Abstract
Background
The mixed-cultivation of different Panax ginseng cultivars can cause adverse effects on stability of yield and quality. K-1 is a superior cultivar with good root shape and stronger disease resistance. DNA markers mined from functional genes are clearly desirable for K-1, as they may associate with major traits and can be used for marker-assisted selection to maintain the high quality of Korean ginseng.
Methods
Five genes encoding pathogenesis-related (PR) proteins of P. ginseng were amplified and compared for polymorphism mining. Primary, secondary, and tertiary structures of PR5 protein were analyzed by ExPASy-ProtParam, PSSpred, and I-TASSER methods, respectively. A coding single nucleotide polymorphism (SNP)–based specific primer was designed for K-1 by introducing a destabilizing mismatch within the 3′ end. Allele-specific polymerase chain reaction (PCR) and real-time allele-specific PCR assays were conducted for molecular discrimination of K-1 from other cultivars and landraces.
Results
A coding SNP leading to the modification of amino acid residue from aspartic acid to asparagine was exploited in PR5 gene of K-1 cultivar. Bioinformatics analysis showed that the modification of amino acid residue changed the secondary and tertiary structures of the PR5 protein. Primer KSR was designed for specific discrimination of K-1 from other ginseng cultivars and landraces. The developed real-time allele-specific PCR assay enabled easier automation and accurate genotyping of K-1 from a large number of ginseng samples.
Conclusion
The SNP marker and the developed real-time allele-specific PCR assay will be useful not only for marker-assisted selection of K-1 cultivar but also for quality control in breeding and seed programs of P. ginseng.
Keywords: Allele-specific polymerase chain reaction, K-1 cultivar, Panax ginseng, Pathogenesis-related protein, Single nucleotide polymorphism
1. Introduction
Panax ginseng Meyer, which belongs to the Panax genus in the Araliaceae family, is one of the most valuable medicinal plants in East Asia. Ginseng root has been used as a tonic to relieve fatigue and stress in a number of herbal remedies for more than 2000 years [1]. Modern science has demonstrated that P. ginseng has a variety of pharmacological effects, including antitumor, antistress, antihyperlipidemi, antidiabetic, antiamnestic, radioprotection, cardiovascular protection, and immunization-enhancing activities [2], [3], [4], [5], [6]. Such pharmacological effects have been found to be principally attributed to ginsenosides. So far, more than 40 naturally occurring ginsenosides have been isolated from ginseng roots, leaves, fruits, and flower buds [7]. Moreover, P. ginseng contains other phytochemical compounds such as polysaccharides, peptides, polyacetylenic alcohols, and fatty acids [6]. For these reasons, ginseng preparations have become the best-selling herbal remedies and functional foods worldwide.
With the growing consumer demand and nearly extinction of wild ginseng, cultivated ginseng has become the chief source of ginseng medicinal materials and products. Korea has been dominant in the cultivation and breeding of P. ginseng. After years of trial and efforts, a number of cultivars with good quality and high yield have been developed and registered in the Korea Seed &Variety Service. A wide range of diversity for morphological and physiological traits exist among different ginseng cultivars, such as stem and berry color, leaf type, root shape, seed yield, content of ginsenosides, disease resistance, and so on [8], [9], [10]. Among these, K-1 is a superior cultivar which has been cultivated from 1984. K-1 has good root shape and thus is suitable for producing red ginseng. Besides, it has productive lateral roots and stronger disease resistance. Nowadays, K-1 cultivar is widely cultivated in many ginseng planting areas such as Buyeo, Gochang, and Chuncheon. Different ginseng cultivars including K-1, however, are frequently mixed-cultivated in ginseng fields due to the lack of a proper cultivar authentication method. Therefore, ginseng products made from different cultivars and mixed seeds are sold in the market, causing infringement of breeder's intellectual property rights and serious effects on stability of yield and quality of ginseng.
The development of DNA molecular markers is one of the most efficient methods for marker-aided selection and cultivar authentication of P. ginseng. In recent years, there have been several efforts for development of DNA markers using random-amplified polymorphic DNA (RAPD) [11], inter simple sequence repeat (ISSR) [12], polymerase chain reaction (PCR)–restriction fragment length polymorphism (RFLP) [13], cleaved amplified polymorphic sequence (CAPS) [14], simple sequence repeat (SSR) [15], and single nucleotide polymorphism (SNP) techniques [16], [17], to maintain the high quality of ginseng cultivars. However, DNA markers exploited in noncoding regions can only be beneficial for cultivar authentication. In contrast, DNA markers mined from functional genes are particularly meaningful because they may associate with major traits of economic value and can be used for marker-assisted selection. Pathogenesis-related (PR) proteins are known to function in higher plants against abiotic and biotic stress, and they are postulated to play roles in prevention of pathogen invasion [18]. In this study, five genes encoding PR proteins of P. ginseng were targeted for polymorphism mining. Finally, a real-time allele-specific PCR assay was developed for high throughput screening of K-1 cultivar from a large number of ginseng samples, based on the coding SNP marker exploited from pathogenesis-related protein 5 (PR5) gene.
2. Materials and methods
2.1. Plant materials, DNA extraction, and first strand cDNA synthesis
Fresh leaves of 14 widely cultivated ginseng cultivars and landraces were collected from fields of different locations in Korea and China (Table 1). Korean ginseng samples were morphologically identified by ginseng taxonomist, Professor Woo-Saeng Kwon. Chinese ginseng landraces were provided by the Institute of Special Animal and Plant Sciences of Chinese Academy of Agricultural Sciences. Voucher specimens of 14 ginseng cultivars and landraces were deposited in the Ginseng Bank, Kyung Hee University. Ginseng leaves were frozen in liquid nitrogen and ground into a fine powder with a mortar and pestle. A genomic DNA purification kit (Exgene Plant SV, GeneAll) was used for DNA extraction of 14 ginseng cultivars and landraces. Total RNA was isolated using an RNeasy mini kit, and first strand cDNA was synthesized from total RNA using a RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher), according to the manufacturer's protocols.
Table 1.
Ginseng plant materials used in this study
| Cultivar or landrace | Voucher | Location | Number of individuals | Accession number of PR51) |
|---|---|---|---|---|
| Chunpoong | GB001 | Gochang, Korea | 10 | MF280378 |
| Yunpoong | GB002 | Gochang, Korea | 10 | MF280374 |
| Gopoong | GB003 | Chuncheon, Korea | 10 | MF280375 |
| Sunpoong | GB004 | Gochang, Korea | 10 | MF280376 |
| Gumpoong | GB005 | Gochang, Korea | 10 | MF280377 |
| K-1 | GBD201 | Gochang, Buyeo, Chuncheon, Korea | 25 | MF280379 |
| Sunwon | GBD048 | Daejeon, Korea | 10 | MF280383 |
| Sunun | GBD043 | Daejeon, Korea | 10 | MF280384 |
| Sunhyang | GBD058 | Daejeon, Korea | 10 | MF280385 |
| Chungsun | GBD073 | Daejeon, Korea | 10 | MF280386 |
| Damaya | GB090 | Jilin, China | 10 | MF280380 |
| Ermaya | GB091 | Jilin, China | 10 | |
| Biantiao | GB092 | Jilin, China | 10 | |
| Huangguo | GB093 | Jilin, China | 10 |
PR5, pathogenesis-related protein 5
The one accession number represents the consensus sequence from the combined individual samples for different cultivar or landraces.
2.2. PCR amplification of five PR genes
Oligonucleotide primers used for amplification of five PR genes were shown in Table 2. PCR was performed in a 20 μL volume mixture consisting of 0.5μM of each primer, 20 ng of cDNA, and 10 μL of 2X EF-Taq PCR PreMix (SolGent, Korea). PCR profiles used were as follows: 1 cycle of 4 min at 94°C, 35 cycles of 30 s at 94°C, 30 s at 58°C, 1 min at 72°C, and a final 7 min extension at 72°C. PCR fragments were analyzed by separation in 1.0% agarose gels containing 0.5% ethidium bromide solution and visualized under a UV lamp.
Table 2.
PR genes and primers used in this study
| PR genes | Primers used for PCR (5′→3′) | Accession numbers |
|---|---|---|
| PR2 (beta-glucanase) | F-CCCCGAACCTTCAATCCCTT R-ACTTTGACTGCTGACTCGGT |
DQ015705 |
| PR3 (chitinase) | F-TTCAATCATCACCAAGCAAAG R-TCCGCAACTTCTACAGTCAA |
FJ790420 |
| PR5 (thaumatin-like protein) | F-GCAGCCACTTTTGACATCCT R-ATCTCTTAAAGGAATATCTCCGA |
GQ452234 |
| PR6 (protease inhibitor) | F-TCTGCGTTTGGGCTCATTAC R- AGCAACCCCATAGCTAGACTT |
GU001147 |
| PR10 (ribonuclease) | F-GCTGTTCAAGGGTTCTTTCC R-GGAATTACGGCATCACCTAT |
GU086324 |
PCR, polymerase chain reaction; PR, pathogenesis-related
2.3. Sequencing and multiple sequence alignment
PCR products of five genes amplified from 14 ginseng cultivars and landraces were purified using a GenoAid PCR Combo Kit (Genotech, Korea), following the manufacturer's instructions. Sequencing was performed in both directions by Sanger's method on an ABI PRISM 3700 DNA sequencer (Applied Biosystems, USA), by using a BigDye Terminator Cycle Sequencing Kit (Applied Biosystems, USA) with the same primers used in PCR. DNA sequences of both directions were assembled with SeqMan software (DNASTAR, USA) to construct a unique sequence for each cultivar or landrace. Multiple sequence alignments were performed using Clustal Omega program [19].
2.4. Bioinformatics analysis
The open reading frame (ORF) of PR5 gene was analyzed by ORF finder in National Center for Biotechnology Information (NCBI). Protein primary structure and properties were analyzed by online ExPASy-ProtParam tool (http://web.expasy.org/protparam/), and secondary structure was predicted by PSSpred online program [20]. I-TASSER method was used for comparative modeling of tertiary structures [21], and CHIMERA program was used for visualization of the three-dimensional structures of PR5 protein.
2.5. Allele-specific PCR
Allele-specific primer KSR was designed for molecular authentication of K-1 cultivar, based on the K-1–specific SNP site exploited from PR5 gene. A destabilizing mismatch (substitution of C for T) was introduced within the second base from the 3′ end, to improve PCR specificity and ensure reliable discrimination for K-1. Molecular authentication of K-1 cultivar was performed by multiplex PCR. A total volume of 20 μL-PCR reaction mixture contained 20 ng of genomic DNA, 0.5μM of PR5F, 0.5μM of PR5R, 0.25μM of KSR, and 10 μL of 2X EF-Taq PCR PreMix (SolGent, Korea). PCR was conducted with the same cycling parameters as mentioned in 2.2.
2.6. Real-time PCR assay for K-1 cultivar
To facilitate rapid identification of K-1 cultivar, a real-time allele-specific PCR assay was conducted on a CFX Connect machine (Bio-Rad, USA). The reaction mixtures consisted of 20 ng genomic DNA, 5μM of primer PR5F, 5μM of primer KSR, and 5 μL of RbTaq qPCR 2X PreMIX (SYBR Green) (Enzynomics, Korea). Chunpoong and H2O were used as negative control and no template control, respectively. The reactions were initiated at 95°C for 5 min, followed by 33 cycles of 15 s at 95°C for denaturation, 20 s at 58°C for annealing, and 20 s at 72°C for extension. The melting step was performed with an increase from 65 to 95°C, rising by 1°C at each step. Endpoint method was used for analyzing of different ginseng samples.
3. Results and discussion
The demand of molecular makers for application in marker-assisted breeding and cultivar discrimination of P. ginseng has greatly increased in recent years. A series of diverse molecular markers including RAPD, PCR-RFLP, sequence-characterized amplified region (SCAR), CAPS, SSR, and SNP have been developed and applied for diversity analysis and genetic identification of ginseng cultivars. Owing to the differences in their principles, each molecular marker technique has its limitations in cultivar discrimination. RAPD and ISSR are difficult to ensure reproducibility due to their susceptibility to experimental conditions; RFLP and CAPS require restriction enzyme digestion of PCR products. SSR markers have proved to be effective for discrimination of P. ginseng cultivars, but a time-consuming silver-stained polyacrylamide gel electrophoresis is usually needed in visualization step because of the small size differences among PCR products amplified from these cultivars [22]. With the development of next generation sequencing technology, a large number of genomic and transcriptomic sequences are available in public databases [23], [24]. SNP markers thus can be identified through comparative sequencing of different cultivars. However, in silico mining of sequences in public databases may result in the discovery of a large amount of heterozygous SNPs and nonallelic SNPs representing paralogous sequences, which are suboptimal for application in marker-assisted breeding. In contrast, SNP mining based on published functional genes is relatively straight forward and meaningful because coding SNP may involve in regulating the activity of the expressed gene product and associate with major traits of economic value. In this study, five genes encoding PR proteins of 10 Korean ginseng cultivars and 4 Chinese ginseng landraces were targeted for polymorphism mining.
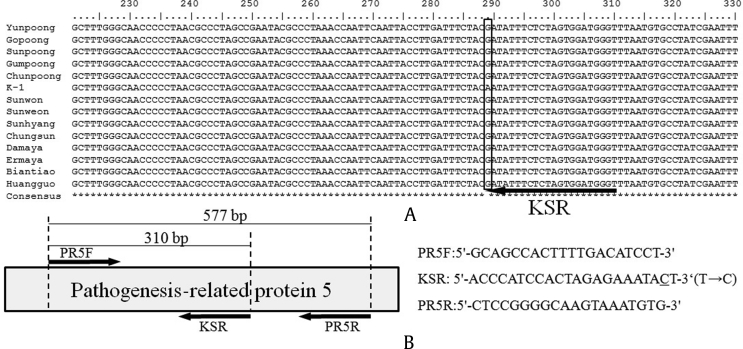
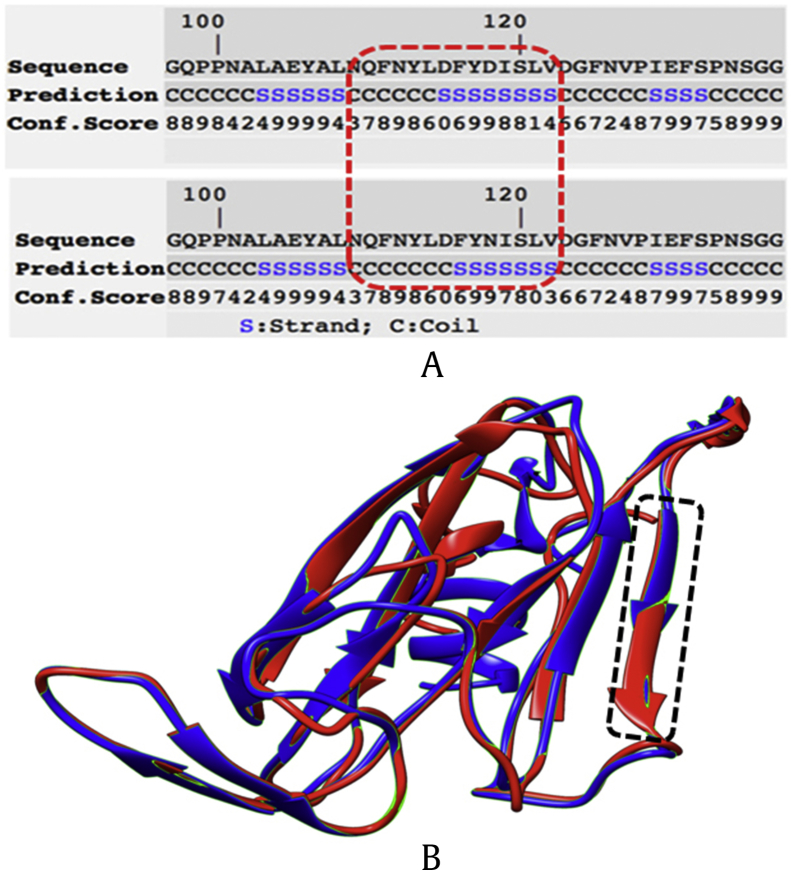
The PCR products amplified from 14 ginseng cultivars and landraces were sequenced and determined to be 577 bp, and the sequences of PR5 gene were registered in GenBank with accession numbers MF280374-MF280386 (Table 1). ORF analysis showed that a 137 AA length of protein is generated from the cDNA sequence of PR5 gene. Multiple sequence alignment result showed that the sequences from 14 cultivars and landraces were almost identical, except that a SNP site specific to K-1 was exploited. As shown in Fig. 1, 13 ginseng cultivars and landraces contain nucleotide G at 289th position, but a base substitution of A for G was observed in K-1 cultivar. This coding SNP is a nonsynonymous substitution leading to the modification of amino acid residue from aspartic acid (D) to asparagine (N). Expasy-Protparam analysis showed that the modification of amino acid residue increased the instability index and isoelectric point of PR5 of K-1. The base substitution also changed the secondary structure including the length of coil and strand, and further changed the tertiary structure of the PR5 protein of K-1 (Fig. 2). Owing to the differences of strand and coil, the protein could play a different role, which may in turn affect the disease resistance of K-1 cultivar.
Fig. 1.
Schematic diagram of positions of SNP and primers used in this study. (A) The locations of SNP and primer KSR. (B) Positions and sequences of primers used in this study. The underlined nucleotide is the destabilizing mismatch introduced intentionally.
SNP, single nucleotide polymorphism.
Fig. 2.
Bioinformatics analysis of the secondary and tertiary structure of PR5. (A) The modification of amino acid residue from D to N changed the secondary structure including the length of coil and strand. (B) The structural changes caused by the target mutation were shown in dotted box.
PR5, pathogenesis-related protein 5.
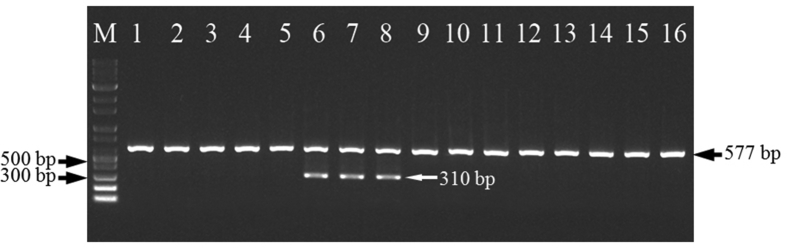
Based on this SNP site, primer KSR was designed for specific discrimination of K-1 from other ginseng cultivars and landraces. The substitution of C for T locating at the second base from the 3′ terminal was the additional mismatch incorporated intentionally (Fig. 1), to achieve absolute allelic specificity. The K-1–specific primer KSR, together with primer pairs PR5F and PR5R, were used in multiplex PCR for molecular identification of K-1 cultivar. The DNAs from K-1 and other ginseng samples, as expected, generated different fragment patterns when they were cross-challenged against the combination of three primers. As shown in Fig. 3, all the ginseng samples yielded the 577 bp-amplicons with primer PR5F and PR5R, which can be used as a positive control showing that PCR process was not problematic. However, only K-1 produced the specific band of 310 bp representing the A allele, which was amplified by PR5F and the specific primer KSR. This indicated that the extra mismatch introduced in primer KSR had a minor effect on the amplification of the specific A allele but produced a dramatic reduction of the nonspecific G allele. The experiment was repeated several times with ginseng samples collected from different locations, and the accuracy and reproducibility of the developed multiplex PCR was further confirmed. We are, therefore, able to draw a conclusion that the SNP marker is effective for specific discrimination of K-1 from other cultivars.
Fig. 3.
Allele-specific PCR profiles of different ginseng cultivars and landraces. Lane M: 1000 bp DNA ladder; lane 1–5: Yunpoong, Gopoong, Sunpoong, Gumpoong, Chunpoong, respectively; lane 6–8: K-1; lane 9–16: Sunwon, Sunun, Sunhyang, Chungsun, Damaya, Ermaya, Biantiao, Huangguo, respectively. Lane 6–8 generated the K-1–specific amplicons of 310 bp.
PCR, polymerase chain reaction.
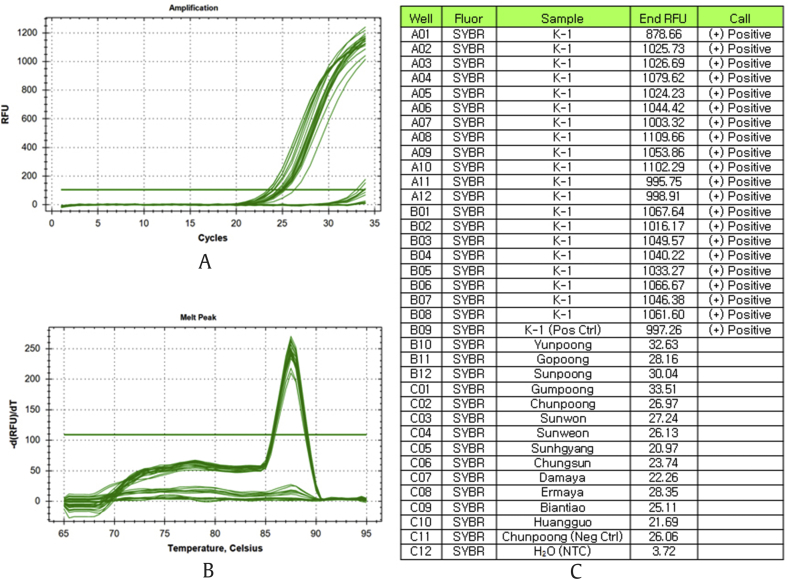
SNP genotyping with allele-specific PCR only needs an agarose gel–based analysis and thus is accessible to minimally equipped molecular biology laboratories. However, the requirement of time-consuming gel-based assays limits its utility in screening a large numbers of samples. To overtake this deficiency, a real-time allele-specific PCR assay with SYBR Green I fluorescent dye was developed. In real-time allele-specific PCR assay, the presence or absence of a detected fluorescent signal indicates whether the specific allele is present in DNA template. The amplification profile, melt peak, and endpoint analysis result were shown in Fig. 4. From the endpoint analysis, we can see that there was no rise in fluorescent signal for cultivars and landraces except K-1, due to the failure in duplex formation which resulted from the mismatches between the allele-specific primer KSR and the binding site in the template DNA. Therefore, K-1 samples could be easily discriminated from other ginseng cultivars and landraces by their positive allele calling signals. The developed assay was tested with 200 K-1 individuals collected from different locations and could identify K-1 with perfect accuracy up to 100 percent. Compared with other nongel–based assays such as TaqMan assay and molecular beacons, the developed real-time allele-specific PCR requires only a plus/minus assay, thus permitting easier automation and accurate genotyping of K-1 cultivar from a large number of ginseng samples.
Fig. 4.
Real-time allele-specific PCR assay of different ginseng cultivars and landraces. (A) Amplification profile. (B) Melt peak. (C) Endpoint analysis. K-1 could be easily discriminated from other cultivars by their positive allele calling signals.
PCR, polymerase chain reaction; RFU, relative fluorescence units.
SNPs represent both natural genetic variation and genetic drift created by breeders during the process of crop improvement and thus have a particularly important application in marker-assisted selection of ginseng cultivars. Dong et al and Chen et al screened a series of disease-resistance–related SNP markers by restriction-site associated DNA sequencing and bred a disease-resistant P. notoginseng cultivar “Miaoxiang Kangqi 1” [25], [26]. Disease-resistance evaluation results showed that SNP could serve as DNA markers for selection of disease-resistant cultivars and accelerate the breeding of new cultivars. In this study, a coding SNP specific to K-1 cultivar was exploited from PR5 gene. Bioinformatics analysis showed that this mutation site might cause the structural changes of PR5 protein, which could in turn affect the disease resistance. Because the biological function of PR5 protein was not clear, the association between this SNP and disease resistance should be further confirmed. Although this coding SNP may or may not be helpful markedly in ginseng resistance breeding, it will show association with the trait of K-1. Therefore, this SNP marker, as well as the developed real-time allele-specific PCR assay, will be useful not only for correct labeling and marker-assisted selection of K-1 cultivar but also for quality control in breeding and seed programs of P. ginseng.
Conflicts of interest
All contributing authors declare no conflicts of interest.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (81503178) and the Korea Institute of Planning & Evaluation for Technology in Food, Agriculture, Forestry & Fisheries (KIPET NO: 317007-3).
References
- 1.Cho J., Han Y., Oh H., Park H., Sung H., Park J. The Society for Korean Ginseng; Seoul: 1995. Understanding of Korean ginseng: Korean ginseng contains various effective components. [Google Scholar]
- 2.Jia L., Zhao Y., Liang X.J. Current evaluation of the millennium phytomedicine- ginseng (II): Collected chemical entities, modern pharmacology, and clinical applications emanated from traditional Chinese medicine. Curr Med Chem. 2009;16:2924–2942. doi: 10.2174/092986709788803204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shin H.R., Kim J.Y., Yun T.K., Morgan G., Vainio H. The cancer-preventive potential of Panax ginseng: a review of human and experimental evidence. Canc Causes Contr. 2000;11:565–576. doi: 10.1023/a:1008980200583. [DOI] [PubMed] [Google Scholar]
- 4.Kim S.H., Park K.S. Effects of Panax ginseng extract on lipid metabolism in humans. Pharmacol Res. 2003;48:511–513. doi: 10.1016/s1043-6618(03)00189-0. [DOI] [PubMed] [Google Scholar]
- 5.Cheng Y., Shen L.H., Zhang J.T. Anti-amnestic and anti-aging effects of ginsenoside Rg1 and Rb1 and its mechanism of action. Acta Pharmacol Sin. 2005;26:143–149. doi: 10.1111/j.1745-7254.2005.00034.x. [DOI] [PubMed] [Google Scholar]
- 6.Choi K.T. Botanical characteristics, pharmacological effects and medicinal components of Korean Panax ginseng C A Meyer. Acta Pharmacol Sin. 2008;29:1109–1118. doi: 10.1111/j.1745-7254.2008.00869.x. [DOI] [PubMed] [Google Scholar]
- 7.Shin B.K., Kwon S.W., Park J.H. Chemical diversity of ginseng saponins from Panax ginseng. J Ginseng Res. 2015;39:287–298. doi: 10.1016/j.jgr.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kwon W.S., Lee M.G., Lee J.H. Characteristics of flowering and fruiting in new varieties and lines of Panax ginseng C.A. Meyer. J Ginseng Res. 2001;25:41–44. [Google Scholar]
- 9.Ahn I.O., Lee S.S., Lee J.H., Lee M.J., Jo B.G. Comparison of ginsenoside contents and pattern similarity between root parts of new cultivars in Panax ginseng C.A. Meyer. J Ginseng Res. 2008;32:15–18. [Google Scholar]
- 10.Lee S.S., Lee J.H., Ahn I.O. Vol. 18. 2005. Characteristics of new cultivars in Panax ginseng C.A. Meyer; pp. 3–18. (Proceedings of the Ginseng Society Conference). [Google Scholar]
- 11.In D.S., Kim Y.C., Bang K.H., Chung J.W., Kim O.T., Hyun D.Y., Cha S.W., Kim T.S., Seong N.S. Genetic relationships of Panax species by RAPD and ISSR analyses. Kor J Med Crop Sci. 2005;13:249–253. [Google Scholar]
- 12.Bang K.H., Lee S.W., Hyun D.Y., Cho J.H., Cha S.W., Seong N.S., Huh M.K. Molecular authentication and genetic polymorphism of Korean ginseng (Panax ginseng C.A. Meyer) by inter-simple sequence repeats (ISSRs) markers. J Life Sci. 2004;14:425–428. [Google Scholar]
- 13.Kim O.T., Bang K.H., In D.S., Lee J.W., Kim Y.C., Shin Y.S., Hyun D.Y., Lee S.S., Cha S.W., Seong N.S. Molecular authentication of ginseng cultivars by comparison of internal transcribed spacer and 5.8 S rDNA sequences. Plant Biotechnol Rep. 2007;1:163–167. [Google Scholar]
- 14.Lee J.W., Bang K.H., Kim Y.C., Seo A.Y., Jo I.H., Lee J.H., Kim O.T., Hyun D.Y., Cha S.W., Cho J.H. CAPS markers using mitochondrial consensus primers for molecular identification of Panax species and Korean ginseng cultivars (Panax ginseng C. A. Meyer) Mol Biol Rep. 2012;39:729–736. doi: 10.1007/s11033-011-0792-4. [DOI] [PubMed] [Google Scholar]
- 15.Kim N.H., Choi H.I., Ahn I.O., Yang T.J. EST-SSR marker sets for practical authentication of all nine registered ginseng cultivars in Korea. J Ginseng Res. 2012;36:298–307. doi: 10.5142/jgr.2012.36.3.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang H., Li G., Kwon W.S., Yang D.C. Development of EST intron-targeting SNP markers for Panax ginseng and their application to cultivar authentication. Int J Mol Sci. 2016;17:884. doi: 10.3390/ijms17060884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim K., Lee S.C., Lee J., Lee H.O., Joh H.J., Kim N.H., Park H.S., Yang T.J. Comprehensive survey of genetic diversity in chloroplast genomes and 45S nrDNAs within Panax ginseng species. PLoS One. 2015;10 doi: 10.1371/journal.pone.0117159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee O.R., Gayathri S., Kim J.Y., In J.G., Kwon W.S., Kim J.H., Yang D.C. Defense genes induced by pathogens and abiotic stresses in Panax ginseng C.A. Meyer. J Ginseng Res. 2011;35:1–11. [Google Scholar]
- 19.Sievers F., Higgins D.G. Clustal Omega, accurate alignment of very large numbers of sequences. Methods Mol Biol. 2014;1079:105–116. doi: 10.1007/978-1-62703-646-7_6. [DOI] [PubMed] [Google Scholar]
- 20.Yan R., Xu D., Yang J., Walker S., Zhang Y. A comparative assessment and analysis of 20 representative sequence alignment methods for protein structure prediction. Sci Rep. 2013;3:2619. doi: 10.1038/srep02619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang J., Yan R., Roy A., Xu D., Poisson J., Zhang Y. The I-TASSER Suite: Protein structure and function prediction. Nat Meth. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jo I.H., Kim Y.C., Kim D.H., Kim K.H., Hyun T.K., Ryu H., Bang K.H. Applications of molecular markers in the discrimination of Panax species and Korean ginseng cultivars (Panax ginseng) J Ginseng Res. 2017;41:444–449. doi: 10.1016/j.jgr.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jayakodi M., Lee S.C., Lee Y.S., Park H.S., Kim N.H., Jang W., Lee H.O., Joh H.J., Yang T.J. Comprehensive analysis of Panax ginseng root transcriptomes. BMC Plant Biol. 2015;15:138. doi: 10.1186/s12870-015-0527-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu J., Chu Y., Liao B., Xiao S., Yin Q., Bai R., Dong L., Li X., Qian J., Zhang J. Panax ginseng genome examination for ginsenoside biosynthesis. GigaScience. 2017;6:1–15. doi: 10.1093/gigascience/gix093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dong L., Chen Z., Wang Y., Wei F., Zhang L., Xu J., Wei G., Wang R., Yang J., Liu W. DNA marker-assisted selection of medicinal plants (I). Breeding research of disease-resistant cultivars of Panax notoginseng. China J Chin Mater Med. 2017;42:56–62. doi: 10.19540/j.cnki.cjcmm.2017.0004. [DOI] [PubMed] [Google Scholar]
- 26.Chen Z., Ma X., Dong L., Zhang L., Wei G., Xiao L., Wang Y., Wei F., Liu W., Yu Y. DNA marker-assisted selection of medicinal plants (III). Evaluation of disease resistance of “Miaoxiang Kangqi 1”—a new cultivar of Panax notoginseng. China J Chin Mater Med. 2017;42:2046–2051. doi: 10.19540/j.cnki.cjcmm.20170519.001. [DOI] [PubMed] [Google Scholar]