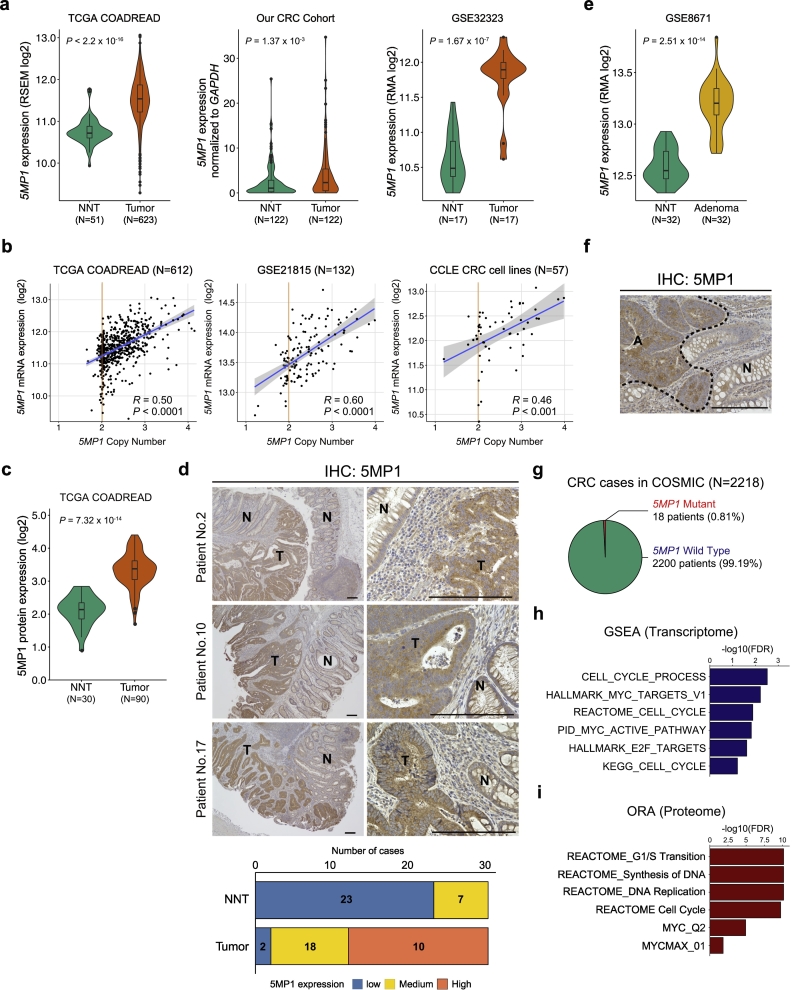
Fig. 1.
A translation initiation regulatory factor 5MP1 on chromosome 7p is amplified in CRC.
(a) Violin plots of 5MP1 mRNA expression levels in CRC tissues and non-neoplastic tissues (NNT) of colorectal mucosa in the TCGA COADREAD dataset (left), our dataset obtained by qRT-PCR (medium), and GSE32323 (right). P represents p-values from two-sided Mann-Whitney U tests.
(b) Correlation between the 5MP1 copy number and the 5MP1 mRNA expression levels in the TCGA COADREAD dataset (left), GSE21815 (medium), and CRC cell lines in Cancer Cell Line Encyclopedia (CCLE) (right). R represents the Pearson correlation coefficient.
(c) Violin plots of 5MP1 protein expression levels in CRC tissues and NNT in the TCGA proteome dataset. P represents p-value from the two-sided Mann-Whitney U test.
(d) Representative images of immunohistochemical staining for 5MP1 in CRC tissues (upper). Proportions of 5MP1 levels in tumor tissues and NNT are shown using a three-stage staining score (lower). T, Tumor; N, NNT; Scale bars, 200 μm.
(e) Violin plots of 5MP1 mRNA expression levels in colorectal adenoma tissues and NNT in GSE8671. P represents p-value from the two-sided Mann-Whitney U test.
(f) Representative image of immunohistochemical staining for 5MP1 in a surgically resected colorectal adenoma tissue. A, Adenoma; N, NNT; Scale bars, 200 μm.
(g) Pie chart of 5MP1 mutation distribution in CRC cases of the COSMIC database (N = 2218).
(h,i) Gene set enrichment analysis (GSEA) of the TCGA COADREAD transcriptome dataset (N = 623) (h) and Overrepresentation enrichment analysis of the TCGA COADREAD proteome dataset (N = 90) (i) showing six significantly enriched gene sets involved in cell cycle progression and c-Myc targets that are positively correlated with 5MP1 expression levels. Threshold of False Discovery Rate (FDR) is <0.25 and < 0.05, respectively.