A Newcastle disease virus was isolated from a chicken from a live bird market in the Mbeya region of Tanzania. Complete genome characterization of the isolate identified it as a member of subgenotype Vd. This is the first complete genome sequence of this subgenotype.
ABSTRACT
A Newcastle disease virus was isolated from a chicken from a live bird market in the Mbeya region of Tanzania. Complete genome characterization of the isolate identified it as a member of subgenotype Vd. This is the first complete genome sequence of this subgenotype.
ANNOUNCEMENT
Newcastle disease virus (NDV), a member of the genus Avulavirus within the family Paramyxoviridae, has a negative-sense single-stranded nonsegmented RNA genome (1). Although it represents a single serotype, NDV has at least 19 different genotypes divided into two distinct classes (2, 3). To date, only partial genomes of subgenotype Vd viruses are available in public databases (4, 5). In this study, we report the first isolation of a subgenotype Vd virus from Tanzania. This virus was isolated from a chicken in 2012, and the obtained sequence represents the first subgenotype Vd complete genome.
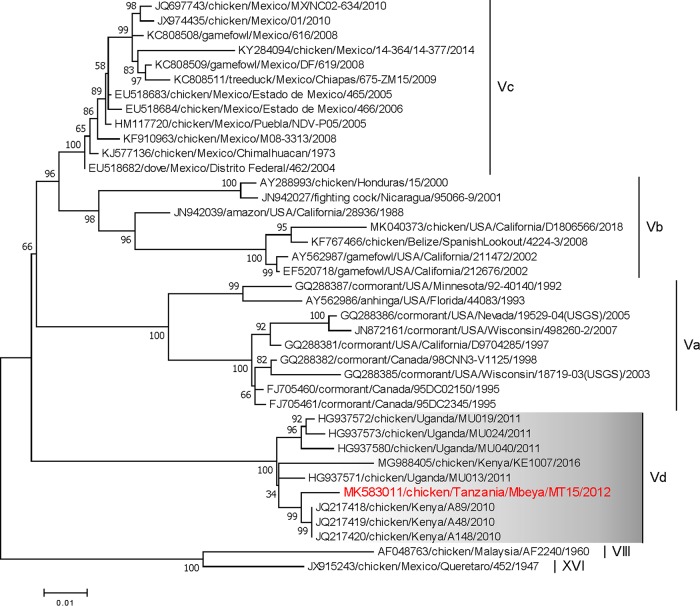
A virulent NDV from a cloacal swab collected from a chicken at a live bird market in the Mbeya region of Tanzania in 2012 was isolated at the Southeast Poultry Research Laboratory of the USDA in Athens, GA. The virus was propagated in 9-day-old specific-pathogen-free embryonating chicken eggs, and the intracerebral pathogenicity index (ICPI) was estimated following standard procedures (6). Viral RNA was isolated from allantoic fluid using the QIAamp viral RNA minikit (Qiagen, USA). The Illumina libraries were prepared using a stranded RNA-Seq library preparation kit (KAPA Biosystems, USA) per the manufacturer’s instructions. The distribution size and concentration of the prepared libraries were checked on a Bioanalyzer 2100, using a high-sensitivity (HS) DNA kit (Agilent Technologies, Germany), and a Qubit fluorometer, using a double-stranded DNA (dsDNA) HS assay kit (Life Technologies, USA), respectively. Next-generation paired-end sequencing (2 × 250 bp) was performed on a MiSeq instrument using the 500-cycle MiSeq reagent kit v.2 (Illumina, USA). Sequence data were assembled using a de novo approach and utilizing MIRA v.3.4.1 (7) within a customized workflow on the Galaxy platform (8), as described previously (9). A total of 805,566 raw paired-end reads were generated. A single NDV contig was assembled. The final consensus was 15,180 nucleotides (nt) long (99.9% genome coverage), was called from 508,118 NDV raw reads using BWA-MEM (10), and had 46.6% GC content. The median read depth coverage of the NDV assembly was 5,508×, and the maximum depth coverage was 9,354×. Six nucleotides missing at both the 5ʹ and 3ʹ ends of the obtained consensus sequence were sequenced utilizing a single 3′-nucleotide tailing reaction of both the genomic RNA and the full-length positive-sense antigenomic RNA, followed by a single reverse transcription reaction targeted to the common polynucleotide tails, as described previously (11). The final genome consensus of the isolated strain, designated chicken/Tanzania/Mbeya/MT15/2012 (here, MT15), comprises 15,192 nucleotides and was annotated using the annotation tool of Geneious 9.1.8. The genome complies with the paramyxovirus “rule of six” (12) and contains six open reading frames (ORFs) (3′-NP-P-M-F-HN-L-5′) of 1,470 nt, 1,188 nt, 1,095 nt, 1,662 nt, 1,716 nt, and 6,615 nt in length, respectively. The ORFs were identified using Geneious and confirmed by alignment with published NDV genomes. A preliminary BLAST search of the complete genome sequence showed 99% nucleotide identity with the genotype V incomplete genomes of chicken/Kenya/A89/2010 (14,888 nt; GenBank accession number JQ217418) and chicken/Kenya/A48/2010 (14,945 nt; GenBank accession number JQ217419). Detailed phylogenetic analysis based on the complete fusion gene (13) classified MT15 as a member of subgenotype Vd, together with isolates from Kenya (obtained in 2010 to 2016) and Uganda (obtained in 2011) (Fig. 1). The MT15 isolate shared 98.3% and 97.6% nucleotide identity with the Kenyan and Ugandan viruses, respectively.
FIG 1.
Phylogenetic analysis of NDV isolates of genotype V; this was based on the complete fusion gene sequences constructed with the maximum likelihood method from the general time-reversible model in MEGA version 7.0. The tree with the highest log likelihood (−7,045.63) is shown. The percentage for which the associated taxa clustered together is shown next to each branch. The initial tree(s) for the heuristic search was obtained automatically by applying neighbor-joining and BioNJ algorithms to a matrix of pairwise distances estimated using the maximum composite likelihood (MCL) approach, and then the topology with the superior log likelihood value was selected. A discrete gamma distribution was used to model evolutionary rate differences among sites (5 categories [+G, parameter = 0.4593]). The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 33.48% of the sites). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 39 nucleotide sequences (sequences from genotypes VIII and XVI are included as an outgroup). All positions containing gaps and missing data were eliminated. There were a total of 1,646 positions in the final data set. The isolate used in this study is shown in red.
According to the World Organisation for Animal Health, an ICPI value of 0.7 or greater or the presence of at least three basic amino acids at the fusion protein cleavage site indicates a virulent form of NDV (6), while values above 1.5 are typical for velogenic viruses (14). Analysis of the deduced amino acid sequence of the fusion protein cleavage site (6, 15) of MT15 showed a polybasic amino acid motif and a phenylalanine at position 117 (112RRQKR↓F117), which is typical for virulent NDV strains. This result was consistent with the ICPI value of 1.86 (6). Velogenic Newcastle disease viruses are important pathogens, and the sequence information provides valuable information for characterizing and tracking the viruses.
Data availability.
The complete genome sequence of chicken/Tanzania/Mbeya/MT15/2012 has been deposited in GenBank under the accession number MK583011. Raw data were deposited in the SRA under accession number SRR9071773, BioSample number SAMN11660474, and BioProject number PRJNA543308.
ACKNOWLEDGMENTS
We acknowledge Timothy Olivier and Suzanne DeBlois for their technical assistance in characterizing these viruses.
The mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
This study was supported by USDA CRIS projects 6040-32000-066 and 6040-32000-072 and the Centers for Excellence in Influenza Research agreement number AAI12004-001-02001.
REFERENCES
- 1.Amarasinghe GK, Aréchiga Ceballos NG, Banyard AC, Basler CF, Bavari S, Bennett AJ, Blasdell KR, Briese T, Bukreyev A, Caì Y, Calisher CH, Campos Lawson C, Chandran K, Chapman CA, Chiu CY, Choi K-S, Collins PL, Dietzgen RG, Dolja VV, Dolnik O, Domier LL, Dürrwald R, Dye JM, Easton AJ, Ebihara H, Echevarría JE, Fooks AR, Formenty PBH, Fouchier RAM, Freuling CM, Ghedin E, Goldberg TL, Hewson R, Horie M, Hyndman TH, Jiāng D, Kityo R, Kobinger GP, Kondō H, Koonin EV, Krupovic M, Kurath G, Lamb RA, Lee B, Leroy EM, Maes P, Maisner A, Marston DA, Mor SK, Müller T, et al. 2018. Taxonomy of the order Mononegavirales: update 2018. Arch Virol 163:2283–2294. doi: 10.1007/s00705-018-3814-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dimitrov KM, Ramey AM, Qiu X, Bahl J, Afonso CL. 2016. Temporal, geographic, and host distribution of avian paramyxovirus 1 (Newcastle disease virus). Infect Genet Evol 39:22–34. doi: 10.1016/j.meegid.2016.01.008. [DOI] [PubMed] [Google Scholar]
- 3.Snoeck CJ, Owoade AA, Couacy-Hymann E, Alkali BR, Okwen MP, Adeyanju AT, Komoyo GF, Nakoune E, Le Faou A, Muller CP. 2013. High genetic diversity of Newcastle disease virus in poultry in West and Central Africa: cocirculation of genotype XIV and newly defined genotypes XVII and XVIII. J Clin Microbiol 51:2250–2260. doi: 10.1128/JCM.00684-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ogali IN, Wamuyu LW, Lichoti JK, Mungube EO, Agwanda B, Ommeh SC. 2018. Molecular characterization of Newcastle disease virus from backyard poultry farms and live bird markets in Kenya. Int J Microbiol 2018:2368597. doi: 10.1155/2018/2368597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Byarugaba DK, Mugimba KK, Omony JB, Okitwi M, Wanyana A, Otim MO, Kirunda H, Nakavuma JL, Teillaud A, Paul MC, Ducatez MF. 2014. High pathogenicity and low genetic evolution of avian paramyxovirus type I (Newcastle disease virus) isolated from live bird markets in Uganda. Virol J 11:173. doi: 10.1186/1743-422X-11-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.OIE. 2012. Newcastle disease (infection with Newcastle disease virus), p 964–983. Manual of diagnostic tests and vaccines for terrestrial animals, 8th ed, vol 1. World Organisation for Animal Health, Paris, France. [Google Scholar]
- 7.Chevreux B, Wetter T, Suhai S. 1999. Genome sequence assembly using trace signals and additional sequence information, p 45–56. In Computer science and biology: proceedings of the German Conference on Bioinformatics, GCB ’99. GCB, Hannover, Germany. [Google Scholar]
- 8.Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, Čech M, Chilton J, Clements D, Coraor N, Eberhard C, Grüning B, Guerler A, Hillman-Jackson J, Von Kuster G, Rasche E, Soranzo N, Turaga N, Taylor J, Nekrutenko A, Goecks J. 2016. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res 44:W3–W10. doi: 10.1093/nar/gkw343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dimitrov KM, Sharma P, Volkening JD, Goraichuk IV, Wajid A, Rehmani SF, Basharat A, Shittu I, Joannis TM, Miller PJ, Afonso CL. 2017. A robust and cost-effective approach to sequence and analyze complete genomes of small RNA viruses. Virol J 14:72. doi: 10.1186/s12985-017-0741-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown PA, Briand FX, Guionie O, Lemaitre E, Courtillon C, Henry A, Jestin V, Eterradossi N. 2013. An alternative method to determine the 5′ extremities of non-segmented, negative sense RNA viral genomes using positive replication intermediate 3′ tailing: application to two members of the Paramyxoviridae family. J Virol Methods 193:121–127. doi: 10.1016/j.jviromet.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 12.Kolakofsky D, Pelet T, Garcin D, Hausmann S, Curran J, Roux L. 1998. Paramyxovirus RNA synthesis and the requirement for hexamer genome length: the rule of six revisited. J Virol 72:891–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diel DG, da Silva LH, Liu H, Wang Z, Miller PJ, Afonso CL. 2012. Genetic diversity of avian paramyxovirus type 1: proposal for a unified nomenclature and classification system of Newcastle disease virus genotypes. Infect Genet Evol 12:1770–1779. doi: 10.1016/j.meegid.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 14.Alexander DSD. 1998. Newcastle disease virus and other avian paramyxoviruses, p 156–163. In Swayne DE, Glisson JR, Jackwood MW, Pearson JE, Reed WM (ed), A laboratory manual for the isolation and identification of avian pathogens, 4th ed. American Association of Avian Pathologists, Kennett Square, PA. [Google Scholar]
- 15.Peeters BP, de Leeuw OS, Koch G, Gielkens AL. 1999. Rescue of Newcastle disease virus from cloned cDNA: evidence that cleavability of the fusion protein is a major determinant for virulence. J Virol 73:5001–5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The complete genome sequence of chicken/Tanzania/Mbeya/MT15/2012 has been deposited in GenBank under the accession number MK583011. Raw data were deposited in the SRA under accession number SRR9071773, BioSample number SAMN11660474, and BioProject number PRJNA543308.