Abstract
The Notch signaling pathway is tightly controlled via post‐transcriptional regulatory mechanisms that promote or terminate pathway activity. In this issue, Carrieri et al [1] show that phosphorylation of the Notch intracellular domain (NICD) by cyclin‐dependent kinases (CDKs) suppresses Notch activity by promoting NICD turnover. These findings link Notch pathway activity to the cell cycle, and the authors propose connections between this regulation and the segmentation clock that times embryonic somitogenesis.
Subject Categories: Development & Differentiation; Post-translational Modifications, Proteolysis & Proteomics; Signal Transduction
Signaling through the Notch pathway regulates a wide variety of developmental decisions across numerous metazoans. The pathway appears elegant in its simplicity, with ligand:receptor interactions promoting NICD release, which acts as a transcription factor to promote expression of Notch target genes (Fig 1A). In spite of a lack of signaling cascades or amplification, the pathway is utilized in a variety of dosage‐sensitive developmental decisions, and pathway dysregulation is a major contributor to human disease and cancer 2. Notch signaling is frequently utilized in systems where neighboring cells adopt distinct fates, including lateral inhibition (e.g., in Drosophila neurogenesis) or boundary formation (e.g., in the vertebrate hindbrain), but the pathway also has crucial roles in stem cell maintenance via asymmetric cell division and is important in several contexts where tight temporal and spatial regulation is critical 3. Perhaps one of the most dramatic of these is somitogenesis in vertebrate embryos, where cells in the presomitic mesoderm (PSM) exhibit synchronized oscillatory expression of genes linked to a “segmentation clock” that has been proposed to time somite production 4. The oscillatory clock period influences the rate of somite formation, and the Notch pathway plays roles in maintaining synchrony in neighboring cells and in promoting robust oscillations in some species. Ties between the level of Notch activity, the stability of Notch pathway components, and the speed of segmentation clock oscillations have been suggested 4.
Figure 1. CDK1/2‐dependent phosphorylation of NICD links the cell cycle and the segmentation clock.
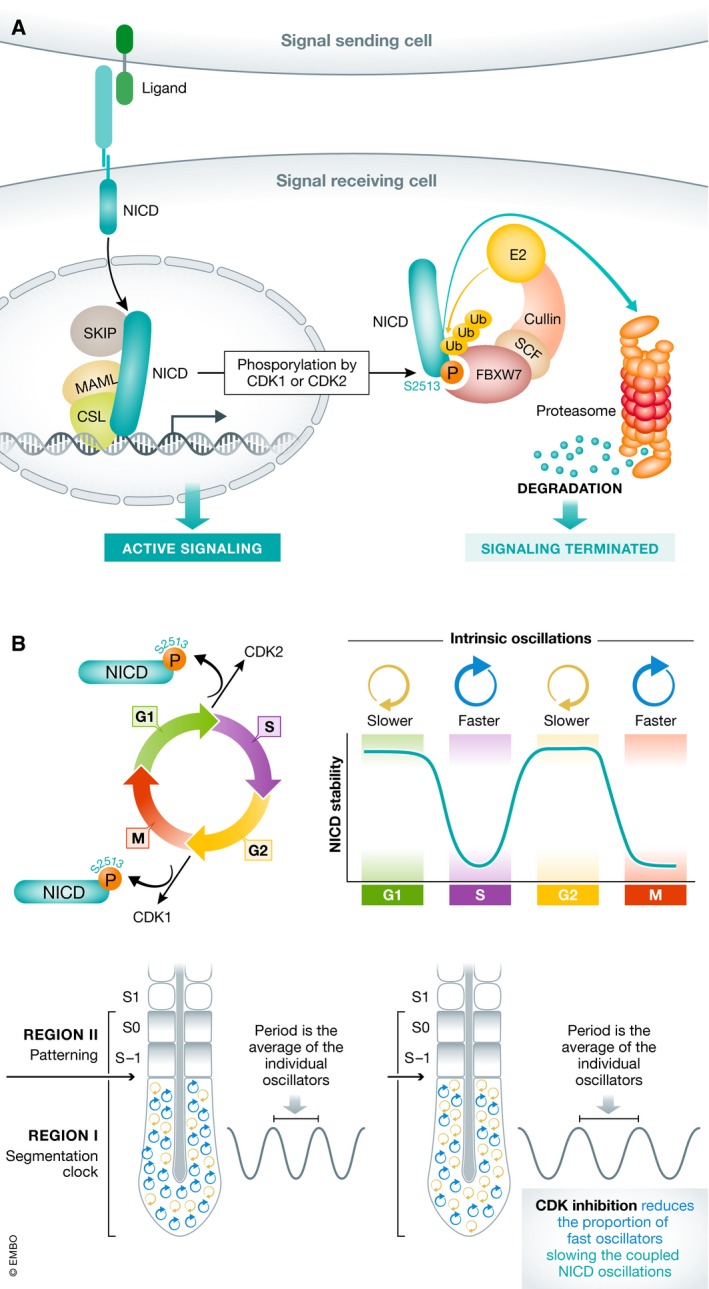
(A) Binding of a ligand on a signal‐sending cell to a receptor on a signal‐receiving cell releases NICD to enter the nucleus of the signal‐receiving cell, bind DNA, and promote active signaling. Phosphorylation of S2513 promotes FBXW7 binding and promotes termination of signal by targeting NICD for SCF E3 ubiquitin ligase‐dependent proteasomal degradation. (B) Peaks of CDK1 or CDK2 activity alter NICD stability in a cell cycle‐dependent manner. If NICD stability influences the intrinsic period of the cell autonomous oscillator, then the cell cycle position of cells in the PSM influences the period of synchronized segmentation clock oscillations. Treatments that block CDK1 activity in the PSM are predicted to reduce the proportion of fast oscillators, slowing the tissue‐level oscillations.
The outcomes of Notch signaling are influenced by both cellular and developmental contexts, and the pathway has pleiotropic effects that require cells to regulate Notch ligands and receptors at all levels. Modes of regulation include the use of protein modifications to fine‐tune ligand:receptor interactions, controlled protein trafficking to influence ligand/receptor presentation, and nuclear interactions that influence signaling outcomes. In the absence of pathway amplification, the duration of the Notch signal is strongly influenced by numerous post‐translational modifications that affect NICD stability 2.
Building on findings from previous work 5, in this issue, Carrieri et al focus on the final steps that shape Notch activity: Signal termination triggered by ubiquitination of the PEST domain by SCF E3 ubiquitin ligase, targeting NICD for proteasomal degradation. In many contexts, recognition of NICD by this complex is mediated by FBXW7, a member of the F‐box protein family that binds phosphorylated NICD 6. The importance of this coordinated signal termination is reflected in numerous cancers where aberrant Notch signaling contributes to disease progression 2. Mutations within the PEST domain of NICD have been observed in many cancers (notably T‐cell acute lymphoblastic leukemia or T‐ALL), most of which result in sustained Notch signaling due to increases in NICD stability. Some T‐ALL lines that are resistant to Notch inhibitors that block NICD production have been identified, and many of these are found to lack Notch1 mutations. Instead, they exhibit inactivating mutations to FBXW7, highlighting the importance of this regulated turnover mechanism 6.
Previous work from several laboratories indicated that phosphorylation of NICD enhances FBXW7 binding, providing a mechanism to fine‐tune NICD half‐life 6. Experiments using small molecule inhibitors demonstrated that inhibition of some CDKs could stabilize NICD, resulting in increased NICD levels and half‐life, and slowing the speed of oscillations in the segmentation clock 5. Here, Carrieri et al utilize a combination of cell culture and in vivo analyses to probe the mechanisms behind these observations. The use of more specific small molecule inhibitors, along with siRNA and genetic perturbations, demonstrates that both CDK1 and CDK2 can phosphorylate NICD and that interference with this modification raises steady‐state NICD levels and increases its half‐life (Fig 1A). Mass spectroscopy analysis identifies a key residue targeted by CDK1 or CDK2 (S2513). Critically, the authors are able to detect interactions between endogenous NICD and FBXW7 and demonstrate that mutations preventing phosphorylation at S2513 block this interaction, suggesting that cell cycle‐specific phosphorylation of NICD could alter Notch pathway activity dependent on the cell cycle phase of the signal‐receiving cell. Supporting this idea, the authors demonstrate that NICD levels vary across the cell cycle, decreasing at G1/S and G2/M when CDK1 and CDK2 are most active (Fig 1B). These findings have implications for diseases associated with increased cell divisions, such as cancer, where Notch signaling is often dysregulated.
Finally, and critically, the authors tie their findings back to temporal regulation of the segmentation clock, demonstrating that NICD levels in the PSM are increased and oscillations are slowed when embryos are treated with specific inhibitors of CDK1 or CDK2, resulting in embryos that form fewer, but larger, somites. This finding is examined in a mathematical model, where cells in the PSM are assumed to act as phase‐coupled oscillators. When cell‐to‐cell coupling is strong, this model gives rise to synchronous oscillations with a period equal to the average of the individual oscillators. When the outcome of CDK inhibition is modeled, the stabilization of NICD and the resultant reduction in the number of fast oscillators produces an increase in the average period of cell autonomous oscillators and slows the period of the clock, recapitulating in vivo results (Fig 1B). Taken together, these findings provide evidence linking the regulated half‐life of NICD with the period of oscillations and are consistent with work from other laboratories where loss of NRARP in mouse embryos led to enhanced NICD levels in the PSM and extended the oscillatory clock period by 5 min 7.
These findings elegantly link two different clocks: the cell cycle and the segmentation clock. Connections between the cell cycle and vertebrate segmentation have a long history 8. Heat shock perturbations of chicken or zebrafish embryos, instead of leading to a single, discrete segmental defect, result in repeated anomalies separated by production of 6–8 normal somites. The intervals between the repeated disruptions correspond fairly well to the length of the cell cycle of PSM cells, and these observations led to biological and mathematical models linking somite production to a “gate” in the cell cycle that promotes cells of a similar age to coordinately engage in changes in gene expression that promote somite formation 8.
While Notch signaling is clearly important in the clock, the detailed mechanisms by which it affects clock function are not entirely clear. In zebrafish, the expression patterns of clock genes or reporter constructs in Notch genetic mutants have suggested that the major role for Notch in the clock is to synchronize oscillations between neighboring PSM cells, converting a field of noisy, cell autonomous oscillators into a synchronized, tissue‐level timing mechanism. Research suggests that increased Notch activity levels accelerate the zebrafish clock, perhaps by increasing coupling strength, while reductions in Notch activity due to loss of ligand expression or to pharmacological perturbations slow oscillations 9. In contrast, data from mammalian embryos indicate that Notch activity is required for segmentation clock function 10, and manipulations that increase Notch signaling seem to slow down the period of oscillation 5, 7. The work by Carrieri et al begins to directly address how Notch activity patterns are shaped in the segmentation clock. Future studies examining these mechanisms across vertebrate evolution are likely to clarify the similarities and distinctions that influence clock function and vertebrate body plans.
EMBO Reports (2019) 20: e48247
See also: FA Carrieri et al (July 2019)
References
- 1. Carrieri FA, Murray PJ, Ditsova D et al (2019) EMBO Rep 20: e46436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Siebel C, Lendahl U (2017) Physiol Rev 97: 1235–1294 [DOI] [PubMed] [Google Scholar]
- 3. Shaya O, Sprinzak D (2011) Curr Opin Genet Dev 21: 732–739 [DOI] [PubMed] [Google Scholar]
- 4. Kageyama R, Shimojo H, Isomura A (2018) Adv Exp Med Biol 1066: 265–277 [DOI] [PubMed] [Google Scholar]
- 5. Wiedermann G, Bone RA, Silva JC et al (2015) Elife 4: 1–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yeh CH, Bellon M, Nicot C (2018) Mol Cancer 17: 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kim W, Matsui T, Yamao M et al (2011) Mol Biol Cell 22: 3541–3549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stern CD, Piatkowska AM (2015) Semin Cell Dev Biol 42: 134–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liao B‐K, Oates AC (2017) Arthropod Struct Dev 46: 429–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ferjentsik Z, Hayashi S, Dale JK et al (2009) PLoS Genet 5: e1000662 [DOI] [PMC free article] [PubMed] [Google Scholar]


