Abstract
Background
Pancreatic ductal adenocarcinoma (PDAC) is an aggressive disease with poor prognosis, and gemcitabine-based chemotherapy remains an effective option for the majority of PDAC patients. Hepatocyte nuclear factor 1α (HNF1A) is a tumor-suppressor in PDAC, but its role in gemcitabine chemoresistance of PDAC has not been clarified.
Methods
The function of HNF1A in gemcitabine was detected by overexpression and knockdown of HNF1A in vitro and in vitro. The regulatory network between HNF1A and ABCB1 was further demonstrated by luciferase assays, deletion/mutation reporter construct assays and CHIP assays.
Findings
Here, we found that HNF1A expression is significantly associated with gemcitabine sensitivity in PDAC cell lines. Moreover, we identified that HNF1A overexpression enhanced gemcitabine sensitivity of PDAC both in vitro and in vitro, while inhibition of HNF1A had the opposite effect. Furthermore, by inhibiting and overexpressing HNF1A, we revealed that HNF1A regulates the expression of MDR genes (ABCB1 and ABCC1) in PDAC cells. Mechanistically, we demonstrated that HNF1A regulates ABCB1 expression through binding to its specific promoter region and suppressing its transcription levels. Finally, the survival analyses revealed the clinical value of HNF1A in stratification of gemcitabine sensitive pancreatic cancer patients.
Interpretation
Our study paved the road for finding novel treatment combinations using conventional cytotoxic agents with functional restoration of the HNF1A protein, individualized treatment through HNF1A staining and improvement of the prognosis of PDAC patients.
Fund
National Natural Science Foundations of China and National Natural Science Foundation of Guangdong Province.
Keywords: Pancreatic ductal adenocarcinoma, HNF1A, Gemcitabine, Chemotherapy resistance, ABCB1
Research in context.
Evidence before this study
Pancreatic ductal adenocarcinoma (PDAC) is an aggressive disease with rapid tumor progression and poor prognosis, which is highly refractory to systemic treatment. A low response rate to gemcitabine is common in the clinic and has been attributed to drug uptake, metabolism and action. Indeed, hepatocyte nuclear factor 1α (HNF1A) has been identified as being associated with the risk of pancreatic cancer and was previously reported as a tumor suppressor in several cancers, including PDAC. Moreover, a recent study showed that stratifying molecular subtypes of PDAC using immunohistochemical methods and HNF1A is associated with significantly different outcomes and responses to chemotherapy. Additionally, gemcitabine resistance in pancreatic cancer cell lines was attributed to the enhanced expression of multidrug resistant genes. Studies on the upstream regulation of ABC transporters are lacking, and the association between HNF1A and ABC transporters in mediating resistance to gemcitabine in PDAC patients has not been elucidated.
Added value of this study
HNF1A expression is significantly associated with gemcitabine sensitivity in PDAC cell lines. By using a gain- and loss-of-functional model, we identified that HNF1A overexpression enhanced gemcitabine sensitivity of PDAC both in vitro and in vivo, while inhibition of HNF1A had the opposite effect. Furthermore, we revealed that HNF1A regulates the expression of multidrug resistance genes (ABCB1 and ABCC1) in PDAC cells. Mechanistically, we demonstrated that HNF1A regulates ABCB1 expression through binding to its specific promoter region and suppressing its transcription levels. Finally, the survival analyses revealed the clinical value of HNF1A in the stratification of gemcitabine sensitive pancreatic cancer patients.
Implications of all the available evidence
A new role of HNF1A in gemcitabine sensitivity of PDAC was identified. HNF1A mediated the resistance of PDAC cells to chemotherapy by directly binding to the specific region of the ABCB1 promoter and regulating its expression. By analyzing of HNF1A staining from PDAC tissues and clinical features, we paved the road for individualized treatment and improved the prognosis of PDAC patients.
Alt-text: Unlabelled Box
1. Introduction
Pancreatic ductal adenocarcinoma (PDAC) remains one of the most lethal diseases with a 5-year survival rate of approximately 5% [1]. More than 80% of PDAC patients lose the opportunity for radical surgery at the time of their diagnosis, which means that chemotherapy is the major treatment for most PDAC patients [2]. Gemcitabine, has been confirmed to be the first effective drug in the treatment of PDAC by inhibiting DNA synthesis and stimulating the apoptosis of cancer cells [3]. Recently, intensified chemotherapy regimens including FOLFIRINOX (fluorouracil, leucovorin, irinotecan, and oxaliplatin) [4], gemcitabine plus Abraxane or albumin-bound paclitaxel [5], and gemcitabine plus capecitabine [6] were shown to significantly improved overall and median progression-free survival. However, the increased toxicity limited the intensified treatment protocol such as FOLFIRINOX to the patients only with a good performance status. Gemcitabine, however, continues to be a part of these therapies (gemcitabine plus Abraxane or albumin-bound paclitaxel, and gemcitabine plus capecitabine). Disappointingly, the low response rate to gemcitabine is common in the clinic, and only <20% of PDAC patients are sensitive to gemcitabine treatment [7]. Thus, the development of a novel therapeutic strategy to explore the mechanisms underlying gemcitabine resistance and enhancement of gemcitabine efficiency in PDAC treatment are still urgently needed.
Gemcitabine, a nucleoside cytidine (pyrimidine), is known to kill cells by incorporating into replicating DNA, thereby inhibiting DNA synthesis and terminating the premature chain. Mechanism of gemcitabine resistance can be attributed to cell plasticity, heterogeneity of the tumor, dense stromal environment, altered metabolism and the regulation of drug influx/efflux [[8], [9], [10]]. However, the exact molecular mechanism underlying gemcitabine resistance in pancreatic cancer has not been fully elucidated. Previous research indicates that hedgehog signaling pathway activating in tumor-associated stromal tissue insensitive pancreatic cancer cells to gemcitabine [10]. Nevertheless, clinical trials combining gemcitabine and hedgehog inhibitor IPI-262 failed to prolong the survival of pancreatic cancer patients. Gemcitabine resistance can be either intrinsic or acquired [11]. One of the principal mechanisms of gemcitabine resistance in pancreatic cancer is the regulation of drug efflux [12].
Multidrug resistance (MDR) is characterized by cross-resistance to chemotherapeutic drugs with target sites. The major mechanisms of gemcitabine chemoresistance are related to drug uptake, metabolism and action. One of the common reasons for MDR in cancer cells is the upregulation of a family of ATP-binding cassette (ABC) transporters [13]. These transporters or drug efflux pumps contribute to MDR by increasing the intracellular drug efflux and reducing the drug accumulation in the cancer cells [13]. Therefore, it is meaningful to clarify the regulatory mechanism of these transporters and target these transporters to restore the sensitivity of chemotherapeutic in pancreatic cancer.
HNF1A, hepatocyte nuclear factor 1 homeobox A, was initially discovered in the liver and subsequently verified to be expressed in several organs including the pancreas, kidney, and intestine [14]. For the roles of HNF1a in pancreas-associated diseases, mutation of HNF1A in the germline has been identified as the cause of maturity onset diabetes of the young 3 (MODY3), which account for 21–64% of all MODY3 cases [15]. In addition, a nonsynonymous variant (rs1169288, I27L) in the HNF1A gene associated with type-2 diabetes risk was reported by a genome wide association study (GWAS) [16]. Interestingly, studies have identified that the HNF1A gene is associated with the risk of pancreatic cancer [17,18], and our previous study and Hoskins et al. previously reported the tumor suppressor role of HNF1A in PDAC [19,20]. As a transcriptional factor, HNF1A has also been shown to play a role not only in the regulation of a series gene involved in drug metabolism but also in the uptake and export of substrates for these enzymes, including the MDR protein family [21]. Moreover, hepatocyte nuclear factor 1 homeobox B (HNF1B), which has a physical interaction and shared pathway with HNF1A, has been reported to contribute to drug resistance in ovarian cancer via its direct interactions with some drug resistance-associated proteins [22]. However, the role of HNF1A in gemcitabine chemoresistance of PDAC has not been clarified.
In this study, we explored the role of HNF1a in the gemcitabine sensitivity of PDAC both in vitro and in vitro. Moreover, we sought to gain a better understanding of the mechanisms by which HNF1A mediates the resistance of PDAC cells to chemotherapy by targeting ABCB1, with the aim of finding novel treatment combinations using conventional cytotoxic agents with functional restoration of HNF1A protein.
2. Materials and methods
2.1. Patients and clinical samples
Between 2012 and 2017, we collected 128 cases of PDAC from the surgical patients in our department. The Hospital's Protection of Human Subjects Committee approved our protocol, and all patients were informed before surgery. All samples were evaluated and histologically diagnosed as PDAC by two certified pathologists (Dr. L Wang and Dr. YJ Zeng, Sun Yat-sen University, Guangdong, China). None of these patients received any preoperative chemotherapy or radiotherapy. The detailed clinical and pathological features of patients are shown in Table 1. The follow-up data were collected completely, and overall survival (OS) was explicitly defined as the time interval beginning on the date of randomization to the date of death or last follow-up evaluation (October 2017), and disease-free time was measured from the date of randomization to the date of the first disease-free failure event occurrence.
Table 1.
Correlation between HNF1A expression and clinicopathologic characteristics of PDAC patients.a
| Characteristics |
N of cases |
HNF1A level |
|||
|---|---|---|---|---|---|
| L | M | H | p-Value | ||
| Total cases | 128 | 70 | 37 | 21 | |
| Gender | 0.920 | ||||
| Male | 75 | 40 | 22 | 13 | |
| Female | 53 | 30 | 15 | 8 | |
| Age | 0.134 | ||||
| <60 | 50 | 22 | 17 | 11 | |
| ≥60 | 78 | 48 | 20 | 10 | |
| Differentiation | 0.230 | ||||
| Well | 20 | 8 | 9 | 3 | |
| Moderate | 80 | 49 | 20 | 11 | |
| Poor | 28 | 13 | 8 | 7 | |
| TNM stage(AJCC)b | 0.004** | ||||
| I | 28 | 11 | 8 | 9 | |
| II | 45 | 20 | 19 | 6 | |
| III | 55 | 39 | 10 | 6 | |
| Lymph-node metastasis | 0.228 | ||||
| Negative | 50 | 32 | 12 | 6 | |
| Positive | 78 | 38 | 25 | 15 | |
| Perineural invasion | 0.012* | ||||
| Negative | 41 | 15 | 15 | 11 | |
| Positive | 87 | 55 | 22 | 10 | |
Abbreviations: N of cases = number of cases; T stage = tumor stage; TNM = tumornode metastasis. L = low, M = moderate, H = high.
Chi-square test, *p < .05, **p < .01.
American Joint Committee on Cancer (AJCC), patients were staged in accordance with the 8th Edition of the AJCC Cancer's' TNM Classification; According to the interpretation of NCCN guidelines for surgical treatment of pancreatic carcinoma, PDAC with stage IV are unresectable.
2.2. Cell culture
Human pancreatic cancer cell lines (BxPC-3, CFPAC-1, L3.6pl, PANC-1, Hs766T, MIA PaCa-2 and Mpanc96) were purchased from American Type Culture Collection (ATCC, Manassas, USA). Cells were maintained in RPMI 1640 (GIBCO-BRL; Invitrogen, CA, USA) or Dulbecco's modified Eagle's medium (DMEM; GIBCO-BRL; Invitrogen, CA, USA) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 mg/ml streptomycin and cultured at 37 °C in humidified air with 5% CO2.
2.3. RNA extraction and quantitative real-time PCR (qRT-PCR)
Total RNA was isolated from cultured cell lines or tissues by TRIzol reagent (Invitrogen, San Diego, CA, USA) following the manufacturer's instructions. The detail procedure was described previously [23]. The results were normalized to the expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH). All the primer sequences were supplied in Table S1. The qRT-PCR data were analyzed and expressed relative to cycle threshold (CT) values. The qRT-PCR results were analyzed and shown as the fold change (2-∆∆CT). For expression in tissues, the levels were first normalized to GAPDH expression as ∆CT and then compared with one of the tissues and converted to the fold change (2-∆∆CT). For the analysis of the expression in cells, the levels were compared with the controls and converted to the fold change (2-∆∆CT). The quantitative PCR for each sample was repeated in triplicate.
2.4. HNF1A overepression and HNF1A inhibition MIA PaCa-2 and PANC-1 clones
MIA PaCa-2 and PANC-1 cell lines were stably transfected with lentiviral vector (pMkO.1-puro vector) via lentiviral transduction (MOI = 3) and selected with 5 μg/ml hygromycin. Human HNF1A cDNA (GeneCopoeia, EX-A1385-M13-10, cDNA clone) and HNF1A shRNA (GeneCopoeia, HSH017954, shRNA clone) or empty vector were subcloned into the pMkO.1-puro vector and selected with 2 μg/ml puromycin. Clones were isolated, expanded and tested for HNF1A expression by qRT-PCT and Western blot analysis. The sequences of shRNA are listed in Table S2.
2.5. Cell proliferation assay
An MTS assay was used to evaluate the effects of gemcitabine after overexpression or inhibition of HNF1A on the proliferation of the PANC-1 and MIA PaCa-2 cell lines. Different groups (HNF1A, HNF1A-I and NC) of PANC-1 and MIA PaCa-2 cells growing on a 6-well plate were collected and 1500 cells were plated into 96-well plates. After treatment with various concentrations of gemcitabine for 48 h, 15 μL of MTS solution was added to each well and incubated at 37 °C for 2 h. Cell numbers were estimated using photometric reading, as described previously [24].
2.6. MTT assay
After gemcitabine treatment for 12 h, a total of 7000 cells were seeded in 96-well plates and treated with increasing amounts of gemcitabine for 48 h or 72 h. Thereafter, 20 μL of 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2 tetrazolium bromide (MTT) (Sigma, Saint louis, USA) (5 mg/ml) was added and incubated for 4 h. The supernatant was replaced with 150 μl of dimethyl sulfoxide (Sigma, St. Louis, USA) and read at 490 nm using a microplate photometer. Every concentration had 5 replicate wells, and each group was assayed in triplicate.
2.7. Colony formation assay
A total of 1000 cells were seeded in 6-well plates and maintained in media containing 10% FBS at 37 °C and treated with gemcitabine, which was replaced every 3 days. Ten days after seeding, colonies were fixed with methanol and stained with 0.1% crystal violet (Sigma-Aldrich, Milwaukee, USA). Visible colonies were then manually counted. Wells were measured in triplicate for each treatment group.
2.8. Cell apoptosis analysis
Standard propidium iodide staining of pancreatic cancer cells using the hypotonic lysis method was used for apoptosis studies with fluorescence activated cell sorting (FACS). All groups were treated with various gemcitabine (1, 5 or 10 μmol/L) for 72 h to induce apoptosis. The cells were then collected via trypsinization, fixed with 70% cold ethanol, mixed with 500 μL of ahypotonic solution (0.1% sodium citrate, 0.1% Triton X-100, 20 μg/ml RNase, and 50 μg/ml propidium iodide), incubated for 30 min and analyzed via flow cytometry.
2.9. Tumor formation assay in a nude mouse model
The athymic BALB/c nude mice (4–6 weeks old) were purchased and maintained at the Laboratory Animal Center of Sun Yat-sen University in a specific pathogen-free environment. Mice were given continuous access to food and water. The animal care and experimental protocols were approved by the Institutional Animal Care and Use Committee and the Institutional Biosafety Committee of Sun Yat-sen University. PANC-1 cells stably transfected with HNF1A vector or control vector were cultured in 6-well plates for 48 h. Then, the cells were collected, washed with PBS and resuspended at 1 × 108 cells/ml. A total of 100 μl of suspended cells was subcutaneously injected into the flank of each nude mouse. Three days after the injection of tumor cells, the tumor growth was evaluated the length and width by electronic calipers in every 3 days interval. The tumor volume was calculated using the following formula: V = (L × W2)/2 (V, volume; L, length diameter; W, width diameter). After one week, these mice were treated with gemcitabine (100 mg/kg body weight) or PBS. The mice were killed at 27 days post injection, and tumors were collected for further study (weight measurement, RNA extraction, and immunohistochemistry (IHC)). Briefly, tumor growth was evaluated by tumor volumes and weights (mean ± standard deviation (SD)), which were measured in mice from the HNF1A (5 mice) or negative control (NC) (5 mice) groups. HNF1A levels were determined by qRT-PCR and Western blotting, and tumor tissues were excised and fixed in 4% paraformaldehyde solution for further staining of Ki67.
2.10. Western blot analysis
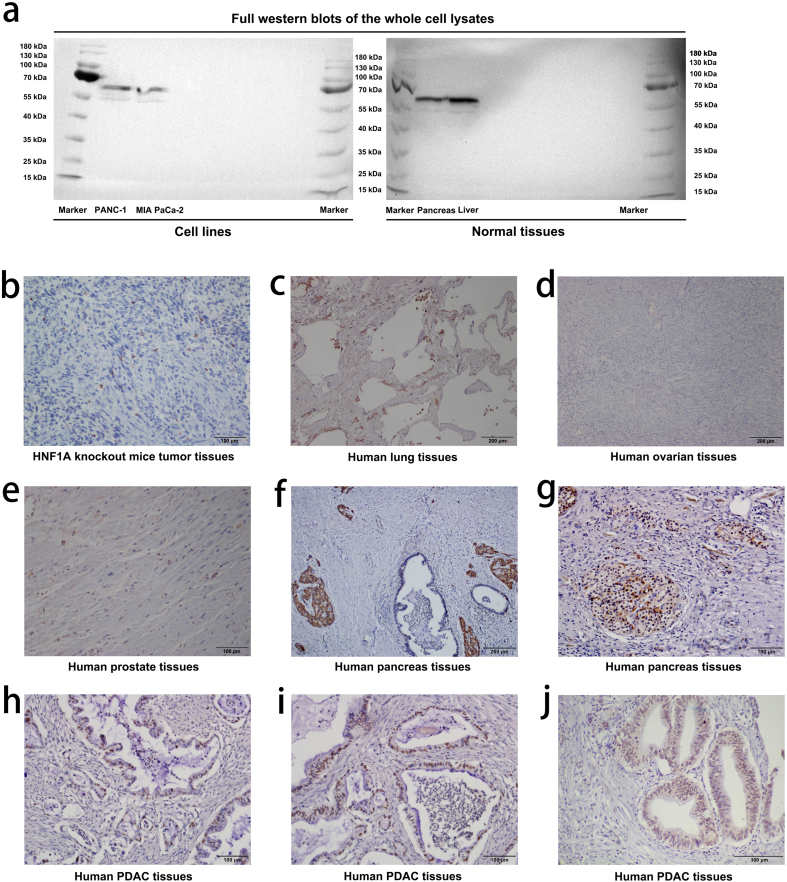
Western blot assay was performed as described previously [23]. Primary antibodies were rabbit anti-human HNF1A antibody (1:1000, #ab96777, Abcam), rabbit anti-human ABCB1 antibody (1:1000, #ab170904, Abcam), rabbit anti-human ABCC1 antibody (1:1000, #ab32574, Abcam), rabbit anti-human ABCC3 antibody (1:1000, #ab226804, Abcam), rabbit anti-human ABCC5 antibody (1:1000, #ab24107, Abcam) and rabbit anti-human GAPDH antibody (1:1000, #ab18162, Abcam). They were then incubated with the following HRP-linked secondary antibody: goat anti-rabbit IgG (1: 10000; Cell Signaling Technology, Boston, USA). The specificity of the antibody ab96777 was shown as full western blots of the whole cell lysates (Fig. S1a).
2.11. Immunofluorescence and immunohistochemistry analysis
For immunofluorescence, cells were fixed in 4% paraformaldehyde according to the manufacturer's instructions and the assay was performed as described previously [23]. The primary antibodies were used as: primary rabbit anti-human HNF1A antibody (1:500, #ab96777, Abcam).
Immunohistochemistry study was performed as described previously [23]. Paraffin-embedded samples of primary carcinomas were immunostained for primary rabbit anti-human HNF1A antibody (1:200, #ab96777, Abcam). To confirm the tissue specificity of antibody ab96777, we performed IHC assays on various samples and found that the expression of HNF1A was absent/low in the HNF1A knockout mice tumor tissues, human lung tissues, human ovarian tissues and human prostate tissues, and was high in the human pancreas islet tissues, and was differentiation in human PDAC tissues [Fig. S1b–j]. These results were consistent with the previous study [25].
The expression levels of HNF1A were scored semiquantitatively based on staining intensity and distribution using the immunoreactive score (IRS) as described elsewhere [26,27]. Concisely, IRS = staining intensity (SI) × percentage of positive cells (PP). SI was assigned as: 0 = negative; 1 = weak; 2 = moderate; and 3 = strong. The percentage of positive pancreatic cancer cells was calculated after at least 10 view fields at 400 × magnification were counted per section. PP was defined as 0 = 0%; 1 = 0–25%; 2 = 25–50%; 3 = 50–75%; and 4 = 75–100%. For categorization of the continuous HNF1A values into low, moderate and high, we chose a commonly used cutoff point for the measurements (total score range 0–12: low, cut point of 0–2; moderate, cut point of 3–6; high, cut point of 8–12). Two pathologists assessed the staining independently, and the discrepancy cases were jointly reevaluated until a consensus was reached.
2.12. Luciferase reporter assay
The ABCB1 promoter (−1,202 DSP in pGL2-basic) is purchased from TranSheep Bio Co. Ltd. (Shanghai, China). A total of 107 cells were transfected with 10 μg of ABCB1 promoter and 1 μg of Renilla (pRL-TK, Promega) as an internal transfection control. Twenty-four hours after transfection, the cells were collected, washed and harvested for firefly/Renilla luciferase assays using Dual-Glo Luciferase.
2.13. Generation of human ABCB1 promoter deletion constructs
The full-length promoter construct (−1202/+118) was generated by digesting with TthIIII. The construct (−278/+118) was generated by blunt end ligation of the resultant promoter fragments and then cloned into the SmaI site of the pGL2basic vector. The other deletion constructs were generated by PCR, digested with restriction enzymes, and then cloned into the MluI and BglII sites of the pGL2-basic reporter vector. The PCR primers used to amplify the deletion construct inserts (−988/+118 and −624/+118) are listed in Table S1. Putative binding site mutagenesis was performed by TranSheep Bio Co. Ltd. (Shanghai, China), and the CTTAACTCTTAAA sequence at position −810 to −798 of the deletion construct inserts (−988/+118) was mutated to GCCGGAAGCCGGC.
2.14. Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assays were performed using EZ-Magna ChIP Chromatin Immunoprecipitation Kit according to the manufacturer's instructions. Briefly, 5 × 106 indicated PANC-1 cells were fixed in 1% formaldehyde for 10 min at room temperature. Fixed cells were collected, lysed and sonicated for 10 cycles of 10 s on/20 s off and 50% AMPL with a Sonics VCX130. Antibodies against HNF1A (5 ul per 1 mg total protein) and rabbit IgG (2 ul per 1 mg total protein) were used for immunoprecipitation. Analysis of the PCR products was performed on the precipitated DNA from a standard 2% (w/v) agarose gel by electrophoresis in Tris-acetate EDTA buffers. The primer sequences used in the ChIP assay are listed in Table S1.
2.15. Statistical analysis
All data were statistically analyzed using SPSS 23.0 (IBM Chicago, IL, USA). The chi-square test (χ2 test) for nonparametric variables and Student's t-test or one-way analysis of variance (ANOVA) for parametric variables were used (two-tailed). Error bars in the experiments indicate standard error of the mean (SEM) or SD for a minimum of three independent experiments. The Kaplan Meier method was used for assessing the differences in patient survival and the log-rank test for a univariate analysis. Cox regression was used for multivariate analyses to assess the relative risk for each factor. Correlation analysis was examined with two-sided Pearson's correlation.
3. Results
3.1. HNF1A expression is significantly associated with gemcitabine sensitivity in PDAC cell lines
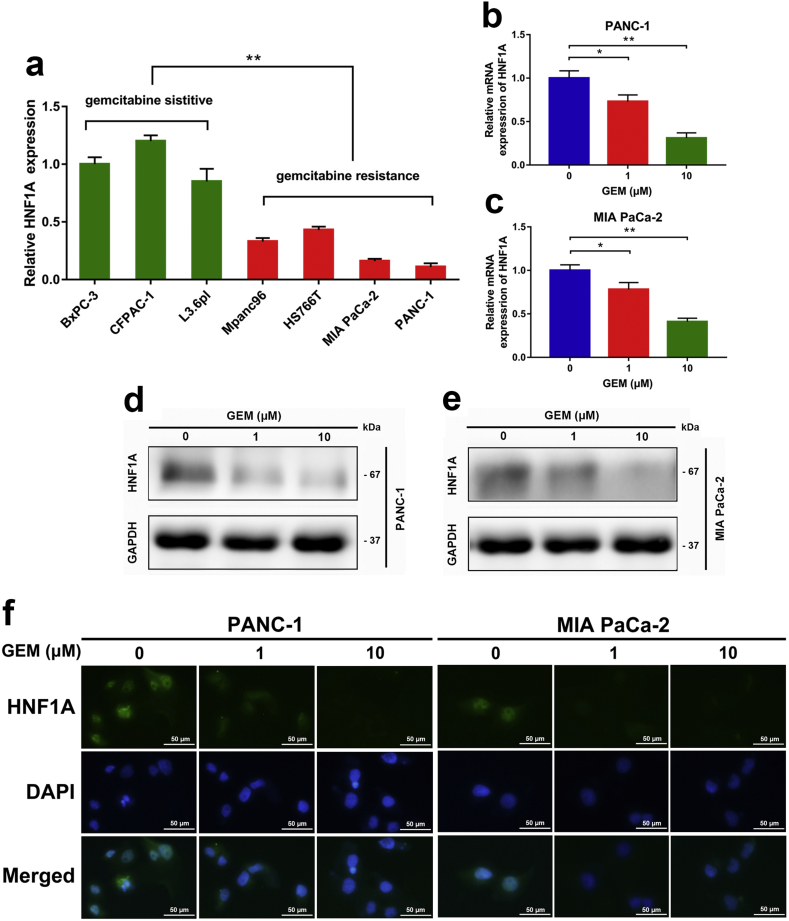
The loss of HNF1A expression has been reported to promote the progression of pancreatic cancer, and HNF1A was identified as a possible tumor suppressor in pancreatic cancer [19,20]. The study found that four cell lines (BxPC-3, SU86.86, CFPAC-1 and L3.6pl) were sensitive to gemcitabine, and five cell lines (PANC-1, Hs766T, MIA PaCa-2, AsPC-1 and Mpanc96) were resistant to gemcitabine based on 50% growth inhibition [24]. To explore the potential role of HNF1A in chemotherapy, we detected the mRNA expression of HNF1A in seven pancreatic cancer cell lines. Data from qRT-PCR analysis showed that the expression of HNF1A in four gemcitabine-resistant cell lines (PANC-1, MIA PaCa-2, Hs766T and Mpanc96) was remarkably lower than that in three gemcitabine-sensitive cell lines (BxPC-3, CFPAC-1 and L3.6pl) (Fig. 1a). Moreover, we evaluated the changes in HNF1A expression in pancreatic cancer cells treated with gemcitabine. In our study, the PANC-1 and MIA PaCa-2 pancreatic cancer cell lines were treated with different concentrations of gemcitabine, and the alterations of HNF1A expression were determined by qRT-PCR, Western blot and immunofluorescence. We found a decreased expression of HNF1A at the mRNA and protein levels after gemcitabine treatment at 1 μM and 10 μM concentrations for 12 h (Fig. 1a–e). Immunofluorescence further demonstrated an inhibition of HNF1A expression in the nucleus after gemcitabine treatment (Fig. 1f).
Fig. 1.
HNF1A expression is significantly associated with gemcitabine sensitivity in PDAC cell lines. (A) HNF1A expression levels were detected in seven pancreatic cancer cell lines by qRT-PCR. Data are shown as fold change (2−∆∆CT) and the mean ± SD from three independent experiments. (B–C) HNF1A expression levels were evaluated by qRT-PCR in PANC-1 and MIA PaCa-2 cell lines after different dose (0, 1, and 10 μm) of gemcitabine treatments for 12 h. (D–E) The protein levels of HNF1A were evaluated by Western Blot in PANC-1 and MIA PaCa-2 cell lines after different dose (0, 1, and 10 μm) of gemcitabine treatments for 12 h. (F) Immunofluorescence staining showed an inhibition of HNF1A expression in nucleus after gemcitabine treatment. The images shown are representative results of three independently repeated experiments and are presented as mean ± SD. Scale bar, 50 μm. *p < .05, **p < .01 (Student's t-test).
3.2. HNF1A enhances the sensitivity of gemcitabine in PDAC cells
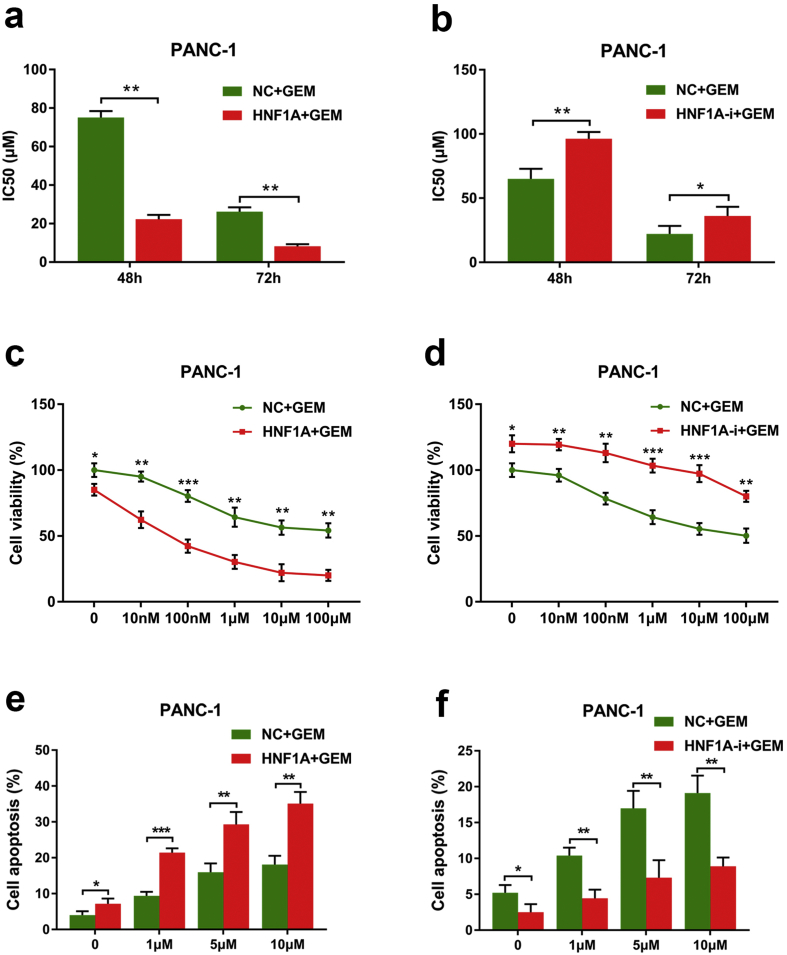
To elucidate the functional role of HNF1A in chemotherapy, we constructed a lentivirus-based expression system to overexpress or deplete HNF1A in two gemcitabine-resistant pancreatic cancer cells. NC, HNF1A-expressing (HNF1A), and HNF1A-shRNA (HNF1A-i) lentivirus were packaged and used to infected PANC-1 and MIA PaCa-2 pancreatic cancer cell lines. After obtaining cell lines that stably overexpressed HNF1A or had depleted HNF1A by puromycin, Western blotting staining showed that the expression of HNF1A was dramatically upregulated following HNF1A-lentivirus infection and significantly reduced by HNF1A-shRNA lentivirus infection in both PANC-1 and MIA PaCa-2 pancreatic cancer cells compared with cells infected with control virus (Fig. S2a–d). To confirm the effect of HNF1A on basal growth of pancreatic cancer cells, we conducted MTS assay and colony formation assay. Consistent with our previous study [19], our results showed that overexpression HNF1A significantly inhibited the proliferation of both PANC-1 and MIA PaCa-2 cells, and HNF1A-i cells showed a higher proliferative capacity than NC cells (Fig. S3). Next, to investigate the effect of HNF1A gemcitabine resistance, we detected the IC50 values of gemcitabine for different time points by MTT assay. We then compared the sensitivity of pancreatic cancer cells to gemcitabine in combination with HNF1A overexpression or inhibition. Our data revealed that the IC50 values of gemcitabine at 48 h and 72 h were both markedly decreased after overexpression of HNF1A followed by gemcitabine treatment (Fig. 2a and S4a), while inhibition of HNF1a significantly increased the IC50 values of gemcitabine (Fig. 2b and S4b). Moreover, we treated the PANC-1 and MIA PaCa-2 cell lines, both of which were resistant to gemcitabine and expressed low levels of HNF1A natively, with different concentrations of gemcitabine in the presence of NC, HNF1A or HNF1A-i. By MTS assay, we found that the proliferation capacity was significantly lower in HNF1A cells (Fig. 2c and S4c), and higher in HNF1A-i cells than in NC cells when treated with no gemcitabine, which indicated a tumor suppressor role of HNF1A in PDAC. Moreover, when treated with various concentrations of gemcitabine (10 nM, 100 nM, 1 μM, 10 μM, 100 μM), the proliferation capacity was much more significantly lower in HNF1A cells (Fig. 2c and S4c), and higher in HNF1A-i cells than in NC cells (Fig. 2d and S4d). In addition, by a fluorescence activated cell sorting assay, we demonstrated that overexpression of HNF1A induced apoptosis in cells treated with gemcitabine at different concentrations (1, 5 or 10 μmol/L) in both PANC-1 and MIA PaCa-2 cells (Fig. 2e and S4e), whereas inhibition of HNF1A had the opposite effect (Fig. 2f and S4f).
Fig. 2.
HNF1A enhances the sensitivity of gemcitabine in PANC-1 cells. (A) MTT assays indicated that the IC50 values of gemcitabine at 48 h and 72 h were all markedly decreased after overexpression of HNF1A followed by gemcitabine treatment. (B) MTT assays indicated that the IC50 values of gemcitabine at 48 h and 72 h were all markedly increased after inhibition of HNF1A followed by gemcitabine treatment. (C) MTS assays indicated that the proliferation capacity was significantly lower in HNF1A-overexpressing cells after treatment with various concentrations of gemcitabine. (D) MTS assays revealed that the proliferation capacity was significantly higher in HNF1A-inhibited cells after treatment with various concentrations of gemcitabine. (E) A flow cytometry assay showed that overexpression of HNF1A induced the apoptosis in cells treatment with gemcitabine at various concentrations (1, 5 or 10 μmol/L). (F) HNF1a inhibition resulted in increased resistance of the cells to treatment with various concentrations (1, 5 or 10 μmol/L) of gemcitabine, resulting in decreased apoptosis rates. The data are shown from 3 parallel experiments and are presented as mean ± SD. *p < .05, **p < .01, ***p < .001 (Student's t-test).
3.3. HNF1A overexpression sensitizes pancreatic xenograft tumors to gemcitabine
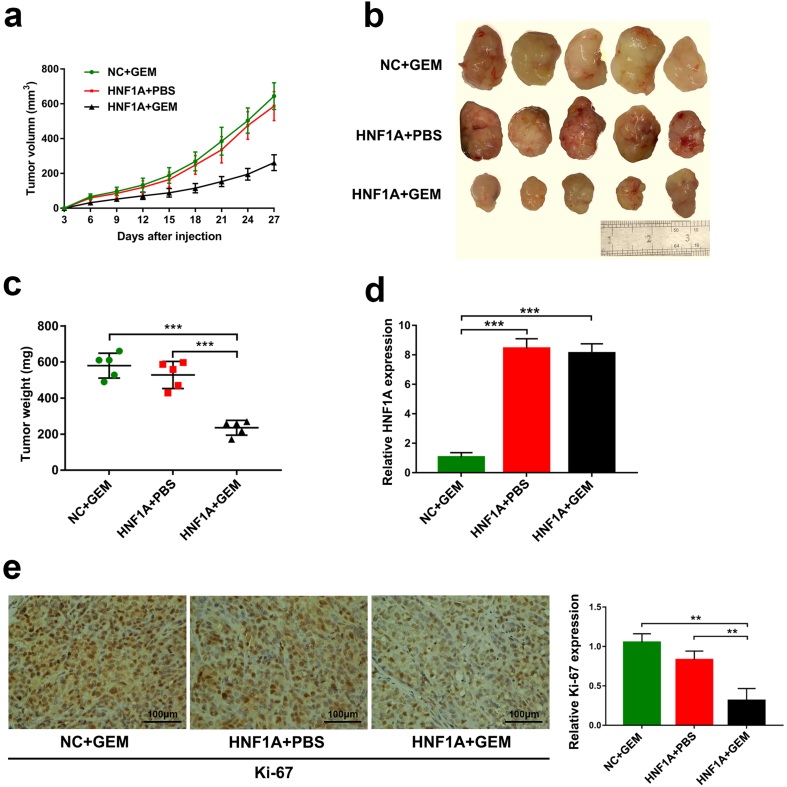
The effect of gemcitabine in combination with HNF1A overexpression on the growth of pancreatic xenograft tumors was further evaluated in vitro. Four weeks after injection, both the tumor size and weight were substantially reduced in the GEM + HNF1A overexpression group compared with the NC + GEM group or the HNF1A + PBS group (Fig. 3a–c). Besides, qRT-PCR analysis revealed the overexpression of HNF1A in the xenograft tumors from the HNF1A + GEM group and HNF1A + PBS group (Fig. 3d). Using immunostaining, we confirmed that tumors from the treatment of the GEM + HNF1A overexpression group exhibited a significantly decreased positive rate of Ki67 (Fig. 3e). Taken together, our data revealed that HNF1A plays a crucial role in gemcitabine chemotherapy for pancreatic cancer both in vitro and in vitro.
Fig. 3.
HNF1A overexpression sensitizes pancreatic xenograft tumors to gemcitabine. (A) After subcutaneous injection of mice with HNF1A-overexpressing cells or NC cells and treatment with gemcitabine or PBS, the tumor growth curves constructed from measurements taken every 3 days revealed growth inhibition in the HNF1A + GEM group. (B) Twenty-seven days after subcutaneous injection, the tumors were harvested from the mice. (C) Tumor weights are shown as the means of tumor weights ± SD when the tumors were removed from mice. *p < .05, **p < .01, ***p < .001 (One-way ANOVA). (D) The HNF1A expression levels were increased in tumor tissues from HNF1A cells compared with negative control cells by qRT-PCR. (E) Representative images (×200) of IHC staining of the tumor. IHC staining showed that overexpression of HNF1A in combination with treatment with gemcitabine resulted in a significantly decreased positive rate of Ki67. Scale bar, 100 μm. Data are representative of three independent experiments and are presented as mean ± SD. **p < .01, ***p < .001 (Student's t-test).
3.4. HNF1A regulates the expression of multidrug resistance genes in PDAC cells
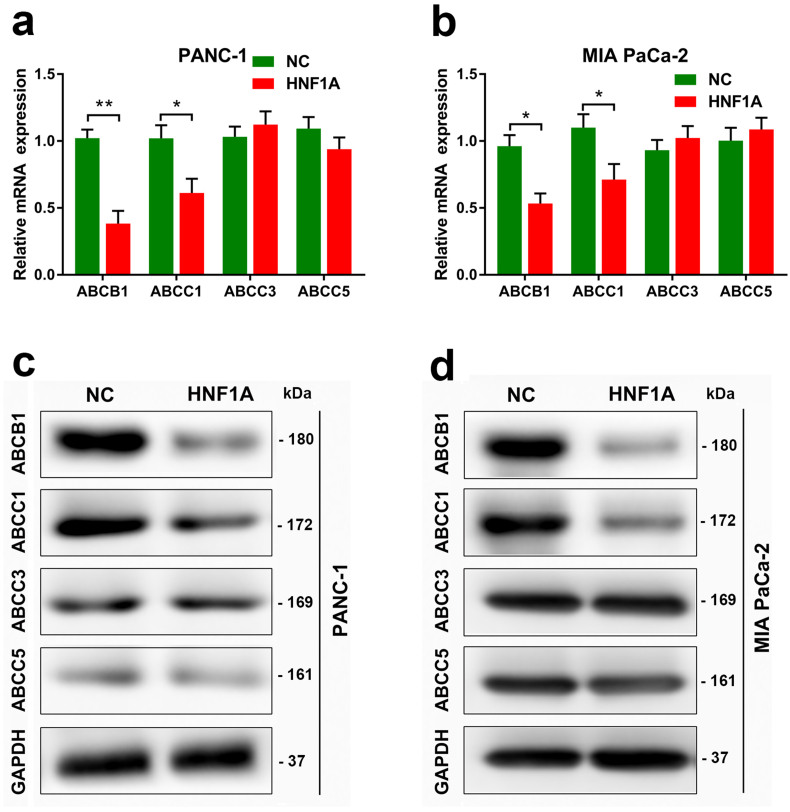
MDR genes contribute to gemcitabine sensitivity in pancreatic cancer cells. The ABCB1, ABCC1, ABCC3 and ABCC5 are well-characterized MDR genes. Using qRT-PCR (Fig. 4a–b) and Western Blot (Fig. 4c–d), we found that the mRNA and protein levels of ABCB1 and ABCC1 were significantly decreased in PANC-1 and MIA PaCa-2 cell lines after overexpression of HNF1A.
Fig. 4.
HNF1A regulates the expression of multidrug resistance genes in PDAC cells. (A-B) Quantitative RT-PCR data showing fold changes (2-∆∆CT) in the mRNA expression level of ABCB1, ABCC1, ABCC3 and ABCC5 genes after overexpression of HNF1A in the PANC-1 and MIA PaCa-2 cell lines. (C-D) Western Blot experiments showing the protein levels of the ABCB1, ABCC1, ABCC3 and ABCC5 genes after overexpression of HNF1A in PANC-1 and MIA PaCa-2 cell lines. The graphs shown are representative results of three repeated assays and are presented as mean ± SD. *p < .05, **p < .01 (Student's t-test).
3.5. HNF1A enhances gemcitabine sensitivity by regulating ABCB1 expression in PDAC cells
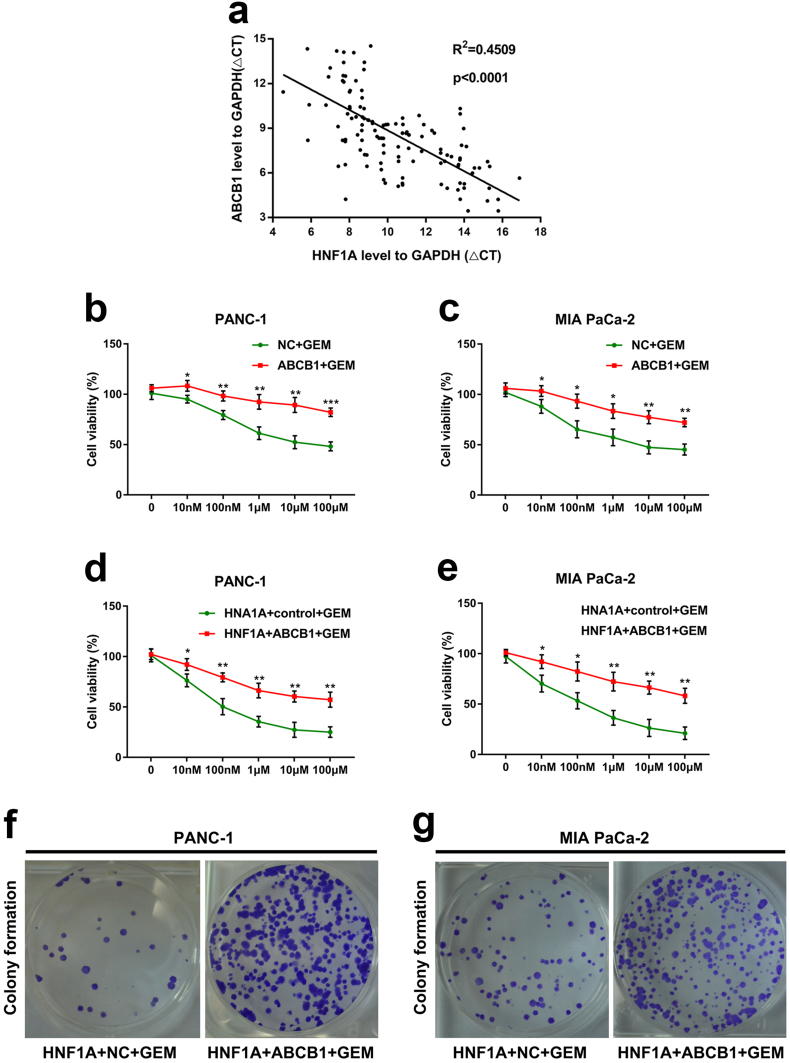
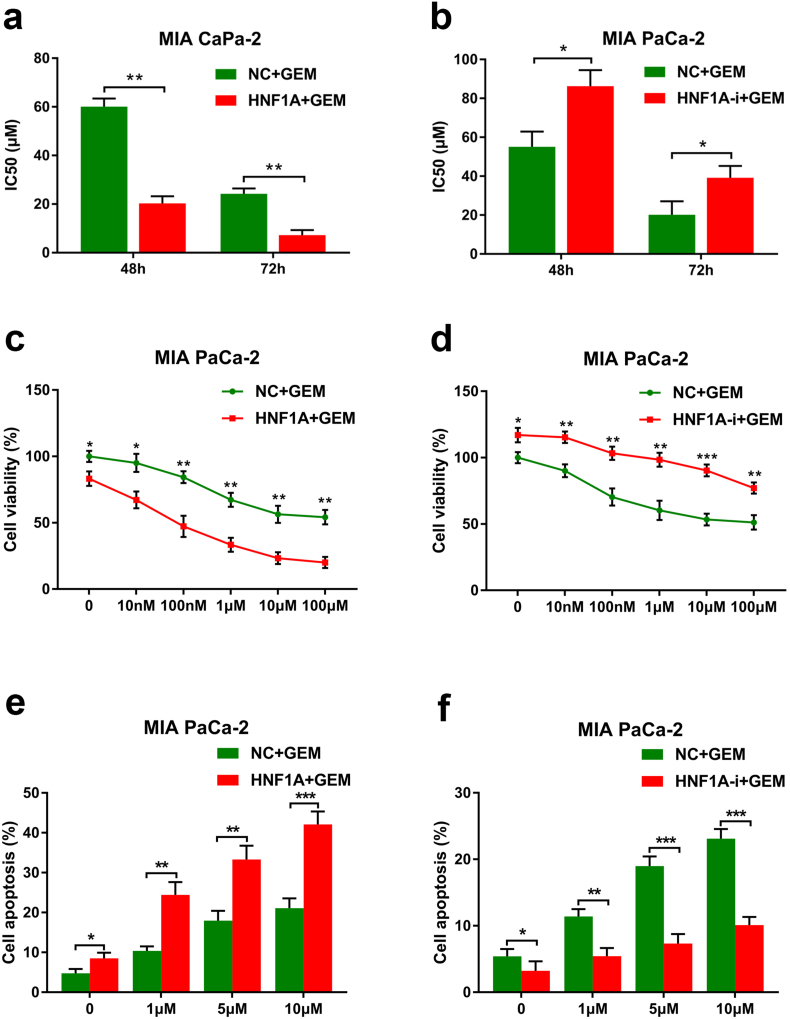
Enhancement of ABCB1 expression is a well-recognized marker for chemotherapy resistance, and its role in gemcitabine resistance in pancreatic cancer has been determined. Therefore, we studied whether ABCB1 participated in tumor chemoresistance induced by HNF1A inhibition. Importantly, by performing a qRT-PCT assay in 128 surgical patients with PDAC in our department, a significantly negative correlation was observed between the ABCB1 mRNA levels and the HNF1A expression levels in pancreatic cancer tissues (R2 = 0.4509, p < .0001, Pearson's correlation analysis, Fig. 5a). Next, the responses of PANC-1 and MIA PaCa-2 cells to gemcitabine decreased after transfection with the ABCB1 vector compared with the control vector (Fig. 5b–c). In addition, the responses of HNF1A-overexpressing PANC-1 and MIA PaCa-2 cells to gemcitabine were also decreased after transfection with the ABCB1 vector compared with the control vector (Fig. 5d–e). Moreover, the cell colony formation assay showed that the enhancement of gemcitabine sensitivity by HNF1A overexpression in PANC-1 and MIA PaCa-2 cells was hindered when combined with ectopic expression of ABCB1 (Fig. 5f–g). These results implied that HNF1A inhibition enhances gemcitabine resistance by inducing ABCB1 expression in pancreatic cancer cells.
Fig. 5.
HNF1A enhances gemcitabine sensitivity by regulating ABCB1 expression in PDAC cells. (A) Pearson's correlation analysis indicated a significantly negative correlation between ABCB1 mRNA levels and HNF1A mRNA levels in pancreatic cancer tissues (n = 128, R2 = 0.4509, p < .0001) (Pearson's correlation analysis). (B–C) The sensitivity of PANC-1 and MIA PaCa-2 cells to gemcitabine was decreased after transfection with the ABCB1 vector compared with the control vector as determined by MTS assay. (D-E) The responses of HNF1A-overexpressing cells to gemcitabine were decreased after transfection with the ABCB1 vector compared with the control vector in both the PANC-1 and MIA PaCa-2 cell lines. (F-G) The cell colony formation assay revealed that the enhancement of gemcitabine sensitivity by HNF1A overexpression in PANC-1 and MIA PaCa-2 cells was hindered when combined with ectopic expression of ABCB1. The graphs shown are representative results of three repeated assays and are presented as mean ± SD. *p < .05, **p < .01, ***p < 0,001 (Student's t-test).
3.6. HNF1A binds to a specific region of the ABCB1 promoter to suppress transcription
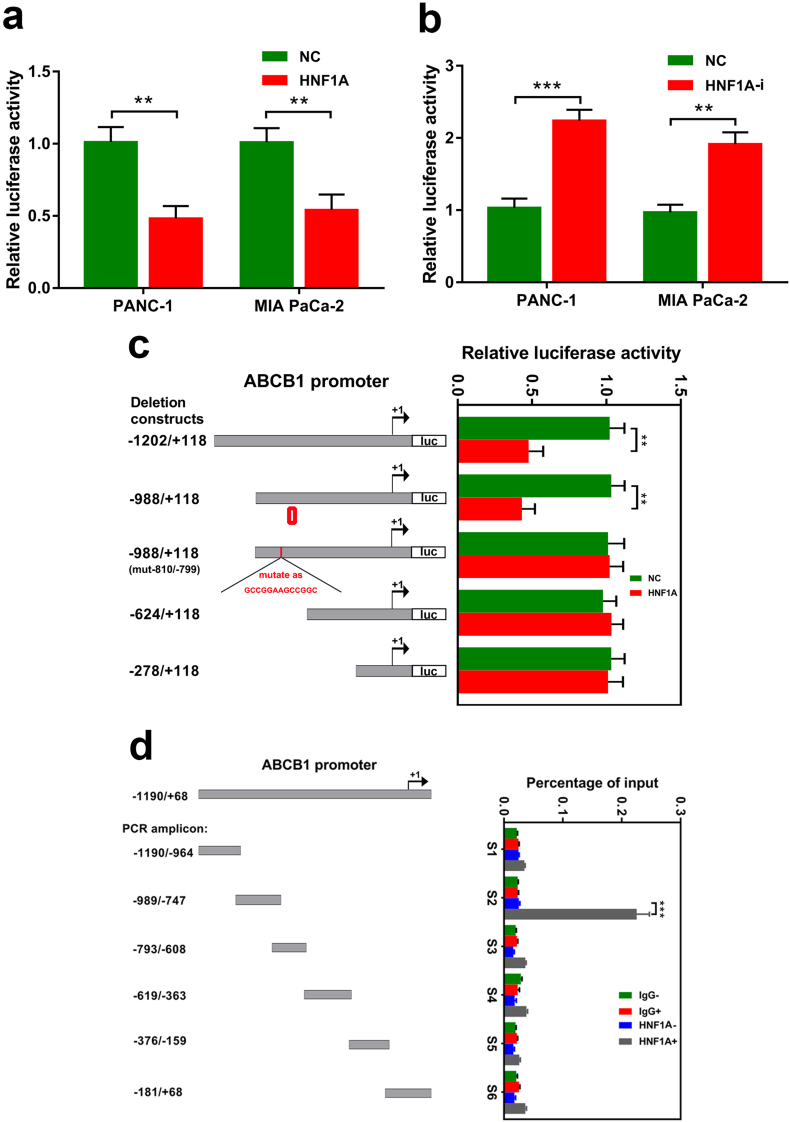
Furthermore, to verify that HNF1A is a crucial transcription factor for ABCB1, using a luciferase assay, we revealed that the transcriptional activation of ABCB1 in PANC-1 cells was inhibited in stably HNF1A overexpressed cells and enhanced in HNF1A depleted cells (Fig. 6a–b). To identify the transcriptional suppression elements in the ABCB1 promoter responsive to HNF1A, we generated and cotransfected a series of deletion/mutation reporter constructs into PANC-1 cells together with either the constitutively active HNF1A or an empty control vector. We found a significant suppressive effect in the HNF1A group compared with the control group after deletion of the 5′ distal region (−1202 bp to −988 bp, Fig. 6c). However, the inhibition was absent when the deletion constructs were −624 bp to +118 bp and −278 bp to +118 bp (Fig. 6c). These results showed that the 5′ upstream region (−988 bp to −624 bp) of the ABCB1 promoter is required for transcriptional suppression by HNF1A. Furthermore, the potential binding sites for HNF1A were identified based on the consensus HNF1 recognition sequence GTTAATnATTAAC [28]. By bioinformatic analysis, a potential HNF1A responsive element was identified at position −810 bp to −798 bp (CTTAACTCTTAAA). Consistent with this idea, the suppressive effect on ABCB1 promoter activity was abrogated when the binding sequence was mutant (Fig. 6c). Finally, we investigated the endogenous promoter occupancy of HNF1A using CHIP analysis. The chromatin was precipitated with a specific HNF1A or IgG antibody, and analyzed by PCR using primer sets that covered the entire length (−1190 bp to +68 bp) of the ABCB1 promoter. Consistent with the cotransfection studies, ChIP analysis showed that HNF1A binding was confined to the specific region of the ABCB1 promoter (−989 bp to −747 sbp, Fig. 6d). Taken together, these results demonstrate that HNF1A regulates ABCB1 expression through binding its specific promoter region and suppressing its transcription.
Fig. 6.
HNF1A binds to the specific region of the ABCB1 promoter to suppress transcription. (A–B) The luciferase assay indicated that the transcriptional activation of ABCB1 in PANC-1 cells was inhibited in stable HNF1A-overexpressing cells and enhanced in HNF1A depletion cells. (C) Comparison of the transcriptional suppressive effect of HNF1A to a series of deletion or mutation constructs of the human ABCB1 promoter. Different human ABCB1 promoter-luciferase reporter constructs together with HNF1A were transiently cotransfected in PANC-1 cells. Twenty-four hours after cotransfection, all samples were harvested for luciferase activity experiments. The graph on the right side reveals the relative luciferase activity of the ABCB1 promoter by HNF1A. (D) ChIP analysis of the human ABCB1 promoter. Protein-DNA complexes from PANC-1 cells were harvested for immunoprecipitation with antibodies against IgG (nonspecific) and HNF1A. After cross-link reversal, the coimmunoprecipitated DNA was amplified by PCR using the indicated primers and resolved in 2% agarose gels. Graphs shown are representative results of three repeated assays and are presented as mean ± SD. **p < .01, ***p < 0,001 (Student's t-test).
3.7. Low levels of HNF1A expression predict chemoresistance in PDAC patients
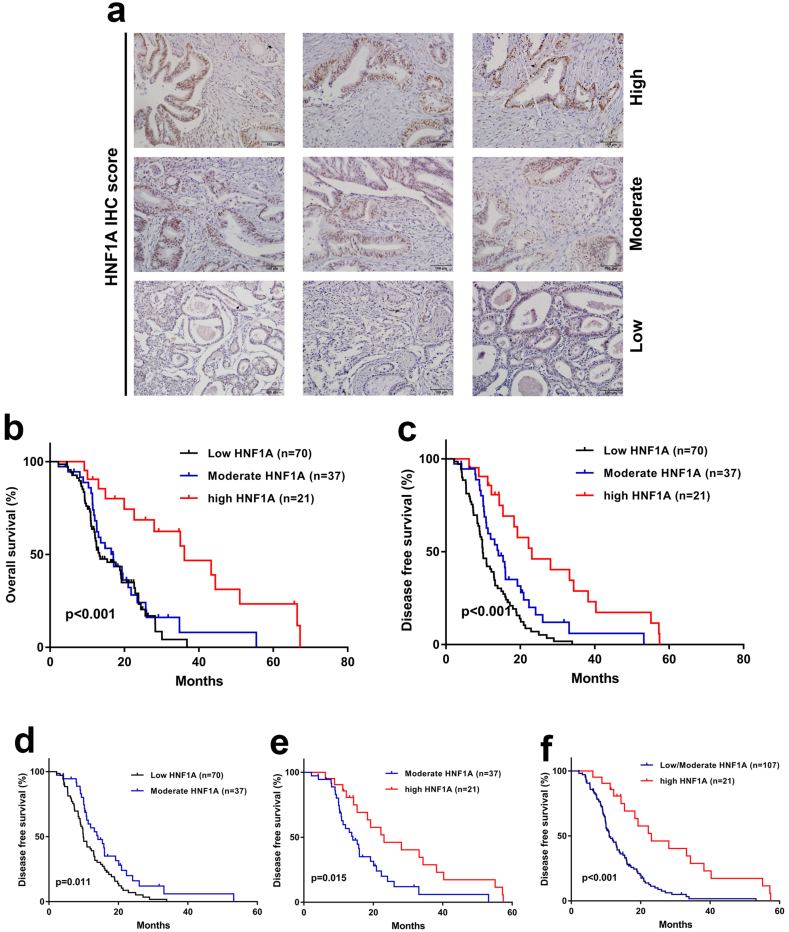
Identifying markers of chemoresistance is important for stratification in clinical trials, especially for pancreatic cancer, whose response rates to its first line agent-gemcitabine, <20%. We next investigated the clinical significance of HNF1A in 128 pancreatic cancer patients, all of whom received regular gemcitabine chemotherapy after radical surgery. PDAC can be stratified into three subtypes according to the staining intensity of HNF1A (Fig. 7a). First, the correlation of HNF1A levels and clinicopathological features was analyzed. We found that HNF1A staining was correlated with TNM stage and perineural invasion (Table 1). By univariate analyses, we found that OS was lower in patients in the moderate/low HNF1A groups than that in the high HNF1A group (Table 2). Moreover, the multivariate analyses showed that HNF1A levels were independently and significantly associated with OS despite adjusting for baseline clinicopathological features (Table 2). In addition, the survival analyses demonstrated an improvement in OS for patients in high HNF1A group compared with the moderate/low HNF1A groups (Fig. 7b). Importantly, the disease-free survival (DFS) analyses confirmed that the high HNF1A group showed the best survival, the low HNF1A group showed the worst survival and the moderate HNF1A group an intermediate DFS (Fig. 7c–f). These data reveal the predictive value of HNF1A in the identification of gemcitabine-sensitive pancreatic cancer patients.
Fig. 7.
Low levels of HNF1A expression predict chemoresistance in PDAC patients. (A) Representative images of HNF1A staining in PDAC tissues (categorization: low, moderate, and high). (B) The Kaplan–Meier analysis of overall survival stratified by the immunoreactive score of HNF1A expression. (C) Kaplan–Meier analysis of disease-free survival stratified by the immunoreactive score of HNF1A expression. (D–F) The disease-free survival analyses confirmed that the high HNF1A group showed the best survival, the low HNF1A group showed the worst survival, and the moderate HNF1A group showed intermediate disease-free survival. The p-values are shown with the use of the log-rank test (two-sided) **p < .01, ***p < .001 (Kaplan-Meier analysis, log-rank test).
Table 2.
Univariate and multivariate analysis of overall survival in PDAC patients (n = 128).
| Variables | Characteristics | Univariate analysis |
Multivariate analysis |
||||
|---|---|---|---|---|---|---|---|
| HR | 95% CI | p value | HR | 95% CI | p value | ||
| Age | <60 (ref) | ||||||
| ≥60 | 1.044 | 0.689–1.581 | 0.839 | ||||
| Gender | Female (ref) | ||||||
| Male | 0.835 | 0.551–1.267 | 0.383 | ||||
| Differentiation | Well (ref) | ||||||
| Moderate | 1.062 | 0.607–1.857 | 0.835 | ||||
| Poor | 1.292 | 0.666–2.506 | 0.440 | ||||
| TNM stage | I (ref) | ||||||
| II | 1.549 | 0.899–2.668 | 0.126 | 1.438 | 0.788–2.623 | 0.238 | |
| III | 2.649 | 1.593–4.406 | <0.001⁎⁎⁎ | 2.887 | 1.549–5.381 | 0.001⁎⁎ | |
| Lymph node metastasis | Negative (ref) Positive |
||||||
| 1.539 | 1.023–2.316 | 0.037⁎ | 1.793 | 1.151–2.794 | 0.010⁎ | ||
| Perineural invasion | Negative (ref) Positive |
||||||
| 1.534 | 1.013–2.323 | 0.038⁎ | 0.787 | 0.458–1.352 | 0.386 | ||
| HNF1A expression | High (ref) | ||||||
| Moderate | 2.637 | 1.423–4.856 | <0.001⁎⁎⁎ | 3.231 | 1.543–6.768 | 0.002⁎⁎ | |
| Low | 2.903 | 1.774–4.751 | <0.001⁎⁎⁎ | 4.109 | 1.978–8.533 | <0.001⁎⁎⁎ | |
Abbreviations: HR = hazard ratio; 95% CI = 95% confidence interval; TNM = tumor node metastasis; T stage = tumor stage; ref. = reference.
Cox regression analysis.
p < .05.
p < .01.
p < .001.
4. Discussion
In the present work, we demonstrated that HNF1A expression was significantly associated with gemcitabine sensitivity in PDAC cell lines. Moreover, overexpression of HNF1A enhanced the sensitivity of gemcitabine in PDAC cells both in vitro and in vitro. Importantly, the mechanism analysis showed that HNF1A suppressed ABCB1 expression by binding to the specific region of the ABCB1 promoter. Interestingly, patients with HNF1A-positive tumors might benefit from gemcitabine chemotherapy.
PDAC remains a clinical challenge and may be ranked as the second leading cause of cancer mortality in 2030 [29]. Less than 20% of pancreatic cancer patients are considered resectable, which offers the only option for a cure. Gemcitabine based single-agent or combinational chemotherapies remain the first-line therapies for most PDAC patients since 1997 [5,7]. However, most patients will face treatment failure due to the development of resistance to chemotherapy. Our previous research and Hoskins et al. reported that HNF1A was involved in the development of different cancers and played a role as a suppressor in pancreatic cancer [19,20]. More recently, Abel et al. reported a novel oncogenic role for HNF1A in pancreatic cancer, particularly in promoting pancreatic cancer stem cell properties [30]. These results were contrast to the data in this study and several previous studies [19,20,31]. Consistent with the opinion of Abel et al., we surmised that HNF1A may function differently in different diseases, even in different molecule subtypes of pancreatic cancer [32]. The cell lines used in Abel et al. came from a low passage PDAC patient primary cell line established from xenografts, and HNF1A was significantly increased in the CD44high/EPCAMhigh subpopulation but not the whole tumors, indicating the different role of HNF1A in different subpopulations of tumors in pancreatic cancer. Supporting the discussing, Yu et al. demonstrated that HNF1A activated PTEN/Akt signaling and then inhibited the proliferation of pancreatic cell lines (including PANC-1, which was used in our research), suggesting a tumor suppressing role in these pancreatic cancer cell lines (SW1990, BxPC-3, AsPC-1, CFPAC-1 and PANC-1) [31]. Recently, Cheng et al. reported that the combination of HNF1A, HNF4A and forkhead box protein A3 (FOXA3) synergistically reprogrammed hepatocellular carcinoma cells to hepatocyte-like cells [33]. In addition, Takashima et al. reproduced the findings above and showed that combinatorial expression of these three transcription factors suppressed the growth of all cell subtypes within the hepatocellular carcinoma cell lines, including cancer stem cells [34]. These studies indicate the intricacy role of HNF1A, even in different cancer stem cells from different carcinomas. For no reason, we should recognize the limitation of the experimental part in our study, is that only two cell lines were used. Interestingly, Muckenhuber et al. reported that stratifying molecular subtypes of PDAC using immunohistochemical methods and HNF1A is associated with significantly differing outcomes and responses to chemotherapy [35]. However, it remains unclear whether HNF1A contributes to gemcitabine chemoresistance in PDAC, and the biological functions and molecular mechanisms of HNF1A in PDAC remain to be elucidated.
In our research, we first focused on the correlation of HNF1A expression and the gemcitabine sensitivity of seven pancreatic cancer cell lines and provided evidence that HNF1A levels were remarkably lower in the gemcitabine-resistant cell lines compared with the gemcitabine-sensitive cell lines. Moreover, treatment with gemcitabine in pancreatic cell lines caused the decreased expression of HNF1A, which indicated that some specific signaling pathways may be involved in the regulation of HNF1A in gemcitabine metabolism.
Therefore, we constructed a gain- and loss-of-functional model to explore the role of HNF1A in gemcitabine resistance in vitro and in vitro. We found that knockdown of HNF1A led to increased IC50 values of gemcitabine and enhanced proliferation in gemcitabine resistant cell lines in vitro. Consistently, overexpression of HNF1A remarkably decreased IC50 values of gemcitabine in vitro, and significantly inhibited the proliferation of gemcitabine resistant pancreatic cell lines both in vitro and in vitro. Actually, the HNF1A/melanoma inhibitory activity (MIA2) axis has been demonstrated to predict the survival benefit to patients with pancreatic cancer who received adjuvant chemotherapy [36]. However, the tumor suppressor function of the HNF1A/MIA2 axis in pancreatic cancer was paradoxical from some data, which even argues for the opposite function [37,38]. In addition, the HNF1A/MIA2 axis was dependent on the germline variant MIA2I141M, whose frequency in PDAC ranged from 0.28 to 0.50 [40]. Based on the indirect findings of previous research, for the first time, we provided evidence that HNF1A plays a crucial role in gemcitabine chemotherapy for pancreatic cancer both in vitro and in vitro.
Most patients with pancreatic cancer will die from the disseminated disease that has developed MDR [13]. A previous study reported a significant upregulation of ABCB4, ABCB11, ABCC1, ABCC3, ABCC5, ABCC10 and ABCG2 at the mRNA level in macrodissected tumors relative to normal tissue, indicating the contribution of ABC transporters to the generally poor treatment response of PDAC [39]. Using support vector machines, Dorman et al. described the development of genomic signatures and revealed a strong association between gemcitabine resistance and ABCB1 copy number in breast cancer cell lines [40]. Nath et al. demonstrated that the gemcitabine resistance in pancreatic cancer cell lines was attributed to the enhanced expression of MDR genes including ABCB1, ABCC1, ABCC3 and ABCC5 [41]. Hong et al. reported that ABCB1 was significantly augmented during the acquisition of gemcitabine resistance in pancreatic cancer cell lines, and the ABC transporter inhibitor verapamil resensitized the resistant cells to gemcitabine in a dose-dependent manner [42]. However, studies on the upstream regulation of ABC transporters are still lacking, and the association between HNF1A and ABC transporters has not been elucidated. We first carried out the upstream regulation of ABC transporters by HNF1A in pancreatic cancer. Overexpression of HNF1A significantly decreased the mRNA and protein levels of ABCB1 and ABCC1 in PDAC cell lines. Among the MDR family, ABCB1 was the first to be explored, and ample evidence suggested that it can confer resistance to cytotoxic and targeted chemotherapy, whereas ABCC1 was first reported in 1992 [43], and was found to mediate resistance to doxorubicin, etoposide and vincristine among others, in cell line models [44]. However, ABCC1 is unlikely to be developed as an anticancer drug due to a lack of convincing evidence to date [13]. Therefore, we delved into the unknown area of the upstream regulation of ABCB1 by HNF1A.
By analyzing our clinical samples from PDAC, we found a significantly negative correlation between HNF1A and ABCB1 mRNA expression levels. Furthermore, the responses of PDAC cell lines to gemcitabine decreased after transfection with ABCB1. In addition, after the overexpression of ABCB1, the enhancement of gemcitabine sensitivity by the HNF1A overexpressing PDAC cell lines was eliminated. Kitanaka et al. reported that the MDR protein 2 (MPR2) promoter combined with HNF1A, and the enhancement of transactivity was observed MODY5 [45]. However, the results from HNF1A knockout mice demonstrated no change in hepatic expression of MPR2 [46]. Interestingly, we identified that the transcriptional activation of ABCB1 in PDAC cells was inhibited in stable HNF1A overexpressing cells and enhanced in HNF1A depleted cells. Moreover, by bioinformatic analyses, specific sequence mutation experiments and ChIP analysis, we confirmed that HNF1A regulates ABCB1 expression through binding to its specific promoter region (−989 bp to −747 bp) and inhibiting its transcription. The majority of HNF1A studies have been conducted in liver and/or pancreatic islet cells. They revealed direct expression regulatory effects of HNF1A that are tissue-specific and, sometimes, opposite effects in liver and islet cells [47]. Previous studies have identified that HNF1A contains a dimerization domain at the N-terminus, a DNA-binding domain in the middle region and a transactivation domain at the C-terminus of the protein [48]. However, in our research, we at first revealed that HNF1A suppressed transcription by directly binding to the specific promoter region of ABCB1, which was, to some extent, indicative of the complexity of the HNF family being involved in the expression of downstream genes and the paradox of the results from different studies [46,49]. Recently, Yao et al. revealed that promoter methylation of ABCB1 may be a valuable indicator of gemcitabine resistant characteristics in PDAC cells [50]. In addition, Suzuki et al. recently showed that some transcription factors, including HNF1A, are involved in the site-specific determination of DNA epigenetic modification in a binding site-directed manner [51]. Whether HNF1A methylates the DNA of ABCB1 promoter requires further exploration. Additionally, the p.Q511L mutation of HNF1A may suppress the transcriptional activity of HNF1A in hepatocellular carcinoma [52], which needed further study in pancreatic cancer and can expand our work.
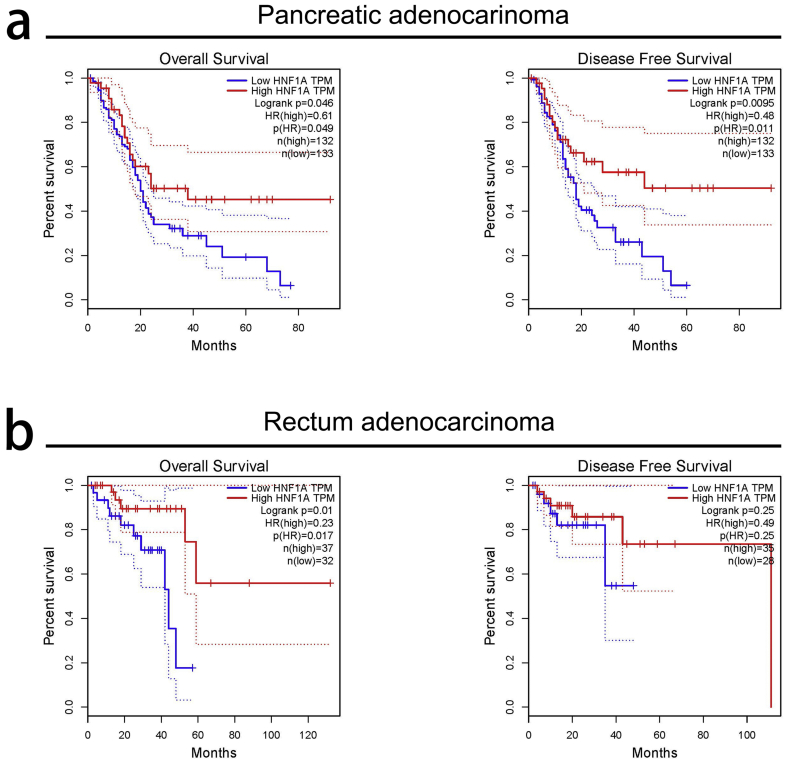
Finally, we provided evidence for the clinical value of HNF1A in the stratification of gemcitabine-sensitive pancreatic cancer patients. We showed that HNF1A staining was significantly associated with TNM stage and perineural invasion. By univariate and multivariate analyses, we revealed that patients with high/moderate HNF1A staining were significantly associated with both OS and DFS compared with low level groups. Importantly, the DFS analyses confirmed that the high HNF1A group showed the best survival, the low HNF1A group showed the worst survival, and the moderate HNF1A group showed an intermediate DFS. Consistently, analyses of TCGA database showed that high HNF1A levels were significantly correlated to the improvement of OS and DFS in pancreatic adenocarcinoma patients (Fig. S5a). In addition, similar results were found in patients with rectum adenocarcinoma (Fig. S5b). However, for the discrepancy of survival data between our research and the paper by Able et al., the different of tumor samples, the experimental methods and the analytical methods should be taken into consideration. Able et al. did not observed a significant association between HNF1A mRNA levels and survival (p = .7017), and they revealed that HNF1A upregulated gene were found to be significantly associated with poor survival. The samples used by Able et al. didn't provide the information about the postoperative chemotherapy, while our samples were collected from 128 pancreatic cancer patients, all of whom received regular gemcitabine chemotherapy after radical surgery. In addition, Able et al. used the HNF1A mRNA level to perform the survival analysis. Considering that pancreatic cancer is composed of a minority of malignant cells within a microenvironment of extracellular matrix, fibroblasts, endothelial cells, and immune cells, the HNF1A mRNA level may be interfered by non-cancer cells in the tumor microenvironment (such as tumor-associated fibroblasts). The immunohistochemistry assays and immunoreactive score (IRS) used in our paper may be more objective and had fewer confounding factors.
Interestingly, Noll et al. reported that HNF1A and KRT81 enable stratification of tumors into different subtypes (substantial differences in OS, and their tumors differed in drug sensitivity) by using IHC [53]. Similar to our results, the mean survival ranged from 43.5 months for HNF1-positive patients to only 16.5 months for the KRT81-positive patient. However, we and others were limited by the number of samples and the retrospective nature of the study; prospective clinical trials are needed to further verify of predictive value of HNF1A.
In conclusion, we determined a new role for HNF1A in the gemcitabine sensitivity of PDAC. We revealed that HNF1A mediated the resistance of PDAC cells to chemotherapy by directly binding to the specific region of the ABCB1 promoter and regulating its expression. HNF1A staining from PDAC tissues and clinical features paved the road for individualized treatment and improved the prognosis of PDAC patients.
The following are the supplementary data related to this article.
Fig. S1.
WB and IHC assays showed the expression of HNF1A in various samples. (a) The specificity of the antibody ab96777 was shown as full western blots of the whole cell lysates. (b) The expression of HNF1A in HNF1A knockout mice tumor tissues. (c) The expression of HNF1A in human lung tissues. (d) The expression of HNF1A in human ovarian tissues. (e) The expression of HNF1A in human prostate tissues. (f-g) The expression of HNF1A was high in human pancreas islet tissues. (h-j) The expression of HNF1A was differentiation in human PDAC tissues.
Fig. S2.
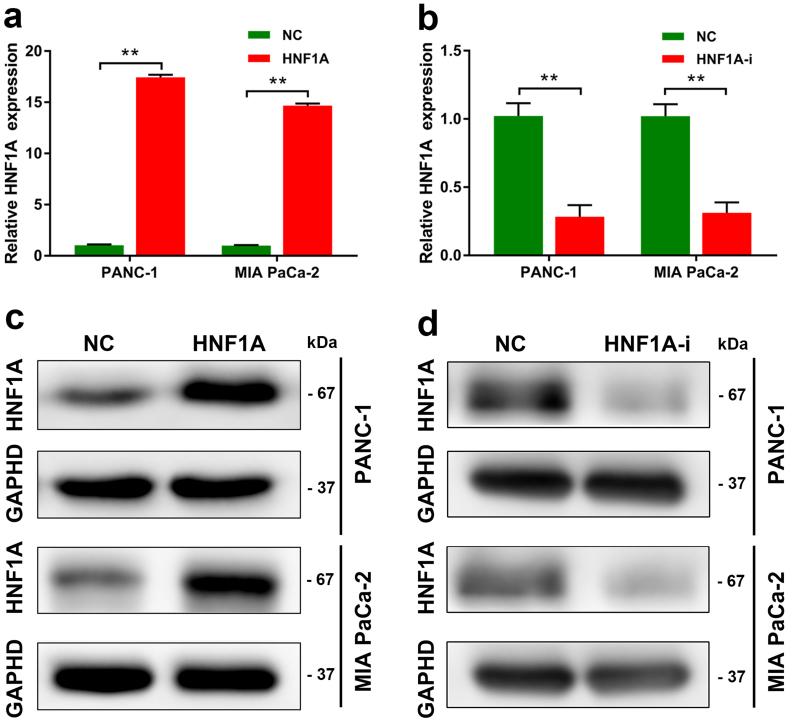
The expression of HNF1A in PDAC cell lines after the construction of lentivirus-based expression system. (a-b) Quantitative RT-PCR data showing the fold changes (2-∆∆CT) in the mRNA expression level of HNF1A after overexpression or inhibition of HNF1A in the PANC-1 and MIA PaCa-2 cell lines. (c-d) Western blot experiments showing the protein levels of HNF1A after overexpression of HNF1A in the PANC-1 and MIA PaCa-2 cell lines. The graphs shown are representative results of three repeated assays and are presented as mean ± SD. **p < .01 (Student's t-test).
Fig. S3.
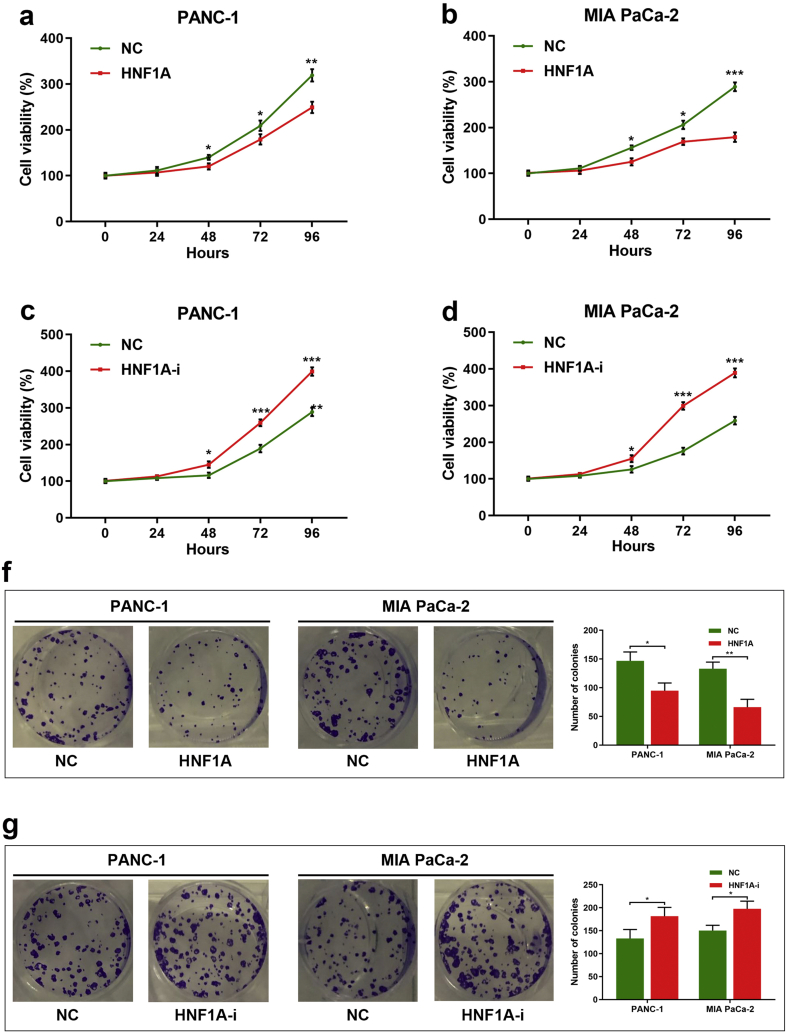
The effect of HNF1A on the basal growth of PANC-1 and MIA PaCa-2 cells. (a-b) MTS assays indicated that the proliferation capacity was significantly inhibited after overexpression of HNF1A in PANC-1 and MIA PaCa-2 cells. (c-d) MTS assays revealed that the proliferation capacity was substantially elevated after inhibition of HNF1A in PANC-1 and MIA PaCa-2 cells. (e) Colony formation assays showed that the proliferation capacity was significantly lower in HNF1A-overexpression PANC-1 and MIA PaCa-2 cells. (f) Colony formation assays showed that the proliferation capacity was significantly higher in HNF1A-inhibition PANC-1 and MIA PaCa-2 cells. Data are representative of three independent experiments and are presented as mean ± SD. *p < .05, **p < .01 (Student's t-test).
Fig. S4.
HNF1A enhances the sensitivity of gemcitabine in MIA PaCa-2 cells. (a) MTT assays indicated that the IC50 values of gemcitabine at 48 h and 72 h were all markedly decreased after overexpression of HNF1A followed by gemcitabine treatment. (b) MTT assays indicated that the IC50 values of gemcitabine at 48 h and 72 h were all markedly increased after inhibiting of HNF1A followed by gemcitabine treatment. (c) MTS assays indicated that the proliferation capacity significantly decreased in HNF1A-overexpressing cells after treatment with various concentrations of gemcitabine. (d) MTS assays revealed that the proliferation capacity significantly increased in HNF1A-inhibited cells after treatment with various concentrations of gemcitabine. (e) The flow cytometry assay showed that overexpression of HNF1A induced apoptosis in cells treated with gemcitabine at various concentrations (1, 5 or 10 μmol/L). (f) HNF1a inhibition resulted in increased resistance of the cells to treatment with various concentrations (1, 5 or 10 μmol/L) of gemcitabine, resulting in decreased apoptosis rates. The data are shown from 3 parallel experiments and presented as mean ± SD. *p < .05, **p < .01 (Student's t-test).
Fig. S5.
Analyses of TCGA database showed that high HNF1A levels predict better survival in pancreatic adenocarcinoma and rectum cancer patients. (a) High HNF1A levels were significantly correlated to the improvement of OS and DFS in pancreatic adenocarcinoma patients. (b) High HNF1A levels were strongly related to the improvement of OS and DFS in patients with rectum adenocarcinoma.
Primers used in PCR.
Oligonucleotide sequences for this study.
Acknowledgments
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (grant number 81571196, 81702417 and 81401689, http://www.nsfc.gov.cn), the National Natural Science Foundation of Guangdong Province (grant number 2017A030313880, http://pro.gdstc.gov.cn).
Conflict of interest
The authors declare no conflict of interest.
Ethics approval and consent to participate
The tissue samples used for the study were approved by the ethics committee of Sun Yat-sen Memorial Hospital of Sun Yat-sen University. Informed consent was obtained from all patients. The protocols for animal experiments were approved by the Institutional Animal Care and Use Committee and Institutional Biosafety Committee of Sun Yat-sen University.
Authors' contributions
Lu YN conceived and conducted most of the experiments, collected the data, analyzed the results and wrote most of the manuscript. Wang Z and Yin PH conceived the idea for the project, coordinated the research and modified the manuscript with Lu YN. Zheng MH did much work to improved quality of our work such as revising the paper meticulously, providing a professional editing, conducting the gain- and loss-of-functional experiments. Lu YN conducted the ChIP analysis, the dual luciferase reporter assay, immunofluorescence, the xenograft tumor assay, and the promoter deletion and mutation assay. Xu DN and Peng JT conducted the Western blotting, IHC, qRT-PCR and medical follow-up. Chen CJ, Chen YQ and Chen HM conducted the MTS assay, the colony formation assay and the flow cytometric assay. All authors approved the final version of the manuscript.
Contributor Information
Yanan Lu, Email: luyanan83@163.com.
Minghui Zheng, Email: zhengmh7@mail.sysu.edu.cn.
Peihong Yin, Email: emperoryin@hotmail.com.
Zhi Wang, Email: wwzz1898@sina.com.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Hidalgo M. Pancreatic cancer. N Engl J Med. 2010;362(17):1605–1617. doi: 10.1056/NEJMra0901557. [DOI] [PubMed] [Google Scholar]
- 3.de Sousa C.L., Monteiro G. Gemcitabine: metabolism and molecular mechanisms of action, sensitivity and chemoresistance in pancreatic cancer. Eur J Pharmacol. 2014;741:8–16. doi: 10.1016/j.ejphar.2014.07.041. [DOI] [PubMed] [Google Scholar]
- 4.Conroy T., Desseigne F., Ychou M., Bouche O., Guimbaud R., Becouarn Y. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med. 2011;364(19):1817–1825. doi: 10.1056/NEJMoa1011923. [DOI] [PubMed] [Google Scholar]
- 5.Von Hoff D.D., Ervin T., Arena F.P., Chiorean E.G., Infante J., Moore M. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl J Med. 2013;369(18):1691–1703. doi: 10.1056/NEJMoa1304369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neoptolemos J.P., Palmer D.H., Ghaneh P., Psarelli E.E., Valle J.W., Halloran C.M. Comparison of adjuvant gemcitabine and capecitabine with gemcitabine monotherapy in patients with resected pancreatic cancer (ESPAC-4): a multicentre, open-label, randomised, phase 3 trial. Lancet. 2017;389(10073):1011–1024. doi: 10.1016/S0140-6736(16)32409-6. [DOI] [PubMed] [Google Scholar]
- 7.Burris H.R., Moore M.J., Andersen J., Green M.R., Rothenberg M.L., Modiano M.R. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. J Clin Oncol. 1997;15(6):2403–2413. doi: 10.1200/JCO.1997.15.6.2403. [DOI] [PubMed] [Google Scholar]
- 8.Shukla S.K., Purohit V., Mehla K., Gunda V., Chaika N.V., Vernucci E. MUC1 and HIF-1alpha signaling crosstalk induces anabolic glucose metabolism to impart gemcitabine resistance to pancreatic cancer. Cancer Cell. 2017;32(1):71–87. doi: 10.1016/j.ccell.2017.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falasca M., Kim M., Casari I. Pancreatic cancer: current research and future directions. BBA Rev Cancer. 2016;1865(2):123–132. doi: 10.1016/j.bbcan.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 10.Olive K.P., Jacobetz M.A., Davidson C.J., Gopinathan A., McIntyre D., Honess D. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324(5933):1457–1461. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jia Y., Gu D., Wan J., Yu B., Zhang X., Chiorean E.G. The role of GLI-SOX2 signaling axis for gemcitabine resistance in pancreatic cancer. Oncogene. 2019;38(10):1764–1777. doi: 10.1038/s41388-018-0553-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deeley R.G., Cole S.P. Function, evolution and structure of multidrug resistance protein (MRP) Semin Cancer Biol. 1997;8(3):193–204. doi: 10.1006/scbi.1997.0070. [DOI] [PubMed] [Google Scholar]
- 13.Robey R.W., Pluchino K.M., Hall M.D., Fojo A.T., Bates S.E., Gottesman M.M. Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat Rev Cancer. 2018;18(7):452–464. doi: 10.1038/s41568-018-0005-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Vries A.G., Bakker-van W.W., Dassel A.C., Losekoot M., Duiker E.W., Gouw A.S. A novel phenotype of a hepatocyte nuclear factor homeobox A (HNF1A) gene mutation, presenting with neonatal cholestasis. J Hepatol. 2015;63(5):1295–1297. doi: 10.1016/j.jhep.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 15.Firdous P., Nissar K., Ali S., Ganai B.A., Shabir U., Hassan T. Genetic testing of maturity-onset diabetes of the young current status and future perspectives. Front Endocrinol (Lausanne) 2018;9:253. doi: 10.3389/fendo.2018.00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reiner A.P., Gross M.D., Carlson C.S., Bielinski S.J., Lange L.A., Fornage M. Common coding variants of the HNF1A gene are associated with multiple cardiovascular risk phenotypes in community-based samples of younger and older European-American adults: the Coronary Artery Risk Development in Young Adults Study and The Cardiovascular Health Study. Circ Cardiovasc Genet. 2009;2(3):244–254. doi: 10.1161/CIRCGENETICS.108.839506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li D., Duell E.J., Yu K., Risch H.A., Olson S.H., Kooperberg C. Pathway analysis of genome-wide association study data highlights pancreatic development genes as susceptibility factors for pancreatic cancer. Carcinogenesis. 2012;33(7):1384–1390. doi: 10.1093/carcin/bgs151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pierce B.L., Ahsan H. Genome-wide “pleiotropy scan” identifies HNF1A region as a novel pancreatic cancer susceptibility locus. Cancer Res. 2011;71(13):4352–4358. doi: 10.1158/0008-5472.CAN-11-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo Z., Li Y., Wang H., Fleming J., Li M., Kang Y. Hepatocyte nuclear factor 1A (HNF1A) as a possible tumor suppressor in pancreatic cancer. Plos One. 2015;10(3) doi: 10.1371/journal.pone.0121082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoskins J.W., Jia J., Flandez M., Parikh H., Xiao W., Collins I. Transcriptome analysis of pancreatic cancer reveals a tumor suppressor function for HNF1A. Carcinogenesis. 2014;35(12):2670–2678. doi: 10.1093/carcin/bgu193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maher J.M., Slitt A.L., Callaghan T.N., Cheng X., Cheung C., Gonzalez F.J. Alterations in transporter expression in liver, kidney, and duodenum after targeted disruption of the transcription factor HNF1alpha. Biochem Pharmacol. 2006;72(4):512–522. doi: 10.1016/j.bcp.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 22.Li J., Zhang Y., Gao Y., Cui Y., Liu H., Li M. Downregulation of HNF1 homeobox B is associated with drug resistance in ovarian cancer. Oncol Rep. 2014;32(3):979–988. doi: 10.3892/or.2014.3297. [DOI] [PubMed] [Google Scholar]
- 23.Zheng S., Chen H., Wang Y., Gao W., Fu Z., Zhou Q. Long non-coding RNA LOC389641 promotes progression of pancreatic ductal adenocarcinoma and increases cell invasion by regulating E-cadherin in a TNFRSF10A-related manner. Cancer Lett. 2016;371(2):354–365. doi: 10.1016/j.canlet.2015.12.010. [DOI] [PubMed] [Google Scholar]
- 24.Arumugam T., Ramachandran V., Fournier K.F., Wang H., Marquis L., Abbruzzese J.L. Epithelial to mesenchymal transition contributes to drug resistance in pancreatic cancer. Cancer Res. 2009;69(14):5820–5828. doi: 10.1158/0008-5472.CAN-08-2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fagerberg L., Hallstrom B.M., Oksvold P., Kampf C., Djureinovic D., Odeberg J. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014;13(2):397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ye H., Zhou Q., Zheng S., Li G., Lin Q., Wei L. Tumor-associated macrophages promote progression and the Warburg effect via CCL18/NF-kB/VCAM-1 pathway in pancreatic ductal adenocarcinoma. Cell Death Dis. 2018;9(5):453. doi: 10.1038/s41419-018-0486-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zlobec I., Terracciano L., Jass J.R., Lugli A. Value of staining intensity in the interpretation of immunohistochemistry for tumor markers in colorectal cancer. Virchows Arch. 2007;451(4):763–769. doi: 10.1007/s00428-007-0466-8. [DOI] [PubMed] [Google Scholar]
- 28.O'Brien V.P., Bokelmann K., Ramirez J., Jobst K., Ratain M.J., Brockmoller J. Hepatocyte nuclear factor 1 regulates the expression of the organic cation transporter 1 via binding to an evolutionary conserved region in intron 1 of the OCT1 gene. J Pharmacol Exp Ther. 2013;347(1):181–192. doi: 10.1124/jpet.113.206359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rahib L., Smith B.D., Aizenberg R., Rosenzweig A.B., Fleshman J.M., Matrisian L.M. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74(11):2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 30.Abel E.V., Goto M., Magnuson B., Abraham S., Ramanathan N., Hotaling E. HNF1A is a novel oncogene that regulates human pancreatic cancer stem cell properties. Elife. 2018;7 doi: 10.7554/eLife.33947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu Y., Liang S., Zhou Y., Li S., Li Y., Liao W. HNF1A/CASC2 regulates pancreatic cancer cell proliferation through PTEN/Akt signaling. J Cell Biochem. 2017;120(3):2816–2827. doi: 10.1002/jcb.26395. [DOI] [PubMed] [Google Scholar]
- 32.Bailey P., Chang D.K., Nones K., Johns A.L., Patch A.M., Gingras M.C. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531(7592):47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 33.Cheng Z., He Z., Cai Y., Zhang C., Fu G., Li H. Conversion of hepatoma cells to hepatocyte-like cells by defined hepatocyte nuclear factors. Cell Res. 2019;29(2):124–135. doi: 10.1038/s41422-018-0111-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takashima Y., Horisawa K., Udono M., Ohkawa Y., Suzuki A. Prolonged inhibition of hepatocellular carcinoma cell proliferation by combinatorial expression of defined transcription factors. Cancer Sci. 2018;109(11):3543–3553. doi: 10.1111/cas.13798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muckenhuber A., Berger A.K., Schlitter A.M., Steiger K., Konukiewitz B., Trumpp A. Pancreatic ductal adenocarcinoma subtyping using the biomarkers hepatocyte nuclear factor-1A and cytokeratin-81 correlates with outcome and treatment response. Clin Cancer Res. 2018;24(2):351–359. doi: 10.1158/1078-0432.CCR-17-2180. [DOI] [PubMed] [Google Scholar]
- 36.Kong B., Wu W., Valkovska N., Jager C., Hong X., Nitsche U. A common genetic variation of melanoma inhibitory activity-2 labels a subtype of pancreatic adenocarcinoma with high endoplasmic reticulum stress levels. Sci Rep. 2015;5:8109. doi: 10.1038/srep08109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones S., Zhang X., Parsons D.W., Lin J.C., Leary R.J., Angenendt P. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321(5897):1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shah R.N., Ibbitt J.C., Alitalo K., Hurst H.C. FGFR4 overexpression in pancreatic cancer is mediated by an intronic enhancer activated by HNF1alpha. Oncogene. 2002;21(54):8251–8261. doi: 10.1038/sj.onc.1206020. [DOI] [PubMed] [Google Scholar]
- 39.Mohelnikova-Duchonova B., Brynychova V., Oliverius M., Honsova E., Kala Z., Muckova K. Differences in transcript levels of ABC transporters between pancreatic adenocarcinoma and nonneoplastic tissues. Pancreas. 2013;42(4):707–716. doi: 10.1097/MPA.0b013e318279b861. [DOI] [PubMed] [Google Scholar]
- 40.Dorman S.N., Baranova K., Knoll J.H., Urquhart B.L., Mariani G., Carcangiu M.L. Genomic signatures for paclitaxel and gemcitabine resistance in breast cancer derived by machine learning. Mol Oncol. 2016;10(1):85–100. doi: 10.1016/j.molonc.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nath S., Daneshvar K., Roy L.D., Grover P., Kidiyoor A., Mosley L. MUC1 induces drug resistance in pancreatic cancer cells via upregulation of multidrug resistance genes. Oncogenesis. 2013;2:e51. doi: 10.1038/oncsis.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hong S.P., Wen J., Bang S., Park S., Song S.Y. CD44-positive cells are responsible for gemcitabine resistance in pancreatic cancer cells. Int J Cancer. 2009;125(10):2323–2331. doi: 10.1002/ijc.24573. [DOI] [PubMed] [Google Scholar]
- 43.Cole S.P., Bhardwaj G., Gerlach J.H., Mackie J.E., Grant C.E., Almquist K.C. Overexpression of a transporter gene in a multidrug-resistant human lung cancer cell line. Science. 1992;258(5088):1650–1654. doi: 10.1126/science.1360704. [DOI] [PubMed] [Google Scholar]
- 44.Mirski S.E., Gerlach J.H., Cole S.P. Multidrug resistance in a human small cell lung cancer cell line selected in adriamycin. Cancer Res. 1987;47(10):2594. [PubMed] [Google Scholar]
- 45.Sachiko K., Utako S., Takashi I. Regulation of human insulin, IGF-I, and multidrug resistance protein 2 promoter activity by hepatocyte nuclear factor (HNF)-1beta and HNF-1alpha and the abnormality of HNF-1beta mutants. J Endocrinol. 2007;192(1):141–147. doi: 10.1677/joe.1.07003. [DOI] [PubMed] [Google Scholar]
- 46.Shih D.Q., Bussen M., Sehayek E., Ananthanarayanan M., Shneider B.L., Suchy F.J. Hepatocyte nuclear factor-1alpha is an essential regulator of bile acid and plasma cholesterol metabolism. Nat Genet. 2001;27(4):375–382. doi: 10.1038/86871. [DOI] [PubMed] [Google Scholar]
- 47.Qadri I., Hu L.J., Iwahashi M., Al-Zuabi S., Quattrochi L.C., Simon F.R. Interaction of hepatocyte nuclear factors in transcriptional regulation of tissue specific hormonal expression of human multidrug resistance-associated protein 2 (abcc2) Toxicol Appl Pharmacol. 2009;234(3):281–292. doi: 10.1016/j.taap.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 48.Chi Y.I., Frantz J.D., Oh B.C., Hansen L., Dhe-Paganon S., Shoelson S.E. Diabetes mutations delineate an atypical POU domain in HNF-1alpha. Mol Cell. 2002;10(5):1129–1137. doi: 10.1016/s1097-2765(02)00704-9. [DOI] [PubMed] [Google Scholar]
- 49.Bristeau A., Catherin A.M., Weiss M.C., Faust D.M. Hormone response of rodent phenylalanine hydroxylase requires HNF1 and the glucocorticoid receptor. Biochem Biophys Res Commun. 2001;287(4):852–858. doi: 10.1006/bbrc.2001.5673. [DOI] [PubMed] [Google Scholar]
- 50.Yao L., Gu J., Mao Y., Zhang X., Wang X., Jin C. Dynamic quantitative detection of ABC transporter family promoter methylation by MS-HRM for predicting MDR in pancreatic cancer. Oncol Lett. 2018;15(4) doi: 10.3892/ol.2018.8041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suzuki T., Maeda S., Furuhata E., Shimizu Y., Nishimura H., Kishima M. A screening system to identify transcription factors that induce binding site-directed DNA demethylation. Epigenetics Chromatin. 2017;10(1):60. doi: 10.1186/s13072-017-0169-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ding C.H., Deng L.F., Chen F., Ding K., Chen W.S., Xie W.F. p.Q511L mutation of HNF1alpha in hepatocellular carcinoma suppresses the transcriptional activity and the anti-tumor effect of HNF1alpha. Biochem Biophys Res Commun. 2018;495(1):86–91. doi: 10.1016/j.bbrc.2017.10.174. [DOI] [PubMed] [Google Scholar]
- 53.Noll E.M., Eisen C., Stenzinger A., Espinet E., Muckenhuber A., Klein C. CYP3A5 mediates basal and acquired therapy resistance in different subtypes of pancreatic ductal adenocarcinoma. Nat Med. 2016;22(3):278–287. doi: 10.1038/nm.4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used in PCR.
Oligonucleotide sequences for this study.