0.19 | 7.65e-2 | 221 | 0.67 | 0.18 | 1.05e-03 |
| E-TABM-36 | mRNA microarray | 41 | 0.80 | 0.18 | 1.57e-2 | 41 | 0.73 | 0.19 | 7.76e-2 | 40 | 0.77 | 0.19 | 0.103 |
When compared with the results in TCGA dataset (360 samples) analysed by Chaudhary et al. (C-index: 0.68 ± 0.07, Brier score: 0.20 ± 0.02, average log-rank p-value: 0.01), our method provided more considerable improvement in prediction accuracy as well as risk-stratification. Worth noticing, in addition to the DL-based approach using only expression data, our method outperformed the three-omics-based procedure by Chaudhary et al. (C-index: 0.69 ± 0.08; Brier score: 0.20 ± 0.02; log-rank p-value: 0.005). Moreover, only 16 genes out of the 100 features proposed by Chaudhary et al. were identified by us, implying the instability of the SG features by DL-based procedure. It could be argued that the comparison was unfair due to 5 less HCC samples in TCGA dataset we used, however, validations in other three datasets with no more than one-unit difference in sample size between Chaudhary et al. and us showed that the PDS-based approach still outperformed the DL-based method (Table 2).
To evaluate the improvement due to PDS without removal of crosstalk effect, we identified 5 pathways which were significant with HCC survival. With these features, we reimplemented the classifier to predict risk subgroups for above four HCC cohorts. Compared with DL-based model, the PDS without crosstalk removal using TCGA HCC samples produced higher C-index (0.79 ± 0.01 vs. 0.70 ± 0.08), low Brier score (0.20 ± 0.01 vs. 0.21 ± 0.02), and more significant average log-rank p-value (8.49e-5 vs. 3.89e-3) on survival difference (Supplementary Table S2). For the LIRI-JP cohort, PDS-based approach without crosstalk correction provided a lower C-index (0.80 vs. 0.84). When applied to NCI cohort, the results showed a moderate improvement in prediction accuracy (C-index: 0.65 vs. 0.62). Similar with the TCGA cohort, PDS-based approach without crosstalk removal offered a much higher C-index (0.79 vs. 0.73) than DL-based method in the E-TABM-36 cohort. Thus, in TCGA cohort and E-TABM-36 cohort, the improvement was mostly due to the PDS-pathway analysis instead of the crosstalk removal. Put the PDS-based approach with crosstalk removal, the PDS-based approach without crosstalk removal, the DL-based approach together for comparison in three validation cohorts: in LIRI-JP cohort, the three approaches performed similarly (C-index: 0.83, 0.80, 0.84) (Supplementary Table S2); for the NCI cohort, the PDS with crosstalk removal, without crosstalk removal, and the DL-based method obtained C-indexes of 0.76, 0.65 and 0.62; for the E-TABM-36 cohort, the three methods produced C-indexes of 0.80, 0.79, and 0.73. The improvement derived from both the PDS-based method and the crosstalk removal, and the relative magnitudes varied among cohorts, and the PDS-based approach with crosstalk correction performs more robustly in all three validation cohorts. Thus, the crosstalk correction procedure is necessary for accurate prediction. Moreover, even without crosstalk correction, our results still outperform the counterpart by deep learning-based approach.
3.3. Validation on independent HCC datasets
To demonstrate that the conclusion would robustly hold for external datasets, we validated the model trained from TCGA dataset on the other three cohorts of HCC, which had expression data from RNA-Seq and microarray platforms. For the LIRI-JP dataset (n = 231), we obtained decent results which were comparable to the counterpart by DL-based approach we applied (Table 2), including high C-index (0.83 vs. 0.84), low Brier score (0.15 vs. 0.15) and more significant log-rank p-value (1.69e-9 vs. 1.07e-7) between the survival subgroups (Fig. 3b). However, the DL-based approach could not offer competent performance in other two datasets. In the NCI cohort (n = 221), the two subgroups identified by our method showed a much higher C-index (0.76 vs. 0.62), low Brier error rate (0.17 vs. 0.19) and more significant log-rank p-value (1.46e-7 vs. 7.653e-2), compared with the results by the SGs from the DL-based approach (Fig. 3c). In addition, we applied this model to a small validation dataset of E-TABM-36 (n = 41) and also obtained preferable results including a higher C-index (0.80 vs. 0.73), a lower Brier score (0.18 vs. 0.19) and a more significant log-rank p-value (0.0157 vs. 0.0776) (Fig. 3d). As to the E-TABM-36 cohort, the DL-based model did not differentiate the survival-risk subgroups significantly; in contrast, we obtained higher C-index and more significant log-rank p-value (Table 2), implying superior survival-risk stratification. When compared with the results analysed by Chaudhary et al., our method provided more considerable improvement in prediction accuracy as well as risk-stratification for all validation datasets (Table 2).
Fig. 3.
Significant survival differences for four cohorts. (a) TCGA cohort, (b) LIRI-JP cohort, (c) NCI cohort, (d) E-TABM-36 cohort. S1: aggressive (higher-risk survival) subtype; S2: moderate (lower-risk survival) subtype.
3.4. The PDS-based methodology outperforms alternative approach
In second approach, we applied univariate Cox-PH analysis on expression data from TCGA cohort, kept the top 100 features and obtained labels with K-means clustering (K = 2) which gave a remarkable log-rank p-value of 5.74e-15. In CV, SVM classifier on TCGA dataset produced higher C-index (0.81 ± 0.03), lower Brier score (0.20 ± 0.02), and significant log-rank p-value (4.16e-6) than our pathway-based model did (Supplementary Table S3). However, validations on external datasets did not yield competent results as in CV. For the LIRI-JP cohort, we obtained a lower C-index of 0.80, a higher Brier score of 0.16, and a less significant log-rank p-value of 4.75e-6 (Supplementary Table S3). When validated on NCI dataset, we reached a lower C-index of 0.56, a higher Brier score of 0.19, and a non-significant log-rank p-value of 0.784 (Supplementary Table S3). In the smallest cohort of E-TABM-36, the model could not stratify the samples into 2 risk groups efficiently.
3.5. Associations of survival subtypes with clinical and genomic features
We tested the associations between the two survival subtypes and clinical features from the TCGA cohort using Fisher exact test and discovered that clinical stage (P = 2.77e-5) and grade (P = 5.55e-6) were both significantly different between two subgroups, as expected. For the LIRI-JP cohort, we also obtained significant result for the stage covariate (P = 1.93e-5). The NCI cohort provided several clinical covariates including stage (P = 0.00150), cirrhosis (P = 0.000475) and multinodular (P = 0.0494), and all these covariates were significantly associated with survival subtypes.
Tumor suppressor gene TP53 is one of the most frequently mutated genes in many cancers and associated with poor prognosis of patients [42]. Using Fisher exact test between two survival subtypes in TCGA cohort, TP53 mutation is significantly more frequent in the aggressive subgroup S1 than the S2 subgroup (P = 8.93e-8; OR = 3.66). Consistently, patients from subtype S1 have much higher risk of TP53 mutation than S2 subtype (P = 1.25e-2; OR = 2.17) in LIRI-JP cohort.
Utilizing DESeq2 package (log2 fold change >1 and FDR <0.05) for differential expression analysis between two HCC subgroups [43], we found 1677 upregulated and 762 downregulated genes in the aggressive subgroup S1 from the TCGA cohort. The upregulated genes included stemness marker gene CD133 (1.16e-12), AFP (P = 4.34e-08), KRT19 (P = 8.32e-14) and tumor marker gene BIRC5 (P = 2.00e-20), the increased expression level of which were identified to be associated with aggressive subtype in HCC [[44], [45], [46], [47]]. Moreover, 29 genes (ADH1B, ADH6, ALDOA, APOC3, AQP9, CDO1, COBLL1, CRAT, CYB5A, CYP4F12, EPHX2, HN1, KHK, LECT2, PAH, RGN, PKLR, PFKFB3, PKM2, PLG, RGS2, SERPINC1, RNASE4, SLC2A2, SLC22A7, SLC38A1, TM4SF1, SULT2A1, SPHK1) in common with a 65-gene signatures in previous study were differentially expressed in similar pattern [1].
3.6. Characterization of survival risk progression and individualized prediction
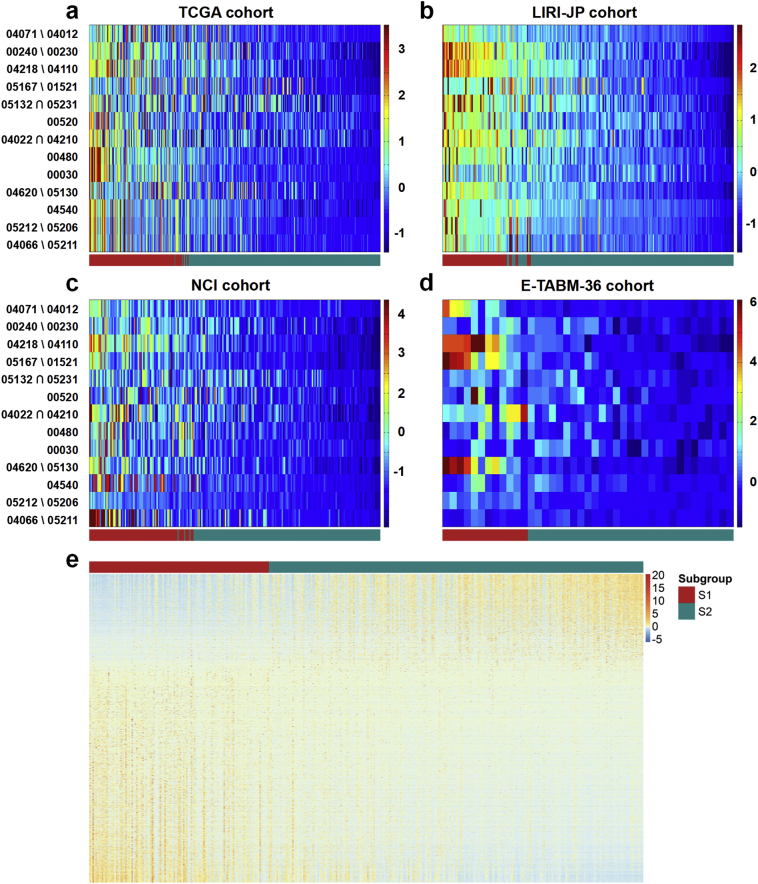
The survival risk of tumor samples is characterized by inherently gradual progression, rather than by clear, abrupt changes between discrete subgroups. For such case supervised classification algorithms, which try to split the samples into distinct groups, thus fails to depict the gradual nature of the phenomenon. In this study, we have utilized SPIN, an unsupervised sorting method, to characterize the gradual changes of survival risk in HCC samples.
Of all samples from four datasets, the ranking profiles by SPIN were not only able to characterize risk progression continuously in HCC samples, but reproduced the stratification of survival-risk subtypes predicted by SVM model aforementioned (Fig. 4 a-d). The deep blue end represented samples which had minimal pathway deregulation from normal liver tissue, while the samples adjacent to the bright red side were far away from the normal tissue in pathway functionality, implying higher risk compared with the samples in the opposite end. To demonstrate it quantitatively, a Cox-PH model built with the ranking and survival outcome was used to infer survival risk for each individual. Similarly, we used C-index to evaluate the fraction of patient pairs whose risk prediction by SPIN ranking were correctly ordered (Table 3). Of note, this metric here was more aggressive than before because it treated these samples as continuous individuals rather than discrete groups, thus more pairs would be evaluated. To compare with the SG features by the DL-based approach, we applied same procedure to these features to predict individualized risk in the same four cohorts. Consistently, we obtained higher accuracy in terms of C-index for all cohorts compared with the SGs (Table 3). According to the order by SPIN, we plotted the expression profile of 2439 differential expression genes mentioned earlier between risk subgroups in TCGA dataset (Fig. 4e), which characterized the progression of HCC in gene expression level.
Fig. 4.
PDS and expression heatmap with samples ranking by SPIN for four cohorts. (a) TCGA cohort, (b) LIRI-JP cohort, (c) NCI cohort, (d) E-TABM-36 cohort. The 13 pathways (rows) are represented with KEGG map ID for clarity, and the samples (columns) are ordered by SPIN methodology and labeled by supervised classification. S1: aggressive (higher-risk survival) subtype; S2: moderate (lower-risk survival) subtype. (e) Differential expressed genes including 1677 upregulated and 762 downregulated in the two subgroups from the TCGA cohort.
Table 3.
Performance of survival risk prediction by Cox-PH model built with SPIN rank and survival data in all four cohorts compared with the counterpart utilizing mRNA features by the DL-based approach.
| Cohort | Samples(N) | PDS-based features |
DL-based features |
||
|---|---|---|---|---|---|
| C-indexa | p-valueb | C-indexa | p-valueb | ||
| TCGA | 355 | 0.70 | 4.31e-17 | 0.66 | 1.49e-9 |
| LIRI-JP | 231 | 0.74 | 3.25e-8 | 0.72 | 1.02e-7 |
| NCI | 221 | 0.65 | 3.85e-7 | 0.63 | 1.91e-5 |
| E-TABM-36 | 41 | 0.68 | 4.03e-4 | 0.62 | 3.67e-2 |
C-index, more pairs would be evaluated than in Table 2.
P-value, for the statistical test if the estimate is significantly different from background (0.5).
3.7. Cancer hallmark analysis
PDS of the 13 pathways identified were found significant associated with survival in HCC patients, then we expected the dominate genes belonging to them to be functionally enriched with tumor etiology. To verify it, we applied PCA analysis for each pathway using the same data with which were used to calculate PDS and obtained 40 common genes in TCGA, LIRI-JP and NCI cohorts after applying the filter of cumulative proportion > 0.8 and loading >0.3. Multiple known HCC therapeutic targets, such as FOXM1, RAF1, TGFA, ENO1, EGF, UAP1L1, GNAS, GPI, IRAK1, MAPK1, MAPK9, IKBKG, MAP2K2, RAC1, CDK1, SQSTM1, TYMS, G6PD, and RAD51, were among the list. The 40-gene list also contained several less-described genes, such as PLCB1, GPX7, HKDC1, MAPK13, RRM2, RALBP1 and GMDS [48], as well as novel HCC markers such as UGDH, UCK2, DUT, NPL, ITPR1, GNG4, RENBP, PPP2R5A, GNB5, WASF1, CYB5R1, HEXA, TKT and TK1.
3.8. Construction of a nomogram using PDS-based features
To make risk prediction more intuitive for clinicians, we constructed a color-based nomogram from the SVM classifier above using the 13 features (Supplementary Fig. S2). The whole samples of TCGA cohort were used, aiming to build a more accurate nomogram, thus, no clinical covariate was embedded due to high missing rate. Moreover, evidence shows that classifier with clinical information integrated produced no improvement compared with the counterpart without clinical covariates in the study of Chaudhary et al., which was also observed in our exploration. To obtain a risk estimate for a patient with this nomogram, the color corresponding to the feature's value needs to be determined using the color bar specific to each feature. This color is transformed to a value with the color legend at the right. Repeating this for each feature and summing the resulting values, yields the score. This score is then converted into the risk estimate by means of the bottom color bar. The importance of the features is represented by means of the brightness of the color: variables with a higher intensity in yellow have a larger impact on the risk prediction. Of note, this nomogram is actually not easy to use as the counterpart based on linear models, because the color-value conversion is not straightforward and accurate by sight. More powerful tools for nomogram construction are desired for nonlinear models like SVM.
4. Discussion
Molecular signatures by statistical models like machine learning trained with high-throughput expression data have long been appreciated and discussed for precision medicine in cancer. However, poor stability and prediction accuracy of SG features have been observed in multiple studies and pose challenges to clinical application. Categories of pathway-based approaches have thus emerged with robust features which are extracted on the basis of molecular mechanisms such as predefined pathways. Pathifier algorithm quantifies pathway deregulation and produces continuous features which can be used to characterize pathway functionality during disease progression. This approach has been utilized in subtypes classification of multiple cancer types with remarkable outcomes, where crosstalk between pathways has never been discussed. A recent procedure filtering SG features with DL framework was applied to prognosis prediction in HCC, a cancer type of high heterogeneity. To demonstrate whether PDS features could outperform these SGs by DL-based approach, we proposed a novel pipeline based on Pathifier with crosstalk accommodated to predict prognosis in the same four HCC datasets. Our pathway-level features led to a substantially better performance of prediction than the SG features by DL-based methodology not only in terms of accuracy but also survival difference between subtypes.
One major advantage of our PDS-based features was in survival risk stratification. Compared with the DL-based approach [15], we found that statistically significant survival difference between aggressive subgroup (S1) and moderate subgroup (S2) was more pronounced in the TCGA cohort of HCC patients in CV. This finding also held in other three HCC datasets (LIRI-JP, NCI and E-TABM-36), especially for the small cohort of E-TABM-36 where the DL-based model could not obtain statistically significant outcome. Furthermore, we also evaluated subtype distinction in terms of other clinical criterions. More importantly, more decent performance in all four HCC cohorts showed the robustness of our methodology (Table 2), whereas the DL-based approach could not offer comparable results in other three datasets than LIRI-JP cohort.
The robustness and accuracy of our classifier is supported not only by the inherent stability of pathway-level features but also by eliminating false positive features as possible. The general philosophy of Pathifier algorithm is to extract coarse-grained variables from expression space of genes belong to each pathway to represent the extent of deregulation from normal behavior [5]. With coarse-grained PDS which is less sensitive to noise, we can thus construct simple models with less features that capture the essential aspects of the problem. On the other hand, to avoid false positive features which may dampen performance of validation on independent datasets, we have adopted the survival-significant pathways/sub-pathways only shared by the three large datasets (TCGA, LIRI-JP and NCI) as HCC-specific features, rather than use significant features from one arbitrary dataset.
More importantly, in addition to classification of survival subtypes, the ranking profiles of HCC samples generated by these features can not only characterize tumor progression continuously among patients, but also provide individualized risk prediction as well as pathway deregulation details for each patient. Governing genes identified in these pathways provides important biological insight in selecting treatment strategies for HCC patients. For example, repeated overexpression of the mammalian transcription factor Forkhead Box M1 (FOXM1) has been implicated in all major hallmarks of cancer [49]. Findings such as these have generated interest in the development of targeted inhibitors, in this case for FOXM1 [50,51].
Though we have developed a pipeline for robust stratification of survival subtypes and accurate prognosis prediction in hepatocellular carcinoma, it has a few limitations. First, similar to Chaudhary et al., we obtain class label of the TCGA HCC samples using whole TCGA dataset. Therefore, when we implement CV on TCGA dataset using SVM model, the C-statistics can be inflated; however, validations on other external datasets produce more unbiased C-statistics. Another limitation is that the sample size of one of the three validation datasets (E-TABM-36) is only 41, which may introduce bias into validation. However, validations on the other two large datasets (LIRI-JP, NCI) with sample size of 232, 221 indicate that our model is generally predictive; in addition, we have applied our approach to a relatively large HCC dataset from GSE54236 (N = 78) [52], and still obtained very good prediction accuracy (C-index = 0.88) as well as drastically different risk subgroups of HCC (log-rank p-value = 1.54e-8). An additional hurdle is that a certain number of normal samples are required to estimate PDS more accurately. Hopefully, we have gained improved result in E-TABM-36 cohort with the aid of normal samples from NCI cohort after batch effect adjustment. In terms of prediction accuracy, it may be argued that the sample size differences contribute to improvements in our prediction model in comparison with the results by Chaudhary et al. Though we have used 5 less samples (355 vs. 360) from TCGA cohort in CV than the DL-based study, validations on the other three datasets with very close sample size (LIRI-JP: 231 vs. 230, NCI: 221 vs. 221, E-TABM: 41 vs.40) to the DL-based study still provide better performance consistently. Furthermore, we have also implemented the DL-based approach with our curated datasets and obtained similar outcomes, indicating the higher accuracy and robustness of our approach.
In summary, the PDS-based features derived from Pathifier with crosstalk accommodated provides an accurate and robust stratification of HCC patients with prognostic significance, with the promise to improve precision therapy with subtype-specific efficacy. The dominant genes identified were well consistent with therapeutic targets of HCC from other independent studies. We also expect that our procedure is applicable to other cancer types with good performance. Validations on other cancer types with large sample size are desired for future research.
Funding sources
The research was supported in part by 2016YFC0902403(Yu) of Chinese Ministry of Science and Technology, and by National Natural Science Foundation of China 11671256(Yu), and also by the University of Michigan and Shanghai Jiao Tong University Collaboration Grant (2017, Yu). The funders did not play a role in manuscript design, data collection, data analysis, data interpretation or writing of the manuscript.
Declaration of interests
The authors declared no conflict of interest.
Author contributions
Z.Y. and B.F. contributed to the study concept and design; Z.Y. and Y.Z. obtained funding and provided the essential materials; B.F., C.L. and Y.Y. obtained the datasets; B.F., Y.Y., Z.T. and Z.Y. analysed and interpreted the data; B.F. and Z.Y. wrote the manuscript. All authors reviewed and approved the final manuscript.
Acknowledgements
None.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.05.010.
Appendix A. Supplementary data
Supplementary material
References
- 1.Kim S.M., Leem S.H., Chu I.S., Park Y.Y., Kim S.C., Kim S.B. Sixty-five gene-based risk score classifier predicts overall survival in hepatocellular carcinoma. Hepatology. 2012;55(5):1443–1452. doi: 10.1002/hep.24813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Veer L.J.V.T., Dai H., De Vijver M.J.V., He Y.D., Hart A.A.M., Mao M. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 3.Paik S., Shak S., Tang G., Kim C., Baker J., Cronin M.T. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2005;351(27):2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 4.Michiels S., Koscielny S., Hill C. Prediction of cancer outcome with microarrays: a multiple random validation strategy. Lancet. 2005;365(9458):488–492. doi: 10.1016/S0140-6736(05)17866-0. [DOI] [PubMed] [Google Scholar]
- 5.Domany E. Using high-throughput transcriptomic data for prognosis: a critical overview and perspectives. Cancer Res. 2014;74(17):4612–4621. doi: 10.1158/0008-5472.CAN-13-3338. [DOI] [PubMed] [Google Scholar]
- 6.Kanehisa M., Sato Y., Kawashima M., Furumichi M., Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:457–462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fabregat A., Sidiropoulos K., Garapati P., Gillespie M., Hausmann K., Haw R. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014;42:472–477. doi: 10.1093/nar/gkv1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harris M.A., Clark J.I., Ireland A., Lomax J., Ashburner M., Foulger R. The gene ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32 doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alcaraz N., List M., Batra R., Vandin F., Ditzel H.J., Baumbach J. De novo pathway-based biomarker identification. Nucleic Acids Res. 2017;45(16) doi: 10.1093/nar/gkx642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vaske C.J., Benz S.C., Sanborn J.Z., Earl D., Szeto C.W., Zhu J. Inference of patient-specific pathway activities from multi-dimensional cancer genomics data using PARADIGM. Bioinformatics. 2010;26(12):237–245. doi: 10.1093/bioinformatics/btq182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drier Y., Sheffer M., Domany E. Pathway-based personalized analysis of cancer. Proc Natl Acad Sci U S A. 2013;110(16):6388–6393. doi: 10.1073/pnas.1219651110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Livshits A., Git A., Fuks G., Caldas C., Domany E. Pathway-based personalized analysis of breast cancer expression data. Mol Oncol. 2015;9(7):1471–1483. doi: 10.1016/j.molonc.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang S., Chong N., Lewis N.E., Jia W., Xie G., Garmire L.X. Novel personalized pathway-based metabolomics models reveal key metabolic pathways for breast cancer diagnosis. Genome Med. 2016;8(1):34. doi: 10.1186/s13073-016-0289-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Llovet J.M., Zucman-Rossi J., Pikarsky E., Sangro B., Schwartz M., Sherman M. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016;2 doi: 10.1038/nrdp.2016.18. [DOI] [PubMed] [Google Scholar]
- 15.Chaudhary K., Poirion O.B., Lu L., Garmire L.X. Deep learning-based multi-omics integration robustly predicts survival in liver cancer. Clin Cancer Res. 2018;24(6):1248–1259. doi: 10.1158/1078-0432.CCR-17-0853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 17.Cheng A., Kang Y., Chen Z., Tsao C.J., Qin S., Kim J.S. Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: a phase III randomised, double-blind, placebo-controlled trial. Lancet Oncol. 2009;10(1):25–34. doi: 10.1016/S1470-2045(08)70285-7. [DOI] [PubMed] [Google Scholar]
- 18.Lee J.S., Chu I., Heo J., Calvisi D.F., Sun Z., Roskams T. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40(3):667–676. doi: 10.1002/hep.20375. [DOI] [PubMed] [Google Scholar]
- 19.Hoshida Y., Nijman S.M.B., Kobayashi M., Chan J.A., Brunet J., Chiang D.Y. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009;69(18):7385–7392. doi: 10.1158/0008-5472.CAN-09-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai J., Li B., Zhu Y., Fang X., Zhu M., Wang M. Prognostic biomarker identification through integrating the gene signatures of hepatocellular carcinoma properties. EBioMedicine. 2017;19:18–30. doi: 10.1016/j.ebiom.2017.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Long J., Wang A., Bai Y., Lin J., Yang X., Wang D. EBioMedicine. 2019. Development and validation of a TP53-associated immune prognostic model for hepatocellular carcinoma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu G., Dong C., Liu L. Integrated multiple "-omics" data reveal subtypes of hepatocellular carcinoma. PLoS One. 2016;11(11) doi: 10.1371/journal.pone.0165457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujimoto A., Furuta M., Totoki Y., Tsunoda T., Kato M., Shiraishi Y. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat Genet. 2016;48(5):500–509. doi: 10.1038/ng.3547. [DOI] [PubMed] [Google Scholar]
- 24.Roessler S., Jia H.L., Budhu A., Forgues M., Ye Q.H., Lee J.S. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 2010;70(24):10202–10212. doi: 10.1158/0008-5472.CAN-10-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsafrir D., Tsafrir I., Ein-Dor L., Zuk O., Notterman D.A., Domany E. Sorting points into neighborhoods (SPIN): data analysis and visualization by ordering distance matrices. Bioinformatics. 2005;21(10):2301–2308. doi: 10.1093/bioinformatics/bti329. [DOI] [PubMed] [Google Scholar]
- 26.Saldana D.F., Feng Y. SIS: an R package for sure independence screening in ultrahigh-dimensional statistical models. J Statis Softw. 2018;83(1):1–25. [Google Scholar]
- 27.Harrell F.E., Lee K.L., Mark D.B. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15(4):361–387. doi: 10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 28.Brier G.W. Verification of forecasts expressed in terms of probability. Mon Weather Rev. 1950;78(1):1–3. [Google Scholar]
- 29.Zhu Y., Qiu P., Ji Y. TCGA-assembler: open-source software for retrieving and processing TCGA data. Nat Methods. 2014;11(6):599–600. doi: 10.1038/nmeth.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boyault S., Rickman D.S., de Reynies A., Balabaud C., Rebouissou S., Jeannot E. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 2007;45(1):42–52. doi: 10.1002/hep.21467. [DOI] [PubMed] [Google Scholar]
- 31.Hastie T., Stuetzle W. Principal curves. J Am Stat Assoc. 1989;84(406):502–516. [Google Scholar]
- 32.Leek J.T., Johnson W.E., Parker H.S., Jaffe A.E., Storey J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28(6):882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cox D.R. Regression models and life-tables. In: Kotz S., Johnson N.L., editors. Breakthroughs in statistics: methodology and distribution. Springer New York; New York, NY: 1992. pp. 527–541. [Google Scholar]
- 34.Meyer D., Dimitriadou E., Hornik K., Weingessel A., Leisch F. R Package Version 1.6–8; TU Wien: 2017. e1071: Misc functions of the department of statistics, probability theory group (formerly: E1071)https://CRAN.R-project.org/package=e1071 [Google Scholar]
- 35.Schroder M.S., Culhane A.C., Quackenbush J., Haibekains B. Survcomp: an R/bioconductor package for performance assessment and comparison of survival models. Bioinformatics. 2011;27(22):3206–3208. doi: 10.1093/bioinformatics/btr511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Therneau T.M. 2015. A package for survival analysis in S. [R package version 2.38] [Google Scholar]
- 37.Belle V.V., Calster B.V., Huffel S.V., Suykens J.A.K., Lisboa P. Explaining support vector machines: a color based Nomogram. PLoS One. 2016;11(10) doi: 10.1371/journal.pone.0164568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Donato M., Xu Z., Tomoiaga A., Granneman J.G., Mackenzie R.G., Bao R. Analysis and correction of crosstalk effects in pathway analysis. Genome Res. 2013;23(11):1885–1893. doi: 10.1101/gr.153551.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jaccard P. E'tude anatomique de bois comprime's. Bull Soc Vaud Sci Nat. 1910;37:547–579. [Google Scholar]
- 40.Thomas D.C., Baurley J.W., Brown E.E., Figueiredo J.C., Goldstein A.M., Hazra A. Approaches to complex pathways in molecular epidemiology: summary of a special conference of the American Association for Cancer Research. Cancer Res. 2008;68(24):10028–10030. doi: 10.1158/0008-5472.CAN-08-1690. [DOI] [PubMed] [Google Scholar]
- 41.Steck H., Krishnapuram B., Dehingoberije C., Lambin P., Raykar V.C., editors. On ranking in survival analysis: bounds on the concordance indexNeural Inform Process Syst. 2007:1209–1216. [Google Scholar]
- 42.Villanueva A., Hoshida Y. Depicting the role of TP53 in hepatocellular carcinoma progression. J Hepatol. 2011;55(3):724–725. doi: 10.1016/j.jhep.2011.03.018. [DOI] [PubMed] [Google Scholar]
- 43.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamashita T., Forgues M., Wang W., Kim J.W., Ye Q., Jia H. EpCAM and α-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer Res. 2008;68(5):1451–1461. doi: 10.1158/0008-5472.CAN-07-6013. [DOI] [PubMed] [Google Scholar]
- 45.Andersen J.B., Loi R., Perra A., Factor V.M., Leddacolumbano G.M., Columbano A. Progenitor-derived hepatocellular carcinoma model in the rat†. Hepatology. 2010;51(4):1401–1409. doi: 10.1002/hep.23488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cao L., Li C., Shen S., Yan Y., Ji W., Wang J. OCT4 increases BIRC5 and CCND1 expression and promotes cancer progression in hepatocellular carcinoma. BMC Cancer. 2013;13(1):82. doi: 10.1186/1471-2407-13-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma S., Chan K.W., Hu L., Lee T.K., Wo J.Y.H., Ng I.O. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology. 2007;132(7):2542–2556. doi: 10.1053/j.gastro.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 48.Ang C., Miura J.T., Gamblin T.C., He R., Xiu J., Millis S.Z. Comprehensive multiplatform biomarker analysis of 350 hepatocellular carcinomas identifies potential novel therapeutic options. J Surg Oncol. 2016;113(1):55–61. doi: 10.1002/jso.24086. [DOI] [PubMed] [Google Scholar]
- 49.Gartel A.L. FOXM1 in cancer: interactions and vulnerabilities. Cancer Res. 2017;77(12):3135–3139. doi: 10.1158/0008-5472.CAN-16-3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gusarova G.A., Wang I., Major M.L., Kalinichenko V.V., Ackerson T., Petrovic V. A cell-penetrating ARF peptide inhibitor of FoxM1 in mouse hepatocellular carcinoma treatment. J Clin Investig. 2007;117(1):99–111. doi: 10.1172/JCI27527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnson P.W.M., Challis R., Chowdhury F., Gao Y., Harvey M., Geldart T. Clinical and biological effects of an agonist anti-CD40 antibody: a cancer research UK phase I study. Clin Cancer Res. 2015;21(6):1321–1328. doi: 10.1158/1078-0432.CCR-14-2355. [DOI] [PubMed] [Google Scholar]
- 52.Villa E., Critelli R., Lei B., Marzocchi G., Camma C., Giannelli G. Neoangiogenesis-related genes are hallmarks of fast-growing hepatocellular carcinomas and worst survival. Results from a prospective study. Gut. 2016;65(5):861–869. doi: 10.1136/gutjnl-2014-308483. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material