Abstract
Background
Evaluating clinical outcome prior to concurrent chemoradiotherapy remains challenging for oesophageal squamous cell carcinoma (OSCC) as traditional prognostic markers are assessed at the completion of treatment. Herein, we investigated the potential of using sub-region radiomics as a novel tumour biomarker in predicting overall survival of OSCC patients treated by concurrent chemoradiotherapy.
Methods
Independent patient cohorts from two hospitals were included for training (n = 87) and validation (n = 46). Radiomics features were extracted from sub-regions clustered from patients' tumour regions using K-means method. The LASSO regression for ‘Cox’ method was used for feature selection. The survival prediction model was constructed based on the sub-region radiomics features using the Cox proportional hazards model. The clinical and biological significance of radiomics features were assessed by correlation analysis of clinical characteristics and copy number alterations(CNAs) in the validation dataset.
Findings
The overall survival prediction model combining with seven sub-regional radiomics features was constructed. The C-indexes of the proposed model were 0.729 (0.656–0.801, 95% CI) and 0.705 (0.628–0.782, 95%CI) in the training and validation cohorts, respectively. The 3-year survival receiver operating characteristic (ROC) curve showed an area under the ROC curve of 0.811 (0.670–0.952, 95%CI) in training and 0.805 (0.638–0.973, 95%CI) in validation. The correlation analysis showed a significant correlation between radiomics features and CNAs.
Interpretation
The proposed sub-regional radiomics model could predict the overall survival risk for patients with OSCC treated by definitive concurrent chemoradiotherapy.
Fund
This work was supported by the Zhejiang Provincial Foundation for Natural Sciences, National Natural Science Foundation of China.
Keywords: Oesophageal cancer, Radiomics, Prognostic model, Radiation therapy, Imaging
Research in context.
Evidence before this study
Currently, predicting the clinical outcome remains challenging for oesophageal squamous cell carcinoma (OSCC) patients treated by concurrent chemoradiotherapy, as traditional prognostic markers are mainly assessed at the completion of treatment. Sub-regional radiomics analysis has been conducted with proven prognostic power for many cancer types. At present, no previous studies have applied sub-region based radiomics analysis for survival prediction of OSCC.
Added value of this study
In this study involving two medical centres, we performed a sub-region cluster within the tumour region on planning CT. A survival prediction model for OSCC was then constructed based on the sub-regional radiomics features. The constructed sub-regional radiomics survival prediction model showed high prognostic value in both the training cohort and validation cohort.
Implications of all the available evidence
The sub-regional radiomics analysis showed potential in predicting survival of OSCC patients treated by definitive concurrent chemoradiotherapy, which may improve personalized treatment and achieve a better outcome for OSCC patients.
Alt-text: Unlabelled Box
1. Introduction
Previous studies revealed that biomarkers such as VEGF, cyclin D1, Ki-67, and squamous cell carcinoma antigen are prognostic for oesophageal squamous cell carcinoma (OSCC) [1]. These biomarkers are all derived from pathological analysis of tumour samples. Yet there remains a large cohort of OSCC patients who are physiologically unresectable or refused surgery. However, personalized treatment regimen and its long-term efficacy significantly rely on bio-physiological assessment of pre-treatment biomarkers. One alternative is through medical imaging assessment using computed tomography (CT), magnetic resonance (MR) and positron emission computed tomography (PET), which all play essential roles in cancer diagnosis and tumour heterogeneity analysis prior to radiotherapy. More specifically, PET/CT was used to predict survival in oesophageal cancer patients treated with chemoradiotherapy [2]. However, there has not been a literature consensus in terms of prognosis power in PET/CT. Atsumi et al. suggested the maximum standardized uptake value (SUVmax) as a strong prognostic marker for primary OSCC [3]. Lindner et al. found that SUVmax only correlates with survival of OSCC patient in case of primary surgery but not if patients received neoadjuvant therapy [4]. However, Myslivecek et al. concluded that the contribution of (18)F-FDG PET/CT cannot be considered as a potential marker for outcome of chemoradiotherapy [5]. The controversy over the use of SUVmax from PET/CT has hampered its utility in clinical practice. Meanwhile, efforts have been focusing on radiomics as a potential solution in providing surrogate information of the tumour microenvironment utilizing quantification methods such as texture analysis [6]. Beukinga et al. found that the combination of texture features extracted from PET and CT can improve the predictive power of tumour response to neoadjuvant chemoradiotherapy (nCRT) in locally advanced OSCC, compared to SUVmax alone [7]. Despite strong clinical indication, the general clinical use of the PET/CT based radiomics features in tumour response prediction is still limited, in part due to the high cost of PET/CT scans for patients especially in rural areas or developing countries. CT scans, on the other hand, are more accessible and recent studies have explored features extracted from CT alone. Hou et al. found that CT-based radiomics features can be used as imaging biomarkers to predict tumour response to chemoradiotherapy in OSCC patients [8]. Five CT-based radiomics features were found to discriminate responders from non-responders (AUCs from 0.686 to 0.727). Ganeshan et al. showed correlation between unenhanced CT textural features (entropy: p-value = .027, r = 0.512 and uniformity: p-value = .027, r = −0.521, Pearson's correlation) and PET mean standardized uptake value (SUVmean) of non-small cell lung cancer (NSCLC) [9]. Ganeshan et al. applied the same methodology towards oesophageal carcinoma and found that entropy and uniformity are correlated with SUVmean (entropy: p-value < .001, r = 0.748 and uniformity: p-value < .001, r = −0.754, Pearson's correlation) and SUVmax (entropy: p-value = .032, r = 0.469 and uniformity: p-value = .029, r = − 0.476, Pearson's correlation) in the corresponding 18F-FDG PET scans [10]. In other words, texture features from CT scans alone may be a prognostic indicator of survival prediction in OSCC. Thus far, OSCC tumour heterogeneity and its quantitative feature analysis has been carried out for the whole tumour volume, without the consideration of intra-tumoral variation in different sub-regions [11,12]. Yet sub-regional radiomics analysis has been conducted with proven prognostic power for NSCLC [13] and hepatocellular carcinoma [14]. To the best of our knowledge, no previous studies have applied sub-region based radiomics analysis for survival prediction of OSCC.
This study aimed to construct and validate a prognostic model for pre-treatment survival prediction of OSCC patients, based on sub-regional radiomics analysis of CT scans. To identify the clinical and molecular basis of quantitative imaging characteristics in this study, we assessed the association of the radiomics features with the clinical factors and copy number alterations (CNAs). We also compared prediction accuracy of the sub-regional radiomics model with the whole-tumour radiomics model and conventional clinical factors.
2. Methods
2.1. Study design
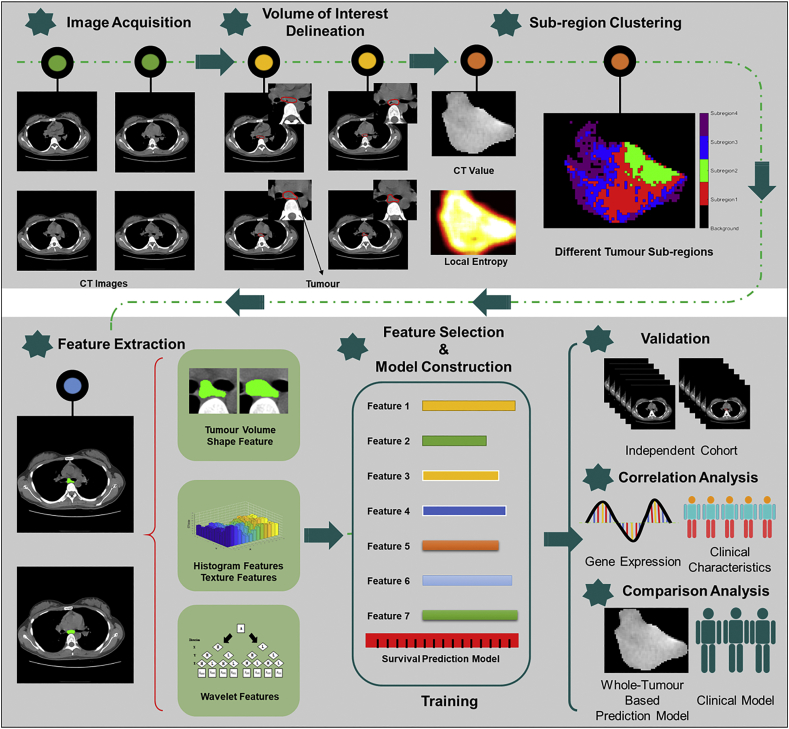
Fig. 1 depicts the schema of the present study, with the following steps: Image Acquisition, Volume of Interest Delineation, Sub-region Clustering, Features Extraction, Feature Selection, Model Construction and Validation. CT image datasets from two institutions were used for training an overall survival prediction model and validation, respectively. More specifically, tumour volumes were delineated on the CT scans of the selected OSCC patients, followed by sub-region clustering and radiomics feature extraction of different sub-regions. Features that showed significant correlation with survival were identified and paired with the corresponding clinical survival outcomes for training and validation using a machine learning method. A survival prediction model was constructed based on the training dataset. The model was then tested using the validation dataset by investigating the underlying genomics correlation of the developed survival prediction model with CNAs and clinical factors. We further validated the developed model by comparison with whole-tumour based radiomics model and conventional clinical factors.
Fig. 1.
Workflow in this study.
2.2. Patients
Two cohorts of OSCC patients from Hangzhou Cancer Hospital and the First Affiliated Hospital of Wenzhou Medical University were retrospectively identified and their CT scans were obtained upon Intuitional Review Board approval from both hospitals. The written informed consent was waived by IRB based on the retrospective nature of this study. This study was constructed following ethical guidelines of World Medical Association (WMA) Declaration of Helsinki. Inclusion criteria are I) histological diagnosis of OSCC and staged according to the 2002 (version 6.0) American Joint Committee on Cancer staging system; II) medically inoperable or refuse to surgery after discussion by the multidisciplinary treatment team; III) Eastern Cooperative Oncology Group Performance Status (ECOG PS) of ≤2; and IV) no evidence of severe organ dysfunction. The exclusion criteria are I) early-stage OSCC; II) evidence of distant metastasis at diagnosis; III) prior administration of surgery or chest radiation or chemotherapy; IV) non-treatment related death; and V) incomplete data on CT images and overall survivals.
All patients received a planning CT scan for treatment planning purpose, followed by radiotherapy treatment with a total dose of 54–60 Gy given in 27–30 fractions (2.0 Gy per fraction, 5 days per week). Treatment plans involved 3-dimension conformal photon fields or Intensity modulated photon fields. The majority of OSCC patients received definitive concurrent chemoradiotherapy based on cisplatin regimen. For patients with advanced ages or poor performance status, radiotherapy alone was delivered to these patients.
Paired CT images and overall survival outcomes of a total of 87 patients diagnosed between May 2012 and November 2016 from Hangzhou Cancer Hospital were used as the training cohort. Same dataset pairs from 46 patients diagnosed between January 2008 and July 2011 from the First Affiliated Hospital of Wenzhou Medical University were used to validate the developed model and to further improve the model by feeding in genomics information with CNAs. Apart from age and sex, patients' clinical characteristics including clinical stage, ECOG PS, and tumour length were collected for both cohorts.
2.3. Image acquisition
The CT images of the training cohort were obtained from the Brilliance Big Bore CT scanner (Philips Electronics, Eindhoven, Netherlands) in Hangzhou Cancer Hospital. The scanning voltage and tube currents were 120 kVp and 406 mAs. The slice thickness ranged from 3 to 5 mm. The CT images in the validation cohort were scanned from the LightSpeed Pro 16 CT (GE Medical Systems, Milwaukee) in the First Affiliated Hospital of Wenzhou Medical College, with 120 kVp and 150 mAs. The slice thickness was 3–8 mm.
2.4. Volume of interest (VOI) delineation and sub-region clustering
The contoured gross tumour volume (GTV) in the radiation treatment planning was used as the volume of interest (VOI) in the training cohort. Because the patients' radiation therapy plans in the validation cohort were not well preserved, the VOIs in the validation cohort were depicted by two radiologists all with more than 10 years of experience. The VOI segmentation was performed using open-source software, ITK-SNAP (http://www.itksnap.org/pmwiki/pmwiki.php). The regions of VOIs in the validation cohort were agreed by both the two radiologists. Air region in the VOI was excluded with a Hounsfield Unit (HU) threshold of zero. VOIs were divided into sub-regions based on the cluster of HU values and local entropy values of CT images. A moving window with the size of 9 × 9 was used in the calculation of local entropy [13] on each slice of CT images. The K-means method was used for clustering sub-regions in this study. The Calinski–Harabasz (CH) value was used as the criterion in selecting the best number of clusters at the patient population level [15]. The number of clusters from 2 to10 were tested in this study.
2.5. Feature extraction
To compensate for the difference in radiomics features caused by different reconstruction slice thicknesses and pixel sizes [16], the voxel sizes of all CT images in this study were reconstructed to 1 × 1 × 5 mm3. The VOIs were normalized to 64 grey levels to compensate for the variation of CT scanners. For each sub-region, region volume, shape, intensity, and texture were quantified as 548 radiomics features using texture analysis and wavelet decomposition method as described in the study of Vallieres et al. [17]. For comparison purpose, 548 radiomics features were also extracted from the whole tumour region for each patient. The radiomics features used in this study were described in Supplementary I. All sub-regional partitioning and radiomics feature extraction were performed on MATLAB 2015b (The Mathworks, Natick, MA).
2.6. Feature selection and survival prediction model construction
To eliminate the potential volume change effect caused by sub-regional analysis on radiomics features, the correlation between the tumour volume and radiomics feature value was assessed using the Pearson correlation method, as compared to the whole tumour region. Radiomics features with a correlation coefficient (CC) of >0.75 were excluded from further study [18]. The redundant features with high internal association (CC > 0.75) of all sub-regional radiomics features were further excluded. The least absolute shrinkage and selection operator (LASSO) for cox algorithm was used to screen features that were highly correlated with survival outcomes [19]. The LASSO algorithm controls the number of selected variables by adjusting the parameter, λ. The objective function of LASSO algorithm is shown in Function 1, where y is truth label, X is the feature matrix, β is the coefficients of features and λ is the tuning parameter. The LASSO method was designed to minimize the objective function. The larger the λ is, the smaller the coefficients of features are. Features with coefficient of larger than zero were selected. To determinate the best λ in the LASSO algorithm, the 8-fold cross-validation strategy was used [20]. The λ resulting the least mean difference between the predicted and actual survival in the cross-validations was used to select the final features. A radiomics score was calculated based on the selected sub-regional radiomics features and corresponding coefficients. The survival prediction model was built based on the radiomics score using the Cox proportional hazards model and a radiomics nomogram was constructed based on this model for visualization purpose.
| (1) |
2.7. Prognostic performance evaluation
The predictive performance of the radiomics score was evaluated in the training cohort and verified in the validation cohort using the concordance index (C-index) [21]. The patients were divided into a high-risk group or low-risk group based on the predicted risk using the proposed model. The receiver operation characteristics (ROC) curve was used to determine the best cut-off risk to stratify patients. The ROC curves for 1-year survival, 2-year survival, and 3-year survival were plotted for the two datasets. The Youden index was used to select the best cut-off value where the sum of sensitivity and specificity is maximized [22]. The optimal cut-off in the training dataset was used to stratify patients in the validation dataset. The Kaplan-Meier survival analysis and Log-rank test were used to compare the differences between the survival curves of the two groups [23].
2.8. Association of sub-regional radiomics features with clinical factors and copy number alteration
The Spearman rank correlation test was used to assess the correlation of sub-regional radiomics features included in the survival model with clinical factors and CNAs in the validation cohort. A corresponding p-value was used to determine the significance of the correlation (p-value < .05). The CNAs of patients in the validation cohort were acquired by whole exon sequencing using the Integrated DNA Technologies (IDT) xGen Exome Research Panel kit for exon capture and sequencing by the Illumina HiSeq 2500. The alignment was performed by bwa 0.7.13 [24]. All CNAs were called by cn.mops [25] package in R language. A total of 1046 CNAs Indexes were analysed. The specific CNAs locations of each CNA index were summarized in the supplementary (Supplementary Table S3).
2.9. Comparison with whole-tumour based radiomics model
The same workflow of feature selection and model construction were applied in building the whole-tumour survival prediction model (WTPM) using the training cohort. Its prediction performance was compared with the sub-regional prediction model (SRPM) using the C-index in both training and validation cohorts.
2.10. Comparison with the clinical factors
The prediction performance of SRPM was also compared with conventional prognostic factors, clinical stage and ECOG PS using the C-index in both training and validation cohorts.
2.11. Statistical analysis
Statistical analysis was done with the R software (R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL: http://www.R-project.org, 2016). The Mann–Whitney U test and chis-q test were used to compare the difference in patient clinical characteristics in the two cohorts. The student t-test was used to compare the C-index of different methods.
3. Results
3.1. Patient characteristics
Patient characteristics in the two cohorts showed consistent demographic distributions, which was summarized in Table 1. The survival analysis showed no significant difference (p-value = .388, Log-rank test) in survival between the two cohorts (Supplementary Fig. S3). Among them, 61 and 37 patients were confirmed deceased during the follow-up period in the training group (70.11%), and the validation group (80.43%), respectively. The median and mean survival time of the training cohort were 13.00 months (10.00–19.00, 95% Confidence Interval [CI]) and 25.66 months (19.85–31.47, 95% CI), respectively. In the validation cohort, median and mean survival time were 12.77 months (9.77–15.43, 95% CI) and 24.21 months (21.27–33.16, 95% CI), respectively.
Table 1.
The patient characteristics in training and validation cohorts.
| Characteristic | Training Cohort (n = 87) | Validation Cohort (n = 46) | p-value |
|---|---|---|---|
| Age, years | .611 | ||
| Median (range) | 64 (45–85) | 61.5(48–75) | |
| Sex | .122 | ||
| Male | 59 | 37 | |
| Female | 28 | 9 | |
| ECOG PS | .73 | ||
| 0–1 | 45 | 26 | |
| 2 | 42 | 20 | |
| T stage | .613 | ||
| T3 | 38 | 18 | |
| T4 | 49 | 28 | |
| N stage | .209 | ||
| N0 | 30 | 11 | |
| N1 | 57 | 35 | |
| Differentiation | .826 | ||
| Well | 17 | 7 | |
| Fairly | 32 | 18 | |
| Poorly | 38 | 21 | |
| Clinical Stage | .068 | ||
| III | 54 | 36 | |
| IVa | 33 | 10 | |
| Treatment modality | .914 | ||
| RT alone | 10 | 5 | |
| Chemoradiotherapy | 77 | 41 | |
| Tumour Length (cm) | .982 | ||
| ≤5 | 49 | 26 | |
| >5 | 38 | 20 |
RT: radiation therapy.
3.2. Sub-region cluster and feature extraction
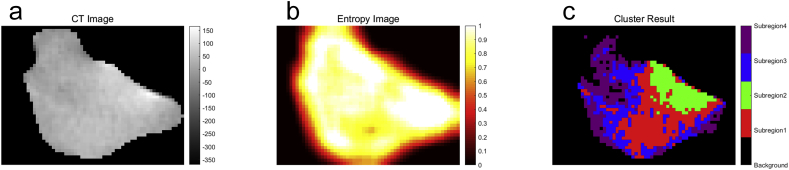
The selection process of the best number of clusters was shown in (Supplementary Fig. S4). When the number of clusters is four, the CH value reached the optimal in the training cohort. Thus, the tumour region was clustered into four sub-regions, as shown in Fig. 2c. Finally, 548 whole-tumour radiomics features and 2192 sub-regional radiomics features were extracted from each patient.
Fig. 2.
(a) The tumour region in the CT images; (b) The local entropy image. (c) The cluster results.
3.3. Feature selection result
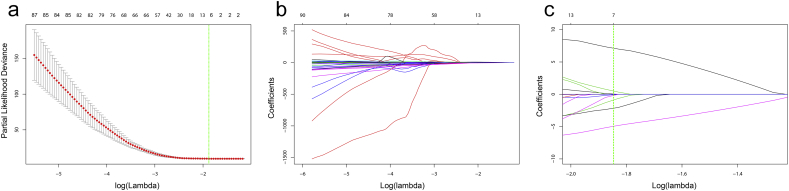
None of radiomics feature were found to have high correlation (CC > 0.75) with the tumour volume. Three hundred and forty features remained after the internal correlation exclusion. The best lambda was set as 0.157851, with log(lambda) = −1.846106, as shown in Fig. 3a. The feature selection process in the LASSO method was shown in Fig. 3b and c. Seven sub-regional radiomics features with non-zero coefficient at the best lambda were selected in the LASSO method. The detailed information of the seven sub-regional features was described in Supplementary Table S1. The corresponding coefficient in LASSO and the p-value in the multivariable analysis for each selected feature were summarized in Supplementary Table S2. Four features are from sub-region 1, two from sub-region 3, and one from sub-region 4. Features Subregion1_LLL_GLCM_inf2h, Subregion1_HHL_Histogram_Uniformity, and Subregion3_LLL_GLCM_inf1h are all independent prognostic factors in the multivariable analysis. The radiomics score was constructed by summing the seven features multiplied with their corresponding coefficient in LASSO.
Fig. 3.
(a) The change of partial likelihood deviance with responding to the change of lambda in the cross-validation process. The green line showed the optimal lambda in the LASSO method with the least partial likelihood deviance. (b) The change of coefficients of each feature in the LASSO method with responding to the change of lambda. (c) The zoomed view of the coefficient change. The green line showed the optimal lambda. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.4. Sub-regional radiomics survival prediction model
With the coefficient of each radiomics feature in the LASSO method, the radiomics score can be formulated as:
| (2) |
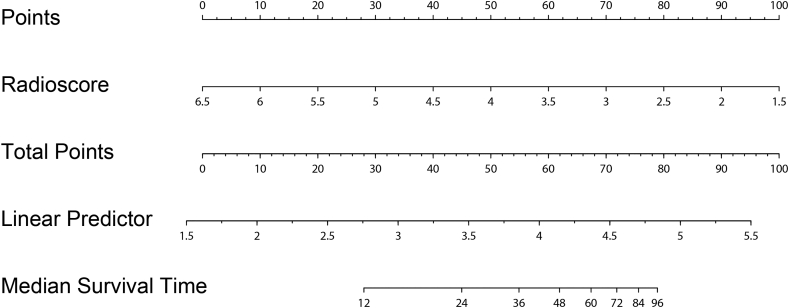
A constant value 22.5 was added to the formula to make positive radiomics score values. The radiomics survival prediction model was constructed based on the radiomics score. The constructed nomogram was shown in Fig. 5.
Fig. 5.
The constructed radiomics nomogram in this study.
The predictive performance of the model was tested in the training set and validation set. The C-index was 0.729 (0.656–0.801, 95% CI) and 0.705 (0.628–0.782, 95%CI) in the two cohorts, respectively. Our evaluation result reflected good prognostic value of the developed model. High C-index value in the validation cohort showed survival prediction feasibility of the model for test cohorts.
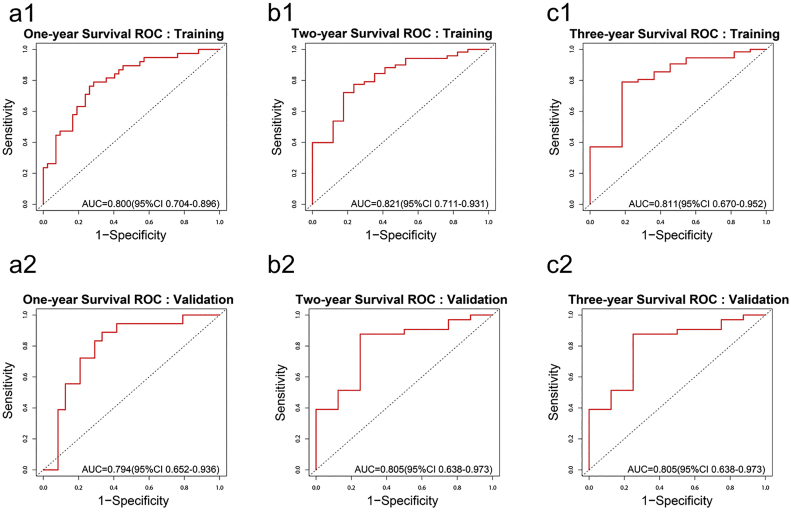
As shown in Fig. 4, the 1-year survival ROC curve showed an AUC of 0.800 (0.704–0.896, 95%CI) in the training cohort and 0.794 (0.652–0.936, 95%CI) in the validation cohort. The stratified high-risk and low-risk group showed significant difference of survival in both the training (p-value < .001, Log-rank test) and validation (p-value < .001, Log-rank test), as shown in Supplementary Fig. S5. The 2-year survival ROC curve showed an AUC of 0.821 (0.711–0.931, 95%CI) in training and 0.805 (0.638–0.973, 95%CI) in validation. The stratified high-risk and low-risk group showed significant difference of survival in both training (p-value < .001, Log-rank test) and validation (p-value = .012, Log-rank test). The 3-year survival ROC curve showed an AUC of 0.811 (0.670–0.952, 95%CI) in training and 0.805 (0.638–0.973, 95%CI) in validation. The stratified high-risk and low-risk group showed significant difference of survival in both the training cohort (p-value < .001, Log-rank test) and validation cohort (p-value < .001, Log-rank test). Results showed clinical utility of the developed model in stratifying high-risk patients regarding 1-year survival, 2-year survival and 3-year survival for treatment strategies adjustment.
Fig. 4.
ROC curves for 1-year (a), 2-year (b) and 3-year (c) survival prediction using developed radiomics survival prediction model in the training cohort (a1, b1, c1) and validation cohort (a2, b2, c2).
3.5. Clinical and biological association
The correlation analysis showed that subregion1_HHL_Uniformity was significantly correlated with tumour length (Fig. S6). Subregion1_HHL_SZE and subregion3_HHH_corrm were significantly correlated with ECOG PS. Subregion3_LLL_inf1h was correlated with both clinical stage and tumour length. Subregion4_Contrast was correlated with the tumour length and ECOG PS.
One hundred and ninety-five CNAs showed non-zero variance among patients. Thirty-one CNAs showed significant correlation with at least one of the selected radiomics features (Fig. S7). The correlation analysis showed the underlying relationship between clinical factors, gene expression and radiography characteristics in oesophageal tumours.
3.6. Comparison with whole-tumour based radiomics model
The WRPM were established by the following radiomics score:
| (3) |
The WRPM showed a C-index of 0.704 (0.637–0.770, 95%CI) in the training cohort and 0.601 (0.531–0.672, 95%CI) in the validation cohort. The C-index of WRPM was lower than that of SRPM, while the statistical analysis showed non-significant difference between the C-indexes of WRPM and SRPM (p-value = .754[Training]; p-value = .982[Validation], t-test).
3.7. Comparison with clinical model
The clinical stage showed a C-index of 0.510 (0.364–0.656, 95%CI) in the training cohort and 0.515 (0.272–0.756, 95%CI) in the validation cohort. The ECOG PS showed a C-index of 0.528 (0.401–0.655, 95%CI) in the training cohort and 0.578 (0.417–0.740, 95%CI) in the validation cohort. Both clinical stage and ECOG PS showed significantly lower C-index than SRPM in the training cohort (p-value = .006 [clinical stage]; p-value = .004 [ECOG PS], t-test). In the validation cohort, no significant difference was observed at 95% confidence interval for clinical stage (p-value = .059, t-test) and ECOG PS (p-value = .061, t-test). When the confidence interval is at 90%,the difference has statistically significance.
4. Discussion
In this study, we performed a sub-region cluster within the tumour region on planning CT. A survival prediction model for OSCC was then constructed based on the sub-regional radiomics features. The constructed sub-regional radiomics survival prediction model showed high prognostic value in both the training cohort (C-index = 0.729 (0.656–0.801, 95% CI)) and validation cohort (C-index = 0.705 (0.628–0.782, 95%CI)). Consistent results on the validation cohort showed utility of this model in independent datasets. The developed model can provide powerful prognostic information to clinicians before radiation therapy and help tailor treatment strategy for patients. For patients with high risk as evaluated by our prediction model the outcome is dismal with chemoradiotherapy and chemoradiotherapy in combination with target drug or immunotherapy are suggested as shown in many clinical trials [[26], [27], [28]].
We also assessed the correlation between sub-regional radiomics features and CNAs or clinical factors. The sub-regional radiomics features showed potential in reflecting gene expression. The SRPM outperformed the WRPM regarding C-index in both datasets, while not statistically significant. The SRPM showed superior prognostic value than conventional clinical factors, clinical stage and ECOG PS and the difference was statistically significant at 90% confidence level. The encouraging prognostic value of CT-based radiomics may shed light on the pre-radiotherapy tumour evaluation and prognosis of OSCC.
The sub-region cluster performed in this study demonstrated the importance of the sub-region analysis in reflecting tumour heterogeneity of OSCC. This result is consistent with previous studies which suggested the prognostic role of sub-regional heterogeneity [12]. The correlation with clinical factors and gene expression showed the potential of sub-regional radiomics features in reflecting tumour biological behaviour. Segal et.al found that genomic activity of human liver cancers can be decoded by non-invasive imaging, namely radiomics features [29]. Combinations of 28 imaging traits can reconstruct 78% of the global gene expression profiles, revealing cell proliferation, liver synthetic function, and patient prognosis. This study showed the potential of radiomics features to reflect gene expression. Also, Hugo et.al suggested that radiomic signature, capturing intratumor heterogeneity, is associated with underlying gene-expression patterns in lung cancer [30]. In the study of Xia et.al, eight sub-regional CT radiomics features were significantly correlated with prognostic gene modules in hepatocellular carcinoma [14]. In the present study, we found that, in oesophageal cancers, 31CNAs showed significant correlation with at least one of the selected radiomics features, which is consistent with previous studies. It may be of interest for future studies to combine radiomics with genomics in the clinical practice and demonstrate the underlying biological pathway for radiomics [31]. The sub-regional radiomics analysis method may better quantify the tumour sub-region which was more correlated with the tumour growth or aggressiveness [12]. A seven-feature based radiomics score was constructed in this study including six wavelet-based radiomics features showing the importance of wavelet decomposition in the radiomics analysis. The wavelet features characterized the heterogeneity at multiple spatial scales which may help quantify the significant radiomics features which were prognostic in OSCC [19,32].
For OSCC patients enrolled in the present study, endoscopic ultrasound (EUS) and CT were both used to for staging. EUS combined with CT has proven to provide excellent sensitivity and specificity in the T stage of oesophageal cancer in a series of studies [33,34]. Currently, the CT-based sub-region radiomics model in this study could apply for OS prediction in these patients and suggestion for subsequent treatment choices.
Additionally, we did whole-exome sequencing for part of the biopsy specimens during our preliminary experiment and the results indicated that CNAs were the only significant predictor for OS in the studied cohort. In literature, Wang et.al demonstrated that other potential biomarkers identified by whole-exome and whole-genome sequencing are also significantly correlated with EC prognosis [35]. For OSCC patients with high risks identified in the current study, definitive concurrent chemoradiotherapy combined with targeted therapy might be a useful treatment option for these patients. Based on our previous phase III study, patients in the definitive concurrent chemoradiotherapy and Erlotinib (EGFR-TKI) group had a median OS time of 24.9 months compared to 20.9 months in non-erlotinib group (p-value = .05) [36].
There are also some limitations in this study. This is a retrospective study, and the validation cohort is of small size. We did not include other imaging types that normally would be used in clinical practice, i.e. PET. Due to the lack of more biological information of patients, we did not further assess the correlation of developed radiomics model with other biological mechanisms for OSCC. In addition, we did not include superficial oesophageal cancer in our study as it is usually treated by endoscopic submucosal dissection in routine practice. Our future research is to explore the radio-genomics model and prospectively validate the proposed model. In addition, we will further assess the tumour change during radiation therapy regarding our developed radiomics score, thus verify the established model.
5. Conclusion
We developed and validated a sub-regional radiomics survival prediction model for OSCC patients. The established model showed high accuracy in improving personalized treatment for OSCC patients. The sub-regional radiomics analysis showed encouraging potential in predicting survival of OSCC treated by radiotherapy and needed to be further confirmed in larger cohorts.
Grants and acknowledgements
The authors would like to thank Dr. Yi Rong for helpful revisement of this manuscript. This work was supported by the Zhejiang Provincial Foundation for Natural Sciences (LZ15H220001: S. Wu), National Natural Science Foundation of China (81672994:S. Wu), Zhejiang Provincial Natural Science Foundation of China (LR16F010001: T. Niu), Zhejiang University Education Foundation ZJU-Stanford Collaboration Fund (T. Niu), National Key Research Plan by the Ministry of Science and Technology of China (2016YFC0104507: T. Niu), Natural Science Foundation of China (81871351: T. Niu), Hangzhou Agriculture and Social Developmental Research Program (20172016A04: S. Wu). The founders had no role in study design, data collection, data analysis, interpretation, writing of the manuscript.
Conflicts of interest
The authors declare no competing financial interests.
Author contributions
Conception and design: T. Niu, S. Wu.
Development of methodology: P. Yang, L. Xu, T. Niu.
Analysis and interpretation of data: P. Yang, X. Wang, L. Yang.
Writing, review, and/or revision of the manuscript: C. Xie, P. Yang, Y. Kuang, T. Niu, S. Wu.
Administrative, technical, or material support: C. Xie, P. Yang, X. Zhang, L. Xu, X. Wang, X. Li, L. Zhang, R. Xie, L. Yang, Z. Jing, H. Zhang, L. Ding, Y. Kuang, T. Niu, S. Wu.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.05.023.
Contributor Information
Tianye Niu, Email: tyniu@zju.edu.cn.
Shixiu Wu, Email: wushixiu@medmail.com.cn.
Appendix A. Supplementary data
Supplementary material
References
- 1.Chen M., Huang J., Zhu Z., Zhang J., Li K. Systematic review and meta-analysis of tumor biomarkers in predicting prognosis in esophageal cancer. BMC Cancer. 2013;13:539. doi: 10.1186/1471-2407-13-539. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chen M, Huang J, Zhu Z, Zhang J, Li K. Systematic review and meta-analysis of tumor biomarkers in predicting prognosis in esophageal cancer. BMC cancer. 2013;13:539. [DOI] [PMC free article] [PubMed]
- 2.Li Y., Lin Q., Luo Z., Zhao L., Zhu L., Sun L. Value of sequential 18F-fluorodeoxyglucose positron emission tomography/computed tomography (FDG PET/CT) in prediction of the overall survival of esophageal cancer patients treated with chemoradiotherapy. Int J Clin Exp Med. 2015;8(7):10947–10955. [PMC free article] [PubMed] [Google Scholar]; Li Y, Lin Q, Luo Z, Zhao L, Zhu L, Sun L, et al. Value of sequential 18F-fluorodeoxyglucose positron emission tomography/computed tomography (FDG PET/CT) in prediction of the overall survival of esophageal cancer patients treated with chemoradiotherapy. International journal of clinical and experimental medicine. 2015;8(7):10947-55. [PMC free article] [PubMed]
- 3.Atsumi K., Nakamura K., Abe K., Hirakawa M., Shioyama Y., Sasaki T. Prediction of outcome with FDG-PET in definitive chemoradiotherapy for esophageal cancer. J Radiat Res. 2013;54(5):890–898. doi: 10.1093/jrr/rrt021. [DOI] [PMC free article] [PubMed] [Google Scholar]; Atsumi K, Nakamura K, Abe K, Hirakawa M, Shioyama Y, Sasaki T, et al. Prediction of outcome with FDG-PET in definitive chemoradiotherapy for esophageal cancer. Journal of Radiation Research. 2013;54(5):890-8. [DOI] [PMC free article] [PubMed]
- 4.Lindner K., Palmes D., Senninger N., Hummel R. PET/CT predicts survival in patients undergoing primary surgery for esophageal cancer. Langenbecks Arch Surg. 2015;400(2):229–235. doi: 10.1007/s00423-014-1264-9. [DOI] [PubMed] [Google Scholar]; Lindner K, Palmes D, Senninger N, Hummel R. PET/CT predicts survival in patients undergoing primary surgery for esophageal cancer. Langenbeck's archives of surgery. 2015;400(2):229-35. [DOI] [PubMed]
- 5.Myslivecek M., Neoral C., Vrba R., Vomackova K., Cincibuch J., Formanek R. The value of (1)(8)F-FDG PET/CT in assessment of metabolic response in esophageal cancer for prediction of histopathological response and survival after preoperative chemoradiotherapy. Biomedical papers of the Medical Faculty of the University Palacky, Olomouc. Czechoslovakia. 2012;156(2):171–179. doi: 10.5507/bp.2011.048. [DOI] [PubMed] [Google Scholar]; .Myslivecek M, Neoral C, Vrba R, Vomackova K, Cincibuch J, Formanek R, et al. The value of (1)(8)F-FDG PET/CT in assessment of metabolic response in esophageal cancer for prediction of histopathological response and survival after preoperative chemoradiotherapy. Biomedical papers of the Medical Faculty of the University Palacky, Olomouc, Czechoslovakia. 2012;156(2):171-9. [DOI] [PubMed]
- 6.Yip C., Landau D., Kozarski R., Ganeshan B., Thomas R., Michaelidou A. Primary esophageal cancer: heterogeneity as potential prognostic biomarker in patients treated with definitive chemotherapy and radiation therapy. Radiology. 2014;270(1):141–148. doi: 10.1148/radiol.13122869. [DOI] [PubMed] [Google Scholar]; Yip C, Landau D, Kozarski R, Ganeshan B, Thomas R, Michaelidou A, et al. Primary esophageal cancer: heterogeneity as potential prognostic biomarker in patients treated with definitive chemotherapy and radiation therapy. Radiology. 2014;270(1):141-8. [DOI] [PubMed]
- 7.Beukinga R.J., Hulshoff J.B., van Dijk L.V., Muijs C.T., Burgerhof J.G.M., Kats-Ugurlu G. Predicting response to neoadjuvant chemoradiotherapy in esophageal cancer with textural features derived from pretreatment (18)F-FDG PET/CT imaging. J Nucl Med. 2017;58(5):723–729. doi: 10.2967/jnumed.116.180299. [DOI] [PubMed] [Google Scholar]; Beukinga RJ, Hulshoff JB, van Dijk LV, Muijs CT, Burgerhof JGM, Kats-Ugurlu G, et al. Predicting Response to Neoadjuvant Chemoradiotherapy in Esophageal Cancer with Textural Features Derived from Pretreatment (18)F-FDG PET/CT Imaging. Journal of nuclear medicine : official publication, Society of Nuclear Medicine. 2017;58(5):723-9. [DOI] [PubMed]
- 8.Hou Z., Ren W., Li S., Liu J., Sun Y., Yan J. Radiomic analysis in contrast-enhanced CT: predict treatment response to chemoradiotherapy in esophageal carcinoma. Oncotarget. 2017;8(61):104444–104454. doi: 10.18632/oncotarget.22304. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hou Z, Ren W, Li S, Liu J, Sun Y, Yan J, et al. Radiomic analysis in contrast-enhanced CT: predict treatment response to chemoradiotherapy in esophageal carcinoma. Oncotarget. 2017;8(61):104444-54. [DOI] [PMC free article] [PubMed]
- 9.Ganeshan B., Abaleke S., Young R.C.D., Chatwin C.R., Miles K.A. Texture analysis of non-small cell lung cancer on unenhanced computed tomography: initial evidence for a relationship with tumour glucose metabolism and stage. Cancer Imaging. 2010;10(1):137–143. doi: 10.1102/1470-7330.2010.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ganeshan B, Abaleke S, Young RCD, Chatwin CR, Miles KA. Texture analysis of non-small cell lung cancer on unenhanced computed tomography: initial evidence for a relationship with tumour glucose metabolism and stage. Cancer Imaging. 2010;10(1):137-43. [DOI] [PMC free article] [PubMed]
- 10.Ganeshan B., Skogen K., Pressney I., Coutroubis D., Miles K. Tumour heterogeneity in oesophageal cancer assessed by CT texture analysis: preliminary evidence of an association with tumour metabolism, stage, and survival. Clin Radiol. 2012;67(2):157–164. doi: 10.1016/j.crad.2011.08.012. [DOI] [PubMed] [Google Scholar]; Ganeshan B, Skogen K, Pressney I, Coutroubis D, Miles K. Tumour heterogeneity in oesophageal cancer assessed by CT texture analysis: Preliminary evidence of an association with tumour metabolism, stage, and survival. Clinical radiology. 2012;67(2):157-64. [DOI] [PubMed]
- 11.Gatenby R.A., Grove O., Gillies R.J. Quantitative imaging in cancer evolution and ecology. Radiology. 2013;269(1):8–15. doi: 10.1148/radiol.13122697. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gatenby RA, Grove O, Gillies RJ. Quantitative imaging in cancer evolution and ecology. Radiology. 2013;269(1):8-15. [DOI] [PMC free article] [PubMed]
- 12.Wu J., Cao G., Sun X., Lee J., Rubin D.L., Napel S. Intratumoral spatial heterogeneity at perfusion MR imaging predicts recurrence-free survival in locally advanced breast cancer treated with neoadjuvant chemotherapy. Radiology. 2018;288(1):26–35. doi: 10.1148/radiol.2018172462. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wu J, Cao G, Sun X, Lee J, Rubin DL, Napel S, et al. Intratumoral Spatial Heterogeneity at Perfusion MR Imaging Predicts Recurrence-free Survival in Locally Advanced Breast Cancer Treated with Neoadjuvant Chemotherapy. Radiology. 2018;288(1):26-35. [DOI] [PMC free article] [PubMed]
- 13.Wu J., Gensheimer M.F., Dong X., Rubin D.L., Napel S., Diehn M. Robust Intratumor partitioning to identify high-risk subregions in lung cancer: a pilot study. Int J Radiat Oncol Biol Phys. 2016;95(5):1504–1512. doi: 10.1016/j.ijrobp.2016.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wu J, Gensheimer MF, Dong X, Rubin DL, Napel S, Diehn M, et al. Robust Intratumor Partitioning to Identify High-Risk Subregions in Lung Cancer: A Pilot Study. International journal of radiation oncology, biology, physics. 2016;95(5):1504-12. [DOI] [PMC free article] [PubMed]
- 14.Xia W., Chen Y., Zhang R., Yan Z., Zhou X., Zhang B. Radiogenomics of hepatocellular carcinoma: multiregion analysis-based identification of prognostic imaging biomarkers by integrating gene data-a preliminary study. Phys Med Biol. 2018;63(3) doi: 10.1088/1361-6560/aaa609. 035044. [DOI] [PubMed] [Google Scholar]; . Xia W, Chen Y, Zhang R, Yan Z, Zhou X, Zhang B, et al. Radiogenomics of hepatocellular carcinoma: multiregion analysis-based identification of prognostic imaging biomarkers by integrating gene data-a preliminary study. Physics in medicine and biology. 2018;63(3):035044. [DOI] [PubMed]
- 15.Liu Y., Li Z., Xiong H., Gao X., Wu J., Wu S. Understanding and enhancement of internal clustering validation measures. IEEE Trans Cybern. 2013;43(3):982–994. doi: 10.1109/TSMCB.2012.2220543. [DOI] [PubMed] [Google Scholar]; Liu Y, Li Z, Xiong H, Gao X, Wu J, Wu S. Understanding and enhancement of internal clustering validation measures. IEEE transactions on cybernetics. 2013;43(3):982-94. [DOI] [PubMed]
- 16.Larue R., van Timmeren J.E., de Jong E.E.C., Feliciani G., Leijenaar R.T.H., Schreurs W.M.J. Influence of gray level discretization on radiomic feature stability for different CT scanners, tube currents and slice thicknesses: a comprehensive phantom study. Acta Oncol. 2017;56(11):1544–1553. doi: 10.1080/0284186X.2017.1351624. [DOI] [PubMed] [Google Scholar]; Larue R, van Timmeren JE, de Jong EEC, Feliciani G, Leijenaar RTH, Schreurs WMJ, et al. Influence of gray level discretization on radiomic feature stability for different CT scanners, tube currents and slice thicknesses: a comprehensive phantom study. Acta oncologica (Stockholm, Sweden). 2017;56(11):1544-53. [DOI] [PubMed]
- 17.Vallieres M., Freeman C.R., Skamene S.R. El Naqa I. a radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities. Phys Med Biol. 2015;60(14):5471–5496. doi: 10.1088/0031-9155/60/14/5471. [DOI] [PubMed] [Google Scholar]; Vallieres M, Freeman CR, Skamene SR, El Naqa I. A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities. Physics in medicine and biology. 2015;60(14):5471-96. [DOI] [PubMed]
- 18.Parada S.A., Eichinger J.K., Dumont G.D., Parada C.A., Greenhouse A.R., Provencher M.T. Accuracy and reliability of a simple calculation for measuring glenoid bone loss on 3-dimensional computed tomography scans. Arthroscopy. 2018;34(1):84–92. doi: 10.1016/j.arthro.2017.07.032. [DOI] [PubMed] [Google Scholar]; Parada SA, Eichinger JK, Dumont GD, Parada CA, Greenhouse AR, Provencher MT, et al. Accuracy and Reliability of a Simple Calculation for Measuring Glenoid Bone Loss on 3-Dimensional Computed Tomography Scans. Arthroscopy: The Journal of Arthroscopic & Related Surgery. 2018;34(1):84-92. [DOI] [PubMed]
- 19.Huang Y., Liu Z., He L., Chen X., Pan D., Ma Z. Radiomics signature: a potential biomarker for the prediction of disease-free survival in early-stage (I or II) non-small cell lung cancer. Radiology. 2016;281(3):947–957. doi: 10.1148/radiol.2016152234. [DOI] [PubMed] [Google Scholar]; Huang Y, Liu Z, He L, Chen X, Pan D, Ma Z, et al. Radiomics Signature: A Potential Biomarker for the Prediction of Disease-Free Survival in Early-Stage (I or II) Non-Small Cell Lung Cancer. Radiology. 2016;281(3):947-57. [DOI] [PubMed]
- 20.Liang W., Xu L., Yang P., Zhang L., Wan D., Huang Q. Novel nomogram for preoperative prediction of early recurrence in intrahepatic cholangiocarcinoma. Front Oncol. 2018;8:360. doi: 10.3389/fonc.2018.00360. [DOI] [PMC free article] [PubMed] [Google Scholar]; Liang W, Xu L, Yang P, Zhang L, Wan D, Huang Q, et al. Novel Nomogram for Preoperative Prediction of Early Recurrence in Intrahepatic Cholangiocarcinoma. Frontiers in oncology. 2018;8:360. [DOI] [PMC free article] [PubMed]
- 21.Park H., Lim Y., Ko E.S., Cho H.-H., Lee J.E., Han B.-K. Radiomics signature on magnetic resonance imaging: association with disease-free survival in patients with invasive breast cancer. Clin Cancer Res. 2018;24(19):4705–4714. doi: 10.1158/1078-0432.CCR-17-3783. [DOI] [PubMed] [Google Scholar]; Park H, Lim Y, Ko ES, Cho H-H, Lee JE, Han B-K, et al. Radiomics Signature on Magnetic Resonance Imaging: Association with Disease-Free Survival in Patients with Invasive Breast Cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2018;24(19):4705-14. [DOI] [PubMed]
- 22.Hu T., Wang S., Huang L., Wang J., Shi D., Li Y. A clinical-radiomics nomogram for the preoperative prediction of lung metastasis in colorectal cancer patients with indeterminate pulmonary nodules. Eur Radiol. 2019;29(1):439–449. doi: 10.1007/s00330-018-5539-3. [DOI] [PubMed] [Google Scholar]; Hu T, Wang S, Huang L, Wang J, Shi D, Li Y, et al. A clinical-radiomics nomogram for the preoperative prediction of lung metastasis in colorectal cancer patients with indeterminate pulmonary nodules. European radiology. 2018. [DOI] [PubMed]
- 23.Wu Y., Xu L., Yang P., Lin N., Huang X., Pan W. Survival prediction in high-grade osteosarcoma using radiomics of diagnostic computed tomography. EBioMedicine. 2018;34:27–34. doi: 10.1016/j.ebiom.2018.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wu Y, Xu L, Yang P, Lin N, Huang X, Pan W, et al. Survival Prediction in High-grade Osteosarcoma Using Radiomics of Diagnostic Computed Tomography. EBioMedicine. 2018;34:27-34. [DOI] [PMC free article] [PubMed]
- 24.Li H., Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]; Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics (Oxford, England). 2009;25(14):1754-60. [DOI] [PMC free article] [PubMed]
- 25.Klambauer G., Schwarzbauer K., Mayr A., Clevert D.A., Mitterecker A., Bodenhofer U. cn.MOPS: mixture of Poissons for discovering copy number variations in next-generation sequencing data with a low false discovery rate. Nucleic Acids Res. 2012;40(9):e69. doi: 10.1093/nar/gks003. [DOI] [PMC free article] [PubMed] [Google Scholar]; Klambauer G, Schwarzbauer K, Mayr A, Clevert DA, Mitterecker A, Bodenhofer U, et al. cn.MOPS: mixture of Poissons for discovering copy number variations in next-generation sequencing data with a low false discovery rate. Nucleic acids research. 2012;40(9):e69. [DOI] [PMC free article] [PubMed]
- 26.Wald O., Smaglo B., Mok H., Groth S.S. Future directions in esophageal cancer therapy. Ann Cardiothorac Surg. 2017;6(2):159–166. doi: 10.21037/acs.2017.02.01. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wald O, Smaglo B, Mok H, Groth SS. Future directions in esophageal cancer therapy. Annals of cardiothoracic surgery. 2017;6(2):159-66. [DOI] [PMC free article] [PubMed]
- 27.Abdo J., Agrawal D.K., Mittal S.K. Targeted” chemotherapy for esophageal cancer. Front Oncol. 2017;7:63. doi: 10.3389/fonc.2017.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]; Abdo J, Agrawal DK, Mittal SK. "Targeted" Chemotherapy for Esophageal Cancer. Frontiers in oncology. 2017;7:63-. [DOI] [PMC free article] [PubMed]
- 28.Bolm L., Kasmann L., Paysen A., Karapetis C., Rades D., Wellner U.F. Multimodal anti-tumor approaches combined with immunotherapy to overcome tumor resistance in esophageal and gastric cancer. Anticancer Res. 2018;38(6):3231–3242. doi: 10.21873/anticanres.12588. [DOI] [PubMed] [Google Scholar]; Bolm L, Kasmann L, Paysen A, Karapetis C, Rades D, Wellner UF, et al. Multimodal Anti-tumor Approaches Combined with Immunotherapy to Overcome Tumor Resistance in Esophageal and Gastric Cancer. Anticancer research. 2018;38(6):3231-42. [DOI] [PubMed]
- 29.Segal E., Sirlin C.B., Ooi C., Adler A.S., Gollub J., Chen X. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat Biotechnol. 2007;25(6):675–680. doi: 10.1038/nbt1306. [DOI] [PubMed] [Google Scholar]; Segal E, Sirlin CB, Ooi C, Adler AS, Gollub J, Chen X, et al. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nature biotechnology. 2007;25(6):675-80. [DOI] [PubMed]
- 30.Aerts H.J., Velazquez E.R., Leijenaar R.T., Parmar C., Grossmann P., Carvalho S. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]; Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nature communications. 2014;5:4006. [DOI] [PMC free article] [PubMed]
- 31.Grossmann P., Stringfield O., El-Hachem N., Bui M.M., Rios Velazquez E., Parmar C. Defining the biological basis of radiomic phenotypes in lung cancer. eLife. 2017:6. doi: 10.7554/eLife.23421. [DOI] [PMC free article] [PubMed] [Google Scholar]; Grossmann P, Stringfield O, El-Hachem N, Bui MM, Rios Velazquez E, Parmar C, et al. Defining the biological basis of radiomic phenotypes in lung cancer. eLife. 2017;6. [DOI] [PMC free article] [PubMed]
- 32.Wilson R., Devaraj A. Radiomics of pulmonary nodules and lung cancer. Transl Lung Cancer Res. 2017;6(1):86–91. doi: 10.21037/tlcr.2017.01.04. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wilson R, Devaraj A. Radiomics of pulmonary nodules and lung cancer. Transl Lung Cancer Res. 2017;6(1):86-91. [DOI] [PMC free article] [PubMed]
- 33.Puli S.R., Reddy J.B., Bechtold M.L., Antillon D., Ibdah J.A., Antillon M.R. Staging accuracy of esophageal cancer by endoscopic ultrasound: a meta-analysis and systematic review. World J Gastroenterol. 2008;14(10):1479–1490. doi: 10.3748/wjg.14.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]; Puli SR, Reddy JB, Bechtold ML, Antillon D, Ibdah JA, Antillon MR. Staging accuracy of esophageal cancer by endoscopic ultrasound: a meta-analysis and systematic review. World journal of gastroenterology. 2008;14(10):1479-90. [DOI] [PMC free article] [PubMed]
- 34.Thosani N., Singh H., Kapadia A., Ochi N., Lee J.H., Ajani J. Diagnostic accuracy of EUS in differentiating mucosal versus submucosal invasion of superficial esophageal cancers: a systematic review and meta-analysis. Gastrointest Endosc. 2012;75(2):242–253. doi: 10.1016/j.gie.2011.09.016. [DOI] [PubMed] [Google Scholar]; Thosani N, Singh H, Kapadia A, Ochi N, Lee JH, Ajani J, et al. Diagnostic accuracy of EUS in differentiating mucosal versus submucosal invasion of superficial esophageal cancers: a systematic review and meta-analysis. Gastrointestinal endoscopy. 2012;75(2):242-53. [DOI] [PubMed]
- 35.Wang X., Li X., Cheng Y., Sun X., Sun X., Self S. Copy number alterations detected by whole-exome and whole-genome sequencing of esophageal adenocarcinoma. Hum Genomics. 2015;9:22. doi: 10.1186/s40246-015-0044-0. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wang X, Li X, Cheng Y, Sun X, Sun X, Self S, et al. Copy number alterations detected by whole-exome and whole-genome sequencing of esophageal adenocarcinoma. Human genomics. 2015;9:22. [DOI] [PMC free article] [PubMed]
- 36.Wu S.X., Wang L.H., Luo H.L., Xie C.Y., Zhang X.B., Hu W. Randomised phase III trial of concurrent chemoradiotherapy with extended nodal irradiation and erlotinib in patients with inoperable oesophageal squamous cell cancer. Eur J Cancer. 2018;93:99–107. doi: 10.1016/j.ejca.2018.01.085. [DOI] [PubMed] [Google Scholar]; .Wu SX, Wang LH, Luo HL, Xie CY, Zhang XB, Hu W, et al. Randomised phase III trial of concurrent chemoradiotherapy with extended nodal irradiation and erlotinib in patients with inoperable oesophageal squamous cell cancer. European journal of cancer (Oxford, England : 1990). 2018;93:99-107. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material