Abstract
Background
Gastric cancer (GC) is the third leading cause of cancer death. Early detection is a key factor to reduce its mortality.
Methods
We retrospectively collected pre- and postoperative serum samples as well as tumour tissues and adjacent normal tissues from 100 GC patients. Serum samples from non-cancerous patients were served as controls (n = 50). A high-throughput protein detection technology, multiplex proximity extension assays (PEA), was applied to measure levels of over 300 proteins. Alteration of each protein was analysed by univariate analysis. Elastic-net logistic regression was performed to select serum proteins into the diagnostic model.
Findings
We identified 19 serum proteins (CEACAM5, CA9, MSLN, CCL20, SCF, TGF-alpha, MMP-1, MMP-10, IGF-1, CDCP1, PPIA, DDAH-1, HMOX-1, FLI1, IL-7, ZBTB-17, APBB1IP, KAZALD-1, and ADAMTS-15) that together distinguish GC cases from controls with a diagnostic sensitivity of 93%, specificity of 100%, and area under receiver operating characteristic curve (AUC) of 0·99 (95% CI: 0·98–1). Moreover, the 19-serum protein signature provided an increased diagnostic capacity in patients at TNM I-II stage (sensitivity 89%, specificity 100%, AUC 0·99) and in patients with high microsatellite instability (MSI) (91%, 98%, and 0·99) compared to individual proteins. These promising results will inspire a large-scale independent cohort study to be pursued for validating the proposed protein signature.
Interpretation
Based on targeted proteomics and elastic-net logistic regression, we identified a 19-serum protein signature which could contribute to clinical GC diagnosis, especially for patients at early stage and those with high MSI.
Fund
This study was supported by a European H2020-Marie Skłodowska-Curie Innovative Training Networks grant (316,929, GastricGlycoExplorer). Funder had no influence on trial design, data evaluation, and interpretation.
Keywords: Gastric cancer, Diagnosis, Biomarker, PEA, Proteomics
Abbreviations: ADAMTS-15, A disintegrin and metalloproteinase with thrombospondin motifs 15; APBB1IP, Amyloid beta A4 precursor protein-binding family B member 1-interacting protein; CA9, Carbonic anhydrase 9; CCL20, C-C motif chemokine 20; CDCP1, CUB domain-containing protein 1; CEACAM5, Carcinoembryonic antigen-related cell adhesion molecule 5; DDAH1, dimethylarginine dimethylaminohydrolase 1; FLI-1, Friend leukemia integration 1 transcription factor; GCNT1, beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N- acetylglucosaminyltransferase; HMOX1, Heme oxygenase 1; IGF1, Insulin-like growth factor I; IL-7, Interleukin-7; KAZALD1, Kazal-type serine protease inhibitor domain containing protein 1; MSLN, Mesothelin; MMP-1, Matrix metalloproteinase-1; MMP-10, Matrix metalloproteinase-10; PPIA, Peptidyl-prolyl cis-trans isomerase A; SCF, Stem cell factor/KIT ligand; TGF alpha, Transforming growth factor alpha; ZBTB-17, Zinc finger and BTB domain-containing protein 17
Research in context.
Evidence before this study
The current clinically used diagnostic biomarkers for gastric cancer (GC) such as carcinoembryonic antigen (CEA), cancer antigen 19-9 (CA19-9), and cancer antigen 72-4 (CA72-4) are neither sufficiently sensitive nor specific. The identification of novel reliable markers and development of new technologies are still urgent for GC detection and hence improving the survival of patients. Large-scale proteome screening in non-invasive biological specimens is a strategy that provides the possibility of discovering new biomarkers. We performed a PubMed database search for studies focused on GC biomarker discovery published until May 2018 using the following terms: “biomarker” AND “proteomic” AND (“gastric cancer” OR “stomach cancer”). The criteria for inclusion were GC diagnostic biomarker studies using any human materials such as serum, plasma, tissue, gastric fluid, urine, circulating cells, or exosomes. Additional studies were identified by surveying the references associated with these primary publications. Studies of biomarkers for prognosis or prediction were not included. Any reports on cell lines or animals were excluded. As a result, almost all studies were based on different types of mass spectrometry or antibody-based tissue microarrays. We have previously developed and optimized in situ proximity ligation assay (PLA) and solid-phase PLA for single protein detection as well as for detection of single protein post-translational modification such as glycosylation and phosphorylation, which have been applied in GC tissue and serum specimens. Here, we extended our research by applying a targeted proteomics approach, multiplex proximity extension assay (PEA), for large scale protein measurement in GC serum and tissue lysate samples in order to identify potential biomarkers for GC diagnosis.
Added value of this study
We identified proteins that are differentially expressed in GC tumour tissues and in sera, as well as changed protein levels before and after surgery. Using a comprehensive multivariate analysis, we identified a nineteen serum protein signature, including the clinically used biomarker CEA, which provided a much greater diagnostic accuracy for GC detection compared to CEA used alone. This protein signature also improved the diagnostic capacity for GC patients at early stage and patients with high microsatellite instability.
Implications of all available evidence
The detection of the nineteen serum protein signature using a mini-panel of multiplex PEA technology or other equivalent approaches would translate into clinical application to improve the GC diagnosis and treatment stratification and bring tangible benefits for GC patients in the future.
Alt-text: Unlabelled Box
1. Introduction
Gastric cancer (GC) is the fifth most commonly diagnosed cancer and the third leading cause of cancer death with over 1,000,000 new cases and an estimated 783,000 deaths in 2018 [1]. Currently, complete surgical resection remains the major curative therapy for gastric cancer. Despite the development of technologies for diagnosis and treatment, most gastric cancer cases in the western world are usually diagnosed at middle or advanced stages, resulting in unsatisfactory treatment results [2,3]. Measurement of tumour protein biomarkers in circulating blood is the most commonly used noninvasive method for early detection of malignant cancers, and is proven to harbor considerable value in screening, diagnosis, monitoring, and prognosis of tumours since some important proteins secreted/released into blood may reflect the quantitative or qualitative changes of the whole body when undergoing any pathological conditions. The currently used gastrointestinal tumour serological protein biomarkers, including carcinoembryonic antigen (CEA), cancer antigen 19-9 (CA19-9), and cancer antigen 72-4 (CA72-4), are insufficient for GC diagnosis since their positive rates in GC patients are <40% and lower than 20% in GC patients at early stage [4,5]. Serum pepsinogen I (PGI) and II (PGII) as well as PGI/PGII ratio are used for GC screening and diagnosis in countries with high or moderate risk, especially in Asia [6]. However, its clinical performance remains controversial and results are different among various ethnicities [7]. Therefore, searching for reliable blood tumour markers for early diagnosis is crucial for early intervention therapy hence for improving the survival of gastric cancer patients.
Technologies developed in recent decades display the ability of large-scale screening of proteins. Most proteomic studies aiming to identify tumour-associated protein markers are based on mass spectrometry (MS) [4,8]. However, the extremely broad range of blood protein concentrations challenges proteomic analyses. Furthermore, many blood proteins in low abundance are likely to be specific at early stages of disease, but their detection with classical MS techniques is impaired by the predominance of high abundant proteins. In addition, MS always requires substantial amount of sample input, limiting its application for many clinical samples where the materials are insufficient. A recently developed technology for multiple proteins detection – multiplex proximity extension assay (PEA; Olink Proteomics™) [9] – enables the simultaneous detection of large-scale proteins with high selectivity and sensitivity with minimal sample volume (as little as 1 μl aliquot), and no requirement for complex sample pre-treatment. A schematic illustration of multiplex PEA is shown in Supplementary Fig. 1. The PEA and proximity ligation assay (PLA) are both proven to be sensitive and specific [10,11]. The specificity is due to the requirement of dual-recognition of a target by a matched pair of DNA-conjugated antibodies. In PEA, upon antigen-antibody binding, the labeled DNA oligonucleotides are brought into close proximity and hybridize to each other. An amplifiable reporter DNA molecule is formed by DNA polymerization, which can subsequently be amplified and quantified by real-time qPCR. The target concentration is proportional to the number of reporter DNA molecules. A limiting factor of most multiplexed immunoassays is the cross-reactivity of antibodies, which restricts the degree of multiplexing to below 10-plex. The design of the DNA-assisted affinity-based PEA excludes the detection of products from unmatched antibody pairs, allowing large-scale of multiplexing without loss of selectivity and sensitivity. The assay sensitivity of multiplex PEA is reported to be comparable with that of standard sandwich single-plex ELISA for each individual protein [9]. Multiplex PEA has been applied for biomarker discovery of many diseases such as cardiovascular diseases, type-1 diabetes, and cancer (publications are listed in https://www.olink.com/data-you-can-trust/publications).
We have previously developed and optimized in situ proximity ligation assay (PLA) and solid-phase PLA for single protein detection as well as for detection of protein post-translational modification such as glycosylation and phosphorylation, which have been applied in GC tissue and serum specimens [12,13]. In the present study, we applied the multiplex PEA technology to measure the levels of over 300 proteins in pre- and post-operative sera from 100 GC patients and control sera from 50 individuals without any type of cancer, as well as in tumour tissues and adjacent normal tissue lysates from the same 100 patients, aiming to identify a set of potential serum protein biomarkers for GC diagnosis.
2. Materials and methods
2.1. Study cohort
This retrospective study was followed the Standards for the Reporting of Diagnosis Accuracy studies (STARD) statement [14]. 100 patients with primary gastric cancer were undergone surgery at the Department of General Surgery and Surgical Oncology in Siena University Hospital, Italy, between July 1990 and February 2010. The subjects were chosen inconsecutively from the hospital surgical database. Sera samples were taken from the 100 patients upon admission to the hospital and one to two weeks after surgery. Fifty control serum samples were collected at the same time from cancer free patients treated in the same surgery department with other gastrointestinal diseases but not related to stomach. Serum samples were aliquoted immediately after centrifugation and stored at −80 °C until tests. Diagnostic criteria of GC were relied on endoscopy and biopsy. Serum levels of CEA, CA19-9, and CA72-4 were achieved from clinically recorded data. Histological classification was followed by Lauren [15] and WHO [16]. TNM classification was determined according to the seventh edition of the cancer staging manual [17]. Curative surgery of the stomach is defined as complete resection of primary cancer with clear surgical margins and adequate lymphadenectomy. Microsatellite instability (MSI) status were accessed as previously described [18]. This study was conducted following the Declaration of Helsinki. All patients were provided with the informed consent and ethical approval was granted by the Institutional Ethics Committee of University of Siena.
2.2. Tissue lysates preparation
Tumour tissues and adjacent normal tissues from the 100 patients were collected shortly after surgery and were immediately snap-frozen in liquid nitrogen and stored in −80 °C. Tissue lysates were prepared as previously described [13]. All tissue samples were cut into thin slices quickly and mixed with lysis buffer (50 mM Tris-HCl, pH 7·4, 150 mM NaCl, 1 mM EDTA, pH 8, 1% Triton X-100, 0.1% sodium deoxycholate, protease inhibitor (Roche Complete Mini)), and zirconium oxide beads (ZrOB20-RNA, 2 mm in diameter, Next Advance, Inc., NY, USA). The ratio of tissue mass: volume of lysis buffer: beads weight was 1:4:2. Homogenization was performed with Bullet Blender device (BBX24B-CE, Next Advance, Inc., NY, USA) according to the manufacturer's recommendation. The homogenized tissues were then centrifuged at 12,000 rpm for 10 min at 4 °C and the supernatant transferred to new tubes. Total protein concentration of tissue lysate was measured by Pierce™ BCA Protein Assay kit (ThermoFisher). All samples were than diluted to 2 mg/ml and the aliquots were frozen at −70 °C before use.
2.3. Protein abundance measurement
Samples from different groups were evenly distributed within a plate but randomly among plates. Assays were done at Uppsala University before accessing patient's clinical information. Protein levels were measured by multiplex PEA (Olink Proteomics™, Sweden) using three commercial panels (Oncology, Inflammation, Cell regulation), and two experimental panels (Cancer, Cellular pathway). Each panel is able to measure 92 proteins in 90 samples simultaneously in as little as 1 μl sample aliquots [9]. Each sample is spiked in four controls to monitor PEA process. Each panel also includes three positive controls and three negative controls, which is used for data normalization and determination of limit of detection (LOD), respectively. Assays were performed following the manufacturer's instructions. In detail, 1 μl of each sample or technical control was incubated with 3 μl incubation mix including antibody probes at 4 °C overnight. The extension mix was added after the antigen-antibody binding, starting the extension and pre-amplification of 17 cycles of PCR (Applied Biosystems 9700, Life Technologies). Samples were diluted 100-fold in this step to minimize the matrix effect and the chance of extension generated by unpaired oligonucleotides. Next, 2·8 μl of each first round PCR product was mixed with 7·2 μl detection reagent from which 5 μl was loaded into the sample wells, while specific pairs of PCR primers were loaded into the primer wells of a microfluid chip (Fluidigm 96.96 Dynamic Array, Fluidigm, CA, USA). The microfluid chip was then primed in Fluidigm IFC controller and loaded for real-time qPCR in Fluidigm Biomark™ thermocycler. Quantification cycle (Cq) value was converted to Normalized Protein eXpression (NPX) by normalizing with extension- and negative controls spiked in each sample. NPX is a unit on log2 scale, where one NPX increase corresponding to a two-fold increase in concentration of the protein. LOD was determined as the NPX value three times the standard deviation beyond its background. Multiplex PEA detects relative protein levels but not absolute concentration. Correlations between NPX values and protein concentrations in mass unit (pg/ml) are available at Olink's website (www.olink.com/products/complete-biomarker-list). The specificity for each panel was determined by carrying out the whole assay in which the test samples were pools of full length recombinant antigens corresponding to every block of 8 proteins from all the 92 proteins in the panel, resulting in signals generated only from the present proteins but not the others (detailed in https://www.olink.com/data-you-can-trust/validation/).
For proteins present in more than one panel, only one was chosen for further analysis. Thus, seven proteins from commercial panels and 18 from experimental panels were removed. The assay reproducibility for the same proteins is displayed in Supplementary Fig. 2, while the reproducibility for sample duplicates is shown in Supplementary Fig. 3.
2.4. Statistics
Data analyses were performed using R software (www.r-project.org) [19].
NPX values were used for analysis since the values tended towards a normal distribution. To minimize the inter-plate variation, samples from different disease groups were evenly distributed throughout the plates, and the inter-plate variation was further normalized for each protein in each plate by adding the Z-score factor calculated as follows: factor = (actual value – median of all samples)/standard deviation. For comparison between GC and controls, NPX values were adjusted if a significant effect (corrected p value <0·05) of age or gender on the protein levels was found by linear regression in both the control and cancer groups. Therefore, 13 proteins measured in serum (CDCP1, CTSV, CXCL9, EPHA1, KIT, OPG, RET, RSPO3, TGFBR2, TNFRSF10B, TRANCE, VEGFR2, WFDC2) were adjusted for age, while no protein was found significantly associated with gender in either group. Principal component analysis (PCA) was applied for an overview of the relationships between variables and the presence of outliers.
Differences between two groups for continuous variables were analysed by non-parametric Mann-Whitney-Wilcoxon test, while for category variables, Chi-square or Fisher's exact test was performed, and ANOVA was applied for comparisons of more than two groups. Differences between before and after surgery were tested using paired Mann-Whitney-Wilcoxon test. Correlation coefficients or co-linearity between each two protein markers were tested by Spearman's rank rho. In order to manage multiple tests errors, P-values were adjusted using the Benjamini-Hochberg procedure using 5% as an acceptable false discovery rate [20].
To evaluate the diagnostic performance, receiver operating characteristic (ROC) curves were constructed, and areas under ROC curve (AUC), optimal cutoff, sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were calculated through R packages pROC [21] and ROCR [22]. The optimum cutoff value was defined by maximizing the Yoden's index (sensitivity+specificity-1).
To explore which combination of analytes would increase the discrimination between cases and controls, elastic-net penalized logistic regression (ENLR) was performed by applying a penalty to the regression coefficients and finding groups of correlated variables. The optimal penalization proportion α was searched via grid search with 10-fold cross-validation and evaluated in terms of the average of misclassification rate, sensitivity, specificity, and AUC. The optimal tuning parameter λ was determined as the mean values of 100 iteratively lambda values minimizing the deviance of the model. Values of regression coefficients were used to access the contribution of individual protein to the case-control discrimination. We estimated the ENLR model through R package glmnet [23] by using 90% of the samples (randomly selected 45 from the control group and 90 from the cancer group) defined as the training set and the remaining 5% samples (5 and 10) as the test set. The entire cross-validation procedure was repeated 10 times to cover all the samples. Proteins with all non-zero coefficients during the 10 times repeated process were selected. The regression coefficients for the selected proteins were then calculated by rerunning ENLR with only these proteins. To further reduce the number of proteins that could be included in the combination model, ROC curves were plotted starting from the first protein with the highest regression coefficient and then compared to the ROC curves generated while adding one more protein at a time. This process was repeated until none had a significant improvement.
2.5. Protein-protein interaction and enrichment analyses
Protein-protein interactions were analysed with the Search Tool for Retrieval of Interacting Genes/proteins (STRING) database (www.string-db.org) [24]. Protein enrichment was performed with FunRich 3.0 (www.funrich.org) software [25].
3. Results
3.1. Patient demographics
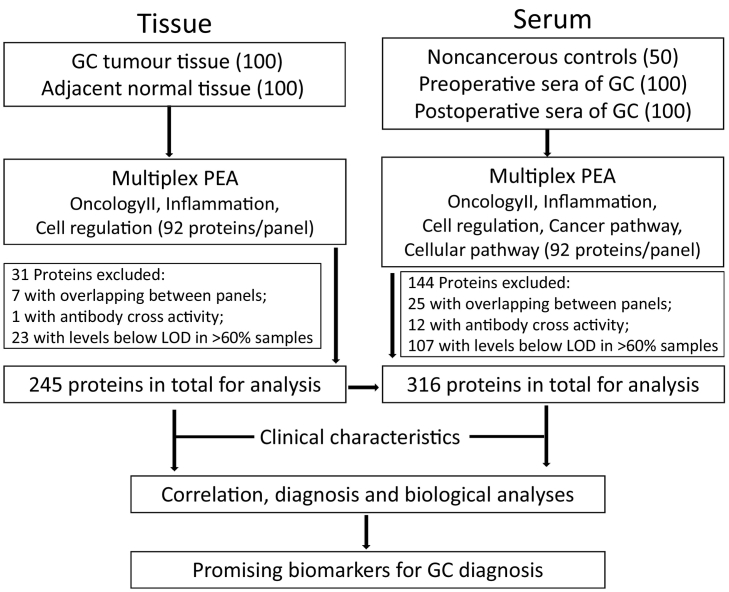
The schematic diagram of this study can be viewed in Fig. 1. Demographic and pathologic characteristics for the 50 non-cancerous control individuals and 100 GC patients are summarized in Table 1 and more detailed information is listed in Supplementary Table 1.
Fig. 1.
A schematic diagram overview of the study. GC, gastric cancer. PEA, proximity extension assay. LOD, limit of detection.
Table 1.
Demographic and pathologic characteristics of 50 control subjects and 100 patients with gastric cancer.
| Variables | No. | Total No. | |
|---|---|---|---|
| Control group | |||
| Age (year) | Mean (range) | 63·5 (25–91) | 50 |
| Gender | F | 38 | 50 |
| M | 12 | ||
| CEA (ng/ml) | Mean (SD) | 1·9 (1·1) | 50 |
| CA19–9 (ng/ml) | Mean (SD) | 20·4 (32·8) | 36 |
| Missing | 14 | ||
| CA72–4 (ng/ml) | Mean (SD) | 2·2 (1·6) | 17 |
| Missing | 33 | ||
| GC group | |||
| Age (year) | Mean (range) | 70·6 (30–92) | 100 |
| Gender | F | 41 | 100 |
| M | 59 | ||
| CEA (ng/ml) | Mean (SD)_Pre | 40·1 (96·1) | 97 |
| Mean (SD)_Post | 13·7 (259·7) | ||
| missing | 3 | ||
| CA19–9 (ng/ml) | Mean (SD)_Pre | 261.6 (1024.4) | 97 |
| Mean (SD)_Post | 93.1 (490.1) | ||
| missing | 3 | ||
| CA72–4 (ng/ml) | Mean (SD)_Pre | 34.3 (143.9) | 90 |
| Mean (SD)_Post | 25.2 (116.5) | ||
| missing | 10 | ||
| Blood type ABO | A | 42 | 94 |
| B | 11 | ||
| O | 38 | ||
| AB | 3 | ||
| Missing | 6 | ||
| Blood type Rh | Rh- | 10 | 94 |
| Rh+ | 84 | ||
| missing | 6 | ||
| Stomach cancer history | No | 84 | 99 |
| Yes | 15 | ||
| missing | 1 | ||
| MSI status | Stable | 63 | 98 |
| High | 35 | ||
| missing | 2 | ||
| Tumour diameter | <50 mm | 30 | 98 |
| ≥50 mm | 68 | ||
| missing | 2 | ||
| TNM stage | I | 8 | 99 |
| II | 20 | ||
| III | 57 | ||
| IV | 14 | ||
| missing | 1 | ||
| Tumour relapse | No | 29 | 65 |
| Yes | 36 | ||
| missing | 35 | ||
MSI: microsatellite instability; EBV: epstein-barr virus.
3.2. Protein detection
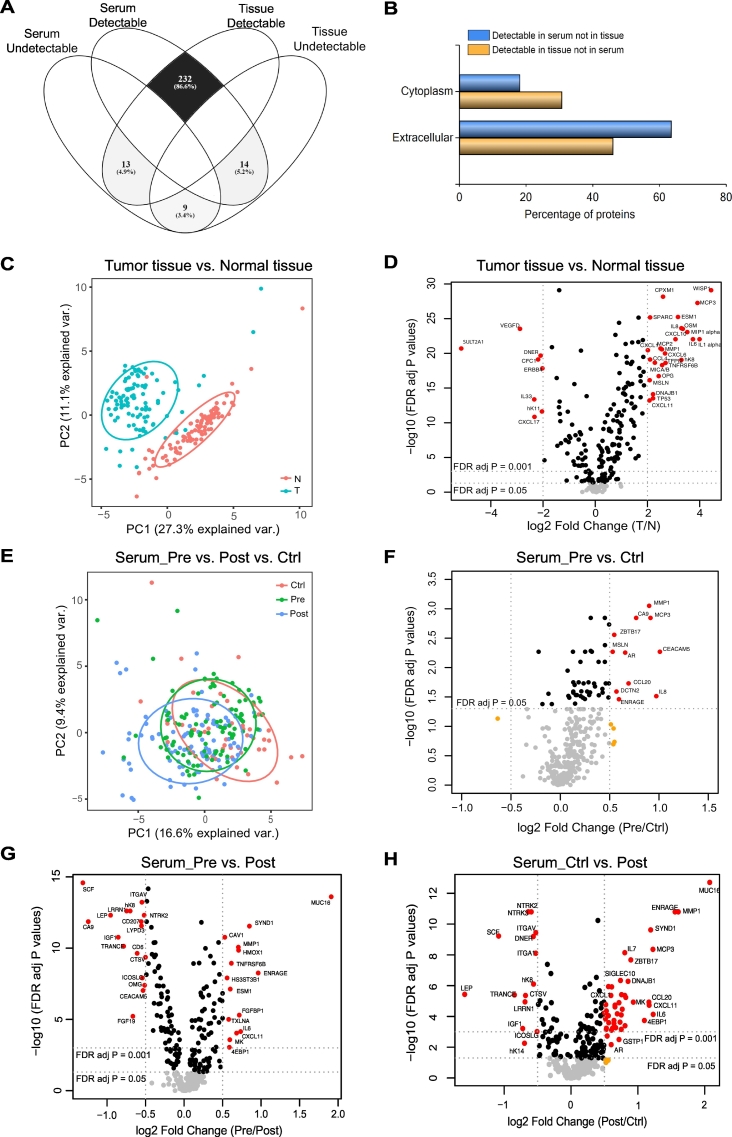
Full names and corresponding UniProtKB accession numbers of all the measured proteins are listed in Supplementary Table 2. Apart from overlapping proteins and proteins with antibody cross activity, proteins levels with NPX values below the LOD in >60% of samples in each disease group were excluded for further analysis (Fig. 1, Supplementary Table 2). This led to 245 proteins for tissue sample analysis and 316 for serum sample analysis while 232 proteins are common between serum and tissue (Fig. 2A). Among the proteins measured in both serum and tissue samples, 13 of those detectable in tissue but not in serum are more cytoplasmic and involved in inflammation, whereas 14 detectable in serum but not in tissue tend to be extracellular proteins and involved in immune-response, as revealed by the functional enrichment analysis tool (FunRich) (Fig. 2B).
Fig. 2.
Multiplex PEA results of proteins measured in GC tissues and serum specimen. (A) Venn diagram showing the number of proteins detectable or undetectable in serum and in tissue specimen. (B) Comparison of subcellular location between proteins detectable in serum but not in tissue and proteins detectable in tissue but not in serum by the FunRich software. The 13 proteins detectable in GC tissue but not in serum are AGR3, ARTN, CAMKK1, IL1A, IL20RA, IL22RA1, IL24, IL33, JUN, LIF, NCLN, NRTN, PAK4. The 14 proteins detectable in GC serum but not in tissue are CRX, DKKL1, FAM19A5, FCRLB, FGF23, IL10, IL10RA, IL2, LYPD1, OPTC, SEZ6L, SLITRK2, TCL1B, WNT9A. (C) Principal component analysis (PCA) plot illustrating the sample distribution of 100 gastric tumour tissues (T, blue) and matched adjacent noncancerous tissues (N, red), based on 245 proteins levels. Each dot represents an individual sample. (D) Volcano plot showing the 245 proteins levels in GC tissues compared to matched non-tumour tissues. The dashed line represents the cutoff line with indicated significance criteria. Points having absolute log fold-change ≥2 and FDR adjusted p-value <0·05 are shown in red, with absolute log fold-change <2 and p-value ≥0·05 are in gray, and the rest are in black. (E) PCA plot illustrating the distribution of 50 serum samples from controls (Ctrl, red), 100 GC preoperative serum samples (Pre, green) and matched 100 postoperative samples (Post, blue), based on 316 proteins levels. (F—H) Volcano plots showing the 316 protein levels in preoperative GC serum samples versus controls (F), between preoperative and postoperative ones (G), and between postoperative samples and controls (H). Points having absolute log fold-change ≥0·5 and FDR adjusted p-value <0·05 are shown in red, with absolute log fold-change <0·5 and p-value <0·05 are in black, with absolute log fold-change <0·5 and p-value ≥0·05 are in gray, and the rest are in orange. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. Cross correlation between each two proteins
As many proteins appear functionally redundant and in order to minimize the risk of selecting a set of biomarkers strongly associated with each other, we investigated the correlations among the proteins. The correlations between each two proteins were assessed by Spearman's rank and the results are illustrated as heatmaps (Supplementary Fig. 4). Among the 245 proteins detected in tumour tissue, 3% (1843/245 × 245) display absolute correlation (ρ) larger than 0·7, and 13.8% (8263/245 × 245) larger than 0·5. Among the 316 proteins measured in serum, only 1% (1026/316 × 316) cross correlation was found with an absolute correlation value larger than 0·7, and 4% (4040/316 × 316) larger than 0·5.
3.4. Proteins differently expressed between GC tumour tissue and adjacent normal tissue
As a first step to evaluate the biological/medical quality of the investigated samples, we performed a PCA of the measured tissue proteins. A clear separation was observed between the 100 tumour tissues and the matched adjacent normal tissues (Fig. 2C). Levels of 200 (81.6%) proteins were significantly altered in tumour tissues compared to normal tissues using the univariate analysis with a false discovery rate (FDR) correction for multiple tests (FDR Padj < 5%): 138 proteins were found to be up-regulated and 62 down-regulated in tumour tissue. The mean (± SD) of each protein in tumour and normal groups, as well as corresponding p-values are detailed in Supplementary Table 3. A volcano plot also illustrates the abundance of proteins changed in tumour tissues compared to corresponding normal tissues (Fig. 2D). Significant associations with clinical variables for proteins expressed in tumour tissues are listed in Table 2, such as MSI status correlated to XPNPEP2, TGFR2, BCR, Flt3L, IFN gamma, SIGLEC6, CPE, GPNMB, TNFRSF19, TWEAK, CBL, and BOC, and some proteins correlated to Borrmann, Ming, or WHO classifications.
Table 2.
Clinical significance of proteins expressed in gastric cancer tumour tissue.
| Variables | Protein | Padj | |
|---|---|---|---|
| Age (mean, range, year) | 70.6 (30–92) | MOG | 0·0002 |
| GFRA2 | 0·0006 | ||
| BOC | 0·02733 | ||
| Gender | |||
| Female | 41 | FASLG | 0·0388 |
| Male | 59 | ||
| MSI status | |||
| Stable | 64 | XPNPEP2 | 0·0002 |
| High | 35 | TGFR2 | 0·0002 |
| BCR | 0·0008 | ||
| Flt3L | 0·002 | ||
| IFN gamma | 0·0039 | ||
| SIGLEC6 | 0·0063 | ||
| CPE | 0·0170 | ||
| GPNMB | 0·0207 | ||
| TNFRSF19 | 0·0266 | ||
| TWEAK | 0·0266 | ||
| CBL | 0·0294 | ||
| BOC | 0·0403 | ||
| Borrman classification | |||
| I | 7 | OMG | 6·11E-17 |
| II | 14 | MOG | 1·59E-05 |
| III | 60 | PROK1 | 0·0113 |
| IV | 18 | ||
| Ming classification | |||
| EXP + MIX (expanding+mixed) | 37 | ZBTB16 | 0·0002 |
| INF (infiltrative) | 40 | NT3 | 0·0008 |
| TWEAK | 0·0072 | ||
| WHO classification | |||
| Poor/Undifferentiated | 38 | OMG | 1·87E-05 |
| Tubular | 34 | MOG | 0·0002 |
| Signet-ring cells & mucinous | 22 | GSAP | 0·0103 |
| Papillary | 2 | IL33 | 0·0415 |
| PROK1 | 0·0453 |
3.5. Proteins differently expressed in sera between the GC and control groups
Blood testing is minimally invasive and as such well-suited to cancer diagnosis. We extended the exploration of the protein profiling also to sera collected before and after surgery from the same 100 GC patients as well as sera from 50 control subjects, in order to develop a diagnostic method more applicable in the clinics.
PCA plot in Fig. 2E illustrates an overview of the distribution of cancer patients before and after operation as well as controls based on the levels of all 316 proteins. The volcano plot (Fig. 2F) displays the protein alterations in the cancer group compared to the control group after univariate analysis. Fifty-four (17·1%) protein differences were found significant (50 increased, 4 decreased) in the cancer group compared to the control group after applying univariate analysis with FDR correction. The mean (± SD) level of each protein in each group, p-values, sensitivity, specificity, PPV, NPV, and AUC are detailed in Supplementary Table 4. The corresponding levels of each of the 54 significantly altered proteins in each group are plotted in Supplementary Fig. 5. Thirty-eight out of the 54 proteins (70.4%) were also significantly elevated in tumour tissues compared to paired normal tissues, while seven proteins were significantly increased in serum but not in tissue comparison (Supplementary Fig. 6). The most significantly increased protein was MMP1 (FDR Padj = 0·0009), having a diagnostic sensitivity of 66%, specificity of 76%, and an AUC of 0·74 (95% CI 0·65–0·82). CEACAM5, also known as CEA, was significantly increased in the sera of GC patients compared to those of the control group, and significantly decreased after surgery. The level of CEA measured by PEA technology is consistent with that measured by standard clinical ELISA method (r = 0·84, Supplementary Fig. 7A), and no significant difference in diagnostic accuracy (CEA_PEA: sensitivity 54%, specificity 76%, AUC 0·68 (0·60–0·77), CEA_ELISA: 52%, 82%, 0·67 (0·59–0·76), P = 0·7, Supplementary Fig. 7B).
Significant associations for pre-operative protein with clinical characteristics were found between loss of heterogeneity of CDH and SMAD4 (p = 0·0005) and between GCNT1 and TNM stage (p = 0·0072).
3.6. Alteration of serum protein levels before and after surgery
To check whether protein abundances can be affected by tumour resection, levels of proteins before operation and one to two weeks after were compared. The univariate analysis with FDR correction for multiple tests showed that 184 of 316 (58·2%) proteins were significantly changed after surgery (88 decreased and 96 increased, Supplementary Table 5). Volcano plots in Fig. 2G-H display the most significantly altered proteins in the comparisons between pre and post groups and between control and post. The biological enrichment analysis by FunRich revealed that the proteins decreased after surgery are predominantly involved in cell adhesion, whereas the proteins increased are tend to be associated with inflammatory response (Supplementary Fig. 8). Notably, the changes for the protein levels after operation were not consistent in all the patients, but following the same trend in most patients as illustrated for each protein and each patient in Supplementary Fig. 9. We then evaluated the clinical associations of proteins differentially changed after surgery, and found associations between CYR61 with LOH of CDH, between GCNT1 with Lauren classification, and proteins associated with tumour site including ERBB2/3/4, GPNMB, ITGAV, ITGB5, LYPD3, NTRK2/3, SEZ6L, XPNPEP2, and TNFRSF19 (Supplementary Table 6).
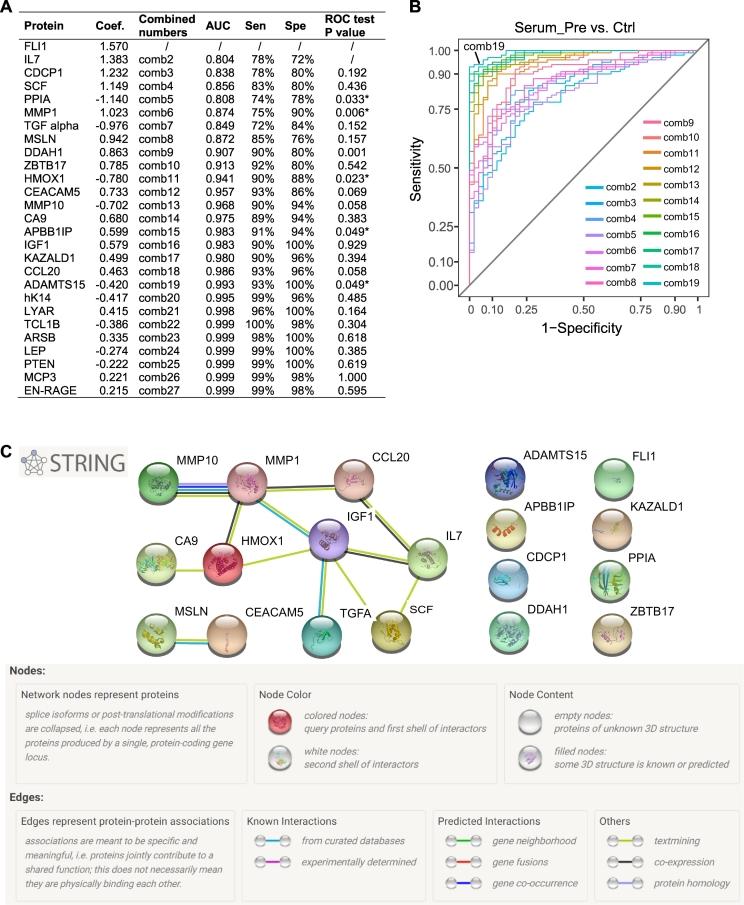
3.7. Optimal proteins combination for distinguishing gastric cancer patients from controls
Multivariate analysis ENLR was performed to further select promising serum biomarkers and generate a diagnostic model for gastric cancer. With cross-validation, the optimal α = 0.4 was chosen for penalty proportion for the model according to the lowest misclassification error and the best accuracy (Supplementary Table 7). After ten-fold cross-validation, 27 proteins were retained to have all non-zero coefficients at each cross-validation step (Fig. 3A). By comparing the ROC curves of different combinations of the proteins that ranked from the largest to the smallest absolute regression coefficients, the combination of the top 19 proteins showed the optimal AUC accuracy number when no significant improvement was found by adding one more protein to the linear combination (Fig. 3A and B). Therefore, the generated diagnostic signature was found to be:
Fig. 3.
Diagnostic capacity for gastric cancer of the identified 19 serum protein signature by elastic-net logistic regression. (A) Diagnostic performances of different protein combinations. Proteins are sorted according to the absolute coefficient from the largest to the smallest. “ROC test P" is the p-value of the comparison of ROC curves generated from successive protein combinations with one more protein added at a time. Coef., coefficient. ROC, receiver operator characteristics. *, p < 0·05. (B) Overlaid ROC curves of each of the combinations from two to 19 serum proteins. Comb, combination. (C) Protein-protein interactions among the 19 proteins assessed with the STRING database.
The sensitivity and specificity of the combined model for distinguishing GC patients from controls were 93% and 100%, respectively, with an AUC of 0.99 (95% CI: 0·98–1). The diagnostic performance of the combined signature is clearly greater than that of each of the 19 proteins and also the clinical used biomarkers CA19–9 and CA72–4 (Table 3). Querying the STRING database confirmed the absence of experimental evidence of protein-protein interactions among those selected 19 proteins. Only MMP1, and MMP10 and other MMP family members may form a complex which activates MMP9; MSLN and CEACAM5 are both attached to cell membrane via glycosylphosphatidylinositol (GPI) anchors (Fig. 3C).
Table 3.
Serum protein biomarkers identified by elastic-net logistic regression (ENLR) for gastric cancer diagnosis.
| Identified proteins | Short name | Ctrl (Mean ± SD) | Pre (Mean ± SD) | Pre vs. Ctrl | FDR Padj | Cut-off | Sen (%) | Spe (%) | AUC (95%CI) | T vs. N | FDR Padj |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Matrix metalloproteinase-1 | MMP1 | 1·56 ± 1·04 | 2·46 ± 1·04 | I | 0·0009 | 2·05 | 66 | 76 | 0·74(0·65–0·82) | I | 2·57E-21 |
| Interleukin-7 | IL7 | 3·3 ± 0·54 | 3·75 ± 0·58 | I | 0·0014 | 3·41 | 73 | 64 | 0·72(0·64–0·81) | – | – |
| Carbonic anhydrase 9 | CA9 | 3·26 ± 1·36 | 4·02 ± 1·21 | I | 0·0014 | 3·40 | 71 | 68 | 0·71(0·62–0·8) | I | 2·82E-05 |
| CUB domain-containing protein 1 | CDCP1 | 3·69 ± 0·63 | 4·19 ± 0·66 | I | 0·0018 | 3·34 | 93 | 40 | 0·71(0·62–0·8) | I | 1·95E-05 |
| Zinc finger and BTB domain-containing protein 17 | ZBTB17 | 1·34 ± 0·69 | 1·88 ± 0·85 | I | 0·0027 | 1·69 | 61 | 76 | 0·7(0·61–0·79) | I | 4·84E-13 |
| dimethylarginine dimethylaminohydrolase 1 | DDAH1 | 1·36 ± 0·51 | 1·82 ± 0·76 | I | 0·0041 | 1·29 | 77 | 54 | 0·69(0·6–0·79) | – | – |
| Friend leukemia integration 1 transcription factor | FLI1 | 0·11 ± 0·33 | 0·42 ± 0·61 | I | 0·0052 | 0·19 | 66 | 66 | 0·69(0·6–0·78) | D | 3·01E-08 |
| Mesothelin | MSLN | 2·58 ± 0·83 | 3·11 ± 0·74 | I | 0·0053 | 2·31 | 87 | 48 | 0·69(0·59–0·78) | I | 7·34E-17 |
| Carcinoembryonic antigen-related cell adhesion molecule 5 | CEACAM5 | 2·42 ± 0·79 | 3·43 ± 1·73 | I | 0·0053 | 2·86 | 54 | 76 | 0·68(0·6–0·77) | I | 0·0229 |
| Kazal-type serine protease inhibitor domain-containing protein 1 | KAZALD1 | 2·78 ± 0·7 | 3·17 ± 0·68 | I | 0·0079 | 3·08 | 56 | 80 | 0·68(0·58–0·77) | D | 1·47E-07 |
| C-C motif chemokine 20 | CCL20 | 6·09 ± 1·3 | 6·78 ± 1·37 | I | 0·0184 | 6·60 | 52 | 76 | 0·66(0·57–0·75) | I | 0·0137 |
| Stem cell factor/KIT ligand | SCF | 7·85 ± 0·81 | 8·08 ± 0·63 | – | – | 7·71 | 83 | 34 | 0·58(0·49–0·68) | I | 1·20E-07 |
| Peptidyl-prolyl cis-trans isomerase A | PPIA | 3·05 ± 1·01 | 2·92 ± 1·05 | – | – | 3·05 | 52 | 56 | 0·48(0·38–0·57) | – | – |
| Transforming growth factor alpha | TGF alpha | 2·43 ± 0·91 | 2·45 ± 1·02 | – | – | 2·48 | 62 | 52 | 0·5(0·4–0·6) | D | 3·57E-05 |
| Heme oxygenase 1 | HMOX1 | 9·91 ± 1·07 | 9·57 ± 0·81 | – | – | 9·96 | 74 | 62 | 0·63(0·52–0·74) | – | – |
| Matrix metalloproteinase-10 | MMP10 | 6·81 ± 0·56 | 6·71 ± 0·61 | – | – | 6·94 | 71 | 40 | 0·54(0·44–0·64) | I | 3·98E-11 |
| Amyloid beta A4 precursor protein-binding family B member 1-interacting protein | APBB1IP | 1·61 ± 0·69 | 1·98 ± 0·98 | – | – | 2·31 | 31 | 88 | 0·6(0·5–0·69) | I | 0·0082 |
| Insulin-like growth factor I | IGF1 | 1·43 ± 1·13 | 1·56 ± 0·88 | – | – | 2·24 | 78 | 34 | 0·49(0·38–0·59) | – | – |
| A disintegrin and metalloproteinase with thrombospondin motifs 15 | ADAMTS15 | 1·78 ± 0·83 | 1·46 ± 0·62 | – | – | 1·94 | 80 | 48 | 0·62(0·52–0·72) | D | 5·86E-13 |
| 19-protein signature | 93 | 100 | 0·99(0·98–1) | ||||||||
| Carcinoembryonic antigen | CEAa | 2·24 ± 0·83 | 43·89 ± 272·52 | I | 2·65 | 52 | 82 | 0·67(0·59–0·76) | |||
| Carbohydrate antigen 19–9 | CA19–9b | 11·31 ± 10·39 | 280·05 ± 1092·91 | I | 37·5 | 42 | 88 | 0·63(0·54–0·73) | |||
| Carbohydrate antigen 72–4 | CA72–4c | 2·22 ± 1·63 | 35·03 ± 145·51 | I | 2·6 | 55 | 88 | 0·72(0·61–0·83) |
I: increase. D: decrease. FDR Padj: P values were tested by non-parametric Mann-Whitney-Wilcoxon and adjusted multiple tests with false discovery rate. CI: confidence interval. Coef.: coefficient calculated by ENLR. T: tumour tissue. N: adjacent normal tissue. –: Proteins not significantly altered.
Cutoff was defined by Yoden's index by maximizing values of sensitivity+specificity-1.
a, b, c,: clinically measured biomarkers. b: 36 controls vs. 97 GC. c: 17 controls vs.90 GC.
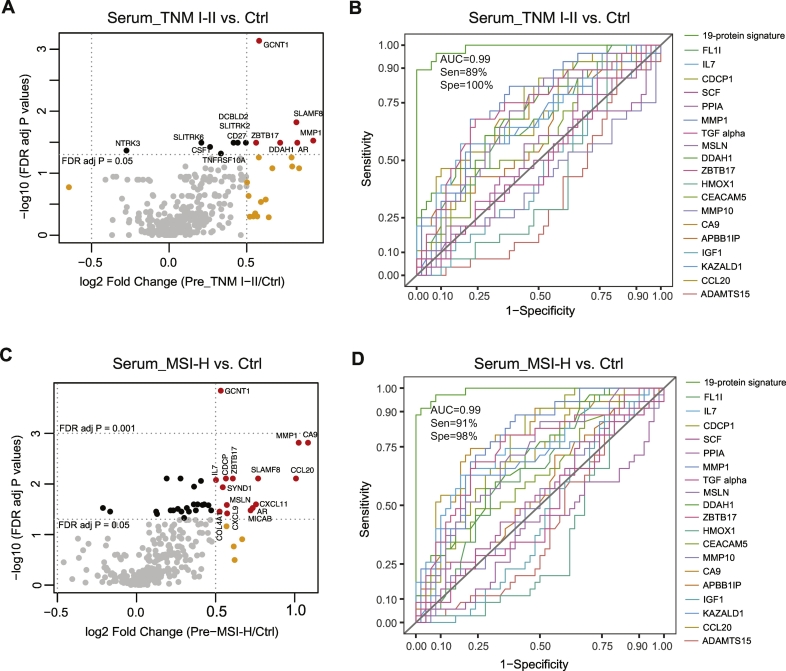
3.8. Proteins significantly altered in sera from GC patients at TNM I-II early stage
Cancer patients at early stage are always difficult to diagnose but early detection is important for successful therapy. Twenty-eight GC patients were diagnosed at the earlier TNM I-II stage in the present cohort. Volcano plot in Fig. 4A illustrates the significantly altered proteins between patients at early stage and controls by univariate analysis. GCNT1 was shown as the most significantly differential protein, and its optimal diagnostic sensitivity, specificity, and AUC of GCNT1 in patients at TNM I-II stage determined by ROC curve were 75%, 86% and 0·82, respectively (Supplementary Fig. 10A and B). PCA plots for both the distribution of tissue and serum samples according to TNM stages as well as volcano plots for protein alterations in different group comparisons in both tissue and serum are demonstrated in Supplementary Fig. 11. With the 19-serum protein signature identified above for the whole cohort, the diagnostic performance for differentiating patients at TNM I-II stage from controls was better than that of each individual protein with an AUC of 0·99 and sensitivity of 89% and specificity of 100% (Fig. 4B), whereas the best score for a single protein was for MMP1 with a sensitivity of 68%, specificity of 78% and AUC of 0·75, and the AUCs for clinically measured biomarkers CEA, CA19–9, and CA72–4 were 0.58, 0.48, and 0.61, respectively.
Fig. 4.
Protein levels in serum samples from GC patients at TNM I-II stage or with high microsatellite instability (MSI) status. (A) Volcano plot showing the comparison of protein levels between patients at TNM I-II stage and controls. (B) ROC curves for the 19-serum protein signature overlaid with each individual protein showing the diagnostic capacity of GC at TNM I-II stage versus controls. (C) Volcano plot showing the comparison of protein levels between patients with MSI-H and controls. (D) ROC curves for the 19-serum protein signature overlaid with each individual protein showing the diagnostic capacity of GC with MSI-H status versus controls. Points in plots A and C having absolute log fold-change ≥0·5 and false discovery rate adjusted p-value <0·05 are shown in red, with absolute log fold-change <0·5 and p-value <0·05 are in black, with absolute log fold-change <0·5 and p-value ≥0·05 are in gray, and the rest are in orange. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.9. Proteins significantly altered in sera from GC patients with high microsatellite instability
One of the leading causes of GC is a defect in DNA mismatch repair, resulting in microsatellite instability. Microsatellite-unstable tumours are hyper-mutated intestinal subtype tumours and have recently been proposed as one of the most robust subgroups in molecular characterization of GC, which occurring in at least 20% of all GC patients and having a better overall prognosis and lower frequency of recurrence [26,27]. The molecular diagnosis of MSI GC is important before treatment, as it triggers different response to chemotherapy, and may require specific surgical treatment such as tailored lymphadenectomy, and it stratifies patients for targeted therapies [28]. In the present cohort, only 35 GC patients were found with high MSI. As shown in Fig. 4C, the volcano plot displays the significantly changed proteins in serum of GC patients with MSI-H status when comparing to the controls by univariate analysis. GCNT1 was also the most significant altered protein in MSI-H versus controls, and the optimal diagnostic sensitivity, specificity, and AUC of GCNT1 for patients with high MSI status were 71%, 86% and 0·82, respectively (Supplementary Fig. 10C and D). Supplementary Fig. 12 illustrates the PCA plots for sample distribution according to MSI status as well as volcano plots for protein alterations in different group comparisons in both tissue and serum. The diagnostic performance of the 19-serum protein signature for differentiating patients with high MSI from controls was significantly better than that of each individual protein with an AUC of 0·99 and sensitivity of 91% and specificity of 98% (Fig. 4D), whereas the best score for a single protein was again for MMP1 with a sensitivity of 88%, specificity of 60% and AUC of 0·78, and the AUCs for clinically measured biomarkers CEA, CA19–9, and CA72–4 were 0.60, 0.63, and 0.79, respectively.
4. Discussion
GC is often either asymptomatic or causing nonspecific symptoms in its early stage, and it shares considerable heterogeneity with distinct morphology [3]. Early detection and diagnosis is crucial to reduce cancer related deaths. In contrast to single tissue biopsy, blood carries information from cancer cells even located at distinct metastatic site, which reflects the overall change of a disease. Upon cancer development and progression, cancer cells secrete proteins into the microenvironment and bloodstream; when tumours grow, the apoptotic and necrotic cells are likely to release numerous proteins into the circulating blood; additionally, cancer systemically elicits an immune response. All of these processes generate circulating proteins as a rich source for biomarkers. Therefore, we measured proteins that are either secreted or located in cytoplasm or involved in immune response.
Applying multiplex PEA technology, we analysed the levels of 316 proteins in serum and 245 in tissue specimens from 100 patients with gastric cancer and 50 controls, and identified a combination of 19 potential serum protein biomarkers that distinguish patients with gastric adenocarcinoma from cancer free controls. The proteins significantly altered in serum may not be significant in tissue, vice versa. The 19-protein signature determined by elastic-net logistic regression enhanced the diagnostic performance for the whole serum cohort to an AUC of 0·99, a sensitivity of 93% and a specificity of 100%. It also satisfactory distinguishes the GC patients at TNM I-II stage and patients with high MSI status from controls.
Most of the 19 proteins are secreted. Apart from the clinically used biomarker CEACAM5, eight proteins have been reported for their clinical significances in both GC blood and tumour tissue, including MMP1 [[29], [30], [31]], MSLN [[32], [33], [34]], CA9 [[35], [36], [37]], CCL20 [[38], [39], [40]], SCF [41,42], TGF alpha [[43], [44], [45]], IGF-1 [46,47], and MMP10 [48]. The first four proteins were also found significantly increased in both GC serum and tumour tissue as analysed by univariate analysis in the present cohort, and SCF, TGF alpha, and MMP10 were significantly altered only in GC tissues, whereas IGF-1 were not changed in neither serum nor tissue.
Up to date, protein dysregulation have been described only in GC tissues but not in GC blood for (1) CDCP1 [49], (2) DDAH1 [50], (3) PPIA [51], and (4) HMOX1 [52,53]. (1) CDCP1 is a transmembrane glycoprotein and its engineered overexpression in gastric cell lines was shown to increase cell migration and invasion [49,54]. Our data revealed that the levels of CDCP1 were significantly increased in GC serum as well as tumour tissues. (2) DDAH1 is an endogenous nitric oxide synthase inhibitor, for which Ye et al. reported that downregulation of DDAH1 was more frequently detected in GC tumour tissues and strongly correlated with aggressive phenotypes and poor prognosis [50]. However, we observed increased DDAH1 levels in GC serum compared to controls, while no difference between GC tumour and adjacent normal tissue in our cohort. (3) PPIA, has been reported to be overexpressed in several types of human cancers including gastric cancer [51,55]. (4) HMOX1 is considered to be the main protein for cytoprotection against various cell stresses, and increased expression of this protein was observed in several types of cancers [56,57]. However, there are no differences in the levels of PPIA and HOMX1 between GC and controls in serum or in tissue in the present study.
Furthermore, we identified six novel proteins associated with GC including (1) IL7, (2) ZBTB17, (3) FLI1, (4) KAZALD1, (5) APBB1IP, and (6) ADAMTS15. (1) A recent bioinformatic analysis based on genome-wide association studies (GWAS) data identified IL7 pathway as one of the three pathways associated with GC risk [58]. (2) ZBTA17, also known as MIZ1, is a transcription factor that forms a complex with Myc and mediates Myc-dependent tumorigenesis [59]. (3) FLI11 is also a transcription factor, and its high level expression as well as translocations were observed in hematopoietic and solid tumours [60]. (4) KAZALD1 is a member of the insulin growth factor-binding protein (IGFBP) superfamily, and hypomethylation of the gene was found in cancer. “The Human Protein Atlas (HPA)” database (https://www.proteinatlas.org) shows a moderate staining (6/10) of KAZALD1 protein in stomach cancer tissues. In the present cohort, the levels of IL7, ZBTB17, FLI1, and KAZALD1 were all significantly increased in serum from GC patients as compared with controls, indicating that they may play roles in GC tumorigenesis and progress. (5) APBB1IP, also known as RIAM (Rap1-GTP-interacting adaptor molecule), appears to function in the signal transduction from Ras activation to actin cytoskeletal remodeling and mediates Rap1-induced adhesion. (6) ADAMTS15 is one of the extracellular metalloproteinases, which functions as a putative tumour suppressor, and has been linked to a number of different cancers such as prostatic, breast and colorectal cancers [61]. The HPA database displays a weak staining for APBB1IP and moderate for ADAMTS15 in 10 GC tissue sections. In our study, ADAMTS15 was also significantly downregulated in GC tissues compared to surrounding normal tissues.
Many of the 19 identified proteins are inflammation related, such as IL7, MMP1, MMP10, CCL20, CDCP1, SCF, and TGFA. As one of the consequences of long-term gastric inflammation is malignances, these markers may play important roles in GC development.
GCNT1 is a glycosyltransferase that adds beta1,6 GlcNAc arm to core 1 O-glycans and forms core 2 O-glycans. Core 2 O-glycans are known to be a particularly good scaffold for sialyl Lewis antigens, which can be recognized by selectin family members and thereby mediate leukocyte rolling and cancer cell metastasis [62]. GCNT1 was significantly increased in sera of GC patients compared to controls in univariate analysis, and was the most significantly increased serum protein in GC patients at TNM I-II stage as well as in high MSI patients. However, there was no change of GCNT1 between tumour and adjacent normal tissues for patients at early stage or with MSI. This indicates that the increase of GCNT1 in serum may be due to inflammation or unleashed immune response. As most blood proteins are glycoproteins and alterations of O-linked glycosylation is related to a majority of cancers, the function role and underline mechanisms of GCNT1 in gastric cancer needs further investigation.
Some very significantly altered serum proteins in univariate analysis, such as GCNT1 and NTRK3, were not selected in the regression model, presumably due to the collinearity of the proteins with others. Conflicting results between univariate analysis and multivariate logistic regression is not unusual. Additionally, different multivariate analysis methods will generate different results. ENLR used in the present study applies automatic variable selection at the same time continuous shrinkage producing a sparse model which minimizes overfitting [63].
The limitations of our study need to be acknowledged. The proteins measured by multiplex PEA are pre-selected, which may exclude at the beginning some promising candidates. Evaluation and development of potential protein biomarkers for GC diagnosis were based on a single cohort; however, the multiple fold cross-validations method provides the power for robust sensitivity and specificity. A large-scale independent cohort of cases and different types of controls, e.g., other gastric diseases and gastrointestinal cancers, will be necessary for validating the proposed protein signature. Furthermore, since the signature is tailored for the European population, it may be challenged by other ethnicities and etiologies. According to the “Phases of biomarker development for early detection of cancer” [64] proposed by Early Detection Research Network (EDRN) of the National Cancer Institute, our study is essentially a cross-sectional and retrospective study. A prospective study is also valuable to further assess the diagnostic/screen value for the proteins combination.
The striking increase or decrease of some protein levels in serum after surgery suggests the role of surveillance of those proteins. To further explore the potential effects, a parallel study which will associate the patients' outcome with protein levels in serum and tissue is ongoing.
Notwithstanding the above limitations, the present study suggests a blood test with a set of serum proteins for GC diagnosis, which may be translated into clinical applications with tangible benefits for GC patients in the future.
Acknowledgments
Acknowledgements
As a part of the GastricGlycoExplorer project, the authors thank all members in the committee. We would like to thank all participants and clinicians who collected the samples.
Funding
The present work was funded by European H2020-Marie Skłodowska-Curie Innovative Training Networks grant agreement N° 316929 (GastricGlycoExplorer). The sponsors of the study had no role in study design, data collection, analysis, interpretation, or writing of the report.
Availability of data and materials
Raw data of the multiplex PEA results are available at the GGE data repository (https://ggerepository.isb-sib.ch/) or can be requested from the corresponding author on reasonable request.
Authors' contributions
MK-M, FR, MG-K, FL, and NGK were involved in the study concept and design, and critical revision of the manuscript for intellectual content. QS processed samples, performed experiments, analysed data, interpreted results, and drafted the manuscript. FO processed samples. KP and FR provided samples and obtained informed consent for all subjects. CW and MG-K analysed data. MK-M was also involved in funding obtain, material support and study supervision. All authors discussed the results and commented on the paper.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.05.044.
Contributor Information
Qiujin Shen, Email: qiujin.shen@igp.uu.se.
Karol Polom, Email: surgoncolclub@gmail.com.
Coralie Williams, Email: c.williams@arianapharma.com.
Felipe Marques Souza de Oliveira, Email: felipe.oliveira@igp.uu.se.
Mariana Guergova-Kuras, Email: mariana.kuras@free.fr.
Frederique Lisacek, Email: frederique.lisacek@sib.swiss.
Niclas G. Karlsson, Email: niclas.karlsson@medkem.gu.se.
Franco Roviello, Email: franco.roviello@unisi.it.
Masood Kamali-Moghaddam, Email: masood.kamali@igp.uu.se.
Appendix A. Supplementary data
Supplementary material
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R., Torre L., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A J Clin. 2018;00:1–31. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]; Bray F, Ferlay J, Soerjomataram I, Siegel R, Torre L, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A J Clin 2018;00:1–31. doi:10.3322/caac.21492. [DOI] [PubMed]
- 2.Pasechnikov V., Chukov S., Fedorov E., Kikuste I., Leja M. Gastric cancer: prevention, screening and early diagnosis. World J Gastroenterol. 2014;20:13842–13862. doi: 10.3748/wjg.v20.i38.13842. [DOI] [PMC free article] [PubMed] [Google Scholar]; Pasechnikov V, Chukov S, Fedorov E, Kikuste I, Leja M. Gastric cancer: Prevention, screening and early diagnosis. World J Gastroenterol 2014;20:13842–62. doi:10.3748/wjg.v20.i38.13842. [DOI] [PMC free article] [PubMed]
- 3.Marrelli D., Polom K., De Manzoni G., Morgagni P., Baiocchi G.L., Roviello F. Multimodal treatment of gastric cancer in the west: where are we going? World J Gastroenterol. 2015;21:7954–7969. doi: 10.3748/wjg.v21.i26.7954. [DOI] [PMC free article] [PubMed] [Google Scholar]; Marrelli D, Polom K, De Manzoni G, Morgagni P, Baiocchi GL, Roviello F. Multimodal treatment of gastric cancer in the west: Where are we going? World J Gastroenterol 2015;21:7954–69. doi:10.3748/wjg.v21.i26.7954. [DOI] [PMC free article] [PubMed]
- 4.Liu W., Yang Q., Liu B., Zhu Z. Serum proteomics for gastric cancer. Clin Chim Acta. 2014;431:179–184. doi: 10.1016/j.cca.2014.02.001. [DOI] [PubMed] [Google Scholar]; Liu W, Yang Q, Liu B, Zhu Z. Serum proteomics for gastric cancer. Clin Chim Acta 2014;431:179–84. doi:10.1016/j.cca.2014.02.001. [DOI] [PubMed]
- 5.Ucar E., Semerci E., Ustun H., Yetim T., Huzmeli C., Gullu M. Prognostic value of preoperative CEA, CA 19-9, CA 72-4, and AFP levels in gastric cancer. Adv Ther. 2008;25:1075–1084. doi: 10.1007/s12325-008-0100-4. [DOI] [PubMed] [Google Scholar]; Ucar E, Semerci E, Ustun H, Yetim T, Huzmeli C, Gullu M. Prognostic value of preoperative CEA, CA 19-9, CA 72-4, and AFP levels in gastric cancer. Adv Ther 2008;25:1075–84. doi:10.1007/s12325-008-0100-4. [DOI] [PubMed]
- 6.Leung W.K., Wu M.S., Kakugawa Y., Kim J.J., Yeoh K.G., Goh K.L. Screening for gastric cancer in Asia: current evidence and practice. Lancet Oncol. 2008;9:279–287. doi: 10.1016/S1470-2045(08)70072-X. [DOI] [PubMed] [Google Scholar]; Leung WK, Wu MS, Kakugawa Y, Kim JJ, Yeoh KG, Goh KL, et al. Screening for gastric cancer in Asia: current evidence and practice. Lancet Oncol 2008;9:279–87. doi:10.1016/S1470-2045(08)70072-X. [DOI] [PubMed]
- 7.Huang Y.K., Yu J.C., Kang W.M., Ma Z.Q., Ye X., Tian S.B. Significance of serum pepsinogens as a biomarker for gastric cancer and atrophic gastritis screening: A systematic review and meta-analysis. PLoS One. 2015;10 doi: 10.1371/journal.pone.0142080. [DOI] [PMC free article] [PubMed] [Google Scholar]; Huang YK, Yu JC, Kang WM, Ma ZQ, Ye X, Tian SB, et al. Significance of serum pepsinogens as a biomarker for gastric cancer and atrophic gastritis screening: A systematic review and meta-analysis. PLoS One 2015;10. doi:10.1371/journal.pone.0142080. [DOI] [PMC free article] [PubMed]
- 8.Kang C., Lee Y., Lee J.E. Recent advances in mass spectrometry-based proteomics of gastric cancer. World J Gastroenterol. 2016;22:8283–8293. doi: 10.3748/wjg.v22.i37.8283. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kang C, Lee Y, Lee JE. Recent advances in mass spectrometry-based proteomics of gastric cancer. World J Gastroenterol 2016;22:8283–93. doi:10.3748/wjg.v22.i37.8283. [DOI] [PMC free article] [PubMed]
- 9.Assarsson E., Lundberg M., Holmquist G., Björkesten J., Thorsen S.B., Ekman D. Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS One. 2014;9 doi: 10.1371/journal.pone.0095192. [DOI] [PMC free article] [PubMed] [Google Scholar]; Assarsson E, Lundberg M, Holmquist G, Björkesten J, Thorsen SB, Ekman D, et al. Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS One 2014;9:e95192. doi:10.1371/journal.pone.0095192. [DOI] [PMC free article] [PubMed]
- 10.Fredriksson S., Gullberg M., Jarvius J., Olsson C., Pietras K., Gústafsdóttir S.M. Protein detection using proximity-dependent DNA ligation assays. Nat Biotechnol. 2002;20:473–477. doi: 10.1038/nbt0502-473. [DOI] [PubMed] [Google Scholar]; Fredriksson S, Gullberg M, Jarvius J, Olsson C, Pietras K, Gústafsdóttir SM, et al. Protein detection using proximity-dependent DNA ligation assays. Nat Biotechnol 2002;20:473–7. doi:10.1038/nbt0502-473. [DOI] [PubMed]
- 11.Lundberg M., Eriksson A., Tran B., Assarsson E., Fredriksson S. Homogeneous antibody-based proximity extension assays provide sensitive and specific detection of low-abundant proteins in human blood. Nucleic Acids Res. 2011;39:e102. doi: 10.1093/nar/gkr424. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lundberg M, Eriksson A, Tran B, Assarsson E, Fredriksson S. Homogeneous antibody-based proximity extension assays provide sensitive and specific detection of low-abundant proteins in human blood. Nucleic Acids Res 2011;39:e102. doi:10.1093/nar/gkr424. [DOI] [PMC free article] [PubMed]
- 12.Conze T., Carvalho A.S., Landegren U., Almeida R., Reis C.A., David L. MUC2 mucin is a major carrier of the cancer-associated sialyl-Tn antigen in intestinal metaplasia and gastric carcinomas. Glycobiology. 2009;20:199–206. doi: 10.1093/glycob/cwp161. [DOI] [PubMed] [Google Scholar]; Conze T, Carvalho AS, Landegren U, Almeida R, Reis CA, David L, et al. MUC2 mucin is a major carrier of the cancer-associated sialyl-Tn antigen in intestinal metaplasia and gastric carcinomas. Glycobiology 2009;20:199–206. doi:10.1093/glycob/cwp161. [DOI] [PubMed]
- 13.de Oliveira F.M.S., Mereiter S., Lönn P., Siart B., Shen Q., Heldin J. Detection of post-translational modifications using solid-phase proximity ligation assay. N Biotechnol. 2018:51–59. doi: 10.1016/j.nbt.2017.10.005. [DOI] [PubMed] [Google Scholar]; Oliveira FMS de, Mereiter S, Lönn P, Siart B, Shen Q, Heldin J, et al. Detection of post-translational modifications using solid-phase proximity ligation assay. N Biotechnol 2018:51–9. doi:10.1016/j.nbt.2017.10.005. [DOI] [PubMed]
- 14.Bossuyt P.M., Reitsma J.B., Bruns D.E., Gatsonis C.A., Glasziou P.P., Irwig L. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. Clin Chem. 2015;61:1446–1452. doi: 10.1373/clinchem.2015.246280. [DOI] [PubMed] [Google Scholar]; Bossuyt PM, Reitsma JB, Bruns DE, Gatsonis CA, Glasziou PP, Irwig L, et al. STARD 2015: An updated list of essential items for reporting diagnostic accuracy studies. Clin Chem 2015;61:1446–52. doi:10.1373/clinchem.2015.246280. [DOI] [PubMed]
- 15.Lauren P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. Acta Pathol Microbiol Scand. 1965;64:31–49. doi: 10.1111/apm.1965.64.1.31. [DOI] [PubMed] [Google Scholar]; Lauren P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. Acta Pathol Microbiol Scand 1965;64:31–49. doi:10.1002/1097-0142(197706)39:6<2475::AID-CNCR2820390626>3.0.CO;2-L. [DOI] [PubMed]
- 16.Watanabe H., Jass J.R., Sobin L.H. Histol. Typing oesophageal gastric tumours. Springer Berlin Heidelberg; Berlin, Heidelberg: 1990. Histological classification of oesophageal tumours; pp. 5–6. [Google Scholar]; Watanabe H, Jass JR, Sobin LH. Histological Classification of Oesophageal Tumours. Histol. Typing Oesophageal Gastric Tumours, Berlin, Heidelberg: Springer Berlin Heidelberg; 1990, p. 5–6. doi:10.1007/978-3-642-83920-7_2.
- 17.Edge S.B. Springer; 2010. American joint committee on Cancer. AJCC cancer staging manual. [DOI] [PubMed] [Google Scholar]; Edge SB, American Joint Committee on Cancer. AJCC cancer staging manual. Springer; 2010. [DOI] [PubMed]
- 18.Corso G., Velho S., Paredes J., Pedrazzani C., Martins D., Milanezi F. Oncogenic mutations in gastric cancer with microsatellite instability. Eur J Cancer. 2011;47:443–451. doi: 10.1016/j.ejca.2010.09.008. [DOI] [PubMed] [Google Scholar]; Corso G, Velho S, Paredes J, Pedrazzani C, Martins D, Milanezi F, et al. Oncogenic mutations in gastric cancer with microsatellite instability. Eur J Cancer 2011;47:443–51. doi:10.1016/j.ejca.2010.09.008. [DOI] [PubMed]
- 19.R core team R: A language and environment for statistical computing. R found stat Comput Vienna, Austria. 2017. http://www.R-project.org/; R core team. R: A language and environment for statistical computing. R Found Stat Comput Vienna, Austria 2017. doi:http://www.R-project.org/.
- 20.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]; Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 1995;57:289–300. doi:10.2307/2346101.
- 21.Robin X., Turck N., Hainard A., Tiberti N., Lisacek F., Sanchez J.C. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinforma. 2011;12 doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]; Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, et al. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 2011;12. doi:10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed]
- 22.Sing T., Sander O., Beerenwinkel N., Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics. 2005;21:3940–3941. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]; Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: Visualizing classifier performance in R. Bioinformatics 2005;21:3940–1. doi:10.1093/bioinformatics/bti623. [DOI] [PubMed]
- 23.Friedman J., Hastie T., Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33 [PMC free article] [PubMed] [Google Scholar]; Friedman J, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw 2010;33. doi:10.18637/jss.v033.i01. [PMC free article] [PubMed]
- 24.Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]; Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015;43:D447–52. doi:10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed]
- 25.Pathan M., Keerthikumar S., Ang C.S., Gangoda L., Quek C.Y.J., Williamson N.A. FunRich: an open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015;15:2597–2601. doi: 10.1002/pmic.201400515. [DOI] [PubMed] [Google Scholar]; Pathan M, Keerthikumar S, Ang CS, Gangoda L, Quek CYJ, Williamson NA, et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics 2015;15:2597–601. doi:10.1002/pmic.201400515. [DOI] [PubMed]
- 26.Cristescu R., Lee J., Nebozhyn M., Kim K.-M., Ting J.C., Wong S.S. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med. 2015;21:449–456. doi: 10.1038/nm.3850. [DOI] [PubMed] [Google Scholar]; Cristescu R, Lee J, Nebozhyn M, Kim K-M, Ting JC, Wong SS, et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med 2015;21:449–56. doi:10.1038/nm.3850. [DOI] [PubMed]
- 27.Cancer T., Atlas G., Bass A.J., Thorsson V.V., Shmulevich I., Reynolds S.M. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cancer T, Atlas G, Bass AJ, Thorsson VV, Shmulevich I, Reynolds SM, et al. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513:202–9. doi:10.1038/nature13480. [DOI] [PMC free article] [PubMed]
- 28.Mereiter S., Polom K., Williams C., Polonia A., Guergova-Kuras M., Karlsson N. The Thomsen-Friedenreich antigen: A highly sensitive and specific predictor of microsatellite instability in gastric Cancer. J Clin Med. 2018 doi: 10.3390/jcm7090256. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mereiter S, Polom K, Williams C, Polonia A, Guergova-Kuras M, Karlsson N, et al. The Thomsen-Friedenreich Antigen: A Highly Sensitive and Specific Predictor of Microsatellite Instability in Gastric Cancer. J Clin Med 2018. doi:10.3390/jcm7090256. [DOI] [PMC free article] [PubMed]
- 29.Kim M., Kim H.J., Choi B.Y., Kim J.-H., Song K.-S., Noh S.-M. Identification of potential serum biomarkers for gastric cancer by a novel computational method, multiple normal tissues corrected differential analysis. Clin Chim Acta. 2012;413:428–433. doi: 10.1016/j.cca.2011.10.026. [DOI] [PubMed] [Google Scholar]; Kim M, Kim HJ, Choi BY, Kim J-H, Song K-S, Noh S-M, et al. Identification of potential serum biomarkers for gastric cancer by a novel computational method, multiple normal tissues corrected differential analysis. Clin Chim Acta 2012;413:428–33. doi:10.1016/j.cca.2011.10.026. [DOI] [PubMed]
- 30.Murray G.I., Duncan M.E., Arbuckle E., Melvin W.T., Fothergill J.E. Matrix metalloproteinases and their inhibitors in gastric cancer. Gut. 1998;43:791–797. doi: 10.1136/gut.43.6.791. [DOI] [PMC free article] [PubMed] [Google Scholar]; Murray GI, Duncan ME, Arbuckle E, Melvin WT, Fothergill JE. Matrix metalloproteinases and their inhibitors in gastric cancer. Gut 1998;43:791–7. doi:10.1136/gut.43.6.791. [DOI] [PMC free article] [PubMed]
- 31.Xu J., E C., Yao Y., Ren S., Wang G., Jin H. Matrix metalloproteinase expression and molecular interaction network analysis in gastric cancer. Oncol Lett. 2016;12:2403–2408. doi: 10.3892/ol.2016.5013. [DOI] [PMC free article] [PubMed] [Google Scholar]; Xu J, E C, Yao Y, Ren S, Wang G, Jin H. Matrix metalloproteinase expression and molecular interaction network analysis in gastric cancer. Oncol Lett 2016;12:2403–8. doi:10.3892/ol.2016.5013. [DOI] [PMC free article] [PubMed]
- 32.Han S.H., Joo M., Kim H., Chang S. Mesothelin expression in gastric adenocarcinoma and its relation to clinical outcomes. J Pathol Transl Med. 2017;51:122–128. doi: 10.4132/jptm.2016.11.18. [DOI] [PMC free article] [PubMed] [Google Scholar]; Han SH, Joo M, Kim H, Chang S. Mesothelin expression in gastric adenocarcinoma and its relation to clinical outcomes. J Pathol Transl Med 2017;51:122–8. doi:10.4132/jptm.2016.11.18. [DOI] [PMC free article] [PubMed]
- 33.B K., I S., A T., U Y., O H., M M. Mesothelin expression correlates with prolonged patient survival in gastric cancer. J Surg Oncol. 2012;105:195–199. doi: 10.1002/jso.22024. [DOI] [PubMed] [Google Scholar]; K. B, S. I, T. A, Y. U, H. O, M. M, et al. Mesothelin expression correlates with prolonged patient survival in gastric cancer. J Surg Oncol 2012;105:195–9. doi:10.1002/jso.22024. [DOI] [PubMed]
- 34.Ito T., Kajino K., Abe M., Sato K., Maekawa H., Sakurada M. ERC/mesothelin is expressed in human gastric cancer tissues and cell lines. Oncol Rep. 2014;31:27–33. doi: 10.3892/or.2013.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ito T, Kajino K, Abe M, Sato K, Maekawa H, Sakurada M, et al. ERC/mesothelin is expressed in human gastric cancer tissues and cell lines. Oncol Rep 2014;31:27–33. doi:10.3892/or.2013.2803. [DOI] [PMC free article] [PubMed]
- 35.Fidan E., Mentese A., Ozdemir F., Deger O., Kavgaci H., Caner Karahan S. Diagnostic and prognostic significance of CA IX and suPAR in gastric cancer. Med Oncol. 2013;30 doi: 10.1007/s12032-013-0540-9. [DOI] [PubMed] [Google Scholar]; Fidan E, Mentese A, Ozdemir F, Deger O, Kavgaci H, Caner Karahan S, et al. Diagnostic and prognostic significance of CA IX and suPAR in gastric cancer. Med Oncol 2013;30. doi:10.1007/s12032-013-0540-9. [DOI] [PubMed]
- 36.Chen J., Röcken C., Hoffmann J., Krüger S., Lendeckel U., Rocco A. Expression of carbonic anhydrase 9 at the invasion front of gastric cancers. Gut. 2005;54:920–927. doi: 10.1136/gut.2004.047340. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chen J, Röcken C, Hoffmann J, Krüger S, Lendeckel U, Rocco A, et al. Expression of carbonic anhydrase 9 at the invasion front of gastric cancers. Gut 2005;54:920–7. doi:10.1136/gut.2004.047340. [DOI] [PMC free article] [PubMed]
- 37.Leppilampi M., Saarnio J., Karttunen T.J., Kivelä J., Pastoreková S., Pastorek J. Carbonic anhydrase isozymes IX and XII in gastric tumors. World J Gastroenterol. 2003;9:1398–1403. doi: 10.3748/wjg.v9.i7.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]; Leppilampi M, Saarnio J, Karttunen TJ, Kivelä J, Pastoreková S, Pastorek J, et al. Carbonic anhydrase isozymes IX and XII in gastric tumors. World J Gastroenterol 2003;9:1398–403. doi:10.3748/wjg.v9.i7.1398. [DOI] [PMC free article] [PubMed]
- 38.Rajkumar T., Vijayalakshmi N., Gopal G., Sabitha K., Shirley S., Raja U.M. Identification and validation of genes involved in gastric tumorigenesis. Cancer Cell Int. 2010;10 doi: 10.1186/1475-2867-10-45. [DOI] [PMC free article] [PubMed] [Google Scholar]; Rajkumar T, Vijayalakshmi N, Gopal G, Sabitha K, Shirley S, Raja UM, et al. Identification and validation of genes involved in gastric tumorigenesis. Cancer Cell Int 2010;10. doi:10.1186/1475-2867-10-45. [DOI] [PMC free article] [PubMed]
- 39.Yoshida A., Isomoto H., Hisatsune J., Nakayama M., Nakashima Y., Matsushima K. Enhanced expression of CCL20 in human helicobacter pylori-associated gastritis. Clin Immunol. 2009;130:290–297. doi: 10.1016/j.clim.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yoshida A, Isomoto H, Hisatsune J, Nakayama M, Nakashima Y, Matsushima K, et al. Enhanced expression of CCL20 in human Helicobacter pylori-associated gastritis. Clin Immunol 2009;130:290–7. doi:10.1016/j.clim.2008.09.016. [DOI] [PMC free article] [PubMed]
- 40.Raja U.M., Gopal G., Shirley S., Ramakrishnan A.S., Rajkumar T. Immunohistochemical expression and localization of cytokines/chemokines/growth factors in gastric cancer. Cytokine. 2017;89:82–90. doi: 10.1016/j.cyto.2016.08.032. [DOI] [PubMed] [Google Scholar]; Raja UM, Gopal G, Shirley S, Ramakrishnan AS, Rajkumar T. Immunohistochemical expression and localization of cytokines/chemokines/growth factors in gastric cancer. Cytokine 2017;89:82–90. doi:10.1016/j.cyto.2016.08.032. [DOI] [PubMed]
- 41.Lim J.B., Kim D.K., Chung H.W. Clinical significance of serum thymus and activation-regulated chemokine in gastric cancer: potential as a serum biomarker. Cancer Sci. 2014;105:1327–1333. doi: 10.1111/cas.12505. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lim JB, Kim DK, Chung HW. Clinical significance of serum thymus and activation-regulated chemokine in gastric cancer: Potential as a serum biomarker. Cancer Sci 2014;105:1327–33. doi:10.1111/cas.12505. [DOI] [PMC free article] [PubMed]
- 42.Zhong B., Li Y., Liu X., Wang D. Association of mast cell infiltration with gastric cancer progression. Oncol Lett. 2017;15:755–764. doi: 10.3892/ol.2017.7380. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhong B, Li Y, Liu X, Wang D. Association of mast cell infiltration with gastric cancer progression. Oncol Lett 2017;15:755–64. doi:10.3892/ol.2017.7380. [DOI] [PMC free article] [PubMed]
- 43.Moskal T., Huang S., Ellis L.M., Fritsche A., Chakrabarty S. Serum levels of transforming growth factor Cancer in gastrointestinal. Cancer Epidemiol Biomarkers Prev. 1995;4:127–131. [PubMed] [Google Scholar]; Moskal T, Huang S, Ellis LM, Fritsche A, Chakrabarty S. Serum Levels of Transforming Growth Factor Cancer in Gastrointestinal. Cancer Epidemiol Biomarkers Prev 1995;4:127–31. [PubMed]
- 44.Choi J., Chul H., Lim H., Ki D. vol. 749. 1999. Detection of transforming growth factor- · in the serum of gastric carcinoma patients; pp. 236–241. [DOI] [PubMed] [Google Scholar]; Choi J, Chul H, Lim H, Ki D. Detection of Transforming Growth Factor- · in the Serum of Gastric Carcinoma Patients 1999;749:236–41. [DOI] [PubMed]
- 45.Konturek P.C., Konturek S.J., Sulekova Z., Meixner H., Karczewska E., Hahn E.G. Expression of hepatocyte growth factor, transforming growth factor alpha, apoptosis related proteins Bax and Bcl-2, and gastrin in human gastric cancer. Aliment Pharmacol Ther. 2001;15:989–999. doi: 10.1046/j.1365-2036.2001.01003.x. [DOI] [PubMed] [Google Scholar]; Konturek PC, Konturek SJ, Sulekova Z, Meixner H, Karczewska E, Hahn EG, et al. Expression of hepatocyte growth factor, transforming growth factor alpha, apoptosis related proteins Bax and Bcl-2, and gastrin in human gastric cancer Aliment Pharmacol Ther. 2001;15:989–99. [DOI] [PubMed]
- 46.Pham T.M., Fujino Y., Kikuchi S., Tamakoshi A., Yatsuya H., Matsuda S. A nested case-control study of stomach cancer and serum insulin-like growth factor (IGF)-1, IGF-2 and IGF-binding protein (IGFBP)-3. Eur J Cancer. 2007;43:1611–1616. doi: 10.1016/j.ejca.2007.04.014. [DOI] [PubMed] [Google Scholar]; Pham TM, Fujino Y, Kikuchi S, Tamakoshi A, Yatsuya H, Matsuda S, et al. A nested case-control study of stomach cancer and serum insulin-like growth factor (IGF)-1, IGF-2 and IGF-binding protein (IGFBP)-3. Eur J Cancer 2007;43:1611–6. doi:10.1016/j.ejca.2007.04.014. [DOI] [PubMed]
- 47.Gu M.-J., Bae Y.-K., Choi J.H. Clinical significance of insulin-growth factor 1 and insulin-growth factor 1 receptor expression in gastrointestinal stromal tumors. Hepatogastroenterology. 2013;60:1383–1386. [PubMed] [Google Scholar]; Gu M-J, Bae Y-K, Choi JH. Clinical significance of insulin-growth factor 1 and insulin-growth factor 1 receptor expression in gastrointestinal stromal tumors. Hepatogastroenterology 2013;60:1383–6. [PubMed]
- 48.Aung P., Oue N., Mitani Y., Nakayama H., Yoshida K., Noguchi T. Systematic search for gastric cancer-specific genes based on SAGE data: melanoma inhibitory activity and matrix metalloproteinase-10 are novel prognostic factors in patients with gastric cancer. Oncogene. 2006;25:2546–2557. doi: 10.1038/sj.onc.1209279. [DOI] [PubMed] [Google Scholar]; Aung P, Oue N, Mitani Y, Nakayama H, Yoshida K, Noguchi T, et al. Systematic search for gastric cancer-specific genes based on SAGE data: melanoma inhibitory activity and matrix metalloproteinase-10 are novel prognostic factors in patients with gastric cancer. Oncogene 2006;25:2546–57. doi:10.1038/sj.onc.1209279. [DOI] [PubMed]
- 49.Uekita T., Tanaka M., Takigahira M., Miyazawa Y., Nakanishi Y., Kanai Y. CUB-domain-containing protein 1 regulates peritoneal dissemination of gastric scirrhous carcinoma. Am J Pathol. 2008;172:1729–1739. doi: 10.2353/ajpath.2008.070981. [DOI] [PMC free article] [PubMed] [Google Scholar]; Uekita T, Tanaka M, Takigahira M, Miyazawa Y, Nakanishi Y, Kanai Y, et al. CUB-domain-containing protein 1 regulates peritoneal dissemination of gastric scirrhous carcinoma. Am J Pathol 2008;172:1729–39. doi:10.2353/ajpath.2008.070981. [DOI] [PMC free article] [PubMed]
- 50.Ye J., Xu J., Li Y., Huang Q., Huang J., Wang J. DDAH1 mediates gastric cancer cell invasion and metastasis via Wnt/β-catenin signaling pathway. Mol Oncol. 2017;11:1208–1224. doi: 10.1002/1878-0261.12089. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ye J, Xu J, Li Y, Huang Q, Huang J, Wang J, et al. DDAH1 mediates gastric cancer cell invasion and metastasis via Wnt/β-catenin signaling pathway. Mol Oncol 2017;11:1208–24. doi:10.1002/1878-0261.12089. [DOI] [PMC free article] [PubMed]
- 51.Bai Z., Ye Y., Liang B., Xu F., Zhang H., Zhang Y. Proteomics-based identification of a group of apoptosis-related proteins and biomarkers in gastric cancer. Int J Oncol. 2011;38:375–383. doi: 10.3892/ijo.2010.873. [DOI] [PubMed] [Google Scholar]; Bai Z, Ye Y, Liang B, Xu F, Zhang H, Zhang Y, et al. Proteomics-based identification of a group of apoptosis-related proteins and biomarkers in gastric cancer. Int J Oncol 2011;38:375–83. doi:10.3892/ijo.2010.873. [DOI] [PubMed]
- 52.Yin Y., Liu Q., Wang B., Chen G., Xu L., Zhou H. Expression and function of heme oxygenase-1 in human gastric cancer. Exp Biol Med (Maywood) 2012;237:362–371. doi: 10.1258/ebm.2011.011193. [DOI] [PubMed] [Google Scholar]; Yin Y, Liu Q, Wang B, Chen G, Xu L, Zhou H. Expression and function of heme oxygenase-1 in human gastric cancer. Exp Biol Med (Maywood) 2012;237:362–71. doi:10.1258/ebm.2011.011193. [DOI] [PubMed]
- 53.Noh S.J., Kim K.M., Jang K.Y. Individual and co-expression patterns of nerve growth factor and heme oxygenase-1 predict shorter survival of gastric carcinoma patients. Diagn Pathol. 2017;12:1–12. doi: 10.1186/s13000-017-0644-1. [DOI] [PMC free article] [PubMed] [Google Scholar]; Noh SJ, Kim KM, Jang KY. Individual and co-expression patterns of nerve growth factor and heme oxygenase-1 predict shorter survival of gastric carcinoma patients. Diagn Pathol 2017;12:1–12. doi:10.1186/s13000-017-0644-1. [DOI] [PMC free article] [PubMed]
- 54.Uekita T., Sakai R. Roles of CUB domain-containing protein 1 signaling in cancer invasion and metastasis. Cancer Sci. 2011;102:1943–1948. doi: 10.1111/j.1349-7006.2011.02052.x. [DOI] [PubMed] [Google Scholar]; Uekita T, Sakai R. Roles of CUB domain-containing protein 1 signaling in cancer invasion and metastasis. Cancer Sci 2011;102:1943–8. doi:10.1111/j.1349-7006.2011.02052.x. [DOI] [PubMed]
- 55.Cheng S., Luo M., Ding C., Peng C., Lv Z., Tong R. Downregulation of Peptidylprolyl isomerase A promotes cell death and enhances doxorubicin-induced apoptosis in hepatocellular carcinoma. Gene. 2016;591:236–244. doi: 10.1016/j.gene.2016.07.020. [DOI] [PubMed] [Google Scholar]; Cheng S, Luo M, Ding C, Peng C, Lv Z, Tong R. Downregulation of Peptidylprolyl isomerase A promotes cell death and enhances doxorubicin-induced apoptosis in hepatocellular carcinoma. Gene 2016;591:236–44. doi:10.1016/j.gene.2016.07.020. [DOI] [PubMed]
- 56.Ahmad A., Zeenat W., Sajad H., Shabir A., Bhat A., Ahmad M. A review on heme oxygenase-1 induction: is it a necessary evil. Inflamm Res. 2018;67:579–588. doi: 10.1007/s00011-018-1151-x. [DOI] [PubMed] [Google Scholar]; Ahmad A, Zeenat W, Sajad H, Shabir A, Bhat A, Ahmad M. A review on heme oxygenase-1 induction: is it a necessary evil. Inflamm Res 2018;67:579–88. doi:10.1007/s00011-018-1151-x. [DOI] [PubMed]
- 57.Sebastián V.P., Salazar G.A., Coronado-arrázola I., Schultz B.M., Vallejos O.P., Berkowitz L. vol. 9. 2018. Heme Oxygenase-1 as a modulator of intestinal inflammation development and progression; pp. 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sebastián VP, Salazar GA, Coronado-arrázola I, Schultz BM, Vallejos OP, Berkowitz L, et al. Heme Oxygenase-1 as a Modulator of Intestinal Inflammation Development and Progression 2018;9:1–12. doi:10.3389/fimmu.2018.01956. [DOI] [PMC free article] [PubMed]
- 58.Yu F., Tian T., Deng B., Wang T., Qi Q., Zhu M. Multi-marker analysis of genomic annotation on gastric cancer GWAS data from Chinese populations. Gastric Cancer. 2018 doi: 10.1007/s10120-018-0841-y. [DOI] [PubMed] [Google Scholar]; Yu F, Tian T, Deng B, Wang T, Qi Q, Zhu M, et al. Multi-marker analysis of genomic annotation on gastric cancer GWAS data from Chinese populations. Gastric Cancer 2018. doi:10.1007/s10120-018-0841-y. [DOI] [PubMed]
- 59.Wiese K.E., Walz S., Von Eyss B., Wolf E., Athineos D., Sansom O. The role of MIZ-1 in MYC-dependent tumorigenesis. Cold Spring Harb Perspect Med. 2013 doi: 10.1101/cshperspect.a014290. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wiese KE, Walz S, Von Eyss B, Wolf E, Athineos D, Sansom O, et al. The role of MIZ-1 in MYC-dependent tumorigenesis. Cold Spring Harb Perspect Med 2013. doi:10.1101/cshperspect.a014290. [DOI] [PMC free article] [PubMed]
- 60.Li Y., Luo H., Liu T., Zacksenhaus E., Ben-David Y. The ets transcription factor Fli-1 in development, cancer and disease. Oncogene. 2015;34:2022–2031. doi: 10.1038/onc.2014.162. [DOI] [PMC free article] [PubMed] [Google Scholar]; Li Y, Luo H, Liu T, Zacksenhaus E, Ben-David Y. The ets transcription factor Fli-1 in development, cancer and disease. Oncogene 2015;34:2022–31. doi:10.1038/onc.2014.162. [DOI] [PMC free article] [PubMed]
- 61.Kumar S., Rao N., Ge R. Emerging roles of ADAMTSS in angiogenesis and cancer. Cancers (Basel) 2012;4:1252–1299. doi: 10.3390/cancers4041252. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kumar S, Rao N, Ge R. Emerging roles of ADAMTSS in angiogenesis and cancer. Cancers (Basel) 2012;4:1252–99. doi:10.3390/cancers4041252. [DOI] [PMC free article] [PubMed]
- 62.Barthel S.R., Gavino J.D., Descheny L., Dimitroff C.J. Targeting selectins and selectin ligands in inflammation and cancer. Expert Opin Ther Targets. 2007 doi: 10.1517/14728222.11.11.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]; Barthel SR, Gavino JD, Descheny L, Dimitroff CJ. Targeting selectins and selectin ligands in inflammation and cancer. Expert Opin Ther Targets 2007. doi:10.1517/14728222.11.11.1473. [DOI] [PMC free article] [PubMed]
- 63.Zou H., Hastie T. Regularization and variable selection via the elastic-net. J R I State Dent Soc. 2005;67:301–320. [Google Scholar]; Zou H, Hastie T. Regularization and variable selection via the elastic-net. J R Stat Soc 2005;67:301–20. doi:10.1111/j.1467-9868.2005.00503.x.
- 64.Pepe M.S., Etzioni R., Feng Z., Potter J.D., Thompson M.L., Thornquist M. Phases of biomarker development for early detection of cancer. J Natl Cancer Inst. 2001;93:1054–1061. doi: 10.1093/jnci/93.14.1054. [11459866] [DOI] [PubMed] [Google Scholar]; Pepe MS, Etzioni R, Feng Z, Potter JD, Thompson ML, Thornquist M, et al. Phases of biomarker development for early detection of cancer. J Natl Cancer Inst 2001;93:1054–61. doi:11459866. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
Raw data of the multiplex PEA results are available at the GGE data repository (https://ggerepository.isb-sib.ch/) or can be requested from the corresponding author on reasonable request.