Abstract
Background
Energy metabolism in carcinogenesis is poorly understood. It is widely accepted the majority of colorectal cancers (CRCs) arise from adenomatous polyps (APs). We aimed to characterize the bioenergetic alterations in APs and CRCs.
Methods
Fifty-six APs, 93 CRCs and adjacent normal mucosae were tested. Oxygen consumption rate (OCR) was measured representing mitochondrial oxidative phosphorylation (OxPhos), and extracellular acidification rate (ECAR)was measured representing glycolysis. Mitochondrial DNA (mtDNA) variants and mutations were studied. Over-expressed metabolic genes in APs were identified by microarray and validated by qRT-PCR, Western blots and immunohistochemistry. Identified genes were knocked down in WiDr and colo205 CRC cell lines, and their expression was analyzed in APs/CRCs with enhanced glycolysis.
Findings
ECAR, not OCR, was significantly increased in APs. While no difference of ECAR was found between CRCs and normal mucosae, OCR was significantly reduced in CRCs. OCR/ECAR ratio was decreased in APs over 1 cm, APs with a villous component and CRCs, indicating their glycolytic tendencies. The number of mtDNA mutations was increased in APs and CRCs, but not correlated with metabolic profiles. Two metabolic genes ALDOB and SLC16A4 were up-regulated in APs. Both ALDOB-knockdown and SLC16A4-knockdown CRC cell lines showed increased basal motichondrial OxPhos and decreased basal glycolysis. Moreover, the increase of mitochondrial ATP-linked respiration and the decrease of glycolytic capacity were showed in SLC16A4-knockdown cells. Finally, APs/CRCs with enhanced glycolysis had increased SLC16A4 expression.
Interpretation
ATP production shifts from OxPhos to glycolysis in the process of AP enlargement and villous transformation. OxPhos defects are present in CRCs but not in APs. APs and CRCs tend to accumulate mtDNA mutations, but these are not correlated with bioenergetic profiles. Finally, the ALDOB and SLC16A4 may contribute to the glycolytic shift in APs/CRCs.
Keywords: Bioenergetics, Mitochondrial oxidative phosphorylation, Glycolysis, Colorectal adenomatous polyp, Colorectal cancer
Research in context.
Evidence before this study
The mitochondrial DNA (mtDNA) mutations and bioenergetic alterations from oxidative phosphorylation (OxPhos) to aerobic glycolysis have both been observed in colorectal cancers (CRCs). It was speculated that mtDNA mutations disrupt electron transport chain proteins, further inhibiting OxPhos and enhancing glycolysis, however, whether mtDNA mutations are associated with the bioenergetic changes in human adenomatous polyps (APs) and CRCs is not clear. Moreover, it has been demonstrated that dynamic bioenergetic changes occur in cancer progression in an ulcerative colitis model. However, no previous study has assessed the bioenergetic alterations in the human adenoma-carcinoma sequence.
Added value of this study
This study demonstrates that mtDNA mutations accumulate in APs and CRCs, but are not associated with their bioenergetics profiles. The OxPhos defects are present in CRCs but not in APs. In APs, ATP production shifts from OxPhos to glycolysis in the process of polyp enlargement and villous transformation. The over-expression of ALDOB and SLC16A4 may contribute to the glycolytic shift in APs/CRCs.
Implications of all the available evidence
The bioenergetic shift from OxPhos to glycolysis occurs during the process of the adenoma-carcinoma sequence, possibly through the enhancement of ALDOB and SLC16A4. The OxPhos defects mainly occur in CRCs. mtDNA mutations accumulate during the adenoma-carcinoma sequence, but these mutations are seemingly not associated with the bioenergetic changes.
Alt-text: Unlabelled Box
1. Introduction
In normal cells under aerobic conditions, energy is mainly generated through mitochondrial oxidative phosphorylation (OxPhos). Over 90 years ago, Otto Warburg discovered that cancer cells secreted more lactate than normal cells under aerobic conditions, indicating their higher glucose usage [1]. Because impaired mitochondrial metabolism and increased glycolysis have been observed in colorectal cancers (CRCs), it has been hypothesized that bioenergetic alterations are involved in CRC carcinogenesis [2].
The majority of CRCs develop from adenomatous polyps (APs) through the chromosomal instability pathway, named the adenoma-carcinoma sequence [3]. Several genomic changes are involved, including the activation of the KRAS proto-oncogene and the inactivation of at least three tumor suppressor genes, APC, p53 and 18q LOH. Although the genetic alterations have been extensively studied in this process [4], the bioenergetic alterations are still unclear.
It was initially believed that aerobic glycolysis in cancer cells was caused by damaged mitochondria [5]. The critical initiating events associated with mitochondrial dysfunction include reactive oxygen species (ROS) formation, glutathione depletion and protein alkylation. The ROS can cause mitochondrial DNA (mtDNA) mutation, stimulation of apoptotic pathways and increased propensity for necrosis due to multi-level failure to synthesize ATP [6]. MtDNA mutations have been found in various tumors, and in some cases, the depression of mitochondrial respiratory function is clearly a consequence of disruptive mtDNA mutations [7,8]. In CRCs, mitochondrial dysfunction has been largely analyzed at the DNA level. By complete genome sequencing, mtDNA mutations have been found in human normal mucosae, APs and CRCs [[9], [10], [11], [12]]. While approximately one-fifth of the somatic substitutions were detected in rRNA genes, the vast majority of somatic substitutions were observed in protein coding genes. It was speculated that the acquisition of somatic mtDNA mutations disrupt electron transport chain proteins, further inhibiting OxPhos [13]. However, it is still not clear whether mtDNA mutations are associated with the changes of OxPhos and glycolysis in CRCs.
Using ulcerative colitis as a model, Ussakli et al. demonstrated the dynamic changes of mitochondria during tumor progression [14]. They showed that cytochrome c oxidase (COX) activity progressively decreased with proximity to dysplasia and was the lowest in tissue adjacent to dysplasia, but was statistically increased in cancer, suggesting that mitochondrial loss contributes to the development of dysplasia but then mitochondrial function is restored in cancer enabling further cell proliferation. However, questions still remain; it is not clear whether COX activity represents the function of mitochondrial OxPhos. Furthermore, the CRCs arising from ulcerative colitis are mainly in the setting of chronic inflammation, probably not representing the bioenergetic changes in all CRC carcinogenesis pathways.
This study aims to answer the fundamental question of whether bioenergetic changes occur during the adenoma-carcinoma sequence. We found that the cellular ATP production shifted from OxPhos to glycolysis in the process of polyp enlargement and villous transformation. The defects of OxPhos were mainly present in CRCs, but not in APs. APs and CRCs had more mtDNA mutations, but the existence of mutations was not correlated with their metabolic profiles. Using microarray analysis, two metabolic-related genes ALDOB (Aldolase B, Fructose-Bisphosphate) and SLC16A4 (solute carrier family 16 member 4) were found to be up-regulated in APs with a villous component and may contribute to the glycolytic shift in APs. The increased expression of ALDOB and SLC16A4 in APs and CRCs suggests they may play a pivotal role in promoting glycolysis.
2. Material and methods
2.1. Patients
The pathologic examination and further functional assays were carried out under informed consents of patients. This study protocol was approved by the Medical Ethics and Human Clinical Trial Committee of the CGMH (IRB NO. 101-0102B). Patients with previous cancer or inflammatory bowel disease history were excluded. Fifty-six APs, 93 CRCs and their adjacent normal mucosae were obtained from patients by either biopsy forceps, polypectomy, endoscopic mucosal resection or surgical resection. Once specimens were obtained, a small piece of tissue was sent for extracellular flux analysis within 2 h. Another small piece of tissue was sent for DNA and RNA analysis. Most of the tissue was fixed in formalin and sent for pathological examination. The clinicopathological data were collected including age, gender, location, size, tumor invasion, regional lymph node involvement and tumor histology.
1109 of patients underwent both fluorine-18-2-fluoro-2-deoxy-d-glucose positron emission tomography and computed tomography (FDG PET/CT) and colonoscopy in a health check-up from Jan 2007 to Dec 2011 were retrospectively reviewed [15]. A total 36 patients had advanced colorectal neoplasm (malignancy, an adenoma ≥ 1 cm, or hsitological evidence of high-grade dysplasia or significant villous component). Six of the 38 neoplasms were also detected by FDG PET/CT. 6 APs and 6 CRCs paraffin embedded blocks were obtained from tissue bank and subjected to immunohistochemical staining.
2.2. Extracellular flux analysis
A Seahorse XF analyzer (Seahorse Bioscience, Billerica, MA) was employed to simultaneously measure the oxygen consumption rates (OCR; representing the function of mitochondrial OxPhos) and extracellular acidification rates (ECAR; representing the function of glycolysis) of tissues over time. OCR and ECAR were reported as absolute rates normalized against measured protein (mMol/min/mg for OCR and milli pH (mpH)/min/mg for ECAR). The metabolic profile of tissues was measured using a Seahorse XF24 analyzer (Seahorse Bioscience) according to the manufacturer's instructions and as previously described [[16], [17], [18]]. Briefly, freshly isolated tissue was rinsed with the assay medium (unbuffered Dulbecco's modified Eagle medium (pH 7.4)) and kept on ice. The tissue was cut into 1 mm slices by hand, and equal amount of specimens were placed in each well of an XF 24 Islet Capture Microplate (Cat No. 101174–100, Seahorse Bioscience). The tissue was then covered with an islet capture screen, which allowed free perfusion while minimizing the tissue movement. Assay medium (500 μl) was then added to each well. The microplate was incubated at 37 °C in a non-CO2 incubator for 30 min. The Seahorse XF24–3 analyzer was then employed to simultaneously measure OCR and ECAR. During the measurement, tissue was not pushed by the sensor. At least three replicates from each tissue were used for the assays. The specimen was transferred from the colonoscopy suite or operation theather to the seahorse platform within two hours.
To test the effects of ALDOB and SLC16A4 gene expression on bioenergetic changes, WiDr and colo205 cells with knockdown of ALDOB or SLC16A4 were further established, respectively. The control and ALDOB- or SLC16A4-inhibited cells were seeded into 24 wells of a XF 24-cell culture microplate at a density of 7 × 104 or 5 × 104 cells/well and incubated in a 5% CO2/air atmosphere at 37 °C for 24 h. OCR and ECAR were then measured under basal conditions.
The Mito Stress test Kit (Seahorse Bioscience, Billerica, MA) was used to reveal key parameters of the metabolic function of ALDOB- and SLC16A4-inhibited cells. OCR was measured under basal conditions and in the presence of the ATP synthase inhibitor oligomycin (1 μM), the mitochondrial uncoupler carbonyl cyanide-4-(trifluoromethoxy)phenyl-hydrazone (FCCP; mitochondrial respiration uncoupler, 0.5 μM), and the respiratory chain inhibitor antimycin A (0.5 μM).
The Glycolysis Stress test Kit (Seahorse Bioscience, Billerica, MA) was used to measure ECAR under various conditions. Three baseline recordings were made, followed by sequential injection of glucose (10 mM), the ATP synthase inhibitor oligomycin (1 μM) and the glycolysis inhibitor 2-deoxy-d-glucose (2-DG; 100 mM). The assays were performed in at least three replicates. All values for OCR and ECAR in cells and tissues were normalized to the protein content of the individual wells.
2.3. The protein measurement of tested tissues/cells
After Seahorse analyzer analysis, screen was removed. Aspirate off the media and wash wells with PBS. Samples were lysed in RIPA buffer with protease inhibitors. The lysate was sonicated on ice and incubated 30 min at 4 °C. After 30 min, the lysate was centrifuged at 12000 rpm for 30 min at 4 °C. The protein concentration was determined by Bradford protein assay.
2.4. mtDNA sequencing
The histology of tissues obtained directly from normal mucosae, APs and CRCs were confirmed by frozen section examination and 1mm3 piece of tissues were sent for mtDNA sequencing. From each tissue, the entire mitochondrial genome was obtained in a two-round amplification method as previously described [9]. The products of polymerase chain reaction (PCR) were sequenced by using BigDye version 3.1 Terminator cycle sequencing chemistries on an ABI-Prism 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA) and compared directly with the revised Cambridge reference sequence (rCRS) by using 4Peaks software (Mekentosj BV, Aalsmeer, The Netherlands).
2.5. RNA extraction, microarray analysis, and real-time Q-PCR
Total RNA was isolated with TRIzol reagent (Invitrogen, Carlsbad, CA). Gene expression profiles of the human APs and adjacent normal mucosae were analyzed with a human gene 2.0 array (Affymetrix, Santa Clara, CA), as previously described [19]. For real-time Q-PCR, first strand cDNA was synthesized with an oligo-T primer using superscript III first strand synthesis kit (Invitrogen, Carlsbad, CA). Gene expression of ALDOB, SLC16A4 and GAPDH mRNA were analyzed with the TaqMan® GENE expression assay (Applied Biosystems, Foster City, CA).
2.6. Protein extraction and Western blot assay
The tissues were homogenized and lysed in ice-cold RIPA lysis buffer (150 mM NaCl, 1.0% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 50 mM Tris [pH 7.5], 1 mM PMSF, 10 μg/ml Leupeptin). The lysates were separated by centrifugation and the supernatants were collected, aliquoted, and stored until analyzed by Western blot. 20 μg of total protein per lane were separated by a 10% SDS-PAGE gel and then transferred to a polyvinylidene fluoride membrane. The membrane was blocked by 5% FBS in 1× Tris-Buffered Saline with 0.1% Tween-20 and then incubated with an anti-ALDOB antibody (dilution 1:1000, Abcam, Cambridge, MA, USA), a anti-SLC16A4 antibody (1:1000 Abcam) and a anti-GAPDH antibody (1:15000, Proteintech, Chicago, IL, USA). The membrane was then incubated with diluted horseradish peroxidase-linked anti-rabbit IgG (1:5000, Cell Signaling Technology, Boston, MA, USA) for 1 h at room temperature. The membranes were washed for 5 min (3×) between each step. The protein-antibody complexes were detected by the chemiluminescent substrate (Cell Signaling), the emitted light captured on an X-ray film, and the intensities of bands were semi-quantified by ImageJ software.
2.7. Immunohistochemistry
Paraffin embedded sections (4 μm thick) were treated for immunohistochemistry as previously described [20]. Primary antibodies anti-ALDOB (1:500 Abcam) and anti-SLC16A4 (1:100, Abcam) were applied for 1 h at room temperature in a humid chamber. Sections were washed three times and then incubated for 40 min at room temperature with appropriate secondary antibodies conjugated to biotin. After washing, sections were incubated for 40 min with a tertiary layer of streptavidin–horseradish peroxidase (1:500, Dako). Peroxidase activity was revealed using 4 mmol/L 3,3-diaminobenzidine as a chromogen in PBS containing 0.2% hydrogen peroxide. Sections were counterstained with hematoxylin, dehydrated through ascending alcohols, cleared in xylene and mounted in mounting medium.
2.8. Establishment of ALDOB-inhibited and SLC16A4-inhibited cell lines
The clones (TRCN0000052508 and TRCN0000038444) of shRNA targeting ALDOB and SLC16A4 were purchased from the National RNA Interference Core Facility (Institute of Molecular Biology, Academia Sinica, Taiwan). Single shRNA plasmids and virus package plasmids (pCMV-△R8.91 and pMD.G) were co-transfected into 293FT cells and the cells were harvested after 72 h of transfection. A pool of stably infected cells was selected in medium containing puromycin.
2.9. Evaluation of the immunohistochemical staining
Expression of ALDOB and SLC16A4 were evaluated with a three-tiered system for the intensity: 1 + (weak), 2+ (moderate), 3+ (strong). Weak, moderate and strong staining was indicated when the staining was clearly visible at x400, x200 and x100 magnification respectively. The stained sections were examined separately by two experienced pathologists who were blinded to the clinical information. If there was a discrepancy in the interpretation, a consensus was reached between the two pathologists by reviewing the slides together.
2.10. Statistics
Data are expressed as mean ± standard error of the mean (SEM). Paired t-test, or unpaired t-test were used, with p < .05 considered statistically significant. Results are presented as means ± SD of three independent experiments.
3. Results
3.1. The clinicopathological factors associated with OxPhos and glycolysis in normal colonic mucosa, AP and CRC
The OCR and the ECAR of colonic APs and adjacent normal mucosae were measured and correlated with clinicopathological features (Table 1). In normal mucosae obtained from patients with APs, OCR was not associated with clinicopathological factors including age, gender, AP location, size and histological type. Similarly, no differences of ECAR were found with most factors except location. The ECAR of the right colonic normal mucosae adjacent to APs was significantly higher than the left colonic normal mucosae adjacent to APs (481.91 ± 63.31 vs 343.86 ± 26.71 mpH/min/mg, P = .027; unpaired t-test). This finding was also observed in normal mucosae obtained from patients with CRCs (Table 2). The ECAR of the normal mucosae adjacent to CRCs was significantly higher in the right colon compared to the left colon (584.46 ± 50.54 vs. 450.98 ± 27.44 mpH/min/mg, P = .013; unpaired t-test), while no significant difference of OCR and ECAR was observed with other clinicopathological factors (age, gender, CEA level, location, tumor size, T stage, lymph node metastasis and differentiation). However, significant differences in both OCR and ECAR were found between normal mucosae adjacent to APs and normal mucosae adjacent to CRCs (Supplementary Table 1). The normal mucosae from CRC patients had a higher OCR (3725.15 ± 164.17 vs. 3001.73 ± 252.63 mMol/min/mg, P = .013, unpaired t-test) and ECAR (494.04 ± 25.41 vs 373.44 ± 25.86 mpH/min/mg, P = .002, unpaired t-test) compared to mucosae adjacent to APs, indicating their demand for higher energy.
Table 1.
Clinicopathological factors of all patients with colorectal polyps included.
| Variable | Patients | Normal mucosa |
Polyp |
||||
|---|---|---|---|---|---|---|---|
| OCR (mMole/min/mg)⁎ | ECAR (mpH/min/mg)⁎ | O/E ratio (mMole/mpH)⁎ | OCR (mMole/min/mg)⁎ | ECAR (mpH/min/mg)⁎ | O/E ratio (mMole/mpH)⁎ | ||
| Total numbers | 56 | 3001.73 ± 252.63 | 373.44 ± 25.86 | 8.73 ± 0.57 | 3080.41 ± 246.16 | 525.38 ± 39.07 | 6.53 ± 0.51 |
| Age | |||||||
| <65 y | 34 | 2963.08 ± 354.52 | 372.43 ± 29.45 | 8.65 ± 0.79 | 2958.15 ± 311.94 | 536.06 ± 55.72 | 5.97 ± 0.45 |
| ≥65 y | 22 | 3061.46 ± 346.33 | 375.01 ± 48.48 | 8.87 ± 0.83 | 3269.36 ± 406.00 | 508.87 ± 51.16 | 7.41 ± 1.08 |
| P value | 0.851 | 0.962 | 0.850 | 0.542 | 0.737 | 0.167 | |
| Gender | |||||||
| Male | 43 | 2889.32 ± 298.97 | 376.35 ± 29.87 | 8.47 ± 0.70 | 2899.38 ± 262.53 | 510.92 ± 42.51 | 6.41 ± 0.61 |
| Female | 13 | 3373.55 ± 458.53 | 363.82 ± 53.46 | 9.59 ± 0.86 | 3679.20 ± 599.59 | 573.22 ± 94.72 | 6.92 ± 0.84 |
| P value | 0.423 | 0.840 | 0.416 | 0.183 | 0.506 | 0.677 | |
| Location | |||||||
| Right | 12 | 3415.88 ± 406.11 | 481.91 ± 63.31 | 8.40 ± 1.28 | 2563.33 ± 599.99 | 411.93 ± 72.43 | 6.45 ± 1.18 |
| Left | 44 | 2888.78 ± 301.67 | 343.86 ± 26.71 | 8.83 ± 0.65 | 3221.43 ± 267.14 | 556.32 ± 44.90 | 6.56 ± 0.57 |
| P value | 0.397 | 0.027 | 0.763 | 0.277 | 0.131 | 0.931 | |
| Size | |||||||
| <10 mm | 6 | 1865.42 ± 449.51 | 237.80 ± 57.84 | 9.96 ± 2.23 | 2464.37 ± 955.10 | 298.87 ± 109.62 | 10.59 ± 3.34 |
| ≥10 mm | 50 | 3138.09 ± 272.42 | 389.72 ± 27.37 | 8.59 ± 0.59 | 3154.34 ± 252.91 | 552.56 ± 40.39 | 6.05 ± 0.37 |
| P value | 0.120 | 0.069 | 0.466 | 0.391 | 0.044 | 0.004 | |
| Histology | |||||||
| Tubular type | 19 | 2504.01 ± 350.03 | 338.22 ± 44.46 | 8.00 ± 0.87 | 2354.36 ± 246.39 | 410.78 ± 55.20 | 7.75 ± 1.24 |
| Tubulo-villous and villous type | 37 | 3257.32 ± 332.81 | 391.53 ± 31.81 | 9.11 ± 0.75 | 3453.25 ± 336.37 | 584.23 ± 49.64 | 5.91 ± 0.41 |
| P value | 0.160 | 0.333 | 0.361 | 0.033 | 0.034 | 0.085 | |
P values were calculated using unpaired t-test. Bold means value <0.05.
Values were presented as Mean ± S.E.M.
Table 2.
Clinicopathological factors of all patients with colorectal cancer included.
| Variable | Patients | Normal mucosa |
Cancer |
||||
|---|---|---|---|---|---|---|---|
| OCR (mMole/min/mg)⁎ | ECAR (mpH/min/mg)⁎ | O/E ratio (mMole/mpH)⁎ | OCR (mMole/min/mg)⁎ | ECAR (mpH/min/mg)⁎ | O/E ratio (mMole/mpH)⁎ | ||
| Total numbers | 93 | 3725.15 ± 164.17 | 494.04 ± 25.41 | 8.24 ± 0.43 | 2986.32 ± 133.49 | 529.46 ± 22.26 | 6.00 ± 0.22 |
| Age | |||||||
| <65 y | 49 | 3706.48 ± 224.11 | 502.14 ± 35.73 | 7.98 ± 0.46 | 3204.57 ± 192.17 | 556.22 ± 30.90 | 6.06 ± 0.25 |
| ≥65 y | 44 | 3745.95 ± 243.79 | 485.02 ± 36.45 | 8.54 ± 0.76 | 2743.27 ± 179.06 | 499.67 ± 31.85 | 5.93 ± 0.37 |
| P value | 0.905 | 0.739 | 0.522 | 0.084 | 0.206 | 0.765 | |
| Gender | |||||||
| Male | 49 | 3593.83 ± 232.78 | 462.84 ± 33.31 | 8.62 ± 0.69 | 3021.03 ± 180.44 | 507.75 ± 30.83 | 6.30 ± 0.32 |
| Female | 44 | 3871.41 ± 231.48 | 528.79 ± 38.57 | 7.82 ± 0.50 | 2947.66 ± 200.09 | 553.65 ± 32.14 | 5.66 ± 0.29 |
| P value | 0.402 | 0.197 | 0.358 | 0.785 | 0.306 | 0.144 | |
| CEA level | |||||||
| < 5 IU/ml | 68 | 3839.04 ± 202.38 | 509.94 ± 31.34 | 8.39 ± 0.55 | 2998.09 ± 159.36 | 534.20 ± 25.86 | 6.09 ± 0.26 |
| ≥ 5 IU/ml | 25 | 3415.37 ± 260.54 | 450.79 ± 40.55 | 7.85 ± 0.65 | 2954.32 ± 247.38 | 516.58 ± 44.47 | 5.75 ± 0.41 |
| P value | 0.255 | 0.305 | 0.585 | 0.885 | 0.728 | 0.485 | |
| Location | |||||||
| Right | 30 | 3886.91 ± 303.81 | 584.46 ± 50.54 | 8.11 ± 0.97 | 2956.53 ± 244.88 | 550.41 ± 40.80 | 5.79 ± 0.38 |
| Left | 63 | 3648.13 ± 195.42 | 450.98 ± 27.44 | 8.30 ± 0.45 | 3000.51 ± 160.20 | 519.49 ± 26.63 | 6.10 ± 0.27 |
| P value | 0.500 | 0.013 | 0.833 | 0.879 | 0.519 | 0.519 | |
| Size | |||||||
| <40 mm | 34 | 3855.09 ± 275.75 | 501.49 ± 44.95 | 8.59 ± 0.70 | 3264.33 ± 244.01 | 573.89 ± 40.65 | 6.22 ± 0.37 |
| ≥40 mm | 59 | 3650.28 ± 205.42 | 489.75 ± 30.84 | 8.04 ± 0.55 | 2826.11 ± 154.33 | 503.86 ± 25.80 | 5.87 ± 0.27 |
| P value | 0.551 | 0.825 | 0.541 | 0.114 | 0.130 | 0.485 | |
| T stage | |||||||
| T1&2 | 20 | 4325.43 ± 289.66 | 574.79 ± 50.68 | 7.40 ± 0.69 | 3277.07 ± 239.82 | 624.18 ± 43.11 | 5.32 ± 0.51 |
| T3&4 | 73 | 3560.70 ± 189.91 | 471.92 ± 28.87 | 8.47 ± 0.52 | 2906.66 ± 156.33 | 503.51 ± 25.07 | 6.18 ± 0.24 |
| P value | 0.055 | 0.096 | 0.314 | 0.256 | 0.025 | 0.107 | |
| LN meta | |||||||
| No | 45 | 3713.32 ± 219.88 | 483.34 ± 31.91 | 8.12 ± 0.57 | 3218.82 ± 162.21 | 556.15 ± 26.68 | 5.94 ± 0.30 |
| Yes | 48 | 3736.24 ± 244.51 | 504.07 ± 39.38 | 8.36 ± 0.65 | 2768.35 ± 205.90 | 504.44 ± 35.02 | 6.06 ± 0.32 |
| P value | 0.945 | 0.686 | 0.783 | 0.092 | 0.248 | 0.786 | |
| Differentiation | |||||||
| Well | 9 | 4586.89 ± 516.83 | 527.70 ± 71.69 | 8.53 ± 1.20 | 3158.92 ± 312.73 | 636.22 ± 62.80 | 5.40 ± 0.86 |
| Moderate & poor | 84 | 3632.82 ± 171.05 | 490.44 ± 27.16 | 8.21 ± 0.46 | 2967.83 ± 144.27 | 518.02 ± 23.47 | 6.06 ± 0.22 |
| P value | 0.086 | 0.667 | 0.827 | 0.674 | 0.117 | 0.374 | |
P values were calculated using unpaired t-test. Bold means value <0.05.
Values were presented as Mean ± S.E.M.
In APs, the OCR and ECAR were significantly different between tubular and tubulo-villous/villous APs (Table 1). APs with a villous component had a higher OCR (3453.25 ± 336.37 vs. 2354.36 ± 246.39 mMol/min/mg, P = .033; unpaired t-test) and ECAR (584.23 ± 49.64 vs. 410.78 ± 55.20 mpH/min/mg, P = .034; unpaired t-test) compared to tubular APs. Moreover, the APs with a size ≥10 mm had a higher ECAR (552.56 ± 40.39 vs 298.87 ± 109.62mpH/min/mg, P = .044; unpaired t-test) and lower OCR/ECAR ratio (6.05 ± 0.37 vs 10.59 ± 3.34, P = .004; unpaired t-test), indicating that large APs tend to use glycolysis for ATP production.
In CRCs, most clinicopathological factors were not associated with OCR and ECAR except T stage (Table 2). ECAR was significantly higher in T1 and T2 stage tumors compared with T3 and T4 stages (624.18 ± 43.11 vs 503.51 ± 25.07 mpH/min/mg, P = .025; unpaired t-test). Surprisingly, there were no significant differences of OCR and ECAR between APs and CRCs (Supplementary Table 1), suggesting that APs and CRCs have the similar bioenergetic profiles and demands for energy.
3.2. The changes of OxPhos and glycolysis in APs and CRCs
The OCR, ECAR and OCR/ECAR ratio were compared between APs/CRCs and adjacent normal mucosae (Fig. 1). While there was no difference in OCR between APs and adjacent normal mucosae (P = .762; paired t-test), the ECAR was significantly higher in APs (P = .001; paired t-test). Moreover, the OCR/ECAR ratio was decreased in APs (P = .001; paired t-test) compared to the adjacent normal mucosae. These results suggest that APs have the same level of OxPhos as the adjacent normal mucosae, but tend to produce more ATP by glycolysis.
Fig. 1.
Analysis of OCR, ECAR and OCR/ECAR ratio between APs, CRCs and their adjacent normal mucosae. (a) OCR of APs and normal mucosae (N = 56, P = .762). (b) OCR of CRCs and normal mucosae (N = 93, P < .001). (c) ECAR of APs and normal mucosae (N = 56, P = .001). (d) ECAR of CRCs and normal mucosae (N = 93, P = .121). (e) OCR/ECAR of APs and normal mucosae (N = 56, P = .001). (f) OCR/ECAR of CRCs and normal mucosae (N = 93, P < .001). Groups were compared by paired t-test. The box and whisker plot shows a 5-number data summary: minimum, 1st quartile, median, 3rd quartile and maximum. The box is divided at the median. The length of the box is the interquartile range. The 1st quartile is the bottom line. The 3rd quartile is the top line.
In CRCs, the OCR was significantly decreased compared to the adjacent normal mucosae (P < .001; paired t-test). While the OCR/ECAR ratio was significantly decreased in CRCs (P < .001; paired t-test), there was no significant change of ECAR between CRCs and normal mucosae (P = .121; paired t-test). These results suggest that the CRCs have defects in OxPhos, and therefore ATP production by CRCs mainly depends on glycolysis.
3.3. The changes of OxPhos and glycolysis in relation to AP/CRC size, histology and stage
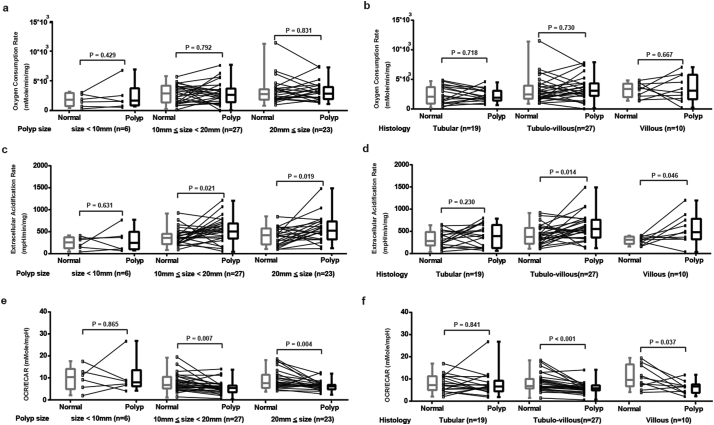
Large APs and APs with a villous component have been shown to have a higher burden of genetic mutations and a greater malignant potential [3,[21], [22], [23], [24], [25]], however, their association with bioenergetic changes are unknown. In order to answer this question, the analysis of OCR, ECAR and OCR/ECAR ratio based on AP size (<1 cm, 1 to 2 cm, and ≥ 2 cm) and histology (tubular, tubulo-villous, and villous) was performed (Fig. 2). Compared to the adjacent normal mucosae, there were no significant changes of OCR in APs of different size and histology (Figure2a and 2b). However, the levels of ECAR were significantly increased in APs of a size ≥1 cm (Fig. 2c) and APs with a villous component (Fig. 2d). The OCR/ECAR ratio was also significantly decreased in APs ≥ 1 cm (Fig. 2e) and APs with a villous component (Fig. 2f). These results suggest that the increase in glycolysis occurs mainly in large APs and APs with a villous component.
Fig. 2.
Subgroup analysis of OCR, ECAR and OCR/ECAR ratio between APs and adjacent normal mucosae. (a) OCR of normal mucosae and different size APs (<1 cm, 1 to 2 cm, and ≥ 2 cm. N = 6, 27 and 23. P = .429, 0.792 and 0.831, respectively). (b) OCR of normal mucosae and APs with different histopathology (tubular, tubulo-villous and villous. N = 19, 27 and 10. P = .718, 0.730 and 0.667, respectively). (c) ECAR of normal mucosae and different size APs (<1 cm, 1 to 2 cm and ≥ 2 cm. N = 6, 27 and 23. P = .631, 0.021 and 0.019, respectively). (d) ECAR of normal mucosae and APs with different histopathology (tubular, tubulo-villous, and villous. N = 19, 27 and 10. P = .230, 0.014 and 0.046, respectively). (e) OCR/ECAR of normal mucosae and different size APs (<1 cm, 1 to 2 cm and ≥ 2 cm. N = 6, 27 and 23. P = .865 0.007 and 0.004, respectively). (f) ECAR of normal mucosae and APs with different histopathology (tubular, tubulo-villous and villous. N = 19, 27 and 10. P = .841, < 0.001 and 0.037, respectively). Groups were compared by paired t-test. The box and whisker plot shows a 5-number data summary: minimum, 1st quartile, median, 3rd quartile and maximum. The box is divided at the median. The length of the box is the interquartile range. The 1st quartile is the bottom line. The 3rd quartile is the top line.
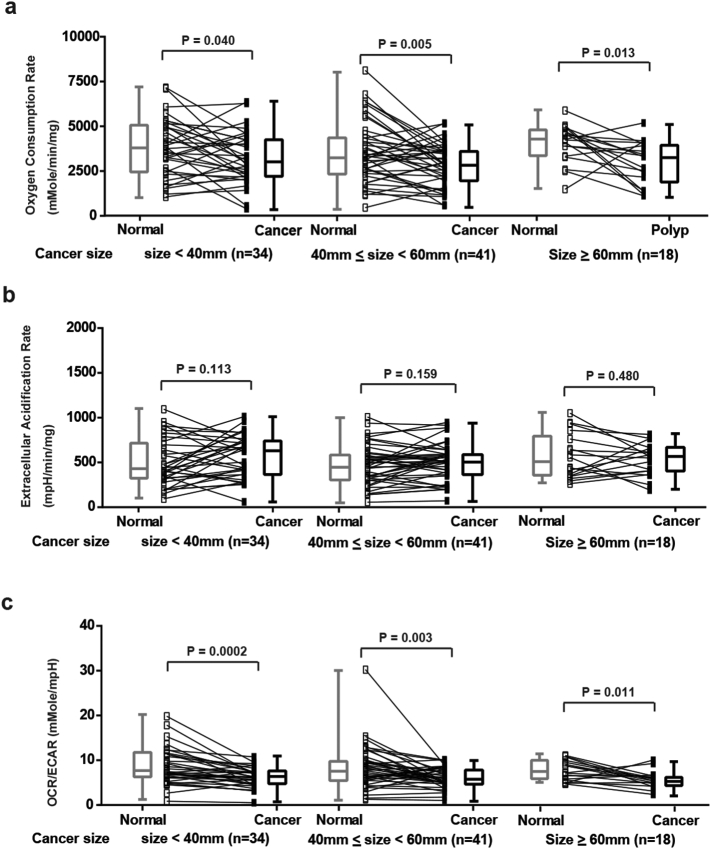
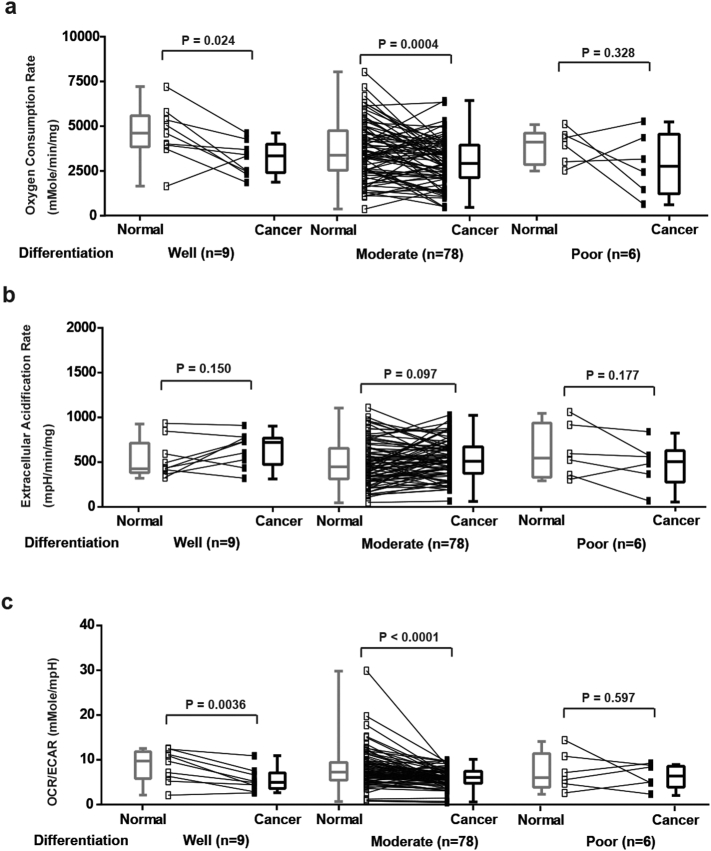
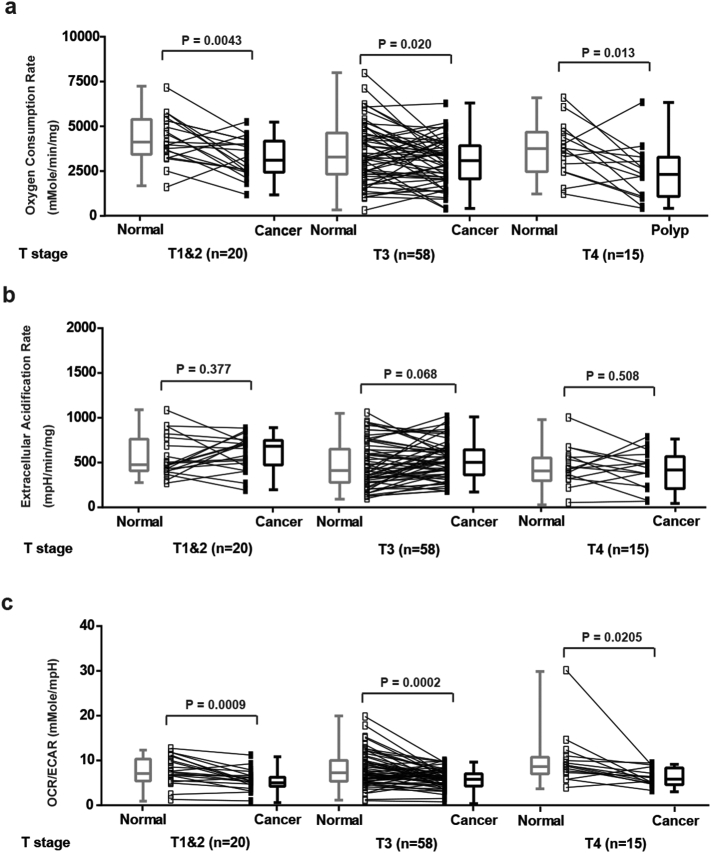
In contrast, the changes of OCR, ECAR and OCR/ECAR ratio observed in CRCs were not associated with either their size (Supplementary Fig. 1), histology (Supplementary Fig. 2) or T stage (Supplementary Fig. 3). These results suggest that the defects in OxPhos in CRCs may occur early in tumor progression.
Supplementary Fig. 1.
Analysis of OCR, ECAR and OCR/ECAR ratio between different sizes of CRCs and adjacent normal mucosae. (a) OCR of normal mucosae and CRCs (<4 cm, 4 to 6 cm and ≥ 6 cm. N = 34, 41 and 18. P = .040, 0.005 and 0.013, respectively). (b) ECAR of normal mucosae and CRCs (<4 cm, 4 to 6 cm and ≥ 6 cm. N = 34, 41 and 18. P = .113, 0.159 and 0.480, respectively). (c) OCR/ECAR of normal mucosae and CRCs (<4 cm, 4 to 6 cm and ≥ 6 cm. N = 34, 41 and 18. P = .0002, 0.003 and 0.011, respectively). Groups were compared by paired t-test. The box and whisker plot shows a 5-number data summary: minimum, 1st quartile, median, 3rd quartile and maximum. The box is divided at the median. The length of the box is the interquartile range. The 1st quartile is the bottom line. The 3rd quartile is the top line.
Supplementary Fig. 2.
Analysis of OCR, ECAR and OCR/ECAR ratio between normal mucosae and CRCs with different degrees of differentiation. (a) OCR of normal mucosae and CRCs (well, moderate and poor. N = 9, 78 and 6. P = .024, 0.0004 and 0.328, respectively). (b) ECAR of normal mucosae and CRCs (well, moderate and poor. N = 9, 78 and 6. P = .150, 0.097 and 0.177, respectively). (c) OCR/ECAR of normal mucosae and CRCs (well, moderate and poor. N = 9, 78 and 6. P = .0036, < 0.0001 and 0.597, respectively). Groups were compared by paired t-test. The box and whisker plot shows a 5-number data summary: minimum, 1st quartile, median, 3rd quartile and maximum. The box is divided at the median. The length of the box is the interquartile range. The 1st quartile is the bottom line. The 3rd quartile is the top line.
Supplementary Fig. 3.
Analysis of OCR, ECAR and OCR/ECAR ratio between normal mucosae and CRCs with different T stages. (a) OCR of normal mucosae and CRCs (T1 & T2, T3 and T4. N = 20, 58 and 15. P = .0043, 0.020 and 0.013, respectively). (b) ECAR of normal mucosae and CRCs (T1 & T2, T3 and T4. N = 20, 58 and 15. P = .377, 0.068 and 0.508, respectively). (c) OCR/ECAR of normal mucosae and CRCs (T1 & T2, T3 and T4. N = 20, 58 and 15. P = .0009, 0.0002 and 0.0205, respectively). Groups were compared by paired t-test. The box and whisker plot shows a 5-number data summary: minimum, 1st quartile, median, 3rd quartile and maximum. The box is divided at the median. The length of the box is the interquartile range. The 1st quartile is the bottom line. The 3rd quartile is the top line.
3.4. The association between bioenergetic changes and variants/mutations in mitochondrial DNA
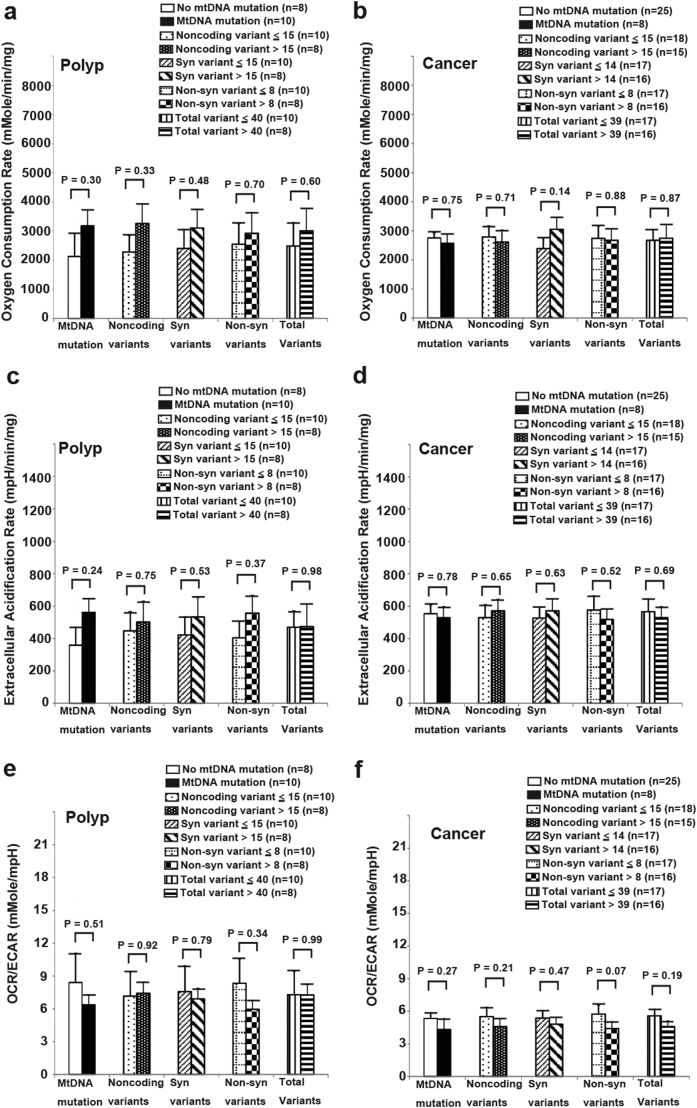
MtDNA variants and mutations are commonly detected in CRCs and APs. It has been hypothesized that the mtDNA variants and mutations may cause the defects in OxPhos. To test this hypothesis, the whole mtDNA genome of 18 APs, 33 CRCs and their adjacent normal mucosae were sequenced using the Sanger method. Because it is difficult to identify all heteroplasmic mtDNA mutations by the Sanger method, only homoplasmic mtDNA variants and mutations were recorded. The number of non-coding variants, synonymous coding variants and non-synonymous coding variants, total variants and the existence of homoplasmic mtDNA mutations in APs, CRCs and their adjacent normal mucosae were analyzed (APs: Supplementary Table 2; CRCs: Supplementary Table 3). The analysis showed that the number and location of mtDNA variants were the same in APs/CRCs and their adjacent normal mucosae from the same patient. However, homoplasmic mtDNA mutations occurred more frequently in APs/CRCs than in the adjacent normal mucosae. In patients with APs, 10 of 18 APs (55.6%) had homoplasmic mtDNA mutations, while only 3 of the 18 adjacent normal mucosae (16.7%) contained homoplasmic mtDNA mutations. In 33 CRCs, 8 tumors had homoplasmic mtDNA mutations (24.2%) while only 3 adjacent normal mucosae (9.1%) contained homoplasmic mtDNA mutations.
Correlations of the number of mtDNA variants and the existence of homoplasmic mtDNA mutations with their bioenergetic profiles were further analyzed. The results showed that the number of mtDNA variants and the existence of homoplasmic mtDNA mutations were not correlated with OCR, ECAR and OCR/ECAR ratio in APs and CRCs (Fig. 3). Similar results were obtained in the adjacent normal mucosae (Supplementary Fig. 4), suggesting that mtDNA variants and homoplasmic mtDNA mutations within APs, CRCs and the adjacent normal mucosae may not be the major cause of bioenergetic changes.
Fig. 3.
Analysis of OCR, ECAR and OCR/ECAR ratio of APs (N = 18) and CRCs (N = 33) divided by the existence of mitochondrial DNA (mtDNA) mutations, the median number of noncoding variants, synonymous variants, non-synonymous variants and total variants. (a) OCR of AP. (b) OCR of CRC. (c) ECAR of AP. (d) ECAR of CRC. (e) OCR/ECAR of AP. (f) OCR/ECAR of CRC. Data and error bars are presented as mean ± SEM. Groups were compared by unpaired t-test.
Supplementary Fig. 4.
Analysis of OCR, ECAR and OCR/ECAR ratio of normal mucosae adjacent to APs (N = 18) and CRCs (N = 33) divided by the existence of mtDNA mutations, the median number of noncoding variants, synonymous variants, non-synonymous variants and total variants. (a) OCR of normal mucosae adjacent to AP. (b) OCR of normal mucosae adjacent to CRC. (c) ECAR of normal mucosae adjacent to AP. (d) ECAR of normal mucosae adjacent to CRC. (e) OCR/ECAR of normal mucosae adjacent to AP. (f) OCR/ECAR of normal mucosae adjacent to CRC. Data and error bars are expressed as mean ± SEM. Groups were compared by unpaired t-test.
3.5. The expression of ALDOB and SLC16A4 may contribute to the increased glycolysis in APs with a villous component
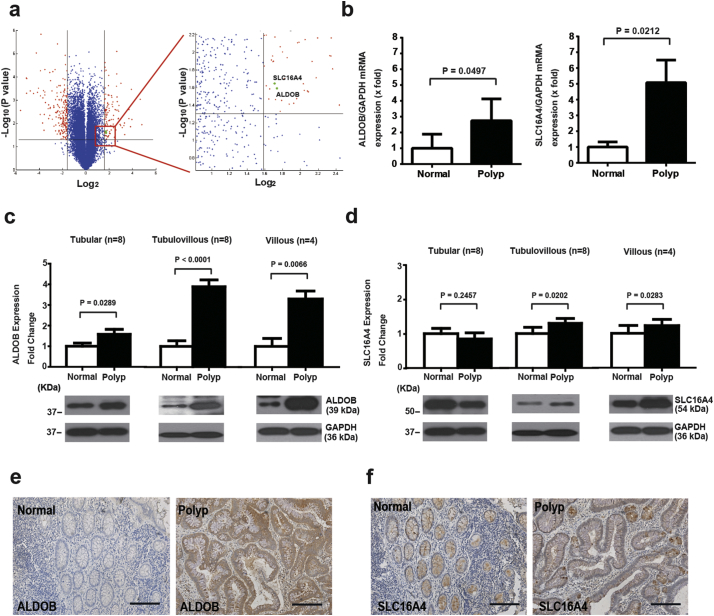
To understand the mechanism of increased glycolysis in APs with a villous component, microarrays were used to analyze differentially expressed genes between the mRNA of 5 APs with a villous component and the adjacent normal mucosae (Fig. 4a). Two metabolic genes, ALDOB and SLC16A4, were up-regulated in APs. This result was further confirmed by real-time PCR showing that the expression of both ALDOB and SLC16A4 mRNA was higher in the APs with a villous component compared with their adjacent normal mucosae (Fig. 4b). To measure their protein expression level in APs, Western blot analysis of ALDOB and SLC16A4 was performed (Fig. 4c, Fig. 4d, and Supplementary Fig. 5). Compared to normal mucosae, the level of ALDOB was significantly increased in tubular, tubulo-villous and villous APs (Fig. 4c). The level of SLC16A4 was significantly increased in tubulo-villous and villous APs, but not in tubular APs (Fig. 4d). Immunohistochemistry showed that both ALDOB and SLC16A4 were expressed in villous APs and their adjacent normal mucosae, but the intensity of ALDOB and SLC16A4 immunostaining in APs was greater (Fig. 4e and f).
Fig. 4.
The analysis of aldolase, fructose-bisphosphate B (ALDOB) and solute carrier family 16 member 4 (SLC16A4) expression between APs and normal mucosae.
(a) Differential Transcripts between normal mucosae and APs with a villous component (N = 5). Two metabolic-related genes (ALDOB and SLC16A4, green dots) were identified from the most differentially expressed transcripts (red dots) based on paired t-test (P < .04) and > 3 fold change. (b) The elevation of ALDOB (N = 8, P = .0497) and SLC16A4 (N = 8, P = .0212) in APs compared to adjacent normal mucosae was confirmed by real-time QPCR. Data and error bars are mean ± SEM. GAPDH was used for normalization purposes. Groups were compared by paired t-test. (c) Western blot analysis of ALDOB in normal mucosae and APs with different histopathology (tubular, tubulo-villous and villous. N = 8, 8 and 4. P = .0289, < 0.0001 and 0.0066, respectively). Data and error bars are mean ± SEM. GAPDH was used for normalization purposes. Groups were compared by paired t-test. (d) Western blot analysis of SLC16A4 in normal mucosae and APs with different histopathology (tubular, tubulo-villous and villous. N = 8, 8 and 4. P = .2457, 0.0202 and 0.0283, respectively). Data and error bars are mean ± SEM. GAPDH was used for normalization purposes. Groups were compared by paired t-test. (e) Immunohistochemical staining of ALDOB in AP and adjacent normal mucosa (200×, scale bar: 100 μm). (f) Immunohistochemical staining of SLC16A4 in AP and adjacent normal mucosa (200×, scale bar: 100 μm). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Supplementary Fig. 5.
The expression levels of ALDOB and SLC16A4 in normal mucosae and APs with different histopathology.
Western blot analysis of ALDOB and SLC16A4 expression in normal mucosae and APs with different histopathology (tubular, tubulovillous and villous). Protein expression was examined in representative pairs of clinical specimens, with GAPDH as the loading control. N: normal mucosae.
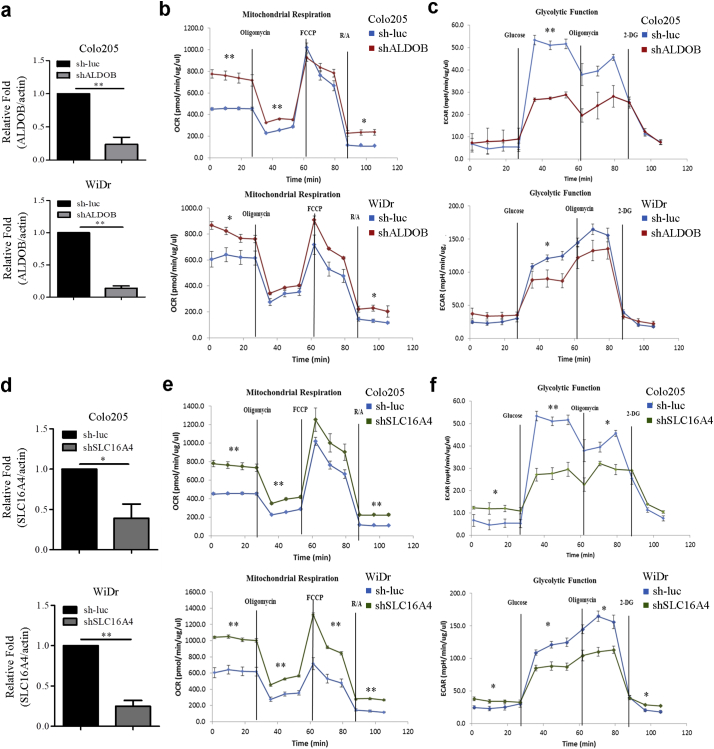
To assess whether ALDOB and SLC16A4 contribute to the bioenergetic changes, the ALDOB and SLC16A4 genes were knocked down in the WiDr and colo205 CRC cell lines separately. The reduced expression of ALDOB and SLC16A4 in the knockdown cell lines was confirmed by qRT-PCR analysis (Fig. 5a & 5d). The mitochondrial OxPhos (Fig. 5b & 5e) and glycolytic activities (Fig. 5c & 5f) of ALDOB-knockdown, SLC16A4-knockdown and control Colo205 and WiDr cell lines were further tested. Both Colo205 and WiDr ALDOB-knockdown cell lines had an increased basal OCR compared to control cell line, but only the Colo205 ALDOB-knockdown cell line had elevated ATP-linked respiration (Fig. 5b). Moreover, both Colo205 and WiDrALDOB-knockdown cell lines showed a significant decrease in glycolysis (Fig. 5c). Thus, the functions of ALDOB are associated with basal motichondrial OxPhos and basal glycolysis. The mitochondrial respiration assay and glycolytic function assays were also performed on Colo205 and WiDr SLC16A4-knockdown cell lines. The results showed that basal OCR and ATP-linked respiration were elevated in both Colo205 and WiDr SLC16A4-knockdown cell lines (Fig. 5e). Moreover, the most significant elevation of maximal respiration was observed in the WiDr SLC16A4-knockdown cell line (Fig. 5e). In the glycolytic function assay, both Colo205 and WiDr SLC16A4-knockdown cell lines showed decreased glycolysis and glycolytic capacity (Fig. 5f). Thus, the functions of SLC16A4 were associated with mitochondrial respiration, basal glycolysis and glycolytic capacity. Overall, these results showed that both ALDOB and SLC16A4 might play an important role in CRC energy metabolism.
Fig. 5.
The bioenergetic functions of ALDOB and SLC16A4 in colorectal cancer cell lines.
(a) qRT-PCR analysis of ALDOB expression in stable (knockdown) cell lines with actin as the loading control. Values were calculated as relative fold change, compared with the control group (sh-luc). Data and error bars are mean ± SEM (N = 3). (b) OCR was performed with the Mito Stress test Kit in ALDOB-depleted Colo205 and WiDr cells at variable time periods. Data and error bars are mean ± SEM (N = 3). (c) ECAR analyses of ALDOB-depleted Colo205 and WiDr cells were performed with the Glycolysis Stress test Kit. Data and error bars are mean ± SEM (N = 3). (d) qRT-PCR analysis of SLC16A4 expression in stable (knockdown) cell lines with actin as the loading control. Values were calculated as relative fold change, compared with the control group (sh-luc). Data and error bars are mean ± SEM (N = 3). (e) OCR was performed with the Mito Stress test Kit in SLC16A4-depleted Colo205 and WiDr cells at variable time periods. Data and error bars are mean ± SEM (N = 3). (f) ECAR analysis of SLC16A4-depleted Colo205 and WiDr cells were performed with the Glycolysis Stress test Kit. Data and error bars are mean ± SEM (N = 3). The ECAR after glucose treatment indicates glycolysis rate. The ECAR after oligomycin treatment indicates glycolysis capacity. Groups were compared by unpaired t-test. (*, p < .05; **, p < .01; ***, p < .001).
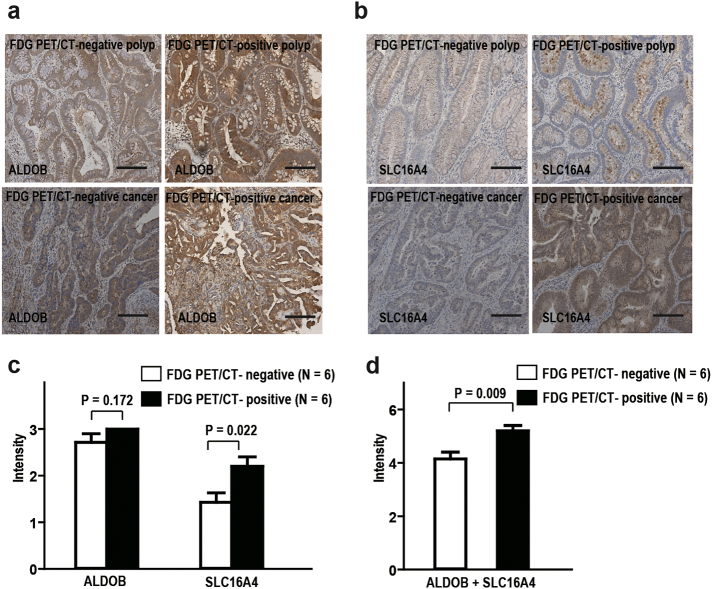
3.6. The FDG PET/CT-positive APs and CRCs have increased SLC16A4 expression
FDG PET/CT is a molecular imaging technology for detecting tumor tissues with increased glycolytic activity. FDG PET/CT positive APs and CRCs have been considered to have a high glycolytic phenotype [26]. To test whether these high glycolytic activity tissues were associated with increased ALDOB and SLC16A4 expression, we further collected tissue from 5 FDG PET/CT positive APs/CRCs (AP, 1; CRCs, 4) and 7 negative APs/CRCs (APs, 5; CRCs, 2). Immunohistochemistry for ALDOB and SLC16A4 was performed (Fig. 6a & 6b), and the staining intensities were scored by independent pathologists (Fig. 6c & 6d). The PET/CT positive group had significantly increased SCL16A4 expression over the negative group (unpaired t-test, P = .022, Fig. 6c), but no difference was seen in ALDOB immunostaining (unpaired t-test, P = .172, Fig. 6c). However, the sum of both staining intensities of same tissues showed that the FDG PET/CT-positive tissues had significantly higher scores than the negative tissues (unpaired t-test, P = .009, Fig. 6d). These results suggest that the FDG PET/CT-positive AP/CRCs may have increased SLC16A4 expression.
Fig. 6.
The expression of ALDOB and SLC16A4 in FDG PET/CT-positive and -negative AP/CRC.
(a) Immunohistochemical staining of ALDOB in FDG PET/CT-positive and -negative AP/CRC (200×, scale bar). (b) Immunohistochemical staining of SLC16A4 in FDG PET/CT-positive and -negative AP/CRC (200×, scale bar). (c) No significant difference of ALDOB intensity between FDG PET/CT-positive AP/CRC and -negative AP/CRC (unpaired t-test, P = .172). The intensity of SLC16A4 expression was increased in FDG PET/CT-positive AP/CRC compared to -negative AP/CRC (unpaired t-test, P = .022). Data and error bars are mean ± SEM (N = 6) (d). The sum of intensity of ALDOB and SLC16A4 expression in each tissue was significantly increased in in FDG PET/CT-positive AP/CRC compared to -negative AP/CRC (unpaired t-test. P = .009). Data and error bars are mean ± SEM (N = 6).
4. Discussion
Aerobic glycolysis, the Warburg effect, has been recognized as a metabolic hallmark of cancer cells. Most CRCs develop from APs through the adenoma-carcinoma sequence. However, the bioenergetic changes during this process are unclear. Ong et al. previously found a depletion of glucose and inositol in APs, suggesting that glycolysis may be critical in early carcinogenesis [27]. Our study further demonstrated that aerobic glycolysis occurs in APs by directly measuring OCR and ECAR. Glycolysis was significantly increased in large APs (size ≥1 cm) and APs with a villous component, suggesting the shift to the Warburg effect appears to commence during AP enlargement and villous transformation.
Mitochondrial dysfunction has been suspected to participate in carcinogenesis [28]. Using ulcerative colitis as a model to study CRC progression, Ussakli et al. showed that the activity of mitochondrial COX progressively decreased with proximity to dysplasia and was the lowest in the mucosa adjacent to dysplasia and in dysplastic mucosa itself, suggesting that mitochondrial loss is associated with the development of dysplasia [14]. Interestingly, COX activity was significantly increased in cancers, indicating that mitochondria function is restored and needed for malignant cells. However, here we show that mitochondrial OxPhos was not altered in APs but was significantly decreased in CRCs, suggesting that mitochondrial dysfunction mainly occurs in cancers. This discrepancy may be due to the different mechanisms of CRC development. Sporadic CRCs are not likely to be initiated by inflammation because most intramural immune cells are recruited after the tumor is formed, but colitis-associated CRCs are often caused by constant inflammation [29]. High levels of ROS are generated by chronic inflammation in patients with inflammatory bowel disease [[30], [31], [32]]. ROS are thought to not only contribute to the development of dysplastic lesions [33,34], but also lead to a reduction of ATP, inhibition of the respiratory chain and mtDNA mutation [35]. Moreover, both sporadic CRCs and colitis-associated CRCs have genetic alterations in the process of carcinogenesis, but the frequency and sequence of these events differs. While defects of p53 commonly occur as early events in colitis-associated CRCs, p53 mutations often occur in later stages of sporadic CRCs [4]. Since p53 defects can impair mitochondrial function at both the DNA and protein level [36,37], it is reasonable to speculate that the early events of mitochondrial dysfunction in colitis-associated dysplasia and the late events of Oxphos defects in sporadic CRCs are linked to p53 mutations. However, further study is needed on this point.
MtDNA mutations have been found in human normal mucosae, APs and CRCs [[9], [10], [11], [12],38]. In our study, we further show that the mtDNA mutations occur more frequently in APs/CRCs than in their adjacent normal mucosae, indicating mtDNA mutations arise during the adenoma-carcinoma sequence. Defects of OxPhos can result from specific mtDNA mutations in human mitochondrial diseases [39], however, whether mtDNA mutations acquired during the carcinogenesis process can cause OxPhos defects is unclear. Defects of OxPhos not directly related to genome damage, e.g., due to insufficient respiration and ROS production, may also occur [40]. Our results show that both OxPhos and glycolysis are not affected by the number of mtDNA variants and the existence of mtDNA mutations, indicating these variants and mutations are not the major cause of the bioenergetic changes in APs and CRCs.
Two metabolic genes, ALDOB and SLC16A4, were over-expressed in APs with a villous component. Moreover, the over-expression of SLC16A4 was shown in FDG PET/CT-positive APs/CRCs, indicating its possible active role in enhancing glycolysis. Aldolase is an important enzyme for glucose and fructose metabolism. In human tissues, three aldolase isoenzymes are expressed in a tissue-dependent manner, and ALDOB is mainly expressed in the liver, kidney and intestine. Besides its metabolic function, aldolase has been considered to participate in carcinogenesis [41]. In lung cancer, aldolase has been shown to promote epithelial-to-mesenchymal transition and cell migration by simultaneously decreasing β-catenin and E-cadherin expression while increasing vimentin and fibronectin expression [42]. Moreover, it has been shown that the Wnt signaling pathway can be activated by aldolase through disrupting the GSK-3β-Axin interaction and targeting Axin to the dishevelled-induced signalosomes [43]. Unlike ALDOB, the SCL16-encoded proteins are monocarboxylate transporters (MCT) catalysing the proton-linked transport of monocarboxylates such as L-lactate, pyruvate and ketone bodies across the plasma membrane [44]. MCT1, MCT4 and MCT5 are expressed in human intestine [45], while MCT1 and MCT4 contribute to the growth and angiogenesis of CRCs [46,47]. However, the role of MCT5 (encoded by SLC16A4 gene) is not clear. Whether ALDOB and SLC16A4 contribute directly to sporadic CRC carcinogenesis is not answered in this study, nevertheless we show increased expression of ALDOB and SLC16A4 in APs with a villous component, and their expression could alter both mitochondrial OxPhos and glycolytic activity.
This is the first report of directly measurement of OCR and ECAR of human APs and CRCs by Seahorse XF analyzer with Islet microplate. Previous studies [[16], [17], [18]] had been demonstrated that muscles and adipose tissue can be directly analyzed using the same method. Because of the 1422 μm height of microchamber of Islet microplate, the tiny tissue could be tested in the Seahorse platform.
The limitation of this study is the cross-sectional design. Ideally, the measurement of OCR and ECAR should be evaluated in prospective studies so that longitudinal information about the progression status of the APs is available. A longitudinal study would allow us to determine whether the APs with increased glycolysis are more prone to develop cancer sooner. However, it is impractical to do this as once discovered, APs are immediately removed. Another limitation of the study is the lack of genetic mutation analysis. Since the accumulation of genetic mutations underlies the adenoma-carcinoma sequence, it is reasonable to speculate that the glycolytic shift occurring in APs may correlate with these genetic alterations; further studies will be performed to clarify this aspect. Indeed, it is not clear from this study whether the metabolic shifting from mitochondrial OxPhos to glycolysis is the cause or the result of CRCs. Nevertheless, we found that ALDOB and SLC16A4 were over-expressed in APs/CRCs. Moreover, the Mito Stress and Glycolysis Stress assays performed with ALDOB-inhibited or SLC16A4-inhibited cell lines showed that ALDOB and SLC16A4 could affect CRC cell mitochondrial OxPhos, basal glycolysis and glycolytic capacities. These results suggest that these two proteins may contribute to the glycolytic shift in APs, and thus may be considered as potential targets for blocking the carcinogenesis process.
In conclusion, this study demonstrated that a shift towards glycolysis occurs with AP enlargement and villous transformation. Mitochondrial OxPhos defects are mainly associated with CRCs. While mtDNA mutations can accumulate during the carcinogenesis process, these mutations may not be the major cause of bioenergetic changes. Finally, ALDOB and SLC16A4 may contribute to the increase of glycolysis seen in APs, indicating their potential roles in bioenergetic alterations during adenoma-carcinoma progression.
The following are the supplementary data related to this article.
Bioenergetic comparison between polyp and cancer and adjacent normal mucosa.
The analysis of mitochondrial DNA variants and mutations in polyps and their adjacent normal mucosa.
The analysis of mitochondrial DNA variants and mutations in colorectal adenocarcinoma and their adjacent normal mucosa.
Acknowledgments
Acknowledgements
We would like to thank the staff members (Yen-Ling Chuang, Tzu-Chi Yu, Chien-Chih Wang, and Hsiu-Min Tseng) of the Liver Research Center, and Genomic Medicine Research Core Labratory in Linkou Chang Gung Memorial Hospital for their technical assistance.
Funding sources
This study was funded by grants from Ministry of Science and Technology (MOST 103-2314-B-182-021; 107-2314-B-182A-025; 106-2321-B-182A-004-MY3) and Chang Gung Medical Research Program, Taiwan (CMRPG3F1401, CMRPG3F1402, CMRPG3F1403, CRRPG3F0061, CRRPG3F0062, CRRPG3F0063, CMRPG3H0241, CMRPG3E2181, CMRPG3B0803, CMRPG3H0721, CMRPG3H0722). The sponsor was not related to the study design in the collection, analysis and interpretation of data.
Delaration of interests
No conflicts of interest were declared.
Author contributions
Lin WR: study concept and design, acquisition of data, drafting of the manuscript; Chiang JM, Hsieh SY and Yu JS: material support, acquisition of data; Lim SN: acquisition of data; Su MY, Chen TH and Huang SW: material support; Chen CW, Tsai CL and Wu RC: analysis and interpretation of data, statistical analysis;
Lin YH: analysis and interpretation of data, drafting of the manuscript; Alison MR, Chiu CT and Yeh CT: critical revision of the manuscript for important intellectual content.
Transcript profiling
The data, including all raw microarray data have been deposited in the Gene Expression Omnibus (GEO) database with accession number GSE81804 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE81804).
References
- 1.Warburg O., Wind F., Negelein E. The metabolism of tumors in the body. J Gen Physiol. Mar 7, 1927;8(6):519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]; Warburg O, Wind F, Negelein E. The Metabolism of Tumors in the Body. J. Gen. Physiol. Mar 7, 1927;8(6):519-30. [DOI] [PMC free article] [PubMed]
- 2.Hagland H.R., Soreide K. Cellular metabolism in colorectal carcinogenesis: influence of lifestyle, gut microbiome and metabolic pathways. Cancer Lett. Jan 28, 2015;356(2 Pt A):273–280. doi: 10.1016/j.canlet.2014.02.026. [DOI] [PubMed] [Google Scholar]; Hagland HR, Soreide K. Cellular metabolism in colorectal carcinogenesis: Influence of lifestyle, gut microbiome and metabolic pathways. Cancer Lett. Jan 28, 2015;356(2 Pt A):273-80. [DOI] [PubMed]
- 3.Vogelstein B., Fearon E.R., Hamilton S.R., Kern S.E., Preisinger A.C., Leppert M. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319(9):525–532. doi: 10.1056/NEJM198809013190901. Sep 1. [DOI] [PubMed] [Google Scholar]; Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, et al. Genetic alterations during colorectal-tumor development. N. Engl. J. Med. 1988 Sep 1;319(9):525-32. [DOI] [PubMed]
- 4.Armaghany T., Wilson J.D., Chu Q., Mills G. Genetic alterations in colorectal cancer. Gastrointest Cancer Res. 2012;5(1):19–27. Jan. [PMC free article] [PubMed] [Google Scholar]; Armaghany T, Wilson JD, Chu Q, Mills G. Genetic alterations in colorectal cancer. Gastrointest. Cancer Res. 2012 Jan;5(1):19-27. [PMC free article] [PubMed]
- 5.Warburg O. On the origin of cancer cells. Science. 1956 Feb24;123(3191):309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]; .Warburg O. On the origin of cancer cells. Science. 1956 Feb24;123(3191):309-14. [DOI] [PubMed]
- 6.Sena L.A., Chandel N.S. Physiological roles of mitochondrial reactive oxygen species. Mol Cell. 2012;48(2):158–167. doi: 10.1016/j.molcel.2012.09.025. Oct 26. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sena LA, Chandel NS. Physiological roles of mitochondrial reactive oxygen species. Mol. Cell. 2012 Oct 26;48(2):158-67. [DOI] [PMC free article] [PubMed]
- 7.Skonieczna K., Malyarchuk B.A., Grzybowski T. The landscape of mitochondrial DNA variation in human colorectal cancer on the background of phylogenetic knowledge. Biochim Biophys Acta. Apr, 2012;1825(2):153–159. doi: 10.1016/j.bbcan.2011.11.004. [DOI] [PubMed] [Google Scholar]; Skonieczna K, Malyarchuk BA, Grzybowski T. The landscape of mitochondrial DNA variation in human colorectal cancer on the background of phylogenetic knowledge. Biochim. Biophys. Acta. Apr, 2012;1825(2):153-9. [DOI] [PubMed]
- 8.Brandon M., Baldi P., Wallace D.C. Mitochondrial mutations in cancer. Oncogene. 2006;25(34):4647–4662. doi: 10.1038/sj.onc.1209607. Aug 7. [DOI] [PubMed] [Google Scholar]; Brandon M, Baldi P, Wallace DC. Mitochondrial mutations in cancer. Oncogene. 2006 Aug 7;25(34):4647-62. [DOI] [PubMed]
- 9.Greaves L.C., Preston S.L., Tadrous P.J., Taylor R.W., Barron M.J., Oukrif D. Mitochondrial DNA mutations are established in human colonic stem cells, and mutated clones expand by crypt fission. Proc Natl Acad Sci U S A. 2006;103(3):714–719. doi: 10.1073/pnas.0505903103. Jan 17. [DOI] [PMC free article] [PubMed] [Google Scholar]; Greaves LC, Preston SL, Tadrous PJ, Taylor RW, Barron MJ, Oukrif D, et al. Mitochondrial DNA mutations are established in human colonic stem cells, and mutated clones expand by crypt fission. Proc. Natl. Acad. Sci. U. S. A. 2006 Jan 17;103(3):714-9. [DOI] [PMC free article] [PubMed]
- 10.He Y., Wu J., Dressman D.C., Iacobuzio-Donahue C., Markowitz S.D., Velculescu V.E. Heteroplasmic mitochondrial DNA mutations in normal and tumour cells. Nature. 2010;464(7288):610–614. doi: 10.1038/nature08802. Mar 25. [DOI] [PMC free article] [PubMed] [Google Scholar]; He Y, Wu J, Dressman DC, Iacobuzio-Donahue C, Markowitz SD, Velculescu VE, et al. Heteroplasmic mitochondrial DNA mutations in normal and tumour cells. Nature. 2010 Mar 25;464(7288):610-4. [DOI] [PMC free article] [PubMed]
- 11.Larman T.C., DePalma S.R., Hadjipanayis A.G., Cancer Genome Atlas Research N, Protopopov A., Zhang J. Spectrum of somatic mitochondrial mutations in five cancers. Proc Natl Acad Sci U S A. Aug 28, 2012;109(35):14087–14091. doi: 10.1073/pnas.1211502109. [DOI] [PMC free article] [PubMed] [Google Scholar]; Larman TC, DePalma SR, Hadjipanayis AG, Cancer Genome Atlas Research N, Protopopov A, Zhang J, et al. Spectrum of somatic mitochondrial mutations in five cancers. Proc. Natl. Acad. Sci. U. S. A. Aug 28, 2012;109(35):14087-91. [DOI] [PMC free article] [PubMed]
- 12.Wang C.Y., Li H., Hao X.D., Liu J., Wang J.X., Wang W.Z. Uncovering the profile of somatic mtDNA mutations in Chinese colorectal cancer patients. PloS One. 2011;6(6) doi: 10.1371/journal.pone.0021613. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wang CY, Li H, Hao XD, Liu J, Wang JX, Wang WZ, et al. Uncovering the profile of somatic mtDNA mutations in Chinese colorectal cancer patients. PloS one. 2011;6(6):e21613. [DOI] [PMC free article] [PubMed]
- 13.Gasparre G., Porcelli A.M., Lenaz G., Romeo G. Relevance of mitochondrial genetics and metabolism in cancer development. Cold Spring Harb Perspect Biol. 2013;5(2) doi: 10.1101/cshperspect.a011411. Feb. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gasparre G, Porcelli AM, Lenaz G, Romeo G. Relevance of mitochondrial genetics and metabolism in cancer development. Cold Spring Harb Perspect Biol. 2013 Feb;5(2). [DOI] [PMC free article] [PubMed]
- 14.Ussakli C.H., Ebaee A., Binkley J., Brentnall T.A., Emond M.J., Rabinovitch P.S. Mitochondria and tumor progression in ulcerative colitis. J Natl Cancer Inst. 2013;105(16):1239–1248. doi: 10.1093/jnci/djt167. Aug 21. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ussakli CH, Ebaee A, Binkley J, Brentnall TA, Emond MJ, Rabinovitch PS, et al. Mitochondria and tumor progression in ulcerative colitis. J. Natl. Cancer Inst. 2013 Aug 21;105(16):1239-48. [DOI] [PMC free article] [PubMed]
- 15.Huang S.W., Hsu C.M., Jeng W.J., Yen T.C., Su M.Y., Chiu C.T. A comparison of positron emission tomography and colonoscopy for the detection of advanced colorectal neoplasms in subjects undergoing a health check-up. PloS One. 2013;8(7) doi: 10.1371/journal.pone.0069111. [DOI] [PMC free article] [PubMed] [Google Scholar]; Huang SW, Hsu CM, Jeng WJ, Yen TC, Su MY, Chiu CT. A comparison of positron emission tomography and colonoscopy for the detection of advanced colorectal neoplasms in subjects undergoing a health check-up. PloS one. 2013;8(7):e69111. [DOI] [PMC free article] [PubMed]
- 16.Chen C.N., Lin S.Y., Liao Y.H., Li Z.J., Wong A.M. Late-onset caloric restriction alters skeletal muscle metabolism by modulating pyruvate metabolism. Am J Physiol Endocrinol Metab. 2015 Jun 1;308(11):E942–E949. doi: 10.1152/ajpendo.00508.2014. [DOI] [PubMed] [Google Scholar]; Chen CN, Lin SY, Liao YH, Li ZJ, Wong AM. Late-onset caloric restriction alters skeletal muscle metabolism by modulating pyruvate metabolism. Am J Physiol Endocrinol Metab. 2015 Jun 1;308(11):E942-9. [DOI] [PubMed]
- 17.Schuh R.A., Jackson K.C., Khairallah R.J., Ward C.W., Spangenburg E.E. Measuring mitochondrial respiration in intact single muscle fibers. Am J Physiol Regul Integr Comp Physiol. 2012;302(6):R712–R719. doi: 10.1152/ajpregu.00229.2011. Mar 15. [DOI] [PMC free article] [PubMed] [Google Scholar]; Schuh RA, Jackson KC, Khairallah RJ, Ward CW, Spangenburg EE. Measuring mitochondrial respiration in intact single muscle fibers. Am J Physiol Regul Integr Comp Physiol. 2012 Mar 15;302(6):R712-9. [DOI] [PMC free article] [PubMed]
- 18.Wang P.W., Kuo H.M., Huang H.T., Chang A.Y., Weng S.W., Tai M.H. Biphasic response of mitochondrial biogenesis to oxidative stress in visceral fat of diet-induced obesity mice. Antioxid Redox Signal. 2014;20(16):2572–2588. doi: 10.1089/ars.2013.5334. Jun 1. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wang PW, Kuo HM, Huang HT, Chang AY, Weng SW, Tai MH, et al. Biphasic response of mitochondrial biogenesis to oxidative stress in visceral fat of diet-induced obesity mice. Antioxid Redox Signal. 2014 Jun 1;20(16):2572-88. [DOI] [PMC free article] [PubMed]
- 19.Tsai C.L., Tsai C.N., Lin C.Y., Chen H.W., Lee Y.S., Chao A. Secreted stress-induced phosphoprotein 1 activates the ALK2-SMAD signaling pathways and promotes cell proliferation of ovarian cancer cells. Cell Rep. 2012;2(2):283–293. doi: 10.1016/j.celrep.2012.07.002. Aug 30. [DOI] [PubMed] [Google Scholar]; Tsai CL, Tsai CN, Lin CY, Chen HW, Lee YS, Chao A, et al. Secreted stress-induced phosphoprotein 1 activates the ALK2-SMAD signaling pathways and promotes cell proliferation of ovarian cancer cells. Cell Rep. 2012 Aug 30;2(2):283-93. [DOI] [PubMed]
- 20.Lin W.R., Lim S.N., McDonald S.A., Graham T., Wright V.L., Peplow C.L. The histogenesis of regenerative nodules in human liver cirrhosis. Hepatology. 2010;51(3):1017–1026. doi: 10.1002/hep.23483. Mar. [DOI] [PubMed] [Google Scholar]; Lin WR, Lim SN, McDonald SA, Graham T, Wright VL, Peplow CL, et al. The histogenesis of regenerative nodules in human liver cirrhosis. Hepatology. 2010 Mar;51(3):1017-26. [DOI] [PubMed]
- 21.Baker S.J., Preisinger A.C., Jessup J.M., Paraskeva C., Markowitz S., Willson J.K. p53 gene mutations occur in combination with 17p allelic deletions as late events in colorectal tumorigenesis. Cancer Res. 1990 Dec 1;50(23):7717–7722. [PubMed] [Google Scholar]; Baker SJ, Preisinger AC, Jessup JM, Paraskeva C, Markowitz S, Willson JK, et al. p53 gene mutations occur in combination with 17p allelic deletions as late events in colorectal tumorigenesis. Cancer Res. 1990 Dec 1;50(23):7717-22. [PubMed]
- 22.Burrell R.A., McClelland S.E., Endesfelder D., Groth P., Weller M.C., Shaikh N. Replication stress links structural and numerical cancer chromosomal instability. Nature. 2013;494(7438):492–496. doi: 10.1038/nature11935. Feb 28. [DOI] [PMC free article] [PubMed] [Google Scholar]; Burrell RA, McClelland SE, Endesfelder D, Groth P, Weller MC, Shaikh N, et al. Replication stress links structural and numerical cancer chromosomal instability. Nature. 2013 Feb 28;494(7438):492-6. [DOI] [PMC free article] [PubMed]
- 23.Jones A.M., Thirlwell C., Howarth K.M., Graham T., Chambers W., Segditsas S. Analysis of copy number changes suggests chromosomal instability in a minority of large colorectal adenomas. J Pathol. 2007;213(3):249–256. doi: 10.1002/path.2234. Nov. [DOI] [PubMed] [Google Scholar]; Jones AM, Thirlwell C, Howarth KM, Graham T, Chambers W, Segditsas S, et al. Analysis of copy number changes suggests chromosomal instability in a minority of large colorectal adenomas. J. Pathol. 2007 Nov;213(3):249-56. [DOI] [PubMed]
- 24.Maltzman T., Knoll K., Martinez M.E., Byers T., Stevens B.R., Marshall J.R. Ki-ras proto-oncogene mutations in sporadic colorectal adenomas: relationship to histologic and clinical characteristics. Gastroenterology. 2001;121(2):302–309. doi: 10.1053/gast.2001.26278. Aug. [DOI] [PubMed] [Google Scholar]; Maltzman T, Knoll K, Martinez ME, Byers T, Stevens BR, Marshall JR, et al. Ki-ras proto-oncogene mutations in sporadic colorectal adenomas: relationship to histologic and clinical characteristics. Gastroenterology. 2001 Aug;121(2):302-9. [DOI] [PubMed]
- 25.O'Brien M.J., Winawer S.J., Zauber A.G., Gottlieb L.S., Sternberg S.S., Diaz B. The National Polyp Study. Patient and polyp characteristics associated with high-grade dysplasia in colorectal adenomas. Gastroenterology. 1990 Feb;98(2):371–379. [PubMed] [Google Scholar]; O'Brien MJ, Winawer SJ, Zauber AG, Gottlieb LS, Sternberg SS, Diaz B, et al. The National Polyp Study. Patient and polyp characteristics associated with high-grade dysplasia in colorectal adenomas. Gastroenterology. 1990 Feb;98(2):371-9. [PubMed]
- 26.Czernin J., Benz M.R., Allen-Auerbach M.S. PET/CT imaging: the incremental value of assessing the glucose metabolic phenotype and the structure of cancers in a single examination. Eur J Radiol. 2010;73(3):470–480. doi: 10.1016/j.ejrad.2009.12.023. Mar. [DOI] [PubMed] [Google Scholar]; Czernin J, Benz MR, Allen-Auerbach MS. PET/CT imaging: The incremental value of assessing the glucose metabolic phenotype and the structure of cancers in a single examination. Eur. J. Radiol. 2010 Mar;73(3):470-80. [DOI] [PubMed]
- 27.Ong E.S., Zou L., Li S., Cheah P.Y., Eu K.W., Ong C.N. Metabolic profiling in colorectal cancer reveals signature metabolic shifts during tumorigenesis. Mol Cell Proteomics. 2010 Feb 10 doi: 10.1074/mcp.M900551-MCP200. ([M900551-MCP200], Paper is Press) [DOI] [PubMed] [Google Scholar]; Ong ES, Zou L, Li S, Cheah PY, Eu KW, Ong CN. Metabolic profiling in colorectal cancer reveals signature metabolic shifts during tumorigenesis. Mol Cell Proteomics. 2010 Feb 10. M900551-MCP200. [DOI] [PubMed]
- 28.Chatterjee A., Dasgupta S., Sidransky D. Mitochondrial subversion in cancer. Cancer Prev Res. 2011;4(5):638–654. doi: 10.1158/1940-6207.CAPR-10-0326. May. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chatterjee A, Dasgupta S, Sidransky D. Mitochondrial subversion in cancer. Cancer Prev. Res. 2011 May;4(5):638-54. [DOI] [PMC free article] [PubMed]
- 29.Romano M., Francesco D.E.F., Zarantonello L., Ruffolo C., Ferraro G.A., Zanus G. From inflammation to Cancer in inflammatory bowel disease: molecular perspectives. Anticancer Res. 2016 Apr;36(4):1447–1460. [PubMed] [Google Scholar]; . Romano M, Francesco DEF, Zarantonello L, Ruffolo C, Ferraro GA, Zanus G, et al. From Inflammation to Cancer in Inflammatory Bowel Disease: Molecular Perspectives. Anticancer Res. 2016 Apr;36(4):1447-60. [PubMed]
- 30.Kruidenier L., Verspaget H.W. Review article: oxidative stress as a pathogenic factor in inflammatory bowel disease—radicals or ridiculous? Aliment Pharmacol Ther. Dec, 2002;16(12):1997–2015. doi: 10.1046/j.1365-2036.2002.01378.x. [DOI] [PubMed] [Google Scholar]; Kruidenier L, Verspaget HW. Review article: oxidative stress as a pathogenic factor in inflammatory bowel disease—radicals or ridiculous? Aliment. Pharmacol. Ther. Dec, 2002;16(12):1997-2015. [DOI] [PubMed]
- 31.Pravda J. Radical induction theory of ulcerative colitis. World J Gastroenterol. Apr 28, 2005;11(16):2371–2384. doi: 10.3748/wjg.v11.i16.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]; Pravda J. Radical induction theory of ulcerative colitis. World J. Gastroenterol. Apr 28, 2005;11(16):2371-84. [DOI] [PMC free article] [PubMed]
- 32.Rezaie A., Parker R.D., Abdollahi M. Oxidative stress and pathogenesis of inflammatory bowel disease: an epiphenomenon or the cause? Dig Dis Sci. 2007;52(9):2015–2021. doi: 10.1007/s10620-006-9622-2. Sep. [DOI] [PubMed] [Google Scholar]; Rezaie A, Parker RD, Abdollahi M. Oxidative stress and pathogenesis of inflammatory bowel disease: an epiphenomenon or the cause? Dig. Dis. Sci. 2007 Sep;52(9):2015-21. [DOI] [PubMed]
- 33.Elson C.O., Sartor R.B., Tennyson G.S., Riddell R.H. Experimental models of inflammatory bowel disease. Gastroenterology. 1995;109(4):1344–1367. doi: 10.1016/0016-5085(95)90599-5. Oct. [DOI] [PubMed] [Google Scholar]; Elson CO, Sartor RB, Tennyson GS, Riddell RH. Experimental models of inflammatory bowel disease. Gastroenterology. 1995 Oct;109(4):1344-67. [DOI] [PubMed]
- 34.Rogler G. Chronic ulcerative colitis and colorectal cancer. Cancer Lett. 2014;345(2):235–241. doi: 10.1016/j.canlet.2013.07.032. Apr 10. [DOI] [PubMed] [Google Scholar]; Rogler G. Chronic ulcerative colitis and colorectal cancer. Cancer Lett. 2014 Apr 10;345(2):235-41. [DOI] [PubMed]
- 35.Du G., Mouithys-Mickalad A., Sluse F.E. Generation of superoxide anion by mitochondria and impairment of their functions during anoxia and reoxygenation in vitro. Free Radic Biol Med. 1998;25(9):1066–1074. doi: 10.1016/s0891-5849(98)00148-8. Dec. [DOI] [PubMed] [Google Scholar]; Du G, Mouithys-Mickalad A, Sluse FE. Generation of superoxide anion by mitochondria and impairment of their functions during anoxia and reoxygenation in vitro. Free Radic. Biol. Med. 1998 Dec;25(9):1066-74. [DOI] [PubMed]
- 36.Park J.H., Zhuang J., Li J., Hwang P.M. p53 as guardian of the mitochondrial genome. FEBS Lett. 2016;590(7):924–934. doi: 10.1002/1873-3468.12061. Apr. [DOI] [PMC free article] [PubMed] [Google Scholar]; Park JH, Zhuang J, Li J, Hwang PM. p53 as guardian of the mitochondrial genome. FEBS Lett. 2016 Apr;590(7):924-34. [DOI] [PMC free article] [PubMed]
- 37.Zhou S., Kachhap S., Singh K.K. Mitochondrial impairment in p53-deficient human cancer cells. Mutagenesis. 2003 May;18(3):287–292. doi: 10.1093/mutage/18.3.287. [DOI] [PubMed] [Google Scholar]; Zhou S, Kachhap S, Singh KK. Mitochondrial impairment in p53-deficient human cancer cells. Mutagenesis. 2003 May;18(3):287-92. [DOI] [PubMed]
- 38.de Araujo L.F., Fonseca A.S., Muys B.R., Placa J.R., Bueno R.B., Lorenzi J.C. Mitochondrial genome instability in colorectal adenoma and adenocarcinoma. Tumour Biol. 2015;36(11):8869–8879. doi: 10.1007/s13277-015-3640-7. Nov. [DOI] [PubMed] [Google Scholar]; de Araujo LF, Fonseca AS, Muys BR, Placa JR, Bueno RB, Lorenzi JC, et al. Mitochondrial genome instability in colorectal adenoma and adenocarcinoma. Tumour Biol. 2015 Nov;36(11):8869-79. [DOI] [PubMed]
- 39.Lightowlers R.N., Taylor R.W., Turnbull D.M. Mutations causing mitochondrial disease: what is new and what challenges remain? Science. 2015;349(6255):1494–1499. doi: 10.1126/science.aac7516. Sep 25. [DOI] [PubMed] [Google Scholar]; Lightowlers RN, Taylor RW, Turnbull DM. Mutations causing mitochondrial disease: What is new and what challenges remain? Science. 2015 Sep 25;349(6255):1494-9. [DOI] [PubMed]
- 40.Berridge M.V., Dong L., Neuzil J. Mitochondrial DNA in tumor initiation, progression, and metastasis: role of horizontal mtDNA transfer. Cancer Res. 2015;75(16):3203–3208. doi: 10.1158/0008-5472.CAN-15-0859. Aug 15. [DOI] [PubMed] [Google Scholar]; Berridge MV, Dong L, Neuzil J. Mitochondrial DNA in Tumor Initiation, Progression, and Metastasis: Role of Horizontal mtDNA Transfer. Cancer Res. 2015 Aug 15;75(16):3203-8. [DOI] [PubMed]
- 41.Lincet H., Icard P. How do glycolytic enzymes favour cancer cell proliferation by nonmetabolic functions? Oncogene. 2015;34(29):3751–3759. doi: 10.1038/onc.2014.320. Jul. [DOI] [PubMed] [Google Scholar]; Lincet H, Icard P. How do glycolytic enzymes favour cancer cell proliferation by nonmetabolic functions? Oncogene. 2015 Jul;34(29):3751-9. [DOI] [PubMed]
- 42.Du S., Guan Z., Hao L., Song Y., Wang L., Gong L. Fructose-bisphosphate aldolase a is a potential metastasis-associated marker of lung squamous cell carcinoma and promotes lung cell tumorigenesis and migration. PloS One. 2014;9(1) doi: 10.1371/journal.pone.0085804. [DOI] [PMC free article] [PubMed] [Google Scholar]; Du S, Guan Z, Hao L, Song Y, Wang L, Gong L, et al. Fructose-bisphosphate aldolase a is a potential metastasis-associated marker of lung squamous cell carcinoma and promotes lung cell tumorigenesis and migration. PloS one. 2014;9(1):e85804. [DOI] [PMC free article] [PubMed]
- 43.Caspi M., Perry G., Skalka N., Meisel S., Firsow A., Amit M. Aldolase positively regulates of the canonical Wnt signaling pathway. Mol Cancer. 2014;13:164. doi: 10.1186/1476-4598-13-164. [DOI] [PMC free article] [PubMed] [Google Scholar]; Caspi M, Perry G, Skalka N, Meisel S, Firsow A, Amit M, et al. Aldolase positively regulates of the canonical Wnt signaling pathway. Mol. Cancer. 2014;13:164. [DOI] [PMC free article] [PubMed]
- 44.Halestrap A.P. The SLC16 gene family - structure, role and regulation in health and disease. Mol Aspects Med. 2013;34(2–3):337–349. doi: 10.1016/j.mam.2012.05.003. Apr-Jun. [DOI] [PubMed] [Google Scholar]; Halestrap AP. The SLC16 gene family - structure, role and regulation in health and disease. Mol. Aspects Med. 2013 Apr-Jun;34(2-3):337-49. [DOI] [PubMed]
- 45.Gill R.K., Saksena S., Alrefai W.A., Sarwar Z., Goldstein J.L., Carroll R.E. Expression and membrane localization of MCT isoforms along the length of the human intestine. Am J Physiol Cell Physiol. 2005;289(4):C846–C852. doi: 10.1152/ajpcell.00112.2005. Oct. [DOI] [PubMed] [Google Scholar]; Gill RK, Saksena S, Alrefai WA, Sarwar Z, Goldstein JL, Carroll RE, et al. Expression and membrane localization of MCT isoforms along the length of the human intestine. Am. J. Physiol. Cell Physiol. 2005 Oct;289(4):C846-52. [DOI] [PubMed]
- 46.Gotanda Y., Akagi Y., Kawahara A., Kinugasa T., Yoshida T., Ryu Y. Expression of monocarboxylate transporter (MCT)-4 in colorectal cancer and its role: MCT4 contributes to the growth of colorectal cancer with vascular endothelial growth factor. Anticancer Res. 2013 Jul;33(7):2941–2947. [PubMed] [Google Scholar]; Gotanda Y, Akagi Y, Kawahara A, Kinugasa T, Yoshida T, Ryu Y, et al. Expression of monocarboxylate transporter (MCT)-4 in colorectal cancer and its role: MCT4 contributes to the growth of colorectal cancer with vascular endothelial growth factor. Anticancer Res. 2013 Jul;33(7):2941-7. [PubMed]
- 47.Vegran F., Boidot R., Michiels C., Sonveaux P., Feron O. Lactate influx through the endothelial cell monocarboxylate transporter MCT1 supports an NF-kappaB/IL-8 pathway that drives tumor angiogenesis. Cancer Res. 2011;71(7):2550–2560. doi: 10.1158/0008-5472.CAN-10-2828. Apr 1. [DOI] [PubMed] [Google Scholar]; Vegran F, Boidot R, Michiels C, Sonveaux P, Feron O. Lactate influx through the endothelial cell monocarboxylate transporter MCT1 supports an NF-kappaB/IL-8 pathway that drives tumor angiogenesis. Cancer Res. 2011 Apr 1;71(7):2550-60. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bioenergetic comparison between polyp and cancer and adjacent normal mucosa.
The analysis of mitochondrial DNA variants and mutations in polyps and their adjacent normal mucosa.
The analysis of mitochondrial DNA variants and mutations in colorectal adenocarcinoma and their adjacent normal mucosa.