Abstract
Background
Myb-binding protein 1A (Mybbp1a) is a nucleolar protein that can regulate rRNA metabolism, the stress response and carcinogenesis. However, the function of Mybbp1a in the progression of hepatocellular carcinoma (HCC) is unclear.
We aimed to determine the role of Mybbp1a in HCC and the underlying mechanism.
Methods
We investigated the function of Mybbp1a in HCC cell models and the xenograft mouse model. The relationship between Mybbp1a and IGFBP5 was found through expression profile chip. The molecular mechanism of Mybbp1a regulating IGFBP5 was proved through CO-IP, CHIP, Bisulfite Sequencing and Pyrosequencing.
Findings
In this study, we observed that Mybbp1a was overexpressed in HCC tissues and associated with the poor prognosis of HCC patients. Suppression of Mybbp1a led to a reduction in the proliferation and migration ability of HCC cells through inhibiting the IGF1/AKT signaling pathway. Further study found that Mybbp1a could form a complex with DNMT1 and induce aberrant hyper-methylation of CpG islands of IGFBP5, which inhibits secretion of IGFBP5 and then activates IGF1/AKT signaling pathway.
Interpretation
These findings extend our understanding of the function of Mybbp1a in the progression of HCC. The newly identified Mybbp1a may provide a novel biomarker for developing potential therapeutic targets of HCC.
Fund
Science Technology Department of Zhejiang Province (No. 2015C03034), National Health and Family Planning Commission of China (No. 2016138643), Innovative Research Groups of National Natural Science Foundation of China (No. 81721091), Major program of National Natural Science Foundation of China (No. 91542205).
Keywords: Myb-binding protein 1A (Mybbp1a), IGF-binding proteins 5(IGFBP5), Hepatocellular carcinoma (HCC), IGF1/AKT pathway, CpG islands methylation
Abbreviations: Mybbp1a, Myb-binding protein 1A; IGFBP5, IGF-binding proteins 5; HCC, Hepatocellular carcinoma; PBS, Phosphate buffer saline; RT-PCR, Real-time polymerase chain reaction; FBS, Fetal bovine serum; IHC, Immunostaining; CCK-8, Cell counting kit-8; Edu, 5-ethynyl-2′-deoxyuridine
Research in context.
Evidence before this study
The mechanism of Mybbp1a in tumor initiation and progression was controversial. It was shown that Mybbp1a acted as a tumor suppressor in breast cancer by enhancing p53-associated anoikis and inhibiting the deacetylation activity of H3K18Ac. However, recent study demonstrated that Mybbp1a could enhance tumor cell proliferation in the early phase of HNSCC and inhibited tumor cell migration and invasion in advanced HNSCC. Moreover, the human protein atlas database showed that Mybbp1a may be favorable prognostic marker in pancreatic cancer, while unfavorable in renal cancer, thyroid cancer and melanoma. In addition, our study found that Mybbp1a was overexpressed in HCC tissues. Based on these, we propose that Mybbp1a may have a special function in the initiation and progression of HCC.
Added value of this study
our study found that Mybbp1a suppressed the transcription of IGFBP5 through the DNMT1-mediated hyper-methylation level of CpG islands in the IGFBP5 CDS site. This biological process led to a loss of the secretion of IGFBP5, which improved the combinative efficiency between IGF1 and IGF1R. Then, phosphorylation of IGF1R induced the activation of the PI3K-AKT pathway, which increased the proliferation rate and migration capacity of HCC cells.
Implications of all the available evidence
These findings extend our understanding of the function of Mybbp1a in the progression of HCC. The newly identified Mybbp1a may provide a novel biomarker for developing potential therapeutic targets of HCC.
Alt-text: Unlabelled Box
1. Introduction
Hepatocellular carcinoma (HCC) is one of the most common solid tumors worldwide, and its incidence and mortality rate have persistently increased in recent years [1,2]. Surgical resection and liver transplantation (LT) are first-line treatments in early HCC. However, many patients are diagnosed with advanced HCC and are not eligible for surgery [3,4]. Furthermore, frequent metastasis and recurrence are the major reasons for the poor prognosis of HCC patients after surgical resection or LT [5]. Potential effective biomarkers for predicting the recurrence and prognosis of HCC are rare. Therefore, the identification of new molecular markers is needed.
MYB binding protein 1a (Mybbp1a) is a nucleolar transcriptional regulator that was first identified because of its ability to specifically bind to the MYB proto-oncogene protein [[6], [7], [8]]. Further studies proved that Mybbp1a also bind to several other transcription factors, including the RelA/p65 subunit of NF-kB, p53, PGC-1a and Prep1, and played an important role in many cellular processes, including the response to nucleolar stress, synthesis of ribosomal DNA and amino acids, and progression of the oxidative phosphorylation respiratory chain [[9], [10], [11], [12], [13], [14], [15], [16]]. However, the function of Mybbp1a in solid tumors is controversial. A recent study proved that Mybbp1a acted as a tumor suppressor protein in breast cancer by enhancing p53-associated anoikis [17].Another study proved that Mybbp1a promoted tumor cell proliferation in the early phase of head and neck squamous cell carcinoma (HNSCC), while inactivation of MYBBP1A induced accelerated tumor cell migration and invasion in advanced tumors [18].Moreover, by integrating prognostic analysis from TCGA database, the human protein atlas database showed that Mybbp1a may be favorable prognostic marker in pancreatic cancer, while unfavorable in renal cancer, thyroid cancer and melanoma.
Thus, more studies are needed to determine the function of Mybbp1a in solid tumors. Here, we demonstrate Mybbp1a is overexpressed in HCC tissues and associated with poor prognosis of HCC patients. Additionally, Mybbp1a could promote the proliferation and migration of HCC cells by activating the IGF1/AKT pathway through inducing CpG-island methylation mediated IGFBP5 silencing.
2. Materials and methods
2.1. Patients and specimens
This study was approved by the ethics committee of the First Affiliated Hospital of Zhejiang University, and informed consent was obtained from all patients. All cancer tissue samples and adjacent non-tumor tissue samples were obtained from 71 patients diagnosed with HCC during September 2012 to December 2013. Clinical data were collected from all these patients including age, gender, size of tumor, number of tumors, vessel invasion, tumor grade and AFP (alpha-fetoprotein). Follow-up information was obtained by reviewing the patient's medical records up to the termination date (January 31, 2016) or death.
2.2. Cell culture
The HCC cell line SK-Hep-1 was purchased from the American Type Culture Collection (Manassas, VA). The liver cancer cell lines Huh7 and HCC-LM3 and the normal liver cell line QSG-7701 were purchased from the Chinese Academy of Science Cell Bank (Shanghai, China). All cell lines were proven by short tandem repeats (STR) and SNP fingerprinting before use. Cells were maintained in MEM or RPMI 1640 medium supplemented with 10% FBS at 37 °C in a humidified atmosphere of 5% CO2. The AKT inhibitor MK2206 was obtained from Selleck Chemicals (Houston, TX) and dissolved in ddH2O for the purpose of this research. IGF1 and IGFBP5 were obtained from Peprotech (Rocky Hill, USA) and dissolved in ddH2O with 3% BSA for research.
2.3. Construction of stably transfected cell lines
The target sequences of Mybbp1a (5′-GCUGGUGAAUGUGCUGAAGAUGGCC-3′),
and IGFBP5 (5′-CTCCCATTCTCCAATGACT-3′) were obtained from Shanghai Genechem Co., Ltd. and used to generate stable Mybbp1a or IGFBP5 knockdown cell lines according to the instructions of manufacturer. Cells were selected by puromycin (4 μg/ml) (Sangon, Shanghai, China) for 72 h, and the transfection efficiency was proved by quantifying EGFP fluorescence, RT-PCR and western blotting. Stably transfected Huh7 and HCC-LM3 cells were maintained in the Modified Eagle medium (MEM, Gibco, Grand Island, NY) containing 10% fetal bovine serum (FBS, Gibco) for further study.
2.4. Real-Time PCR and western blot
Real-Time PCR and western blot were conducted as previously descripted [[19], [20], [21]]. The primers were as follows: Mybbp1a, Forward 5′-GCATCCTGCAGCAGATACAA-3′, Reverse 5′-CAGCTTCACCGACTTCATCA-3′; IGFBP5, Forward 5′-TGACCGCAAAGGATTCTACAAG-3′, Reverse 5′-CGTCAACGTACTCCATGCCT-3′; and GAPDH, Forward 5′-GAATGGGGAAGGTGAAGGTCG-3′, Reverse 5′-GGGGTCATTGATGGCAACAATA-3′. The antibodies are shown in the Supplement Table 1.
2.5. Immunohistochemistry
Immunostaining (IHC) was performed on formalin-fixed, paraffin-embedded clinical tissue specimens. The expression levels of Mybbp1a in HCC tissues and neighbor tissues were detected as the previously descripted [19]. The IHC score was assessed by two independent investigators. The method for the IHC score was as follows: 0, negative; 1, <25% positive tumor cells; 2, 25–50% positive tumor cells; 3, 50%–75% positive tumor cells; and 4, >75% positive tumor cells. The intensity of the dye color was graded as 0 (no color), 1 (light yellow), 2 (light brown), or 3 (brown). The staining index was calculated as follows: staining index = staining intensity × tumor cell staining grade. High Mybbp1a expression levels were defined as a staining index score ≥ 6, while low expression was defined as a staining index < 6.
2.6. Cell proliferation and migration experiments in vitro
HCC cell lines with Mybbp1a knockdown, Mybbp1a + IGFBP5 knockdown or no knockdown (negative control) were used for cell proliferation and migration assays as previously described [20,21].
2.7. Immunofluorescence staining
The HCC Huh7-shMybbp1a, Huh7-NC, HCC-LM3-shMybbp1a and HCC-LM3-NC cells were fixed with 4% paraformaldehyde for 15 min and then permeabilized and simultaneously blocked with 0.1% Triton-X 100 and 1% BSA for 30 min. Cells were incubated sequentially with the Phalloidin-iFluor 647 conjugate (AAT Bioquest®, Inc., CA) for 1 h at room temperature and then with DAPI for 5 min. Finally, the cells were examined with a laser confocal microscope (FV1000, Olympus).
2.8. ELISA assay
The Human IGFBP-5 ELISA Kit was purchased from Ray Biotech, Inc. (ELH-IGFBP5-1, P24593), and the experiment was conducted according to the guidelines.
2.9. Tumor xenograft and lung metastasis
All experimental protocols were approved by the Animal Experimental Ethics Committee of the First Affiliated Hospital of Zhejiang University School of Medicine. The methods were carried out in accordance with the approved guidelines. Cells (5 × 106) were resuspended in 100 μl of PBS and injected subcutaneously into the lateral flanks of immunodeficient mice. Tumor volumes were measured every three days and calculated using the following equation: V (cm3) = width2 (cm2) × length (cm)/2. The mice were sacrificed about five weeks after the injection. In addition, the nude mice were each injected via the tail vein with Mybbp1a-shRNA or Mybbp1a-NC (1 × 106 cells). Mice were sacrificed 40 days after the tail vein injection, and the lungs were embedded in paraffin and sliced for HE staining to identify metastasis.
2.10. Expression microarrays
Huh7 cells were transfected with shMybbp1a and negative control respectively. After RNA preparation using RNeasy Protect Midi Kit (Qiagen, Valencia, CA), hybridization was conducted using Affymetrix whole-genome microarrays (hgu133Plus2) according to the manufacturer's instructions. Raw data were normalized using the RMA algorithm in R statistical computing environment (www.r-project.org) using the default parameters. The relative fold changes were calculated according to negative control.
2.11. Bisulfite sequencing and pyrosequencing
Genomic DNA of cells was extracted using the standard phenol–chloroform technique followed by proteinase K treatment to prevent protein contamination. Bisulfite conversion was performed using an EpiTect Bisulfite Kit (QIAGEN) according to the manufacturer's instruction.The primers for IGFBP5 amplification were as follows: forward 5′-ATGTGTTTTTTTAGTTTTTTGGGTTG-3′; reverse 5′-ACAAAATCCTCCAAAACTCCAATT-3′.
For pyrosequencing, the amplified DNA products were detected by PyroMark Q96 ID (QIAGEN). The primers for IGFBP5 amplification were as follows: forward 5′-TTGGGTTGAGAGTTGGTTAAGGA-3′, reverse 5′-ACCCCAACTCCAAACCACTTACTAC-3′. The primer for sequencing was as follow: 5′-TGAGAGAGTTTAGTGTGTTAGTTAG-3′.
2.12. Co-immunoprecipitation
Co-immunoprecipitation was performed as previously described [22].
2.13. Chromatin Immunoprecipitations-PCR
Chromatin Immunoprecipitations-PCR was conducted by Shanghai Kangchen Bio-tech, and the IGFBP5 primers was showed in the Supplement Table 2.
2.14. Statistical analysis
Data are presented as the means ± standard deviation (SD). For comparisons, Student's t-test and Fisher's exact test were performed, as appropriate. Relapse-free -survival and overall-survival analysis were evaluated by the Kaplan-Meier method, and differences were assessed using the log-rank test. Cox multivariate regression analysis was used to identify independent prognostic factors. All analyses were performed with SPSS 18.0 software (SPSS Inc., Chicago, IL). Differences were considered significant at P < .05.
3. Results
3.1. Mybbp1a is overexpressed in HCC tissues and associated with a poor prognosis of HCC patients
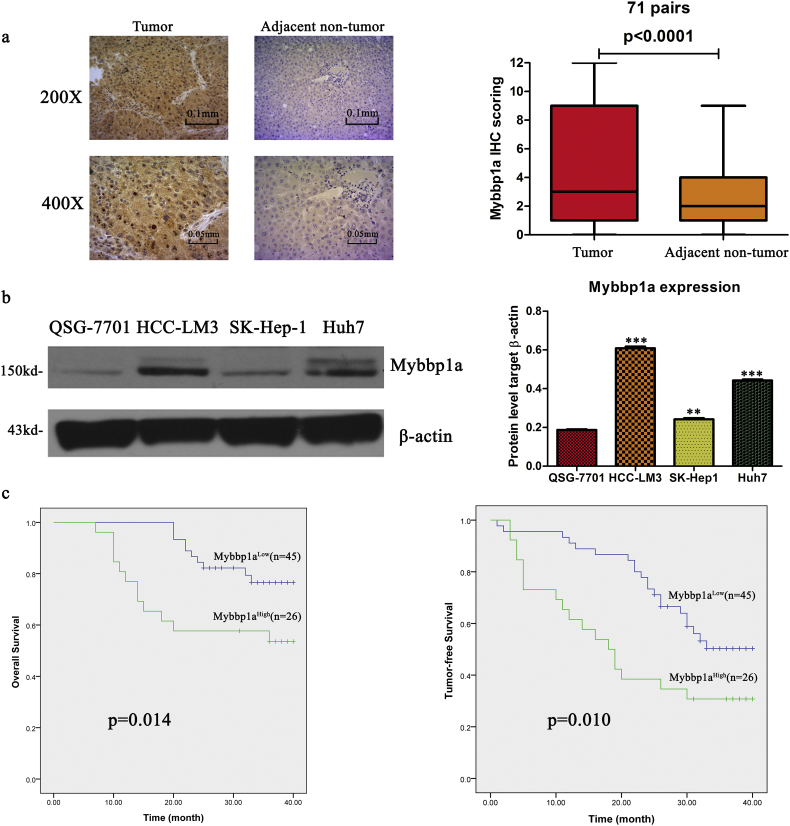
To investigate the clinical significance of Mybbp1a expression, we measured its expression in 71 paired HCC tissues and adjacent non-tumor tissues. Immunohistochemical (IHC) assays showed that the average expression level of Mybbp1a was significantly higher in HCC tissues than that in adjacent non-tumor tissues (Fig. 1a). We also observed that the expression of Mybbp1a was higher in HCC cell lines (SK-Hep-1, Huh7, HCC-LM3) than that in normal liver cells (QSG-7701) (Fig. 1b). Kaplan-Meier analysis showed that higher expression of Mybbp1a was associated with shorter overall survival and tumor-free survival in HCC patients (P = .014 and P = .010, respectively) (Fig. 1c). This result was validated by liver cancer survival analysis in human protein atlas database (www.proteinatlas.org) (Supplement Fig. 1). Cox multivariate regression analysis indicated that high expression of Mybbp1a was an independent and significant factor for overall and tumor-free survival (Supplement Table 2). Taken together, these data indicated that Mybbp1a was overexpressed in HCC and that its overexpression predicted a poor prognosis in HCC patients and might contribute to the progression of HCC.
Fig. 1.
Mybbp1a was overexpressed in HCC tissues and associated with poor prognosis of HCC patients.
(a) Showed the expression of Mybbp1a in HCC tissues and adjacent non-tumor tissues (200×, scale bar = 0.1 mm;400×, scale bar = 0.05 mm), and the results were quantified in the pie chart, (p < .0001, student's t-test). (b) Showed the expression level of Mybbp1a in HCC cell lines and normal liver cells, (**p < .005, ***p < .0005, student's t-test). (c) The overall and relapse-free survival of HCC patients between Mybbp1a high-expression group and Mybbp1a low-expression group were analyzed by Kaplan-meier analysis, (p = .014 and p = .010 respectively, Log Rank (Mantel-Cox)).
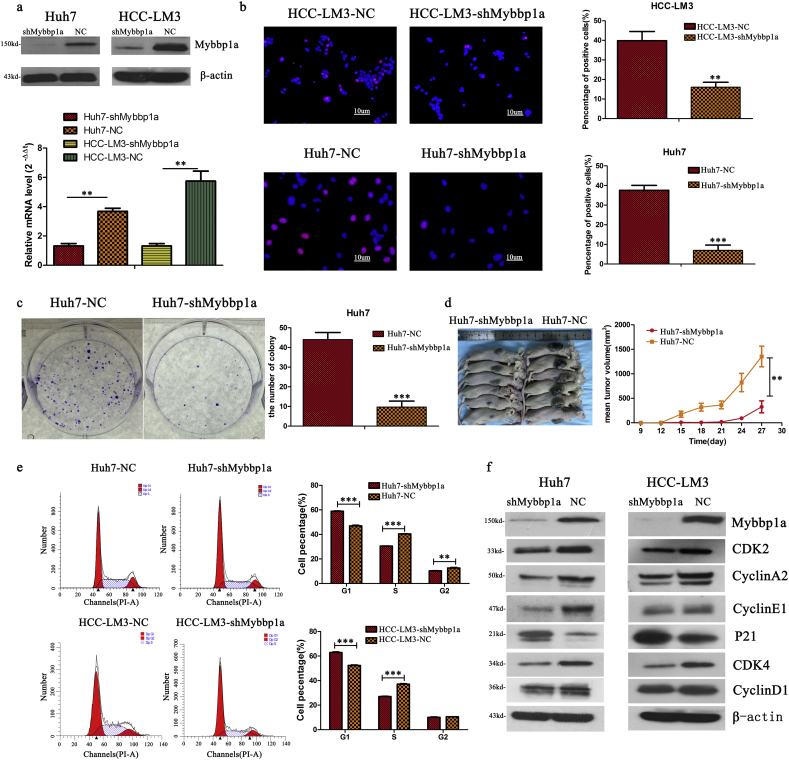
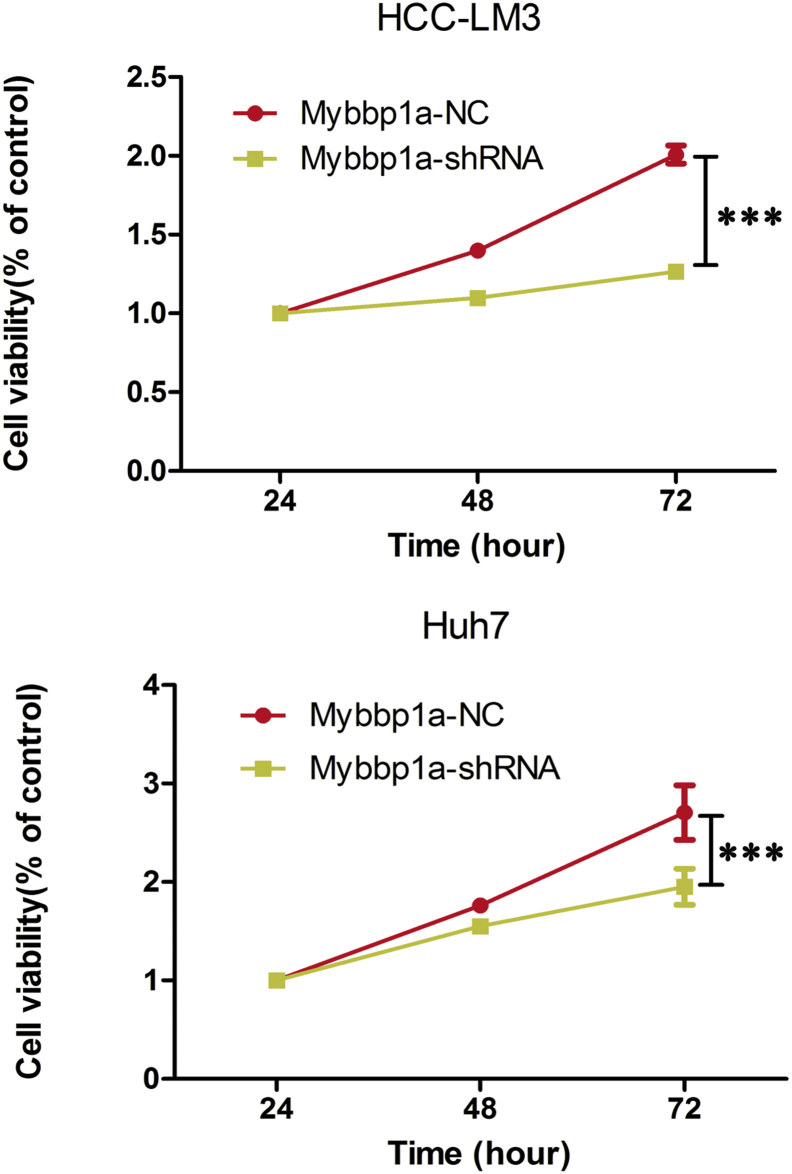
3.2. Suppression of Mybbp1a inhibits the proliferation of HCC cells via G1/S transition
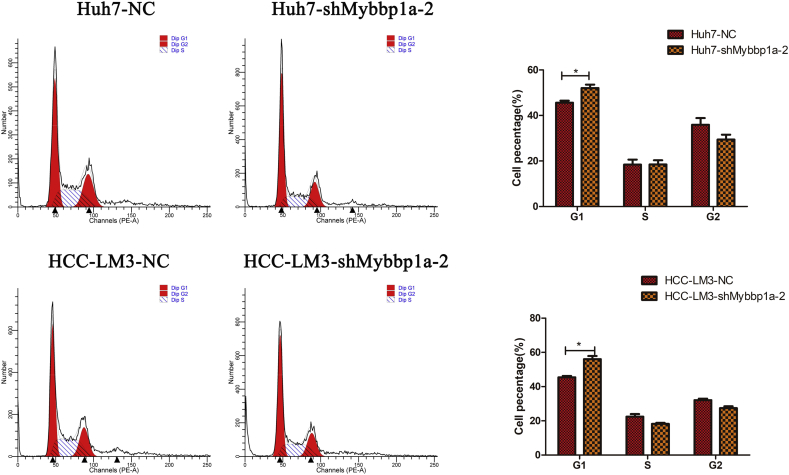
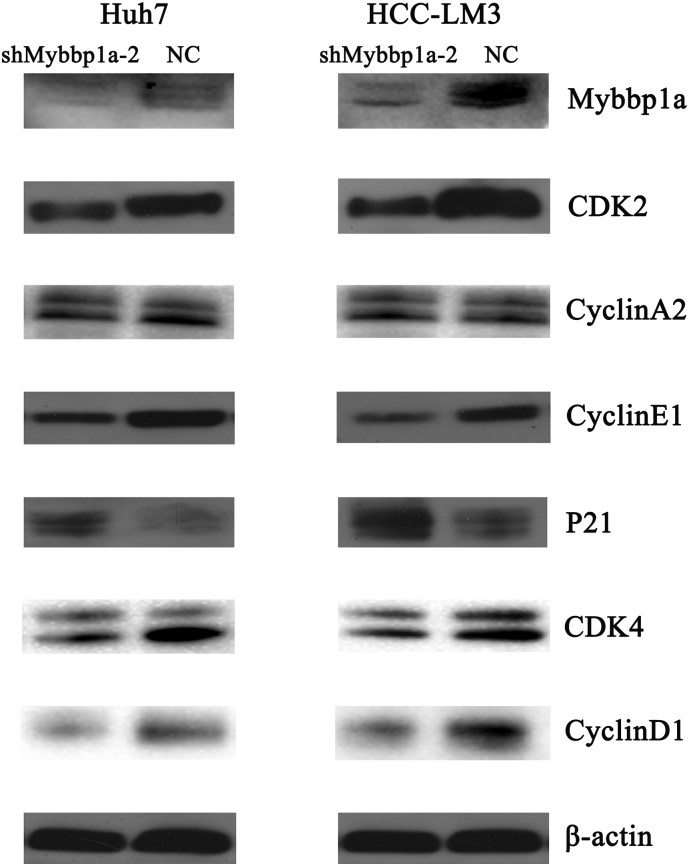
According to the expression level of Mybbp1a in HCC cell lines, we stably transfected Mybbp1a-shRNA into the Huh7 and HCC-LM3 cell lines, and the transfection efficiency was proven at both the protein and mRNA levels (Fig. 2a). Loss of function experiments showed that suppression of Mybbp1a led to lower cell viability and a lower proliferation rate in both Huh7 and HCC-LM3 cells (Supplement Fig. 2, Fig. 2b). Consistently, tumor formation assays showed that suppression of Mybbp1a significantly inhibited the tumorigenesis of HCC cells both in vitro and in vivo (Fig. 2c–d, Supplement Fig. 3). Flow cytometry analysis revealed that knockdown of Mybbp1a induced G1 phase arrest (Fig. 2e, Supplement Fig. 4). We also detected several G1 phase-associated proteins and discovered that suppression of Mybbp1a induced lower expression of CDK2, cyclin A2, CDK4 and cyclin E1, but higher expression of p21 (Fig. 2f, Supplement Fig. 5). These results indicated that Mybbp1a played an important role in the proliferation and regulation of HCC cells.
Fig. 2.
Suppression of Mybbp1a inhibited proliferation of HCC cells via G1/S transition. (a) The transfection efficiency of shMybbp1a and negative control (NC) in Huh7 and HCC-LM3 cells was verified both at protein and mRNA levels, (**p < .005, student's t-test). (b) The Edu assay was used to detect proliferation rate of HCC cells with shMybbp1a or negative control (NC), (scale bar = 10um) (***p < .0005, **p < .005, student's t-test). (c–d) The tumorigenic ability of Huh7-shMybbp1a and Huh7-NC was compared both in vitro and in vivo, (**p < .005, ***p < .0005, student's t-test). (e) The cell cycle was detected by flow cytometry, and the results were analyzed by ModFIT software, (***p < .0005, **p < .005, student's t-test). (f) The expression of G1 phase associated proteins was detected by western blot, and β-actin was as the internal control.
Supplementary Fig. 2.
Showed the results of CCK-8 assay in HCC cells with shMybbp1a or negative control (NC), and the results of 72 h were used for statistical analysis, (***P < .0005, one-way ANOVA).
Supplementary Fig. 3.
showed the results of colony formation assay in HCC cells with shMybbp1a-2 or negative control (NC).
Supplementary Fig. 4.
showed the results of Flow cycle analysis in HCC cells with shMybbp1a-2 or negative control (NC), and the result was statistical analyzed, (*p < .05, student's t-test).
Supplementary Fig. 5.
showed the expression change of G1 phase associated proteins in HCC cells with shMybbp1a-2 or negative control (NC).
3.3. Suppression of Mybbp1a inhibits the migration ability of HCC cells in vitro and in vivo
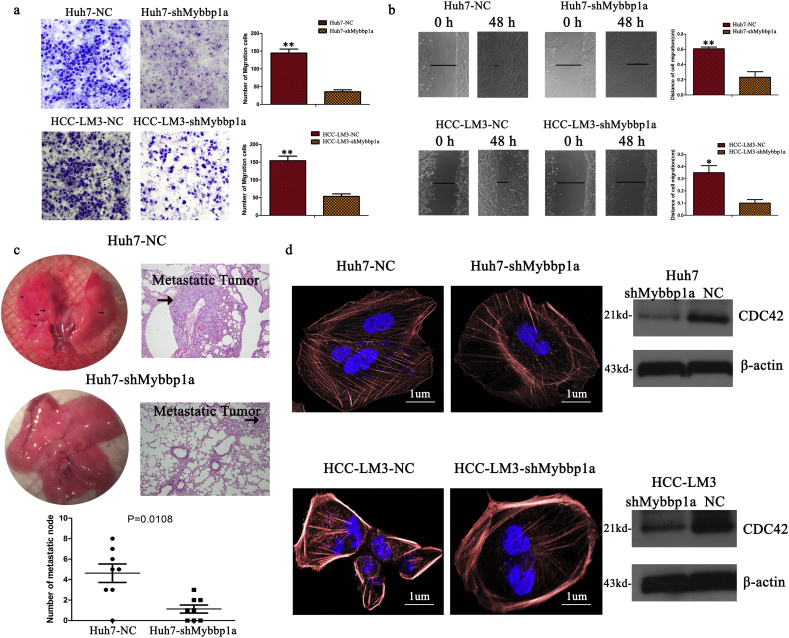
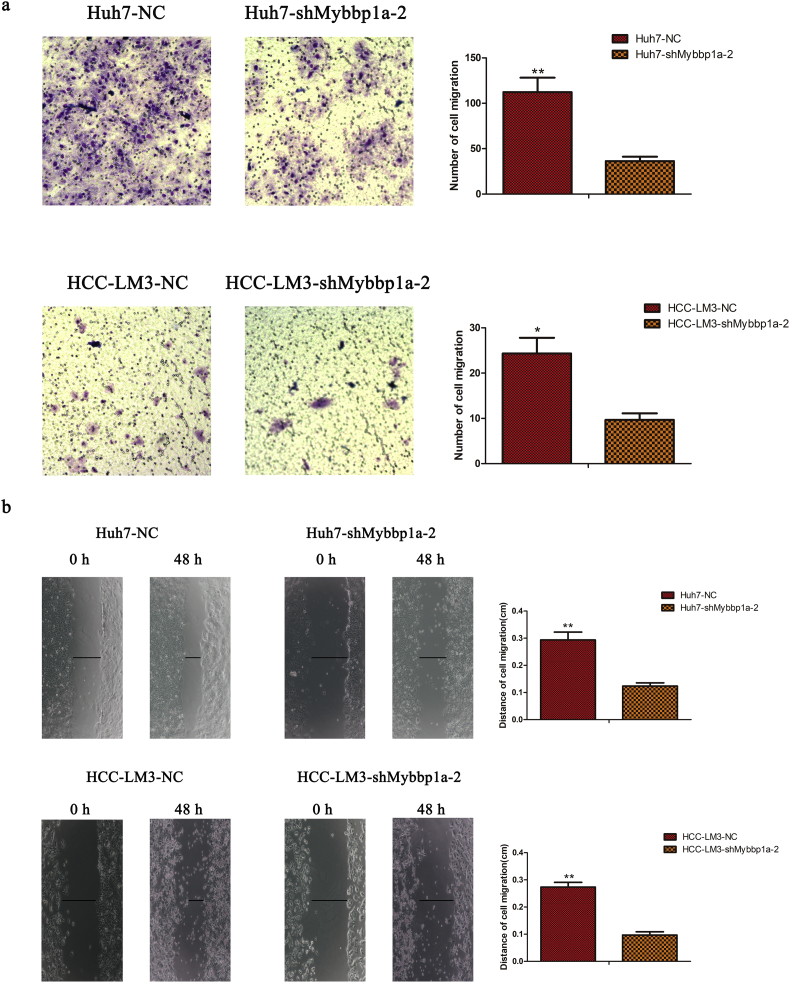
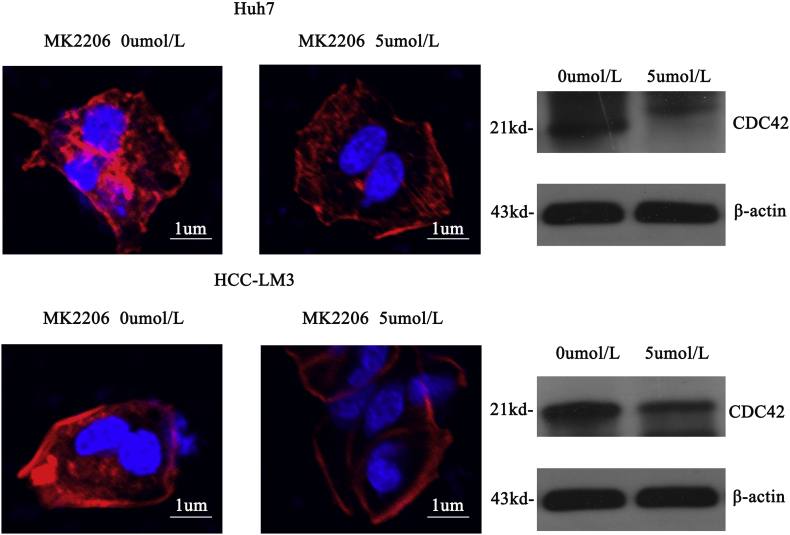
Next, we evaluated whether Mybbp1a played a role in the motility of HCC cells. The results of transwell and wound-healing assays showed that suppression of Mybbp1a led to a lower migration ability than that of the negative control (Fig. 3a-b, Supplement Fig. 6). Consistently, a pulmonary metastasis model showed that the Huh7-shNC group displayed more lung metastasis foci and larger metastatic tumors than those of the Huh7-shMybbp1a group (Fig. 3c). Then, we used phalloidin to observe the cell cytoskeleton in HCC cells and found that the numbers of microfilaments and microtubules were lower in the Huh7-shMybbp1a group than those in the negative control. Meanwhile, we observed that CDC42, which is an important molecule in cell cytoskeleton regulation, significantly decreased under the conditions of Mybbp1a suppression (Fig. 3d). These findings showed that Mybbp1a depletion may inhibit cell migration by regulating CDC42-associated cell cytoskeleton synthesis.
Fig. 3.
Suppression of Mybbp1a inhibited migration capacity of HCC cells. (a) Cell migration capacity was studied by transwell assay and the results were quantified, (**p < .005, student's t-test). (b)Showed the result of wound healing assay; the distance was evaluated with pictures taken at 0 h, 6 h, 24 h and 48 h, (**p < .005, *p < .05, student's t-test). (c) The lung metastasis experiment was conducted with Huh7-shMybbp1a and negative control (NC), the HE photos of metastatic tumor in lung were shown, and the number of metastatic nodes was quantified, (p = .0108, student's t-test). (d) The cytoskeleton of HCC cells with shMybbp1a and negative control (NC) under treatment of phalloidin was shown (scale bar = 1um), and CDC42 was detected by western blotting, and β-actin was as the internal control.
Supplementary Fig. 6.
Showed the results of transwell assay and wound healing assay in HCC cells with shMybbp1a-2 or negative control (NC), and the result was statistical analyzed, (*p < .05, **p < .005, student's t-test).
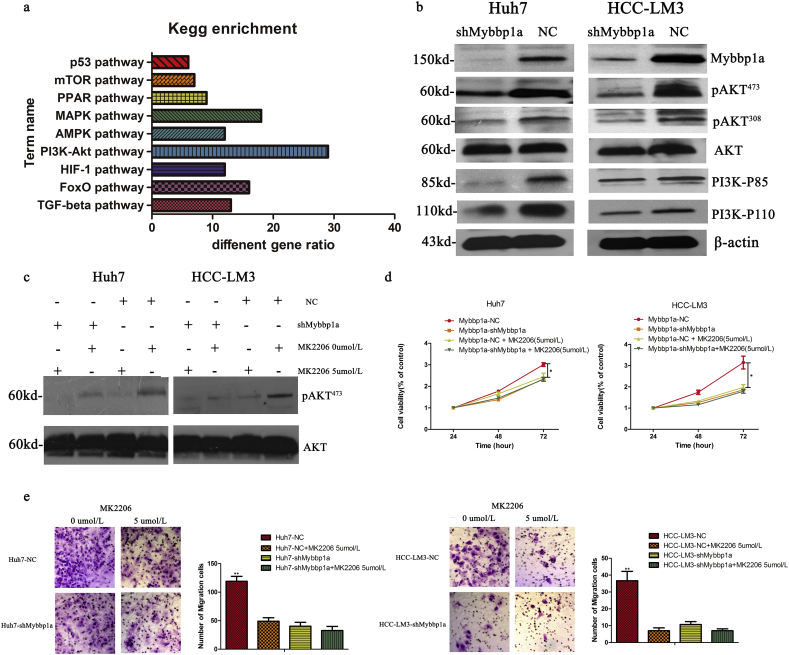
3.4. Mybbp1a promotes the proliferation and migration of HCC cells via PI3K/AKT pathway
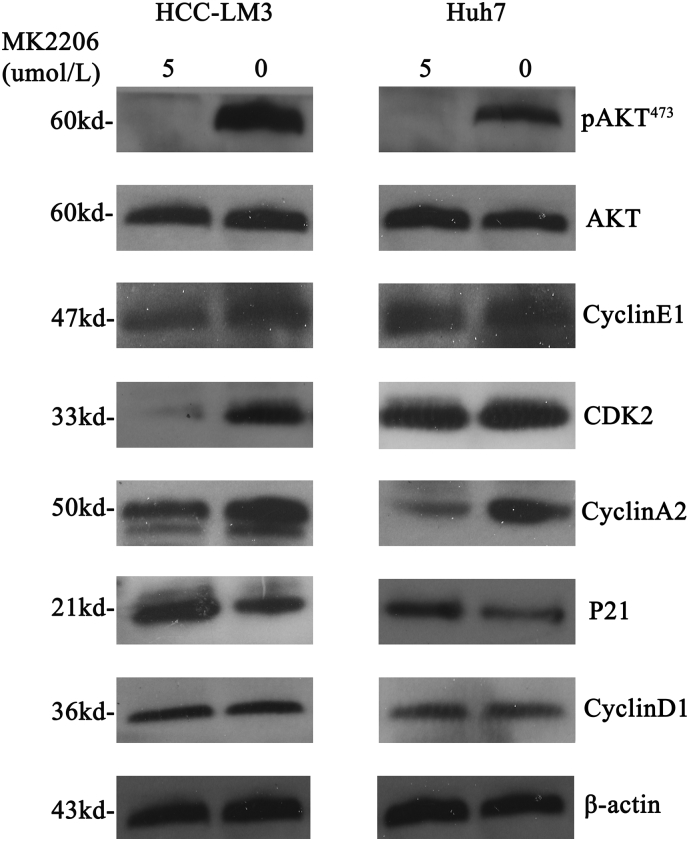
To further explore the mechanism of Mybbp1a in regulating the proliferation and migration of HCC cells, we used Affymetrix whole-genome microarrays (hgu133Plus2) to investigate the differential genes and pathways between the Huh7-shMybbp1a and Huh7-NC groups. KEGG enrichment analysis showed that the PI3K/AKT signaling pathway was significantly changed among the classic biological regulatory pathways (Fig. 4a). Then, we detected the expression of pAKT473, pAKT308, PI3K-P110 and PI3K-P85 in Huh7 and HCC-LM3 cells and found that all of these molecules were significantly decreased under the condition of Mybbp1a suppression (Fig. 4b). To further demonstrate our findings, we used MK2206-2HCL (an inhibitor of AKT) to conduct a remedy experiment, and the inhibition efficiency was validated by western blot analysis (Fig. 4c). Then, we detected cell viability and migration ability in HCC-shMybbp1a and HCC-NC cells with or without 5 μmol/l MK2206-2HCL by the CCK-8 and transwell assays. The results showed that, except for the negative control (NC) group, the cell viability of the other groups was similar at 72 h (Fig. 4d). Consistently, this phenomenon also appeared in a cell migration experiment (Fig. 4e). In addition, we also demonstrated that inhibition of AKT activation induced a change in the expression of G1 phase-associated proteins and cytoskeletal synthesis reduction (Supplementary Fig. 7, Supplementary Fig. 8). Thus, our study proved that Mybbp1a regulated the proliferation and migration of HCC cells by promoting the activation of the PI3K/AKT signaling pathway.
Fig. 4.
Mybbp1a regulated proliferation and migration of HCC cells through PI3K/AKT pathway. (a) The number of differential genes in classic signaling pathways was showed. (b) The expression change of pAKT473, pAKT308, PI3K-P110 and PI3K-P85 in HCC cells was shown, and β-actin was as the internal control. (c) Expression of pAKT473and AKT were detected under different concentration of MK-2206 2HCL in HCC cells with shMybbp1a or negative control (NC). (d) Cell viability of HCC cells with shMybbp1a or negative control (NC) under different concentration of MK-2206 2HCL were shown, and the results of 72 h were used for statistical analysis,(*p < .05,one-way ANOVA). (e) Cell migration capacity of HCC cells with shMybbp1a or negative control (NC) under different concentration of MK-2206 2HCL were shown,(**p < .005, student's t-test).
Supplementary Fig. 7.
Cell G1 phase associated proteins were detected under different concentration of MK2206(0, 5 μmol/l), pAKT473 was used to verify the efficiency of the drug.
Supplementary Fig. 8.
Cell cytoskeleton was detected under different concentration of MK2206(0, 5 μmol/l) (scale bar = 1um), Cytoskeletal synthesis protein CDC42 was detected.
3.5. IGFBP5 is negatively regulated by Mybbp1a in HCC cells
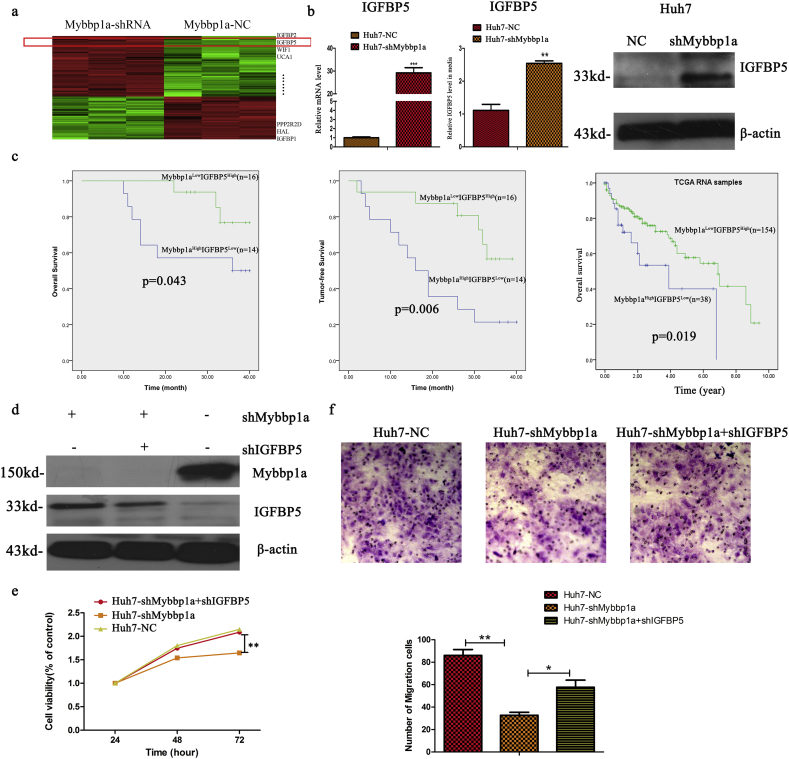
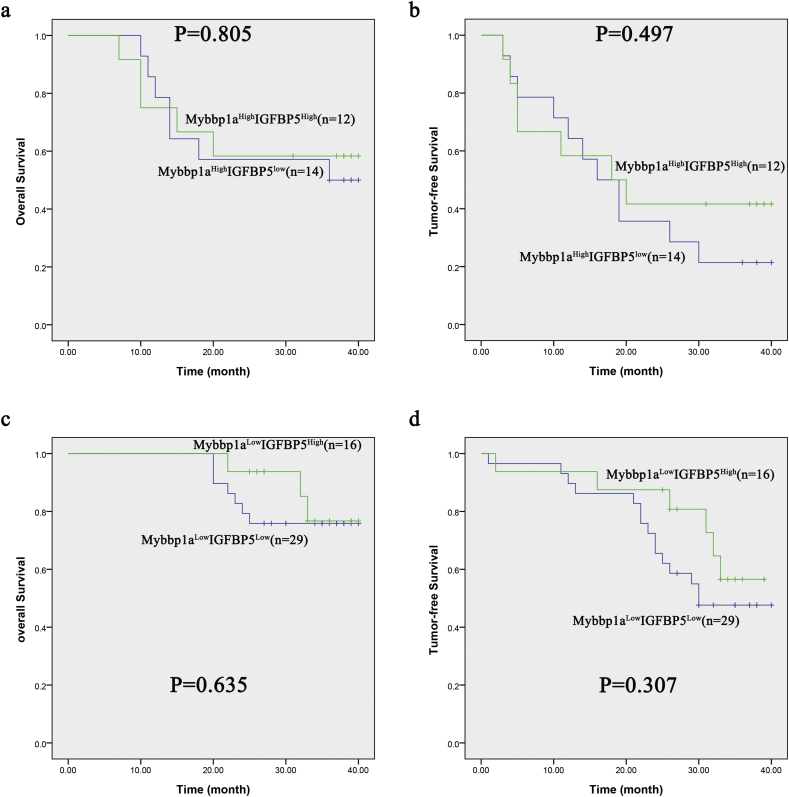
To further investigate the mechanism of Mybbp1a in the activation of the PI3K/AKT pathway, we analyzed the differential gene expression in our Affymetrix whole-genome microarrays. We found that IGFBP1, HAL, PPP2R2D, UCA1, WIF1, IGFBP5 and IGFBP2 had large fold changes under Mybbp1a suppression (Fig. 5a). A recent study proved that IGFBP5 was a potent inhibitor of both the signaling and functional outputs of IGF-1, which plays a vital role in the activation of the PI3K/AKT pathway [23]. Thus, we selected IGFBP5 for further study. First, we validated that IGFBP5 was significantly increased among the mRNA, intracellular protein and extracellular protein levels under Mybbp1a knockdown in Huh7 cells (Fig. 5b).Then, we analyzed patients with Mybbp1aHighIGFBP5Low, Mybbp1aLowIGFBP5High, Mybbp1aHighIGFBP5High or Mybbp1aLowIGFBP5Low, the results showed that the overall and tumor-free survival of patients with Mybbp1aHighIGFBP5Low was shorter than those with Mybbp1aLowIGFBP5High (Fig. 5c, Supplement Fig. 9).In addition, we constructed three stably transfected cell lines, Huh7-shMybbp1a, Huh7-shMybbp1a + shIGFBP5 and a negative control, and the transfection efficiency was verified by western blotting (Fig. 5d). The CCK-8 assay showed that simultaneous knockdown of Mybbp1a and IGFBP5 reversed the decreased viability of cells induced by Mybbp1a knockdown alone (Fig. 5e). Consistently, the results of the transwell assay were similar to those of the cell viability experiment (Fig. 5f). Based on the above findings, we demonstrated that IGFBP5 was negatively regulated by Mybbp1a in HCC cells.
Fig. 5.
IGFBP5 was negatively regulated by Mybbp1a. (a) The differentially expressed genes from the transcriptome chip were shown. (b) The mRNA and protein levels of IGFBP5 between Huh7-shMybbp1a and Huh7-NC cells were detected, (**P < .005, ***p < .0005, student's t-test). (c) Kaplan-meier analysis was used to compare prognosis between two group patients (our HCC cohort and TCGA HCC cohort), including Mybbp1ahighIGFBP5low and Mybbp1alowIGFBP5high group, (p = .043,0.006 and 0.019 respectively, Log Rank (Mantel-Cox)) (d) Transfection efficiency of Huh7-shMybbp1a, Huh7-shMybbp1a + shIGFBP5 and Huh7-NC was verified by western blot. (e) Cell viability of HCC cells with shMybbp1a, shMybbp1a + shIGFBP5 or negative control (NC) was detected, and the results of 72 h were used for statistical analysis (**p < .005, one-way ANOVA). (f) Cell migration capacity of HCC cells with shMybbp1a, shMybbp1a + shIGFBP5 or negative control (NC) was detected, (*p < .05, **p < .005, student's t-test).
Supplementary Fig. 9.
Showed the results of Kaplan-meier analysis among Mybbp1aHighIGFBP5Low, Mybbp1aLowIGFBP5High, Mybbp1aHighIGFBP5High and Mybbp1aLowIGFBP5Low, (p = .805, p = .497, p = .635 and p = .307 respectively, Log Rank (Mantel-Cox)).
3.6. Mybbp1a promotes the activation of the IGF1/AKT signaling pathway by inhibiting the secretion of IGFBP5
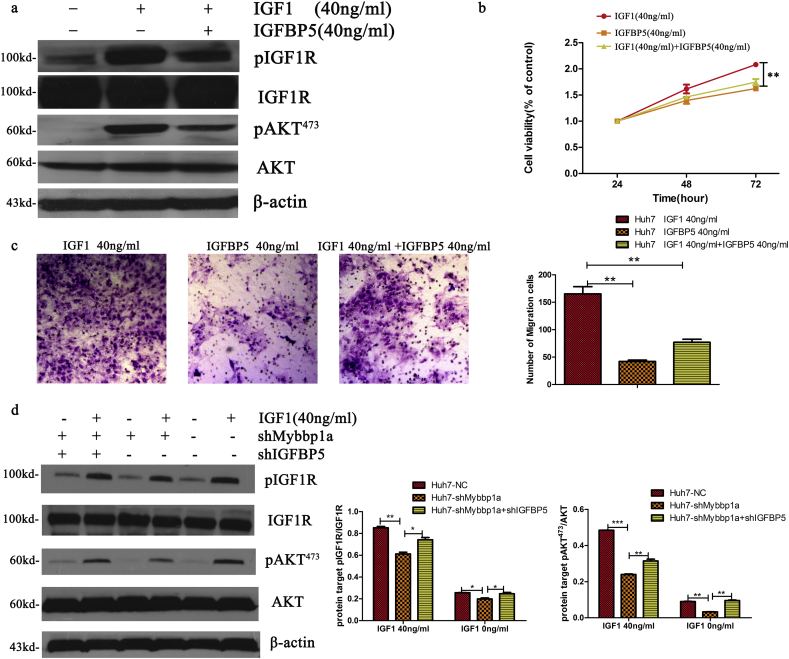
The secretion of IGFBP5 inhibited the activation of IGF1R by competitive binding with IGF1 in the extracellular space [23]. Thus, we hypothesized that the inactivation of the PI3K-AKT pathway was mainly due to the inhibition of the IGF1 pathway in the case of Mybbp1a knockdown. We first validated that the phosphorylation of IGF1R and AKT by IGF1 (Peprotech, Product number 100-11) was inhibited after adding the recombinant protein IGFBP5 (Peprotech, Product number100-05) (Fig. 6a). Then, the CCK-8 assay showed that the cell viability of that IGF1 group was higher than that of IGFBP5 and IGF1 + IGFBP5 groups (Fig. 6b). Similarly, this phenomenon appeared in the cell migration experiment (Fig. 6c). Moreover, in comparison with no IGF1 treatment, we validated that the expression of pIGF1R and pAKT473 both increased under the condition of 40 ng/ml IGF1 in Huh7 cells with shMybbp1a, shMybbp1a + shIGFBP5 or a negative control. However, the expression of pIGF1R and pAKT473 in cells with shMybbp1a + shIGFBP5 and negative controls was higher than that of those with shMybbp1a (Fig. 6d). Taken together, these findings showed that suppression of Mybbp1a led to the inactivation of the IGF1/AKT pathway by inducing the secretion of IGFBP5.
Fig. 6.
Mybbp1a promoted activation of IGF1/IGF1R/PI3K/AKT through inhibiting secretion of IGFBP5. (a) The expression level of pAKT473, AKT, pIGF1R and pIGF1R were detected in Huh7 cells treated with IGF1 (40 ng/ml), IGF1 + IGFBP5 (40 ng/ml) or the control. (b) Cell viability of Huh7 cells treated with IGF1 (40 ng/ml), IGF1 + IGFBP5 (40 ng/ml) or the control was detected, and the results of 72 h were used for statistical analysis, (**P < .005, one-way ANOVA). (c) Cell migration capacity of Huh7 cells treated with IGF1 (40 ng/ml), IGF1 + IGFBP5 (40 ng/ml) or the control was detected, (**p < .005, student's t-test). (d) Expression level of pAKT473, AKT, pIGF1R and pIGF1R were detected after treatment with IGF1 0 ng/ml or 40 ng/ml. The results were quantified by Image J, (*P < .05, **P < .005 and ***P < .0005, student's t-test).
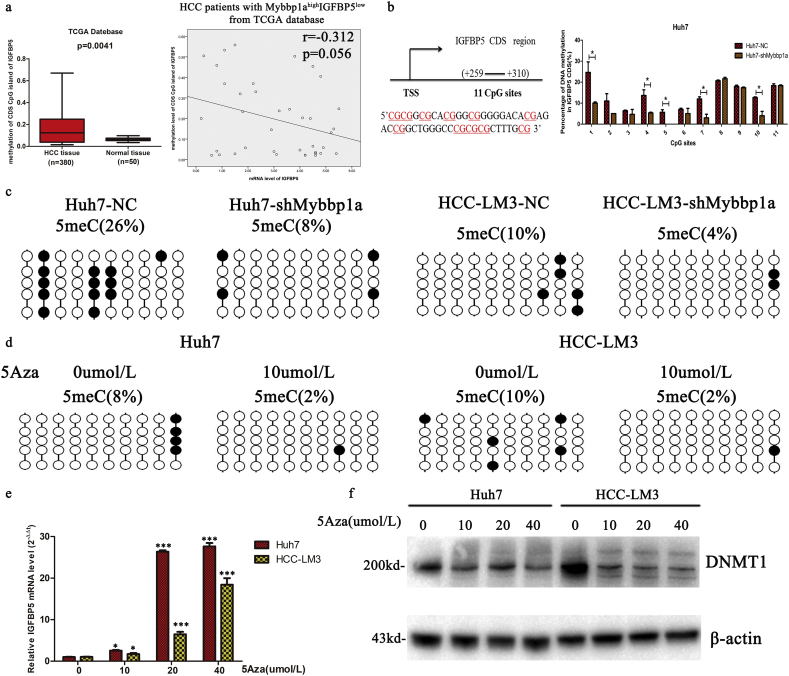
3.7. Mybbp1a inhibits the transcription of IGFBP5 by promoting CpG island methylation
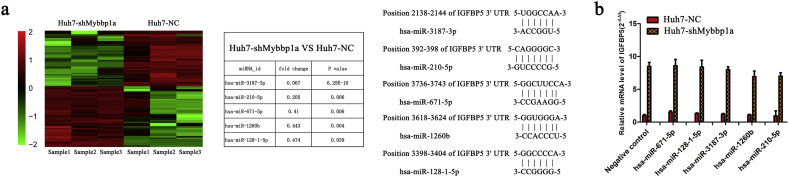
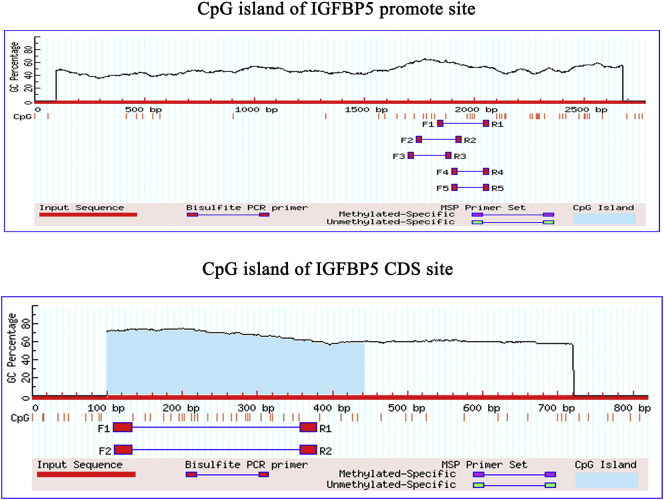
As the suppression of Mybbp1a significantly increased the mRNA level of IGFBP5, we sought to determine how Mybbp1a affected the expression of IGFBP5, through transcriptional level or post-transcriptional level. First, we conducted microRNA sequencing to search for candidate microRNA which affected expression of IGFBP5 in post-transcriptional level. Considering that the mRNA level of IGFBP5 was upregulated under suppression of Mybbp1a, we selected miRNAs that were decreased in the Huh7-shMybbp1a group. Based on the above findings, five miRNAs (miR-3187-3p, miR-210-5p, miR-671-5p, miR-1260b and miR-128-1-5p) were found (Supplement Fig. 10a). However, there was no significant change in the IGFBP5 mRNA level under treatment with miRNA mimics in Huh7 cells (Supplement Fig. 10b). Bioinformatics analysis (MethPrimer 2.0) showed that the CpG islands of IGFBP5 were not in the promoter site, while found one in the CDS (coding sequence) site (Supplement Fig. 11).Then we found that the methylation of CDS CpG island of IGFBP5 in HCC tissues was higher than that in normal tissues, and mRNA level of IGFBP5 was negative correlated with methylation level of CDS CpG island of IGFBP5 in HCC patients with Mybbp1ahighIGFBP5low (Fig. 7a), we thought that Mybbp1a may regulate the mRNA level of IGFBP5 at the transcriptional level through CpG island methylation. By analyzing the methylation level of 40 CpGs in the IGFBP5 CDS site through Bisulfite Sequencing PCR, we observed that the CpG methylation level of the negative control was higher than that of the shMybbp1a group, ranging from 26 CpG to 36 CpG. Then, we quantified the methylation percentage of these 11 CpGs (+259–+310) by pyrosequencing. The results showed that the methylation levels at position 1, 4, 5, 7, and 10 were significantly decreased under Mybbp1a suppression (Fig. 7b). As the results of Bisulfite Sequencing PCR showed that the methylation levels of position 7–11 were significantly different between the NC group and shMybbp1a group in HCC cell lines (Fig. 7c); thus, we selected these five CpG sites for further study. Then, we used Azacitidine (5Aza) to treat the HCC cell lines, and the results of Bisulfite Sequencing PCR showed that the methylation levels of position 7–11 were decreased (Fig. 7d). In addition, we found that the mRNA level of IGFBP5 was significantly increased after treatment with 5Aza (0, 10, 20, 40 μmol/l) (Fig. 7e). The inhibition efficiency of 5Aza was verified by detecting the expression of DNMT1 under different concentrations of 5Aza (Fig. 7f). Therefore, our study demonstrated that Mybbp1a inhibited the transcription of IGFBP5 by promoting the methylation of CpG islands in the CDS site.
Supplementary Fig. 10.
(a) The heat map and candidate miRNAs from the miRNA sequencing were shown, and complementary pairing sequence between miRNAs 3′UTR and IGFBP5 mRNA were obtained from TargetScan database. (b) The mRNA level of IGFBP5 under treatment of miRNAs mimics was detected and quantified.
Supplementary Fig. 11.
The CpG islands of IGFBP5 promote site and CDS site were analyzed by Methprimer.
Fig. 7.
Mybbp1a inhibits transcription of IGFBP5 via promoting CpG islands methylation. (a) CpG island methylation level of IGFBP5 in HCC tissue and normal tissue was obtained from TCGA database, and relationship between CpG island methylation level and mRNA level of IGFBP5 was analyzed, (p = .0041, student's t-test and p = .056, Pearson) (b) CpG sites of IGFBP5 was showed, TSS indicates transcription start site. And the percentages of DNA methylation in specific sites of the IGFBP5 CDS were analyzed by pyrosequencing, (*p < .05, student's t-test) (c–d) The percentages of DNA methylation in specific sites of the IGFBP5 CDS were analyzed by bisulfite sequencing PCR in HCC stable cell line with/without Mybbp1a knockdown, or HCC cell lines treated with 5Aza (0,10 μmol/l) for 48 h. (e) The mRNA level of IGFBP5 was detected under different concentration of 5Aza (0, 10, 20, 40 μmol/l) after 48 h, (*p < .05, ***p < .0005, student's t-test). (f) Expression of DNMT1 under different concentration of 5Aza (0, 10, 20, 40 μmol/l) for 48 h was detected.
3.8. Mybbp1a promotes the methylation of CpG islands of IGFBP5 through DNMT1
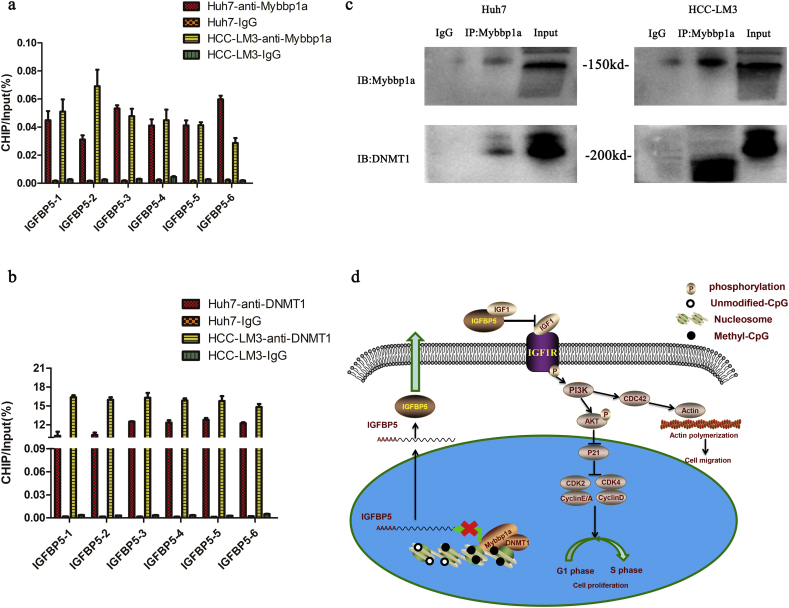
To demonstrate the mechanism of Mybbp1a's regulation of the methylation of CpG islands of IGFBP5, we conducted Chip-PCR analysis to determine whether Mybbp1a and DNMT1 could combine with the CDS site of IGFBP5. We designed 6 primers according to the CDS site of IGFBP5, and the results showed that all of the primers had PCR products (Fig. 8a–b). In addition, the result of Co-IP showed that Mybbp1a could also bind to DNMT1 in HCC cell lines (Fig. 8c). Thus, our study proved that Mybbp1a was able to bind to the CDS site of IGFBP5 and then induce methylation of the CpG site by recruiting DNMT1.
Fig. 8.
Mybbp1a promoting methylation of CpG island of IGFBP5 through DNMT1. (a–b) Chromatin immunoprecipitation (ChIP) was used to enrichment IGFBP5 CDS region by Mybbp1a or DNMT1 antibody, and PCR was used to verify the efficiency according to the Input. (c) Co-immunoprecipitation assay confirms the protein binding between Mybbp1a and DNMT1. (d) The molecular model depicted the mechanism of Mybbp1a as an oncogene in HCC.
4. Discussion
MYB binding protein 1a (Mybbp1a), a nucleolar transcriptional regulator, was first identified as a binding protein with the MYB proto-oncogene protein [6,7]. Further studies demonstrated that Mybbp1a played a vital role in response to P53-associated nucleolar stress, the synthesis of ribosomal DNA and amino acids, and the progression of the oxidative phosphorylation respiratory chain [[9], [10], [11], [12], [13], [14], [15], [16],24]. However, the mechanism of Mybbp1a in tumor initiation and progression was controversial. It was shown that Mybbp1a acted as a tumor suppressor in breast cancer by enhancing p53-associated anoikis and inhibiting the deacetylation activity of H3K18Ac [17,25]. However, a recent study demonstrated that Mybbp1a enhanced tumor cell proliferation in the early phase of HNSCC and inhibited tumor cell migration and invasion in advanced HNSCC [18]. In addition, we found that Mybbp1a was overexpressed in HCC tissues by the Human Protein Atlas database. Based on these, we propose that Mybbp1a may have a special function in the initiation and progression of HCC.
We first demonstrated that Mybbp1a was overexpressed in HCC tissues and that the high expression of Mybbp1a was associated with poor overall and tumor-free survival. Then, loss-of-function experiments showed that suppression of Mybbp1a inhibited the proliferation and migration of HCC cells through G1 phase arrest and suppression of CDC42-mediated cytoskeleton synthesis. To further illustrate this phenomenon, we used transcriptome chip analysis in HCC cells and found that the expression of PI3K/AKT pathway-related genes had significantly changed. As the activation of PI3K/AKT promoted the progression of the cell cycle and cytoskeleton synthesis [[26], [27], [28], [29], [30]]. And our study validated that suppression of Mybbp1a decreased the expression of pAKT308, pAKT473, PI3K-P110 and PI3K-P85. In addition, the cell viability and migration capacities of HCC cells with Mybbp1a knockdown were similar to those of HCC cells under the condition of AKT inhibitors. Thus, we proved that Mybbp1a regulated the proliferation and migration of HCC cells via the PI3K-AKT pathway.
To further illustrate the mechanism of Mybbp1a on the activation of the PI3K-AKT pathway, we analyzed the differentially expressed genes on the transcriptome chip and found that the expression of IGFBP5 was significantly increased. IGFBP5, one of the IGF-binding proteins (IGFBP-1 to -6), is a secreted protein that inhibits the IGF1/IGF1R/PI3K/AKT axis through a competitive combination with circulating IGF1 [31]. Recent studies reported that IGFBP5 was a tumor suppressor in many solid tumors, including breast cancer [32], head and neck cancer [33], osteosarcoma [34], uterine leiomyoma [35], melanoma [36], colorectal cancer [37], gastrointestinal stromal tumors [38] and papillary thyroid carcinoma [39]. Based on these data, we hypothesized that the deactivation of the PI3K/AKT pathway under the suppression of Mybbp1a might be due to the increased secretion of IGFBP5.
We first validated that the expression of IGFBP5 was increased both at the mRNA and protein levels under Mybbp1a knockdown. Then, we found that HCC patients with Mybbp1aHighIGFBP5Low had shorter overall and tumor-free survival than those with Mybbp1aLowIGFBP5High. CCK-8 and transwell assays showed that the cell viability and migration capacity of HCC cells with Mybbp1a suppression were reversed when suppressing both Mybbp1a and IGFBP5. In addition, we demonstrated that IGFBP5 inhibited the IGF1-mediated activation of the PI3K-AKT pathway in HCC cells. The expression of pIGF1R and pAKT473 in HCC cells with shMybbp1a was lower than that of with shMybbp1a + shIGFBP5 or negative controls under the condition of 40 ng/ml IGF1. In summary, we demonstrated that suppression of Mybbp1a inhibited the activation of the IGF1/IGF1R/PI3K/AKT axis through inducing the secretion of IGFBP5.
Thus, we sought to determine how the suppression of Mybbp1a could increase the mRNA level of IGFBP5 in HCC cells, through inhibiting mRNA degradation or promoting mRNA synthesis. First, we thought that suppression of Mybbp1a might induce changes in the expression of IGFBP5-related miRNAs. miRNAs are a class of highly conserved short RNAs of 21–23 nucleotides that induce the degradation of mRNAs by binding to the 3′untranslated region (3′UTR) of their targeted mRNAs [40,41]. Then, we performed miRNA sequencing in HCC cells with shMybbp1a or the negative control and found five candidate miRNAs (miR-3187-3p, miR-210-5p, miR-671-5p, miR-1260b and miR-128-1-5p). However, the mimics of these miRNAs could not significantly decrease the mRNA level of IGFBP5 in HCC cells with shMybbp1a. Then, we tried to find a breakthrough from DNA methylation modification, which is the major epigenetic modification that involves alterations of the chromatin structure. Increasing evidence demonstrated that aberrant methylation of CpG islands in the tumor suppressor gene led to transcriptional silencing in cancer [42,43]. Based on these data, we analyzed the sequence of IGFBP5, including the promoter region, CDS site, 5′ UTR Exons, 3′ UTR Exons and Introns, and found that there were CpG islands in the CDS site of IGFBP5. Then, the results of Bisulfite Sequencing and Pyrosequencing showed that the methylation levels of specific sites significantly decreased under Mybbp1a suppression. In addition, we found that Mybbp1a was able to specifically bind to DNMT1 and the CDS site of IGFBP5. Thus, we demonstrated that Mybbp1a inhibited the secretion of IGFBP5 by increasing the methylation level of CpG islands of the CDS site by recruiting DNMT1.
In conclusion, our study found that Mybbp1a suppressed the transcription of IGFBP5 through the DNMT1-mediated hyper-methylation level of CpG islands in the IGFBP5 CDS site. This biological process led to a loss of the secretion of IGFBP5, which improved the combinative efficiency between IGF1 and IGF1R. Then, phosphorylation of IGF1R induced the activation of the PI3K-AKT pathway, which increased the proliferation rate and migration capacity of HCC cells (Fig. 8d). Though the most of the published studies reported the negative role of Mybbp1a in various cancer types, however, high heterogeneity of tumors from different tissue sources indicated that Mybbp1a may act as different role during progression of different solid tumors. Therefore, our findings raised an intriguing hypothesis that Mybbp1a might serve as a “switch” to regulate the secretion of IGFBP5 to cater to the needs of HCC progression.
However, our study only demonstrated Mybbp1a could affect the methylation of CpG island in CDS site of IGFBP5, further studies were needed to prove whether Mybbp1a could regulate other CpG islands methylation of IGFBP5 or regulate the transcription of IGFBP5 by other pathways?
The following are the supplementary data related to this article.
Supplementary Fig. 1.
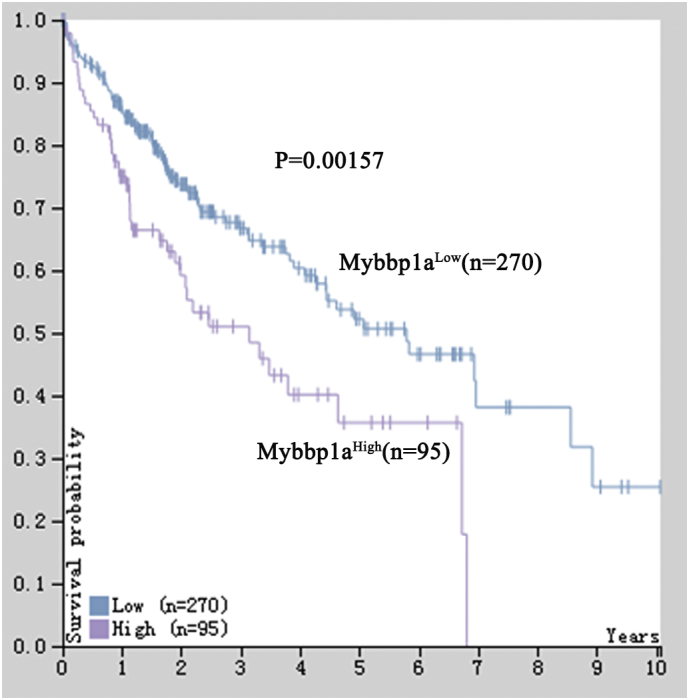
Showed the results of Kaplan-meier analysis between Mybbp1a high-expression and low-expression, the follow-up data was supplied by TCGA database, (p = .00157, Log Rank (Mantel-Cox)).
Supplementary tables
Author contributions
Xiaoyu Weng, Zhen Lv, Jian Wu and Shusen Zheng designed the study. Xiaoyu Weng, Jingbang Wu, Junru Chen, Bin He, Cheng Zhang, Rongliang Tong, Wendi Hu and Linping Cao, Chaofeng Ding and Diyu Chen performed the experiments. Xiaoyu Weng, Zhen Lv and Jian Wu drafted the manuscript. Xiaoyu Weng, Zhen Lv and Chuanhui Peng analyzed the results and made pictures.
Acknowledgments
Acknowledgement
This study was supported by grant from Science Technology Department of Zhejiang Province (No. 2015C03034), National Health and Family Planning Commission of China (No. 2016138643), Innovative Research Groups of National Natural Science Foundation of China (No. 81721091), Major program of National Natural Science Foundation of China (No. 91542205). These funding sources had no role in the study design, data collection, data analysis, interpretation, or writing of this manuscript.
Consent for publications
I have obtained consent to publish from the participant to report individual patient data.
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding authors on reasonable request.
Ethics approval and consent to participate
This study was approved by the ethics committee of the First Affiliated Hospital of Zhejiang University, and informed consent was obtained from all patients. All experimental protocols were approved by the Animal Experimental Ethics Committee of the First Affiliated Hospital of Zhejiang University School of Medicine.
Conflicts of interest statement
The authors do not have a commercial or other association with pharmaceutical companies or other parties that might pose a conflict of interest.
Contributor Information
Jian Wu, Email: drwujian@zju.edu.cn.
Shusen Zheng, Email: shusenzheng@zju.edu.cn.
References
- 1.Finn R.S. Current and future treatment strategies for patients with advanced hepatocellular carcinoma: role of mTOR inhibition. Liver Cancer. 2012;1(3–4):247–256. doi: 10.1159/000343839. [DOI] [PMC free article] [PubMed] [Google Scholar]; Finn RS. Current and Future Treatment Strategies for Patients with Advanced Hepatocellular Carcinoma: Role of mTOR Inhibition. Liver cancer. 2012;1(3-4):247-56. [DOI] [PMC free article] [PubMed]
- 2.Cheng J., Xie H.-Y., Xu X., Wu J., Wei X., Su R. NDRG1 as a biomarker for metastasis, recurrence and of poor prognosis in hepatocellular carcinoma. Cancer Lett. 2011;310(1):35–45. doi: 10.1016/j.canlet.2011.06.001. [DOI] [PubMed] [Google Scholar]; Cheng J, Xie H-Y, Xu X, Wu J, Wei X, Su R, et al. NDRG1 as a biomarker for metastasis, recurrence and of poor prognosis in hepatocellular carcinoma. Cancer letters. 2011; 310(1):35-45. [DOI] [PubMed]
- 3.Abou-Alfa G.K. Sorafenib use in hepatocellular carcinoma: more questions than answers. Hepatology. 2014;60(1):8–15. doi: 10.1002/hep.27044. [DOI] [PubMed] [Google Scholar]; Abou-Alfa GK. Sorafenib use in hepatocellular carcinoma: More questions than answers. Hepatology. 2014. [DOI] [PubMed]
- 4.Desai J.R., Ochoa S., Prins P.A., He A.R. Systemic therapy for advanced hepatocellular carcinoma: an update. J Gastroint Oncol. 2017;8(2):243–255. doi: 10.21037/jgo.2017.02.01. [DOI] [PMC free article] [PubMed] [Google Scholar]; Desai JR, Ochoa S, Prins PA, He AR. Systemic therapy for advanced hepatocellular carcinoma: an update. Journal of gastrointestinal oncology. 2017; 8(2):243-55. [DOI] [PMC free article] [PubMed]
- 5.Chow A.K., Ng L., Lam C.S., Wong S.K., Wan T.M., Cheng N.S. The enhanced metastatic potential of hepatocellular carcinoma (HCC) cells with sorafenib resistance. Plo S one. 2013;8(11) doi: 10.1371/journal.pone.0078675. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chow AK, Ng L, Lam CS, Wong SK, Wan TM, Cheng NS, et al. The Enhanced metastatic potential of hepatocellular carcinoma (HCC) cells with sorafenib resistance. Plo S one. 2013; 8(11): e78675. [DOI] [PMC free article] [PubMed]
- 6.Tavner F.J., Simpson R., Tashiro S., Favier D., Jenkins N.A., Gilbert D.J. Molecular cloning reveals that the p 160 Myb-binding protein is a novel, predominantly nucleolar protein which may play a role in transactivation by Myb. Mol Cell Biol. 1998;18(2):989–1002. doi: 10.1128/mcb.18.2.989. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tavner FJ, Simpson R, Tashiro S, Favier D, Jenkins NA, Gilbert DJ, et al. Molecular Cloning Reveals that the p 160 Myb-Binding Protein Is a Novel, Predominantly Nucleolar Protein Which May Play a Role in Transactivation by Myb. Molecular and Cellular Biology. 1998; 18(2): 989-1002. [DOI] [PMC free article] [PubMed]
- 7.Keough R.A., Macmillan E.M., Lutwyche J.K., Gardner J.M., Tavner F.J., Jans D.A. Myb-binding protein 1a is a nucleocytoplasmic shuttling protein that utilizes CRM1-dependent and independent nuclear export pathways. Exp Cell Res. 2003;289(1):108–123. doi: 10.1016/s0014-4827(03)00262-3. [DOI] [PubMed] [Google Scholar]; Keough RA, Macmillan EM, Lutwyche JK, Gardner JM, Tavner FJ, Jans DA, et al. Myb-binding protein 1a is a nucleocytoplasmic shuttling protein that utilizes CRM1-dependent and independent nuclear export pathways. Experimental cell research. 2003; 289(1): 108-23. [DOI] [PubMed]
- 8.Keough R., Woollatt E., Crawford J., Sutherland G.R., Plummer S., Casey G. Molecular cloning and chromosomal mapping of the human homologue of MYB binding protein (P 160) 1A (MYBBP1A) to 17p13.3. Genomics. 1999;62(3):483–489. doi: 10.1006/geno.1999.6035. [DOI] [PubMed] [Google Scholar]; Keough R, Woollatt E, Crawford J, Sutherland GR, Plummer S, Casey G, et al. Molecular Cloning and Chromosomal Mapping of the Human Homologue of MYB Binding Protein (P 160) 1A (MYBBP1A) to 17p13.3. Genomics. 1999; 62(3): 483-9. [DOI] [PubMed]
- 9.Fan M., Rhee J., St-Pierre J., Handschin C., Puigserver P., Lin J. Suppression of mitochondrial respiration through recruitment of p 160 myb binding protein to PGC-1α: modulation by p 38 MAPK. Genes Dev. 2004;18(3):278–289. doi: 10.1101/gad.1152204. [DOI] [PMC free article] [PubMed] [Google Scholar]; Fan M, Rhee J, St-Pierre J, Handschin C, Puigserver P, Lin J, et al. Suppression of mitochondrial respiration through recruitment of p 160 myb binding protein to PGC-1α: modulation by p 38 MAPK. Genes & development. 2004; 18(3):278-89. [DOI] [PMC free article] [PubMed]
- 10.Owen H.R., Elser M., Cheung E., Gersbach M., Kraus W.L., Hottiger M.O. MYBBP1a is a novel repressor of NF-κB. J Mol Biol. 2007;366(3):725–736. doi: 10.1016/j.jmb.2006.11.099. [DOI] [PubMed] [Google Scholar]; Owen HR, Elser M, Cheung E, Gersbach M, Kraus WL, Hottiger MO. MYBBP1a is a Novel Repressor of NF-κB. Journal of Molecular Biology. 2007; 366(3): 725-36. [DOI] [PubMed]
- 11.Yamauchi T., Keough R.A., Gonda T.J., Ishii S. Ribosomal stress induces processing of Mybbp 1a and its translocation from the nucleolus to the nucleoplasm. Genes Cells. 2008;13(1):27–39. doi: 10.1111/j.1365-2443.2007.01148.x. [DOI] [PubMed] [Google Scholar]; Yamauchi T, Keough RA, Gonda TJ, Ishii S. Ribosomal stress induces processing of Mybbp 1a and its translocation from the nucleolus to the nucleoplasm. Genes to Cells. 2008; 13(1):27-39. [DOI] [PubMed]
- 12.Olausson K.H., Nistér M., Lindström M.S. p53-dependent and -independent nucleolar stress responses. Cells. 2012;1(4):774–798. doi: 10.3390/cells1040774. [DOI] [PMC free article] [PubMed] [Google Scholar]; Olausson KH, Nistér M, Lindström MS. p53 -Dependent and -Independent Nucleolar Stress Responses. Cells. 2012;1(4):774-98. [DOI] [PMC free article] [PubMed]
- 13.Hochstatter J., Hölzel M., Rohrmoser M., Schermelleh L., Leonhardt H., Keough R. Myb-binding protein 1a (Mybbp1a) regulates levels and processing of pre-ribosomal RNA. J Biol Chem. 2012;287(29):24365–24377. doi: 10.1074/jbc.M111.303719. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hochstatter J, Hölzel M, Rohrmoser M, Schermelleh L, Leonhardt H, Keough R, et al. Myb-binding Protein 1a (Mybbp1a) Regulates Levels and Processing of Pre-ribosomal RNA. The Journal of biological chemistry. 2012;287(29):24365-77. [DOI] [PMC free article] [PubMed]
- 14.Tan B.C.-M., Yang C.-C., Hsieh C.-L., Chou Y.-H., Zhong C.-Z., Yung B.Y.-M. Epigeneitc silencing of ribosomal RNA genes by Mybbp1a. J Biomed Sci. 2012;19(1):57. doi: 10.1186/1423-0127-19-57. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tan BC-M, Yang C-C, Hsieh C-L, Chou Y-H, Zhong C-Z, Yung BY-M, et al. Epigeneitc silencing of ribosomal RNA genes by Mybbp1a. Journal of Biomedical Science. 2012;19(1):57-. [DOI] [PMC free article] [PubMed]
- 15.Kumazawa T., Nishimura K., Katagiri N., Hashimoto S., Hayashi Y., Kimura K. Gradual reduction in rRNA transcription triggers p53 acetylation and apoptosis via MYBBP1A. Sci Rep. 2015;5 doi: 10.1038/srep10854. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kumazawa T, Nishimura K, Katagiri N, Hashimoto S, Hayashi Y, Kimura K. Gradual reduction in rRNA transcription triggers p53 acetylation and apoptosis via MYBBP1A. Scientific reports. 2015;5:10854. [DOI] [PMC free article] [PubMed]
- 16.Jones L.C., Okino S.T., Gonda T.J., Whitlock J.P. Myb-binding protein 1a augments AhR-dependent gene expression. J Biol Chem. 2002;277(25):22515–22519. doi: 10.1074/jbc.M200740200. [DOI] [PubMed] [Google Scholar]; Jones LC, Okino ST, Gonda TJ, Whitlock JP. Myb-binding Protein 1a Augments AhR-dependent Gene Expression. Journal of Biological Chemistry. 2002;277(25):22515-9. [DOI] [PubMed]
- 17.Akaogi K., Ono W., Hayashi Y., Kishimoto H., Yanagisawa J. MYBBP1A suppresses breast cancer tumorigenesis by enhancing the p53 dependent anoikis. BMC Cancer. 2013;13:65. doi: 10.1186/1471-2407-13-65. [DOI] [PMC free article] [PubMed] [Google Scholar]; Akaogi K, Ono W, Hayashi Y, Kishimoto H, Yanagisawa J. MYBBP1A suppresses breast cancer tumorigenesis by enhancing the p53 dependent anoikis. BMC cancer. 2013;13:65-. [DOI] [PMC free article] [PubMed]
- 18.Acuña Sanhueza G.A., Faller L., George B., Koffler J., Misetic V., Flechtenmacher C. Opposing function of MYBBP1A in proliferation and migration of head and neck squamous cell carcinoma cells. BMC Cancer. 2012;12:72. doi: 10.1186/1471-2407-12-72. [DOI] [PMC free article] [PubMed] [Google Scholar]; Acuña Sanhueza GA, Faller L, George B, Koffler J, Misetic V, Flechtenmacher C, et al. Opposing function of MYBBP1A in proliferation and migration of head and neck squamous cell carcinoma cells. BMC cancer. 2012;12:72-. [DOI] [PMC free article] [PubMed]
- 19.Lv Z., Weng X., Du C., Zhang C., Xiao H., Cai X. Downregulation of HDAC6 promotes angiogenesis in hepatocellular carcinoma cells and predicts poor prognosis in liver transplantation patients. Mol Carcinog. 2016;55(5):1024–1033. doi: 10.1002/mc.22345. [DOI] [PubMed] [Google Scholar]; Lv Z, Weng X, Du C, Zhang C, Xiao H, Cai X, et al. Downregulation of HDAC6 promotes angiogenesis in hepatocellular carcinoma cells and predicts poor prognosis in liver transplantation patients. Molecular carcinogenesis. 2016;55(5):1024-33. [DOI] [PubMed]
- 20.Xiao H., Tong R., Yang B., Lv Z., Du C., Peng C. TAZ regulates cell proliferation and sensitivity to vitamin D3 in intrahepatic cholangiocarcinoma. Cancer Lett. 2016;381(2):370–379. doi: 10.1016/j.canlet.2016.08.013. [DOI] [PubMed] [Google Scholar]; Xiao H, Tong R, Yang B, Lv Z, Du C, Peng C, et al. TAZ regulates cell proliferation and sensitivity to vitamin D3 in intrahepatic cholangiocarcinoma. Cancer letters. 2016;381(2):370-9. [DOI] [PubMed]
- 21.Tong R., Yang B., Xiao H., Peng C., Hu W., Weng X. KCTD11 inhibits growth and metastasis of hepatocellular carcinoma through activating Hippo signaling. Oncotarget. 2017;8(23):37717–37729. doi: 10.18632/oncotarget.17145. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tong R, Yang B, Xiao H, Peng C, Hu W, Weng X, et al. KCTD11 inhibits growth and metastasis of hepatocellular carcinoma through activating Hippo signaling. Oncotarget. 2017;8(23):37717-29. [DOI] [PMC free article] [PubMed]
- 22.Xiao H.T.R., Ding C., Lv Z., Du C., Peng C., Cheng S. γ-H2AX promotes hepatocellular carcinoma angiogenesis via EGFR/HIF-1α/VEGF pathways under hypoxic condition. Oncotarget. 2015;6:2180–2192. doi: 10.18632/oncotarget.2942. [DOI] [PMC free article] [PubMed] [Google Scholar]; Xiao H TR, Ding C, Lv Z, Du C, Peng C, Cheng S, Xie H, Zhou L, Wu J, Zheng S. γ-H2AX promotes hepatocellular carcinoma angiogenesis via EGFR/HIF-1α/VEGF pathways under hypoxic condition. Oncotarget. 2015;6:2180–92. [DOI] [PMC free article] [PubMed]
- 23.Ding M., Bruick R.K., Yu Y. Secreted IGFBP5 mediates mTORC1-dependent feedback inhibition of IGF-1 signaling. Nat Cell Biol. 2016;18(3):319–327. doi: 10.1038/ncb3311. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ding M, Bruick RK, Yu Y. Secreted IGFBP5 Mediates mTORC1-Dependent Feedback Inhibition of IGF-1 Signaling. Nature cell biology. 2016;18(3):319-27. [DOI] [PMC free article] [PubMed]
- 24.Oriente F., Fernandez Diaz L.C., Miele C., Iovino S., Mori S., Diaz V.M. Prep1 deficiency induces protection from diabetes and increased insulin sensitivity through a p160-mediated mechanism. Mol Cell Biol. 2008;28(18):5634–5645. doi: 10.1128/MCB.00117-08. [DOI] [PMC free article] [PubMed] [Google Scholar]; Oriente F, Fernandez Diaz LC, Miele C, Iovino S, Mori S, Diaz VM, et al. Prep1 Deficiency Induces Protection from Diabetes and Increased Insulin Sensitivity through a p160-Mediated Mechanism. Molecular and Cellular Biology. 2008;28(18):5634-45. [DOI] [PMC free article] [PubMed]
- 25.Karim M.F., Yoshizawa T., Sato Y., Sawa T., Tomizawa K., Akaike T. Inhibition of H3K18 deacetylation of Sirt7 by Myb-binding protein 1a (Mybbp1a) Biochem Biophys Res Commun. 2013;441(1):157–163. doi: 10.1016/j.bbrc.2013.10.020. [DOI] [PubMed] [Google Scholar]; Karim MF, Yoshizawa T, Sato Y, Sawa T, Tomizawa K, Akaike T, et al. Inhibition of H3K18 deacetylation of Sirt7 by Myb-binding protein 1a (Mybbp1a). Biochemical and biophysical research communications. 2013;441(1):157-63. [DOI] [PubMed]
- 26.Georgakilas A.G., Martin O.A., Bonner W.M. p21: a two-faced genome guardian. Trends Mol Med. 2017;23(4):310–319. doi: 10.1016/j.molmed.2017.02.001. [DOI] [PubMed] [Google Scholar]; Georgakilas AG, Martin OA, Bonner WM. p21: A Two-Faced Genome Guardian. Trends in Molecular Medicine. 2017;23(4):310-9. [DOI] [PubMed]
- 27.Massague J. G1 cell-cycle control and cancer. Nature. 2004;432(7015):298–306. doi: 10.1038/nature03094. [DOI] [PubMed] [Google Scholar]; Massague J. G1 cell-cycle control and cancer. Nature. 2004;432(7015):298-306. [DOI] [PubMed]
- 28.Jiang B.-H., Liu L.-Z. PI3K/PTEN signaling in angiogenesis and tumorigenesis. Adv Cancer Res. 2009;102:19–65. doi: 10.1016/S0065-230X(09)02002-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jiang B-H, Liu L-Z. PI3K/PTEN Signaling in Angiogenesis and Tumorigenesis. Advances in cancer research. 2009;102:19-65. [DOI] [PMC free article] [PubMed]
- 29.Chu Julia Y., Dransfield I., Rossi Adriano G., Vermeren S. Non-canonical PI3K-Cdc42-Pak-Mek-Erk signaling promotes immune-complex-induced apoptosis in human neutrophils. Cell Rep. 2016;17(2):374–386. doi: 10.1016/j.celrep.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chu Julia Y, Dransfield I, Rossi Adriano G, Vermeren S. Non-canonical PI3K-Cdc42-Pak-Mek-Erk Signaling Promotes Immune-Complex-Induced Apoptosis in Human Neutrophils. Cell reports. 2016;17(2):374-86. [DOI] [PMC free article] [PubMed]
- 30.Di Blasio L., Gagliardi P.A., Puliafito A., Primo L. Serine/threonine kinase 3-phosphoinositide-dependent protein kinase-1 (PDK1) as a key regulator of cell migration and cancer dissemination. Cancers. 2017;9(3):25. doi: 10.3390/cancers9030025. [DOI] [PMC free article] [PubMed] [Google Scholar]; Di Blasio L, Gagliardi PA, Puliafito A, Primo L. Serine/Threonine Kinase 3-Phosphoinositide-Dependent Protein Kinase-1 (PDK1) as a Key Regulator of Cell Migration and Cancer Dissemination. Cancers. 2017;9(3):25. [DOI] [PMC free article] [PubMed]
- 31.Schneider M.R., Wolf E., Hoeflich A., Lahm H. IGF-binding protein-5: flexible player in the IGF system and effector on its own. J Endocrinol. 2002;172(3):423–440. doi: 10.1677/joe.0.1720423. [DOI] [PubMed] [Google Scholar]; Schneider MR, Wolf E, Hoeflich A, Lahm H. IGF-binding protein-5: flexible player in the IGF system and effector on its own. Journal of Endocrinology. 2002;172(3):423-40. [DOI] [PubMed]
- 32.Akkiprik M., Hu L., Sahin A., Hao X., Zhang W. The subcellular localization of IGFBP5 affects its cell growth and migration functions in breast cancer. BMC Cancer. 2009;9:103. doi: 10.1186/1471-2407-9-103. [DOI] [PMC free article] [PubMed] [Google Scholar]; Akkiprik M, Hu L, Sahin A, Hao X, Zhang W. The subcellular localization of IGFBP5 affects its cell growth and migration functions in breast cancer. BMC cancer. 2009;9:103-. [DOI] [PMC free article] [PubMed]
- 33.Niu J., Huang Y.-J., Wei S., Liu Z., Wang L.-E., Chang S. Association between a functional polymorphism (-1195T>C) in the IGFBP5 promoter and head and neck cancer risk. Head Neck. 2011;33(5):650–660. doi: 10.1002/hed.21514. [DOI] [PMC free article] [PubMed] [Google Scholar]; Niu J, Huang Y-J, Wei S, Liu Z, Wang L-E, Chang S, et al. Association between a Functional Polymorphism (-1195T>C) in the IGFBP5 Promoter and Head and Neck Cancer Risk. Head & neck. 2011;33(5):650-60. [DOI] [PMC free article] [PubMed]
- 34.Su Y., Wagner E.R., Luo Q., Huang J., Chen L., He B.C. Insulin-like growth factor binding protein 5 suppresses tumor growth and metastasis of human osteosarcoma. Oncogene. 2011;30(37):3907–3917. doi: 10.1038/onc.2011.97. [DOI] [PubMed] [Google Scholar]; Su Y, Wagner ER, Luo Q, Huang J, Chen L, He BC, et al. Insulin-like growth factor binding protein 5 suppresses tumor growth and metastasis of human osteosarcoma. Oncogene. 2011;30(37):3907-17. [DOI] [PubMed]
- 35.Ling J., Jiang L., Zhang C., Dai J., Wu Q., Tan J. Upregulation of miR-197 inhibits cell proliferation by directly targeting IGFBP5 in human uterine leiomyoma cells. In Vitro Cell Dev Biol Anim. 2015;51(8):835–842. doi: 10.1007/s11626-015-9887-x. [DOI] [PubMed] [Google Scholar]; Ling J, Jiang L, Zhang C, Dai J, Wu Q, Tan J. Upregulation of miR-197 inhibits cell proliferation by directly targeting IGFBP5 in human uterine leiomyoma cells. In Vitro Cellular & Developmental Biology - Animal. 2015;51(8):835-42. [DOI] [PubMed]
- 36.Wang J., Ding N., Li Y., Cheng H., Wang D., Yang Q. Insulin-like growth factor binding protein 5 (IGFBP5) functions as a tumor suppressor in human melanoma cells. Oncotarget. 2015;6(24):20636–20649. doi: 10.18632/oncotarget.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wang J, Ding N, Li Y, Cheng H, Wang D, Yang Q, et al. Insulin-like growth factor binding protein 5 (IGFBP5) functions as a tumor suppressor in human melanoma cells. Oncotarget. 2015;6(24):20636-49. [DOI] [PMC free article] [PubMed]
- 37.Yu L., Lu Y., Han X., Zhao W., Li J., Mao J. microRNA-140-5p inhibits colorectal cancer invasion and metastasis by targeting ADAMTS5 and IGFBP5. Stem Cell Res Ther. 2016;7:180. doi: 10.1186/s13287-016-0438-5. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]; Yu L, Lu Y, Han X, Zhao W, Li J, Mao J, et al. microRNA -140-5p inhibits colorectal cancer invasion and metastasis by targeting ADAMTS5 and IGFBP5. Stem cell research & therapy. 2016;7:180. [DOI] [PMC free article] [PubMed] [Retracted]
- 38.Simon S., Grabellus F., Ferrera L., Galietta L., Schwindenhammer B., Mühlenberg T. DOG1 regulates growth and IGFBP-5 in gastrointestinal stromal tumors. Cancer Res. 2013;73(12):3661–3670. doi: 10.1158/0008-5472.CAN-12-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]; Simon S, Grabellus F, Ferrera L, Galietta L, Schwindenhammer B, Mühlenberg T, et al. DOG1 regulates growth and IGFBP-5 in gastrointestinal stromal tumors. Cancer research. 2013;73(12):3661-70. [DOI] [PMC free article] [PubMed]
- 39.Liu L., Wang J., Li X., Ma J., Shi C., Zhu H. miR-204-5p suppresses cell proliferation by inhibiting IGFBP5 in papillary thyroid carcinoma. Biochem Biophys Res Commun. 2015;457(4):621–626. doi: 10.1016/j.bbrc.2015.01.037. [DOI] [PubMed] [Google Scholar]; Liu L, Wang J, Li X, Ma J, Shi C, Zhu H, et al. miR-204-5p suppresses cell proliferation by inhibiting IGFBP5 in papillary thyroid carcinoma. Biochemical and biophysical research communications. 2015;457(4):621-6. [DOI] [PubMed]
- 40.He L.H.G. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;(5):522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]; He L HG. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004(5):522-31. [DOI] [PubMed]
- 41.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; DP Bartel. MicroRNAs: target recognition and regulatory functions. Cell. 2009(136):215–33. [DOI] [PMC free article] [PubMed]
- 42.Miremadi A., Oestergaard M.Z., Paul P.D., Caldas C. Cancer genetics of epigenetic genes. Hum Mol Genet. 2007;16:28–49. doi: 10.1093/hmg/ddm021. [DOI] [PubMed] [Google Scholar]; Miremadi A, Oestergaard MZ, Paul PD, Caldas C. Cancer genetics of epigenetic genes. Human molecular genetics. 2007(16):28-49. [DOI] [PubMed]
- 43.Widschwendter M., Jones Peter A. The potential prognostic, predictive, and therapeutic values of DNA methylation in cancer. Clin Cancer Res. 2002;(8):17–21. [PubMed] [Google Scholar]; Widschwendter M, Peter A. Jones. The potential prognostic, predictive, and therapeutic values of DNA methylation in cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2002(8):17-21. [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding authors on reasonable request.