Abstract
LIM domain containing 2 (LIMD2) is a small LIM-only protein that has been demonstrated to promote tumor progression; however, the expression and function of LIMD2 in non-small cell lung cancer (NSCLC) has not previously been reported. In the present study, reverse transcription-quantitative PCR and western blot analysis were conducted to examine the mRNA and protein expression levels of LIMD2. Cell Counting Kit-8, Transwell and wound-healing assays were performed in order to examine cell proliferation, invasion and migration, respectively. The data revealed that the LIMD2 expression levels were significantly increased in NSCLC tissues and cell lines, compared with adjacent non-tumor tissues and normal lung epithelial cells, respectively. In addition, the high expression of LIMD2 was significantly associated with lymph node metastasis, distant metastasis and advanced clinical stage in NSCLC. The patients with NSCLC with a high expression of LIMD2 exhibited shorter survival times than those with low LIMD2 expression. The knockdown of LIMD2 caused remarkable decreases in NSCLC cell proliferation, migration and invasion. Bioinformatics analysis and luciferase reporter gene assay data further confirmed that LIMD2 was a direct target gene of microRNA-124 (miR-124), a well-known tumor suppressor in NSCLC. The expression of LIMD2 was negatively regulated by miR-124 in NSCLC cells. In addition, miR-124 was downregulated in NSCLC tissues compared with adjacent non-tumor tissues, and an inverse correlation was observed between the expression of LIMD2 and miR-124 in NSCLC tissues. In conclusion, the present study demonstrates that LIMD2 serves an oncogenic role in NSCLC, suggesting that it may be used as a potential therapeutic target for the treatment of NSCLC.
Keywords: non-small cell lung cancer, LIM domain containing 2, oncogene, microRNA-124
Introduction
Lung cancer is the most common malignant tumor worldwide and is the leading cause of cancer-associated mortality each year (1–3). Non-small cell lung cancer (NSCLC) accounts for ~85% of all lung cancer cases, which primarily consist of adenocarcinomas and squamous cell carcinomas (1–3). Due to high recurrence and metastasis, the outcomes of the currently available treatment strategies are unsatisfactory, and the 5-year survival rate is <16% (1–3). A thorough understanding of the molecular mechanisms underlying NSCLC progression will enable the development of novel diagnostic and therapeutic targets (4,5).
LIM domain-containing proteins shuttle between the cytoplasm and the nucleus and bind to partners in both compartments, often coupling changes in gene expression with extracellular cues (6). LIM domain-containing 2 (LIMD2) is an important member of the LIM domain-containing protein family (7). Initially, LIMD2 was reported to be a biomarker of the lymph node metastasis of papillary thyroid carcinoma (7). In addition, LIMD2 has been demonstrated to be overexpressed in metastatic lesions and regulates cell motility and tumor progression by directly binding to and activating integrin-linked kinases (6). However, the expression pattern and function of LIMD2 in NSCLC has, to the best of our knowledge, not been reported.
MicroRNAs (miRs), are a class of small non-coding RNAs containing 22–25 nucleotides, and are important regulators of gene expression that function by directly binding to the complementary sequences of 3′unstranslated regions (UTR) in target mRNAs, causing RNA degradation or translational repression (8–10). By altering the expression of their target genes, miRs participate in a variety of biological processes, including cell proliferation, apoptosis, differentiation and tumorigenesis (8,11). Furthermore, numerous miRs have been reported to be deregulated and exert pivotal functions in NSCLC, including miR-186 (12), miR-148 (13), miR-92 (14), miR-599 (15), as well as miR-124 (16). For instance, a previous study has demonstrated that miR-124 is significantly downregulated in patients with gefitinib-resistant NSCLC and cell lines (16). The depletion of miR-124 induces gefitinib resistance, and the overexpression of miR-124 sensitizes gefitinib-resistant cells to gefitinib (16). However, the relationship between miR-124 and LIMD2 in NSCLC has not previously been elucidated.
The aim of the present study was to explore the clinical significance of LIMD2 expression in NSCLC and to explore the functions of LIMD2 in regulating the malignant phenotypes of NSCLC cells. Additionally, the regulatory mechanism underlying LIMD2 expression in NSCLC cells were examined.
Materials and methods
Tissues samples
The present study was approved by the Ethics Committee of Nanfang Hospital of Southern Medical University (Guangzhou, China). A total of 52 NSCLC tissues and their matched adjacent non-tumor tissues were collected at our hospital between September 2011 and March 2013. These 52 patients with NSCLC included 41 males and 11 females, aged 38–74 years with mean age of 62.7 years. Written informed consent was obtained from each patient. The inclusion/exclusion criteria were that no patient received any chemotherapy or radiotherapy prior to surgical resection. The NSCLC tissues and the adjacent non-tumor tissues were confirmed by the pathologists at Nanfang Hospital, and the uniform distance between NSCLC tissues and adjacent tissues was 5 cm. All tissues were immediately snap-frozen in liquid nitrogen and stored at −80°C. The clinical information of the patients is summarized in Table I.
Table I.
Correlation of LIMD2 expression with clinicopathologic characteristics in non-small cell lung cancer.
| Variable | Cases (n=52) | Low expression (n=26) | High expression (n=26) | P-value |
|---|---|---|---|---|
| Age (years) | 0.41416 | |||
| <60 | 15 | 6 | 9 | |
| ≥60 | 37 | 20 | 17 | |
| Sex | 0.499 | |||
| Male | 41 | 22 | 19 | |
| Female | 11 | 4 | 7 | |
| Smoking | 0.258 | |||
| Yes | 31 | 18 | 13 | |
| No | 21 | 8 | 13 | |
| TNM stage | 0.011a | |||
| I–II | 24 | 17 | 7 | |
| III–IV | 28 | 9 | 19 | |
| Differentiation | 0.093 | |||
| Well and moderate | 23 | 15 | 8 | |
| Poor | 29 | 11 | 18 | |
| Lymph node metastasis | 0.001a | |||
| Yes | 27 | 7 | 20 | |
| No | 25 | 19 | 6 |
P<0.05. LIMD2, LIM domain containing 2.
Cell culture
The normal human bronchial epithelial cell line BEAS-2B, and human NSCLC cell lines A549, H522, H1650 and H1975 were purchased from the Chinese Academy of Sciences Cell Bank. These cells were cultured in DMEM supplemented with 10% FBS (both from Thermo Fisher Scientific, Inc.) and incubated in a humidified atmosphere containing 5% CO2 at 37°C.
Cell transfection
For cell transfections, A549 and H1975 cells (5×105 cells per well) were seeded into 6-well plates. Cells were transfected with 100 nM negative control (NC) small interfering RNA (siRNA; Santa Cruz Biotechnology, Inc.), 100 nM LIMD2-specific siRNA (Santa Cruz Biotechnology, Inc.), 100 nM miR-NC mimics, 100 nM miR-124 mimics, 100 nM NC inhibitor or 100 nM miR-124 inhibitor (all from Guangzhou FulenGen, Co., Ltd., Guangzhou, China), using Lipofectamine 2000 (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from the cultured cells or tissues using TRIzol (Thermo Fisher Scientific, Inc.). Total RNA was reverse transcribed to cDNA by using a PrimeScript® RT reagent kit (Takara Biotechnology Co., Ltd.), according to the manufacturer's protocol. PCR performed with the SYBR premix real-time PCR reagent (Takara Biotechnology Co., Ltd.) using the ABI7500 real-time PCR system (Thermo Fisher Scientific, Inc.). The thermocycling conditions were 95°C for 5 min, followed by 35 cycles of 95°C for 30 sec, 60°C for 30 sec and 72°C for 30 sec. GAPDH and U6 were used as the internal controls for LIMD2 and miR-124, respectively. Relative quantification was performed using the comparative 2−ΔΔCq method (17). The primers for LIMD2 were, forward, 5′-TGCCAGAAGACCGTGTACC-3′ and reverse, 5′-TTTGCAGTAGAACTCCCCGTG-3′. The primers for GAPDH were, forward, 5′-CTGGGCTACACTGAGCACC-3′ and reverse, 5′-AAGTGGTCGTTGAGGGCAATG-3′. The primers for U6 (cat. no. HmiRQP9001) and miR-124 (cat. no. HmiRQP0074) were purchased from Guangzhou FulenGen, Co., Ltd., and these sequences were not supplied by the manufacturer.
Western blot analysis
Total protein was isolated from tissues or cell lines using radioimmunoprecipitation assay lysis buffer (Beyotime Biotechnology of Biotechnology, Haimen, China). The lysates were centrifuged at 12,000 × g for 5 min at 4°C, and the supernatant was collected. The protein concentration was determined using a BCA kit (Beyotime Institute of Biotechnology), according to the manufacturer's protocol. The proteins (50 µg per lane) were separated by SDS-PAGE on 10% gels and then transferred onto polyvinylidene fluoride membranes (EMD Millipore). Following blocking with 5% skim milk in TBS-Tween at room temperature for 3 h, the membrane was incubated with rabbit anti-human LIMD2 antibody (1:500; cat. no. ab205375; Abcam) or rabbit anti-human GAPDH antibody (1:500; cat. no. ab9485; Abcam) at room temperature for 3 h. Following three washes with TBS-Tween, the membrane was incubated with horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (1:5,000; ab6721; Abcam) at room temperature for 40 min. The protein bands were detected using Pierce ECL Western Blotting Substrate kit (Thermo Fisher Scientific, Inc.). Densitometric analysis was performed using ImageJ software (version 1.48; National Institutes of Health).
Cell proliferation assay
Cell Counting Kit-8 (CCK-8) assays were conducted to assess cell proliferation. Transfected cells were seeded into 96-well plates (5,000 cells per well). Following incubation at 37°C in 5% CO2 for 0, 24, 48 and 72 h, 10 µl CCK-8 (Beyotime Institute of Biotechnology) was added into each well. Following incubation at 37°C for 30 min, the absorbance at a wavelength of 450 nm was determined.
Cell migration assay
Transfected cells in DMEM supplemented with 10% FBS were seeded in 6-well plates (100,000 cells per well). Following incubation at 37°C in 5% CO2 for 48 h, wounds were created by scratching the cell surface with a 10 µl pipette tip. Following two washes with PBS, the cells were cultured at 37°C in a humidified incubator containing 5% CO2. At 0 and 24 h, the scratches were images under an inverted light microscope (CKX41; Olympus Corporation).
Cell invasion assay
The cell invasion assay was performed in a Transwell chamber (24-well, 8-mm pore size; Corning, Inc.) that was precoated with Matrigel (BD Biosciences). The transfected cells (100,000 cells) in DMEM were seeded in the upper chambers, and DMEM supplemented with 10% FBS was added to the bottom chambers. Following incubation at 37°C for 24 h, the non-invaded cells on the upper surface of the membrane were removed with cotton swabs. The invaded cells were stained with 0.5% crystal violet at room temperature for 5 min, and counted under an inverted light microscope.
Bioinformatics analysis and luciferase reporter gene assay
TargetScan online software (version 7.2; http://www.targetscan.org/) was used to analyze the targeting relationship between miR-124 and LIMD2. The wild-type (WT) or mutant (MT) LIMD2 3′UTR containing the miR-124 targeting sequence was inserted into the pMIR-REPORT™ miRNA Expression Reporter Vector system (Ambion; Thermo Fisher Scientific, Inc.). Subsequently, the WT or MT luciferase reporter gene plasmid was then co-transfected with miR-NC or miR-124 mimics into H1975 and A549 cells using Lipofectamine 2000 (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Following transfection for 48 h, the luciferase activities were measured using a Dual-Luciferase® Reporter assay kit (Promega Corporation, Madison, WI, USA) on Multiskan™ GO Microplate Spectrophotometer (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The ratio of firefly luciferase activity to Renilla luciferase activity was determined. The vector used to express Renilla luciferase was the pMIR-REPORT™ miRNA Expression Reporter Vector system (Thermo Fisher Scientific, Inc.).
Statistical analysis
All data are presented as the mean ± standard deviation. All statistical calculations were performed using SPSS 20.0 (IBM Corp.). Statistical analysis between two groups was performed using Student's t-test. Statistical analysis between more than two groups was performed using one-way ANOVA followed by Tukey's post hoc test. Survival analysis was conducted using the Kaplan-Meier method with the log-rank test. Spearman correlation analysis was used to analyze the correlation between miR-124 and LIMD2 mRNA expression in NSCLC tissues. P<0.05 was considered to indicate a statistically significant difference.
Results
Upregulation of LIMD2 is associated with NSCLC progression
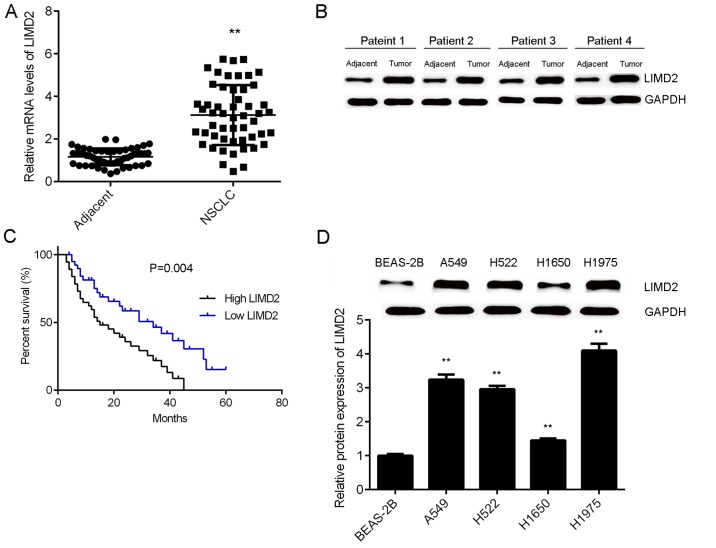
To reveal the role of LIMD2 in NSCLC, the present study initially examined the mRNA and protein expression of LIMD2 in NSCLC tissues and adjacent non-tumor tissues via RT-qPCR and western blotting. As illustrated in Fig. 1A and B, the expression levels of LIMD2 were significantly higher in NSCLC tissues (only epithelial tissues), than in their matched adjacent non-tumor tissues. Based on the mean expression value of LIMD2, these patients were divided into a high LIMD2 expression group and low LIMD2 expression group. Further investigation revealed that the high expression of LIMD2 was significantly associated with lymph node metastasis, distant metastasis and advanced clinical stage in NSCLC (Table I). In addition, it was observed that patients with NSCLC that had high LIMD2 expression exhibited worse prognoses (Fig. 1C). Furthermore, the expression levels of LIMD2 were significantly increased in the NSCLC cell lines (A549, H522, H1650 and H1975) compared with the normal human lung epithelial cells (BEAS-2B; Fig. 1D). Taken together, these findings suggest that the upregulation of LIMD2 may participate in the malignant progression of NSCLC.
Figure 1.
Upregulation of LIMD2 is associated with the progression of NSCLC. LIMD2 (A) mRNA and (B) protein was upregulated in NSCLC tissues compared with adjacent non-tumor tissues. *P<0.01 vs. adjacent. (C) The NSCLC patients with high LIMD2 expression exhibited shorter survival times than those with a low expression of LIMD2. (D) LIMD2 was upregulated in NSCLC cell lines compared to normal human lung epithelial BEAS-2B cells. **P<0.01 vs. BEAS-2B. NCSLC, non-small cell lung cancer; LIMD2, LIM domain containing 2.
Knockdown of LIMD2 inhibits the malignant phenotypes of NSCLC cells
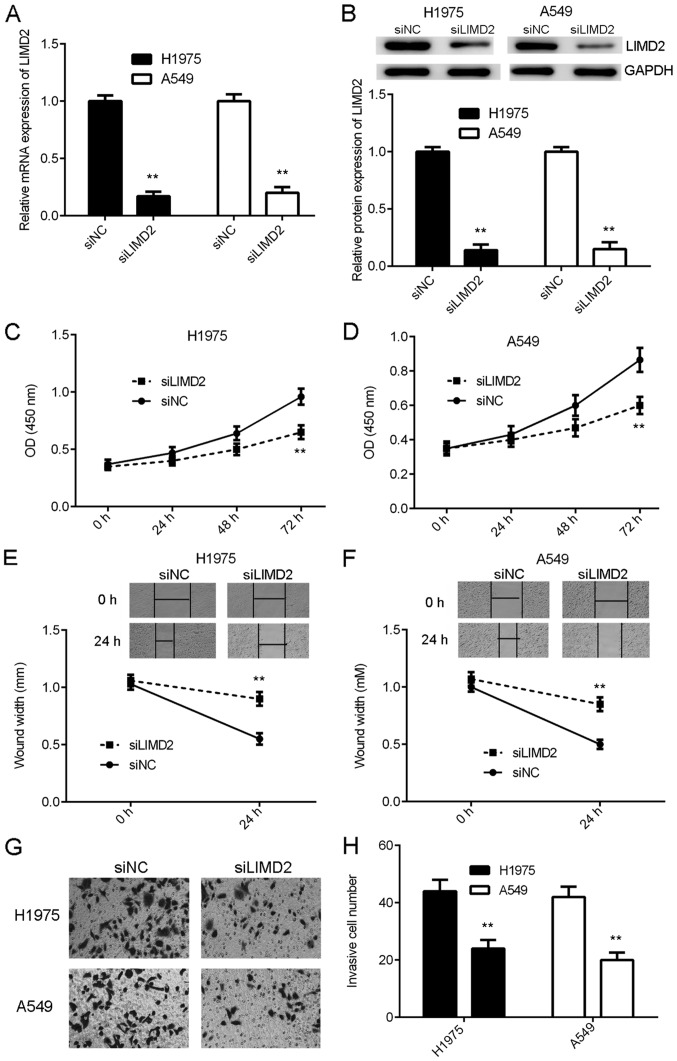
As LIMD2 was significantly upregulated in NSCLC, H1975 and A549 cells were transfected with LIMD2 siRNA to knockdown its expression. Transfection with NC siRNA was used as the control group. Following transfection, the mRNA and protein expression of LIMD2 were significantly reduced in the siLIMD2 group, compared with the siNC group (Fig. 2A and B). The present study then examined the effects of LIMD2 downregulation on NSCLC cell proliferation, migration and invasion. As indicated in Fig. 2C-H, knockdown of LIMD2 markedly inhibited the proliferation, migration and invasion of H1975 and A549 cells, compared with siNC. Therefore, the knockdown of LIMD2 inhibited the malignant phenotype of NSCLC cells in vitro.
Figure 2.
Knockdown of LIMD2 inhibits the malignant phenotypes of non-small cell lung cancer cells. H1975 and A549 cells were transfected with siLIMD2 or siNC. Following transfection, (A) reverse transcription-quantitative PCR and (B) western blotting were used to examine the mRNA and protein expression levels of LIMD2. Cell Counting Kit-8 for (C) H1975 and (D) A549, wound healing for (E) H1975 and (F) A549 and Transwell assays for (G) H1975 and (H) A549 cells were used to examine cell proliferation, migration and invasion, respectively. Magnification for wound healing assay was ×40. Magnification for Transwell assays was ×400. **P<0.01 vs. siNC. LIMD2, LIM domain containing 2; si, small interfering RNA; NC, negative control; OD, optical density.
LIMD2 is a target gene of miR-124
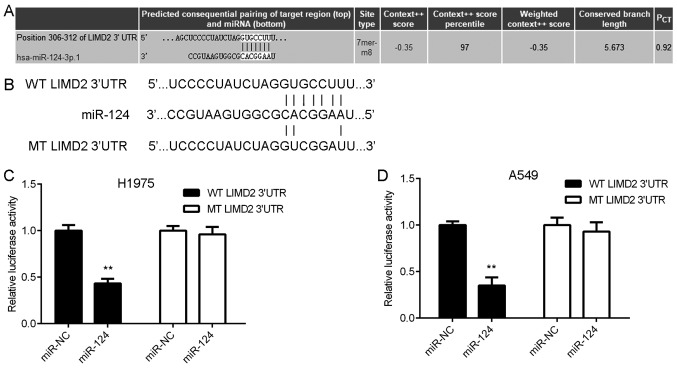
The present study subsequently attempted to examine the regulatory mechanism underlying the expression of LIMD2 in NSCLC using bioinformatics analysis. The TargetScan online software demonstrated that LIMD2 was a potential target of miR-124 (Fig. 3A). To verify this bioinformatics prediction, WT and MT LIMD2 luciferase reporter plasmids were generated (Fig. 3B) and then luciferase reporter gene assays were performed using H1975 and A549 cells. As shown in Fig. 3C and D, the overexpression of miR-124 led to a significant reduction in the luciferase activity of the cells transfected with the WT LIMD2 3′UTR reporter plasmid, yet did not affect the luciferase activity of the cells transfected with the MT LIMD2 3′UTR reporter plasmid. These findings suggest that LIMD2 is a target gene of miR-124 in NSCLC cells.
Figure 3.
LIMD2 is a target gene of miR-124 (A) TargetScan online software revealed that LIMD2 was a potential target of miR-124. (B) WT and MT LIMD2 luciferase reporter plasmids were generated. Luciferase reporter gene assay data showed that overexpression of miR-124 led to a significant reduction in the luciferase activity of (C) H1975 and (D) A549 cells transfected with the WT LIMD2 3′UTR reporter plasmid, but did not affect the luciferase activity of cells transfected with the MT LIMD2 3′UTR reporter plasmid. **P<0.01 vs. miR-NC. LIMD2, LIM domain containing 2; UTR, untranslated region; miR, microRNA; WT, wild-type; MT, mutant; NC, negative control.
LIMD2 is negatively regulated by miR-124 in NSCLC cells
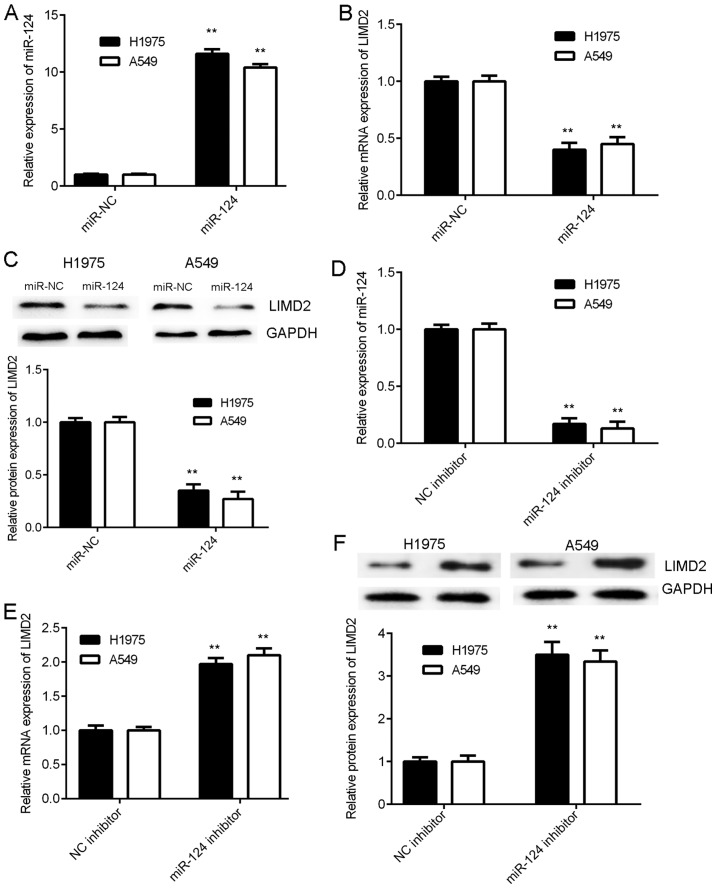
As miRs generally negatively regulate the expression of their target genes, the present study examined the effects of miR-124 on the expression of LIMD2 in NSCLC cells. H1975 and A549 cells were initially transfected with an miR-124 mimic or miR-NC. The RT-qPCR data revealed that the expression levels of miR-124 were significantly increased in the miR-124 group compared with the miR-NC group (Fig. 4A). As illustrated in Fig. 4B and C, the mRNA and protein levels of LIMD2 were significantly reduced in the miR-124 group compared with the miR-NC group. Therefore, the overexpression of miR-124 resulted in the marked downregulation of LIMD2 expression in NSCLC cells. To further confirm these findings, H1975 and A549 cells were transfected with miR-124 inhibitor or NC inhibitor. Following transfection, the expression levels of miR-124 were significantly increased in the miR-124 inhibitor group compared with the NC inhibitor group (Fig. 4D). In addition, a significant increase was observed in the mRNA and protein expression of LIMD2 in the miR-124 inhibitor group, when compared with the NC inhibitor group (Fig. 4E and F). Taken together, the above data indicate that LIMD2 is negatively regulated by miR-124 in NSCLC cells.
Figure 4.
LIMD2 is negatively regulated by miR-124 in non-small cell lung cancer cells. H1975 and A549 cells were transfected with miR-124 mimic or miR-NC. Following transfection, (A) RT-qPCR was performed to examine miR-124 expression. (B) RT-qPCR and (C) western blotting were performed to examine the mRNA and protein expression of LIMD2, respectively. **P<0.01 vs. miR-NC. Then, H1975 and A549 cells were transfected with miR-124 inhibitor or NC inhibitor. Following transfection, (D) RT-qPCR was performed to examine miR-124 expression. (E) RT-qPCR and (F) western blotting were performed to examine the mRNA and protein expression of LIMD2, respectively. **P<0.01 vs. NC inhibitor. RT-qPCR, reverse transcription-quantitative PCR; miR, microRNA; NC, negative control; LIMD2, LIM domain containing 2.
Downregulation of miR-124 is inversely correlated with the upregulation of LIMD2 in NSCLC tissues
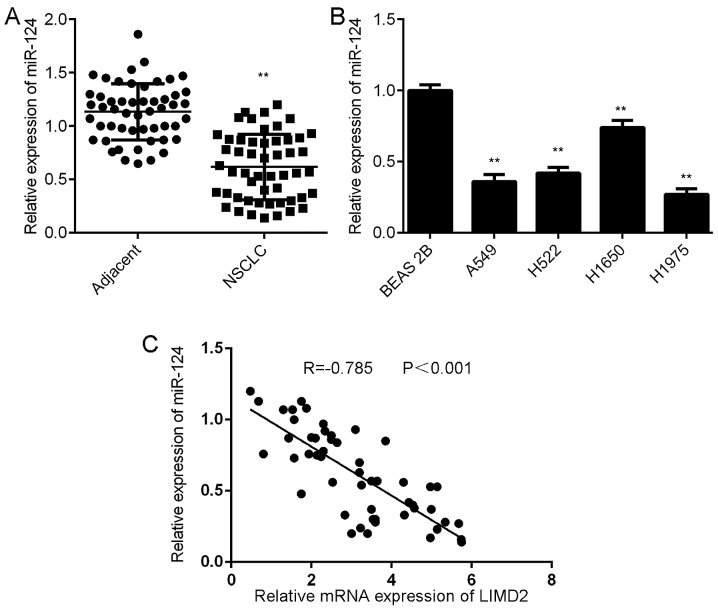
Further investigation revealed that the expression levels of miR-124 were significantly lower in NSCLC tissues than in their matched adjacent non-tumor tissues (Fig. 5A). Consistently, miR-124 was also downregulated in the NSCLC cell lines compared with BEAS-2B cells (Fig. 5B). Notably, an inverse correlation was observed between miR-124 and LIMD2 expression in the NSCLC tissues (Fig. 5C). These findings suggest that the reduced expression of miR-124 may contribute to the increased expression of LIMD2 in NSCLC tissues.
Figure 5.
Reduced expression of miR-124 is associated with increased expression of LIMD2 in NSCLC tissues. (A) RT-qPCR was performed to examine the expression of miR-124 in NSCLC tissues and their matched adjacent non-tumor tissues. **P<0.01 vs. adjacent. (B) RT-qPCR was performed to examine the expression of miR-124 in NSCLC cell lines compared with BEAS-2B cells. **P<0.01 vs. BEAS-2B. (C) An inverse correlation was observed between miR-124 and LIMD2 expression in NSCLC tissues. NCSLC, non-small cell lung cancer; RT-qPCR, reverse transcription-quantitative PCR; miR, microRNA; LIMD2, LIM domain containing 2.
Discussion
The results of the present study revealed that the expression levels of LIMD2 were significantly increased in NSCLC tissues and cell lines, compared with adjacent non-tumor tissues and normal lung epithelial cells. In addition, the high expression of LIMD2 was demonstrated to be significantly associated with lymph node metastasis, distant metastasis and advanced clinical stage in NSCLC. The patients with NSCLC that had a high expression of LIMD2 expression exhibited shorter survival times than those with low LIMD2 expression. The knockdown of LIMD2 resulted in marked decreases in the proliferation, migration and invasion of NSCLC cells. Bioinformatics analysis and luciferase reporter gene assay data further confirmed that LIMD2 was a direct target gene of miR-124, which is a well-established tumor suppressor in NSCLC (16). The expression of LIMD2 was negatively regulated by miR-124 in NSCLC cells. Furthermore, miR-124 was downregulated in NSCLC tissues when compared with adjacent non-tumor tissues, and an inverse correlation was observed between the expression of LIMD2 and miR-124 in NSCLC tissues.
LIM domain-containing proteins have been demonstrated to exert different functions, including the regulation of gene expression, cell adhesion and motility, and these proteins have also been revealed to participate in the development and progression of human cancers (18). LIMD2, a member of the LIM domain-containing protein family, has previously been demonstrated to promote tumor cell metastasis (6). Inhibition of the expression of LIMD2 can inhibit tumor cell motility and invasiveness in thyroid cancer (7). However, whether LIMD2 exerts a promoting function in NSCLC remains unknown. In the present study, the expression of LIMD2 in NSCLC tissues and matched adjacent non-tumor tissues was measured using RT-qPCR and western blot analysis, and it was observed that LIMD2 was significantly upregulated in NSCLC tissues compared with adjacent non-tumor tissues. Further investigation demonstrated that the increased expression of LIMD2 was significantly associated with NSCLC progression as well as the poor prognosis of patients. These findings suggest that LIMD2 may function as an oncogene in NSCLC. To further clarify the exact function of LIMD2 in NSCLC, the present study used two common NSCLC cell lines H1975 and A549 to perform in vitro experiments. The findings of these experiments revealed that the knockdown of LIMD2 significantly reduced the proliferation, migration and invasion of H1975 and A549 cells. These findings suggest that targeting LIMD2 may be a promising strategy for the treatment of NSCLC.
The present study further examined the regulatory mechanism underlying LIMD2 upregulation in NSCLC. As miRs are key regulators of gene expression (10), a bioinformatics prediction was performed to analyze the potential miRs that can directly target LIMD2. Among the predicted miRs, miR-124 was selected for subsequent investigation, as miR-124 has been demonstrated to serve a tumor-suppressive role in NSCLC (19,20). The downregulation of miR-204 in plasma and tissues is associated with a poor prognosis in patients with NSCLC (19,20). Additionally, certain target genes of miR-124 have been identified in NSCLC (20). For example, the study of Li et al (20) reported that miR-124 inhibited NSCLC cell proliferation by targeting STAT3. Lin et al (21) demonstrated that miR-124 served a suppressive role in NSCLC by targeting cadherin 2 and regulating the epithelial-mesenchymal transition. Yang et al (22) reported that miR-204 suppressed NSCLC cell invasion and migration by targeting Janus kinase 2. In addition, CD164, LIM homeobox 2, STAT3 and snail family transcriptional repressor 2 have been identified as target genes of miR-124 in NSCLC (16,21–23). In the present study, a luciferase reporter gene assay data confirmed that LIMD2 was a direct target gene of miR-124, and the expression of LIMD2 was negatively mediated by miR-124 in NSCLC. In addition, the increased expression of LIMD2 was inversely correlated with the decreased expression of miR-124 in NSCLC tissues, suggesting that the downregulation of miR-124 may contribute to the upregulation of LIMD2 in NSCLC.
To the best of our knowledge, the present study is the first to report that the overexpression of LIMD2 promotes the progression of NSCLC and that the knockdown of LIMD2 may inhibit the malignant phenotypes of NSCLC cells. These findings suggest that LIMD2 may be used as a potential therapeutic target for NSCLC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
SQ collected sample tissues and performed clinical experiments. XX, YT and PH performed cell experiments and analyzed data. FZ and YX were responsible for study design wrote the manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Nanfang Hospital of Southern Medical University, Guangzhou, China. Written informed consents were obtained.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 4.Landi L, Cappuzzo F. Pharmacotherapy targeting the EGFR oncogene in NSCLC. Expert Opin Pharmacother. 2014;15:2293–2305. doi: 10.1517/14656566.2014.957179. [DOI] [PubMed] [Google Scholar]
- 5.Markou A, Sourvinou I, Vorkas PA, Yousef GM, Lianidou E. Clinical evaluation of microRNA expression profiling in non small cell lung cancer. Lung Cancer. 2013;81:388–396. doi: 10.1016/j.lungcan.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 6.Peng H, Talebzadeh-Farrooji M, Osborne MJ, Prokop JW, McDonald PC, Karar J, Hou Z, He M, Kebebew E, Orntoft T, et al. LIMD2 is a small LIM-only protein overexpressed in metastatic lesions that regulates cell motility and tumor progression by directly binding to and activating the integrin-linked kinase. Cancer Res. 2014;74:1390–1403. doi: 10.1158/0008-5472.CAN-13-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cerutti JM, Oler G, Michaluart P, Jr, Delcelo R, Beaty RM, Shoemaker J, Riggins GJ. Molecular profiling of matched samples identifies biomarkers of papillary thyroid carcinoma lymph node metastasis. Cancer Res. 2007;67:7885–7892. doi: 10.1158/0008-5472.CAN-06-4771. [DOI] [PubMed] [Google Scholar]
- 8.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 9.Moss EG. MicroRNAs: Hidden in the genome. Curr Biol. 2002;12:R138–R140. doi: 10.1016/S0960-9822(02)00708-X. [DOI] [PubMed] [Google Scholar]
- 10.John B, Enright AJ, Aravin A, Tuschl T, Sande C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 12.Huang T, Wang G, Yang L, Peng B, Wen Y, Ding G, Wang Z. MiR-186 inhibits proliferation, migration, and invasion of non-small cell lung cancer cells by downregulating Yin Yang 1. Cancer Biomark. 2017;21:221–228. doi: 10.3233/CBM-170670. [DOI] [PubMed] [Google Scholar]
- 13.He M, Xue Y. MicroRNA-148a suppresses proliferation and invasion potential of non-small cell lung carcinomas via regulation of STAT3. Onco Targets Ther. 2017;10:1353–1361. doi: 10.2147/OTT.S123518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ren P, Gong F, Zhang Y, Jiang J, Zhang H. MicroRNA-92a promotes growth, metastasis, and chemoresistance in non-small cell lung cancer cells by targeting PTEN. Tumour Biol. 2016;37:3215–3225. doi: 10.1007/s13277-015-4150-3. [DOI] [PubMed] [Google Scholar]
- 15.Tian W, Wang G, Liu Y, Huang Z, Zhang C, Ning K, Yu C, Shen Y, Wang M, Li Y, et al. The miR-599 promotes non-small cell lung cancer cell invasion via SATB2. Biochem Biophys Res Commun. 2017;485:35–40. doi: 10.1016/j.bbrc.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 16.Hu FY, Cao XN, Xu QZ, Huang Z, Zhang C, Ning K, Yu C, Shen Y, Wang M, Li Y, et al. iR-124 modulates gefitinib resistance through SNAI2 and STAT3 in non-small cell lung cancer. J Huazhong Univ Sci Technolog Med Sci. 2016;36:839–845. doi: 10.1007/s11596-016-1672-x. [DOI] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Matthews JM, Lester K, Joseph S, Curtis DJ. LIM-domain-only proteins in cancer. Nat Rev Cancer. 2013;13:111–122. doi: 10.1038/nrc3418. [DOI] [PubMed] [Google Scholar]
- 19.Guo W, Zhang Y, Shi Y, Xi J, Fan H, Xu S. Decreased expression of miR-204 in plasma is associated with a poor prognosis in patients with non-small cell lung cancer. Int J Mol Med. 2015;36:1720–1726. doi: 10.3892/ijmm.2015.2388. [DOI] [PubMed] [Google Scholar]
- 20.Li X, Yu Z, Li Y, Liu S, Gao C, Hou X, Yao R, Cui L. The tumor suppressor miR-124 inhibits cell proliferation by targeting STAT3 and functions as a prognostic marker for postoperative NSCLC patients. Int J Oncol. 2015;46:798–808. doi: 10.3892/ijo.2014.2786. [DOI] [PubMed] [Google Scholar]
- 21.Lin J, Xu K, Wei J, Heimberger AB, Roth JA, Ji L. MicroRNA-124 suppresses tumor cell proliferation and invasion by targeting CD164 signaling pathway in non-small cell lung cancer. J Gene Ther. 2016;2:6. doi: 10.13188/2381-3326.1000006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Q, Wan L, Xiao C, Hu H, Wang L, Zhao J, Lei Z, Zhang HT. Inhibition of LHX2 by miR-124 suppresses cellular migration and invasion in non-small cell lung cancer. Oncol Lett. 2017;14:3429–3436. doi: 10.3892/ol.2017.6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang M, Meng B, Liu Y, Yu J, Chen Q, Liu Y. MiR-124 inhibits growth and enhances radiation-induced apoptosis in non-small cell lung cancer by inhibiting STAT3. Cell Physiol Biochem. 2017;44:2017–2028. doi: 10.1159/000485907. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request.