Abstract
Chemotherapy resistance is a major obstacle to the effective treatment of patients with gastric cancer (GC). Mounting evidence has indicated that the dysregulation of microRNAs (miRNAs) is associated with the sensitivity of cancer cells to chemotherapy. However, the mechanisms underlying miRNA-mediated chemoresistance in GC cells remain to be elucidated. The present study aimed to identify functional miRNAs that may regulate the sensitivity of human GC cells to cisplatin (DDP) treatment. miRNA microarray analysis was used to identify differentially expressed miRNAs between the human cisplatin-sensitive GC cell line SGC7901 and the corresponding cisplatin-resistant cell line SGC7901/DDP. miRNA (miR)-362-5p, which is associated with numerous types of tumors, was identified to be downregulated in the SGC7901/DDP cell line. However, the biological role of miR-362-5p in SGC7901/DDP cells remains to be explored. The expression level of miR-362-5p was demonstrated to be reduced in SGC7901/DDP cells compared with SGC7901 cells by reverse transcription-quantitative PCR. Upregulation of miR-362-5p significantly increased cisplatin sensitivity and cisplatin-induced apoptosis, whereas downregulation of miR-362-5p attenuated these effects. Databases predicted that suppressor of zeste 12 protein (SUZ12) may function as a target of miR-362-5p. In addition, the mRNA and protein expression levels of SUZ12 in SGC7901/DDP cells were significantly higher compared with SGC7901 cells and negatively associated with miR-362-5p expression. MTT and western blot analysis assays confirmed that knockdown of SUZ12 enhanced cisplatin sensitivity and decreased NF-κB/p65 protein levels in SGC7901/DDP cells. In addition, upregulation of miR-362-5p in SGC7901/DDP cells decreased the protein expression level of SUZ12, whereas downregulation of miR-362-5p increased the SUZ12 expression level. The results of the present study suggested that dysregulated miR-362-5p may target SUZ12 to promote the development of cisplatin resistance and attenuate cisplatin-induced apoptosis. Therefore, miR-362-5p upregulation combined with cisplatin treatment may serve as a promising therapeutic strategy for patients with cisplatin-resistant GC.
Keywords: cisplatin resistance, gastric cancer, microRNA-362-5p, suppressor of zeste 12 protein, drug therapy
Introduction
Gastric cancer (GC) is one of the major malignancies worldwide (1). The overall 5-year survival rate of patients with GC remains poor (2). The poor prognosis of patients with this disease results from late diagnosis and a poor response to available therapies. Cisplatin (DDP)-based chemotherapy is the main treatment strategy for GC (3). Although combined drug therapy pre- and post-surgery has increased overall survival, drug resistance continues to be a major clinical obstacle to therapeutic efficacy (4). Drug resistance is categorized into two types: i) Intrinsic resistance, in which tumors are resistant to the drug prior to treatment, thereby preventing the drug from effectively treating the tumor even with early diagnosis; ii) acquired resistance, in which resistance to the drug is formed after prolonged cycles of chemotherapy despite an initial positive reaction. Cisplatin typically kills tumor cells by inducing apoptosis; however, it has been revealed that many solid tumors are resistant to drug-induced apoptosis (5). Therefore, it is essential to identify novel molecular markers for improving the sensitivity of GC cells to cisplatin.
MicroRNAs (miRNAs) are small non-coding RNAs (19–24 nucleotides in length) that regulate gene expression at the post-transcriptional level through base pairing with the 3′-untranslated region (3′-UTR) of target mRNAs, resulting in translational repression or degradation of the mRNAs (6,7). An increasing number of miRNAs have been demonstrated to be implicated in the mechanism of drug resistance and the genes or pathways involved in drug resistance (8). Upregulation of miR-101 has been revealed to enhance cellular sensitivity to cisplatin treatment by promoting the apoptosis-inducing effects of cisplatin in GC (9). Previous studies demonstrated that miR-362-5p serves a critical role in malignant tumors. By collecting seven publicly available and independent renal cell carcinoma miRNA expression profiling datasets, Ying et al (10) revealed that hsa-miR-362-5p is downregulated in renal carcinoma. Ni et al (11) have demonstrated that miR-362-5p targets the cylindromatosis gene, thereby promoting hepatocellular carcinoma cell growth and metastasis. It has also been demonstrated that upregulation of miR-362-5p significantly accelerates proliferation, migration and invasion of human breast cancer MCF7 cells (12). However, the biological role of miR-362-5p in SGC7901/DDP cells remains to be explored.
Suppressor of zeste 12 protein (SUZ12) is a core component of polycomb repressive complex 2 (PRC2), which epigenetically silences gene transcription (13). In addition to SUZ12, PRC2 contains the embryonic ectoderm development protein and the catalytic subunit enhancer of zeste 2 polycomb repressive complex 2 (14), which is involved in the pathogenesis of GC (15). Amplification and overexpression of SUZ12 have been observed in multiple tumor types, such as GC, ovarian cancer and non-small cell lung cancer (16–18). Moreover, there is evidence that SUZ12 serves an important role in GC by acting as an oncogene (16). However, the role of SUZ12 in the cisplatin resistance of GC cells has yet to be investigated.
The present study investigated the potential function of miR-362-5p in vitro and aimed to further understand its underlying mechanism in cisplatin-resistant GC cells. In addition, this study defined the molecular mechanism of SUZ12. These findings may provide novel insights into the tumor biology of GC.
Materials and methods
Cell lines and culture
The human GC cell line SGC7901 and the corresponding cisplatin-resistant cell line SGC7901/DDP were obtained from Nanjing KeyGen Biotechnology Company. Cells were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.), containing 10% FBS (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 mg/ml streptomycin. To maintain the cisplatin-resistant phenotype of SGC7901/DDP cells, cisplatin (800 ng/ml; Jiangsu Hansoh Pharmaceutical Group Co., Ltd.) was added to the medium. The cells were maintained in a humidified incubator with an atmosphere of 5% CO2 at 37°C.
Total RNA extraction and quality control
Total RNA was extracted from cells (~1×107 cells) using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. RNA levels were measured using a Nanodrop 2000 spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific, Inc.) at UV absorbances of 260, 280 and 230 nm. All RNA samples used met pre-determined quality control standards (A260/A230 >2.0; A260/A280 >1.8).
miRNA microarray analysis
miRNAs from ~1×107 cells were extracted using the miRVana™ miRNA isolation kit (cat. no. AM1560, Ambion; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The miRNAs extracted from three matched pairs of SGC7901 and SGC7901/DDP cells were labeled and hybridized with an Affymetrix GeneChip miRNA 4.0 Array (Affymetrix; Thermo Fisher Scientific, Inc.), which contained miRNAs from the miRBase v20 database (http://www.mirbase.org/). The microarray was scanned by CapitalBio Technology Corporation. The data were normalized and analyzed using GeneSpring 13.0 software (Agilent Technologies, Inc.). Student's t-test was used for analysis between two groups of data. Differential expression of miRNA was defined as a difference of >2-fold in miRNA expression that was statistically significant at P<0.05. Cluster analysis and graphical presentation of the data were performed using Cluster 3.0 software (developed by Michael Eisen, updated by Michiel de Hoon, University of Tokyo, Human Genome Center).
Prediction of miRNA target genes
The mature miRNA sequences were acquired from miRBase (http://www.mirbase.org/index.shtml). Potential miRNA targets were predicted using microRNA.org (http://www.microrna.org/microrna/getMirnaForm.do) and miRDB (http://mirdb.org).
Reverse transcription-quantitative PCR (RT-qPCR)
To detect the expression of miR-362-5p, RT-qPCR was performed using Hairpin-it™ microRNAs (GenePharma) and U6 snRNA Normalization RT-qPCR Quantitation kits (GenePharma) according to the manufacturer's instructions. U6 was selected as the internal control for miRNA-362-5p expression. The primers of miR-362-5p and U6 were purchased from GenePharma and the sequences were as follows: miR-362-5p RT primer 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACTCAC-3′, PCR primers, forward 5′-GTCACGAAATCCTTGGAACCTAG-3′ and reverse 5′-TATGGTTGTTCTCGTCTCCTTCTC-3′; and U6 RT primer 5′-CGCTTCACGAATTTGCGTGTCAT-3′, PCR primers, forward 5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse 5′-CGCTTCACGAATTTGCGTGTCAT-3′. The miRNA RT reactions were performed as follows: 25°C for 30 min, 42°C for 30 min, 85°C for 5 min and save at 4°C The PCR reactions were performed as follows: Prevariant at 95°C for 3 min, cyclic reaction at 95°C for 12 sec and 62°C for 40 sec and for 40 cycles. For analysis of SUZ12 mRNA expression, total RNA was reverse-transcribed using a PrimeScript™ RT Reagent kit (Takara Bio, Inc.). RT-qPCR was performed using SYBR® Premix Ex Taq™ II (Takara Bio, Inc.). β-actin was used as the internal control. The PCR primers of mRNA were purchased from Sangon Biotech Co., Ltd. and the sequences of primers were as follows: SUZ12, forward 5′-ATTCATCGCCAACCTGGATT-3′, reverse 5′-TGGCCTGCACACAAGAATATG-3′; β-actin, forward 5′-ATTCCTATGTGGGCGACGAG-3′, reverse 5′-AGAGGCGTACAGGGATAGCA-3′. The mRNA RT reactions were performed as follows: 37°C for 15 min, 85°C for 5 sec and save at 4°C The PCR reactions were performed as follows: Prevariant at 95°C for 30 sec, cyclic reaction at 95°C for 5 sec and 60°C for 30 sec and for 40 cycles. All reactions were performed in triplicate on a LightCycler 96 System (Roche Applied Science). The relative expression levels of miR-362-5p and SUZ12 mRNA were calculated using the 2−ΔΔCq method (19) and were normalized to the U6 and β-actin expression levels, respectively.
Lentiviral transfection, stable cell line establishment and siRNA transfection
The lentiviral vector carrying miR-362-5p mimic-green fluorescent protein (GFP)-puromycin, the negative control-GFP-puromycin or miR-362-5p inhibitor small interfering RNA (siRNA)-GFP-puromycin and the corresponding viruses [(1×108 plaques forming units (pfu)] were constructed by GenePharma. Prior to transfection, ~2×105 cells were plated into six-well plates. When the cells were in the logarithmic growth phase, they were then infected with lentivirus with a multiplicity of infection of 10 pfu/cell. RPMI-1640 medium without penicillin and streptomycin was added to the wells for cell culture. Cells were transfected for 24 h at 37°C, and the culture medium containing lentivirus was removed from the wells and fresh complete medium was added for continued cell culture. At 72 h, the expression of GFP was observed under a fluorescence microscope to determine transfection efficiency. As the lentiviral vectors also carried the gene expressing a puromycin resistance protein, purified cells were obtained by screening with puromycin. RT-qPCR was performed to verify the efficiency of miR-362-5p upregulation or downregulation. The siRNA specific for SUZ12 (si-SUZ12) and negative control siRNA (si-NC) were synthesized by GenePharma. The sequences of siRNAs were as follows: si-SUZ12 5′-CUGCCUCCAUUCGAAACAUTTAUGUUUCGAAUGGAGGCAGTT-3′ and si-NC 5′-UUCUCCGAACGUGUCACGUTTACGUGACACGUUCGGAGAATT-3′. Prior to transfection, ~5×105 cells were seeded in 6-well plates. After 24 h incubation at 37°C, cells were transfected with si-SUZ12 (100 pmol) or si-NC (100 pmol) using 5 µl Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.) and maintained at 37°C in a humidified incubator containing 5% CO2 for 48 h.
Western blotting
Cells (~1×107) were washed twice with PBS and lysed in RIPA buffer (Thermo Fisher Scientific, Inc.) supplemented with protease inhibitors (Thermo Fisher Scientific, Inc.) on ice for 30 min. Cell lysates were centrifuged for 15 min (13,000 × g; 4°C), and the supernatant was collected. Protein concentrations were quantified by the bicinchoninic acid assay method (Thermo Fisher Scientific, Inc.). Proteins (60 µg) were separated by 10% SDS-PAGE. Subsequently, the proteins were transferred to a PVDF membrane (Merck KGaA) and blocked with 5% skimmed milk for 4 h at 4°C The membrane was incubated with antibodies against SUZ12 (cat. no. ab175187; 1:1,000; Abcam), caspase-3 (cat. no ab184787; 1:1,000; Abcam), cleaved-poly-(ADP-ribose) polymerase (PARP; cat. no 5625S; 1:1,000; Cell Signaling Technology, Inc.), NF-κB (cat. no. 8242S; 1:1,000; Cell Signaling Technology, Inc.), β-actin (cat. no. 4970S; 1:1,000; Cell Signaling Technology, Inc.) or GAPDH (cat. no. 5174S; 1:1,000; Cell Signaling Technology, Inc.) at 4°C overnight. Following three washes with TBS + Tween-20 (0.05%, v/v), the membrane was incubated with horseradish peroxidase-conjugated secondary antibodies at 4°C for 4 h. Protein bands were visualized with an enhanced chemiluminescence reagent kit (Thermo Fisher Scientific, Inc.) using Fine Do X6 imaging system (Tanon Science and Technology Co., Ltd.). Protein expression levels were semi-quantified using the Tanon 3500/3500R gel imaging system (Tanon Science and Technology Co., Ltd.) and normalized to the expression of the internal control GAPDH or β-actin.
Cell viability assay
Cells were plated into 96-well plates (5×103 cells/well). At 24 h post-transfection, the cells were treated with a range of concentrations of cisplatin at 37°C. At 48 h post-transfection, 20 µl MTT (5 mg/ml; Sigma-Aldrich; Merck KGaA) was added to each well, followed by culturing for 4 h in an incubator with an atmosphere of 5% CO2 at 37°C. To dissolve the crystals, 150 µl DMSO (Sigma-Aldrich; Merck KGaA) was added to each well, and the plates were agitated lightly for 10 min. The absorbance of the samples at 490 nm was measured with an ELx800 spectrophotometer (BioTek Instruments, Inc.). The median inhibitory concentration (IC50) for cisplatin was estimated based on the cell viability using SPSS 16.0 software. All reactions were performed in triplicate.
Apoptosis assay
The apoptotic rates of SGC7901/DDP and SGC7901 cells were evaluated by flow cytometry (FCM) with an Annexin V-FITC/propidium iodide (PI) Cell Apoptosis Detection kit (BestBio). SGC7901/DDP and SGC7901 cells (~1×106 cells) were treated with cisplatin at a final concentration of 0.8 and 0.3 µg/ml, respectively. Following 48 h of treatment at 37°C the cells were collected and washed twice with ice-cold PBS. Cells were resuspended with 400 µl of 1X binding buffer and maintained at a final density of approximately 1×106 cells/ml. Annexin V-FITC (5 µl) was added to the suspension, which was incubated in the dark for 15 min at room temperature. Following the addition of PI (10 µl) to the suspension, cells were incubated for an additional 5 min in the dark at room temperature. Subsequently, cell apoptosis was assessed (early + late) using a Gallios flow cytometer (Beckman Coulter, Inc.).
Statistical analysis
All experiments were repeated at least three times. Statistical analyses were performed using SPSS 16.0 software (SPSS, Inc.). Data are expressed as the mean ± standard deviation. Comparison between two groups was analyzed by t-test. ANOVA was used for comparison between multiple groups followed by the Least Significant Difference post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-362-5p screening in cisplatin-sensitive and cisplatin-resistant GC cells
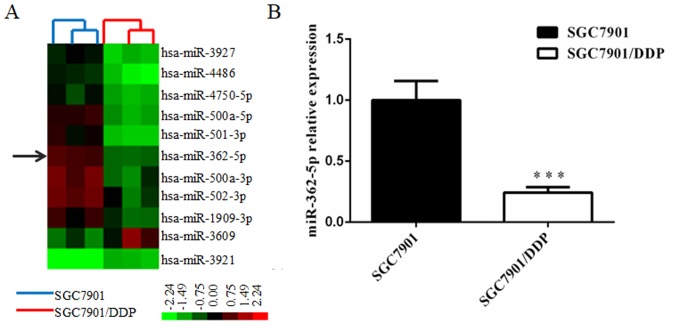
The differential miRNA expression profiles between SGC7901/DDP cells and SGC7901 cells were obtained by miRNA microarray analysis. The Affymetrix miRNA GeneChip 4.0 was used to scan and quantify the signal intensity of probes of 1,316 human mature miRNAs on the chips for the two cell lines. The results indicated that 48 miRNAs were significantly differentially expressed (>2-fold) in SGC7901/DDP cells relative to the parental cells, including 19 upregulated miRNAs and 29 downregulated miRNAs. miR-362-5p was revealed to be downregulated in SGC7901/DDP cells and was selected for further research to determine its function in the cisplatin resistance of GC cells. Cluster analysis was performed with Cluster 3.0 software (Fig. 1A). To study the potential effects of miRNA-362-5p on the cisplatin resistance of GC, the difference in miR-362-5p expression between SGC7901 cells and SGC7901/DDP cells was determined by RT-qPCR. The results revealed that the expression level of miR-362-5p was significantly lower in SGC7901/DDP cells compared with expression in SGC7901cells (P<0.01; Fig. 1B). These results suggested that reduced expression of miR-362-5p may be associated with cisplatin resistance in SGC7901/DDP cells.
Figure 1.
miR-362-5p expression in cisplatin-resistant SGC7901/DDP cells is lower compared with cisplatin-sensitive SGC7901 cells. (A) Microarray assay of miRNA expression levels in gastric cancer cells. Hierarchical clustering of miRNA expression levels in SGC7901/DDP cells relative to SGC7901 cells. Red represents high relative expression; green represents low relative expression. Upregulated and downregulated miRNAs were defined by P<0.05 and >2-fold difference in expression relative to the control cells. The arrow indicates miR-362-5p. (B) Reverse transcription-quantitative PCR analysis demonstrated that miR-362-5p was significantly downregulated in the SGC7901/DDP cells compared with SGC7901 cells. n=3; ***P<0.001 vs. SGC7901; miRNA, microRNA; miR-362-5p, microRNA-362-5p.
Overexpression of miR-362-5p reverses cisplatin resistance in SGC7901/DDP cells
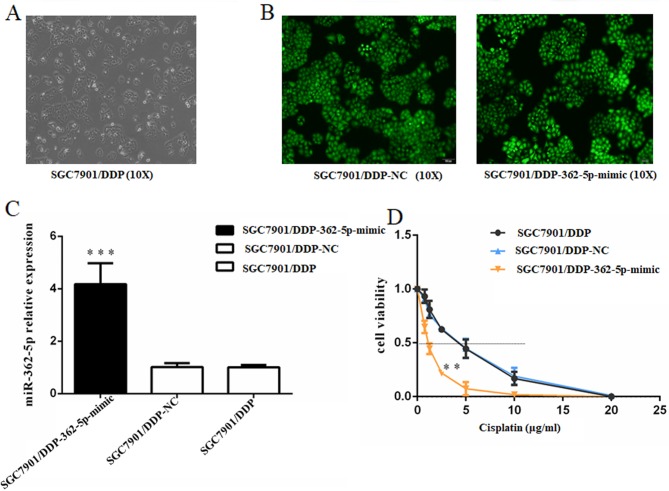
To further determine the association between cisplatin resistance and the regulation of miR-362-5p expression, the effects of miR-362-5p upregulation on cisplatin resistance in cisplatin-resistant SGC7901/DDP cell lines were investigated. Lentiviral plasmids carrying miR-362-5p mimic or negative control miRNA were transfected into SGC7901/DDP cells. After screening with puromycin, the GFP-positive rate of the cells subjected to lentiviral transfection was >90% based on fluorescence microscopy (Fig. 2A and B). RT-qPCR was performed to verify the transfection efficiency in the constructed cell lines, and the results demonstrated that miR-362-5p expression levels were significantly increased (P<0.01; Fig. 2C). After stable cell line establishment, the cells were treated with different concentrations of cisplatin for 48 h. The IC50 for cisplatin was estimated based on cell viability. The SGC7901/DDP cells transfected with miR-362-5p mimic exhibited a notably lower survival status compared with control SGC7901/DDP-NC and SGC7901/DDP cells (IC50, 1.609±0.332 vs. 5.133±0.569 and 5.161±0.641 µg/ml, respectively; P<0.01; Fig. 2D). These results indicated that upregulation of miR-362-5p may enhance the cisplatin sensitivity of cisplatin-resistant GC cells.
Figure 2.
Upregulation of miR-362-5p reverses the cisplatin resistance of SGC7901/DDP cells. (A) Light microscopy (magnification, ×100) and (B) lentiviral transfection assay of SGC7901/DDP cells (magnification, ×100). Cells that were successfully transfected expressed green fluorescent protein. (C) Reverse transcription-quantitative PCR analysis demonstrated that the expression level of miR-362-5p was markedly increased in SGC7901/DDP cells transfected with miR-362-5p mimic compared with NC-transfected and untransfected cells. (D) Viability of cells treated with cisplatin for 48 h. MTT assays were conducted on SGC7901/DDP cells treated with various concentrations of cisplatin (0, 0.75, 1.25, 2.5, 5, 10 or 20 µg/ml). The IC50 for cisplatin was estimated based on the cell viability. The IC50 for cisplatin of the miR-362-5p mimic-transfected cells was notably lower compared with the controls. n=3; ***P<0.001 vs. SGC7901/DDP-NC and SGC7901/DDP; **P<0.01 vs. SGC7901/DDP-NC and SGC7901/DDP; IC50, median inhibitory concentration; miR-362-5p, microRNA-362-5p; NC, negative control.
Inhibition of miR-362-5p enhances the cisplatin resistance of SGC7901 cells
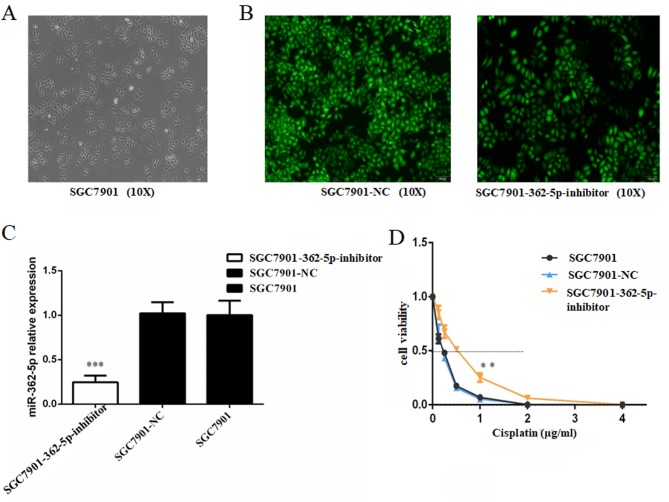
Based on the aforementioned results, the effects of miR-362-5p inhibition on cisplatin-induced cytotoxicity in cisplatin-sensitive SGC7901 cell line was further investigated. SGC7901 cells were transfected with lentiviral plasmids carrying miR-362-5p inhibitor or its negative control. The transfected cells were screened with puromycin and then observed for fluorescence (Fig. 3A and B). The data from RT-qPCR revealed that the miR-362-5p inhibitor significantly decreased the expression level of miR-362-5p in SGC7901 cells compare with negative control and blank control (P<0.001; Fig. 3C), which confirmed that the cells were successfully transfected. Subsequently, a range of concentrations of cisplatin was applied to the cultures and the IC50 for cisplatin was estimated based on cell viability. Cisplatin sensitivity was significantly attenuated in SGC7901 cells transfected with the miR-362-5p inhibitor compared with the control SGC7901-NC and SGC7901 cells (IC50, 0.676±0.042 vs. 0.286±0.025 and 0.300±0.009 µg/ml, respectively; P<0.01; Fig. 3D). These data verified that downregulation of miR-362-5p enhances the cisplatin resistance of cisplatin-sensitive GC cell lines.
Figure 3.
Inhibition of miR-362-5p reduces the cisplatin sensitivity of SGC7901 cells. (A) Light microscopy (magnification, ×100) and (B) lentiviral plasmid transfection assay of SGC7901 cells (magnification, ×100). Cells that were successfully transfected expressed green fluorescent protein. (C) Reverse transcription-quantitative PCR analysis demonstrated that the expression level of miR-362-5p was markedly decreased in SGC7901 cells transfected with miR-362-5p inhibitor compared with the control groups. (D) Viability of cells treated with cisplatin for 48 h. MTT assays were conducted on SGC7901 cells treated with various concentrations of cisplatin (0, 0.125, 0.25, 0.5, 1.0, 2.0 or 4.0 µg/ml). The IC50 for cisplatin was estimated based on the cell viability. The IC50 for cisplatin of SGC7901 cells transfected with miR-362-5p inhibitor was notably higher compared with the control groups (P<0.01). n=3; **P<0.01, ***P<0.001; IC50, median inhibitory concentration; miR-362-5p, microRNA-362-5p; NC, negative control.
miR-362-5p modulates cisplatin resistance by targeting SUZ12 in GC cells
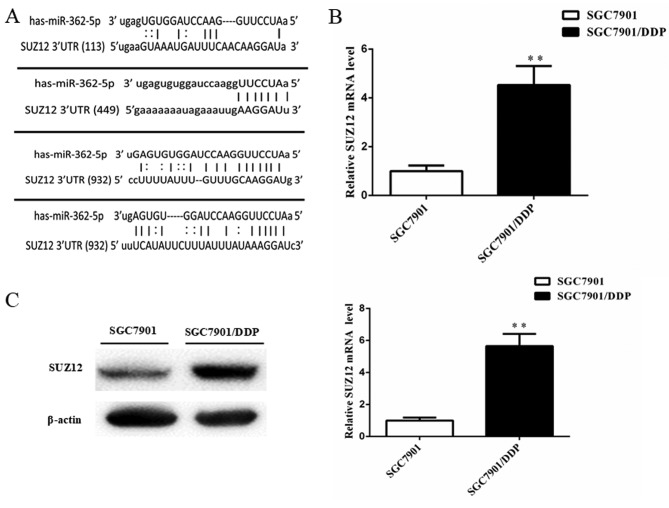
To explore the mechanism of miR-362-5p-mediated cisplatin resistance in human GC cells, the bioinformatics algorithms in the microRNA.org and miRDB databases were used to search for possible targets of miR-362-5p. The databases predicted SUZ12 as a target of miR-362-5p. The 3′-UTR of SUZ12 mRNA contains four miR-362-5p target sites at nucleotide positions 113–139, 449–472, 932–954 and 1,196-1,222 (Fig. 4A). A previous study has demonstrated that SUZ12 acts as an oncogene in GC cells (16). However, whether SUZ12 is involved in the cisplatin resistance of GC cells is not fully understood. The mRNA and protein expression levels of SUZ12 in SGC7901/DDP cells were markedly higher compared with the SGC7901 cells based on RT-qPCR and western blot analysis (Fig. 4B and C), which suggested that there may be an association between miRNA-362-5p and SUZ12 in cisplatin-resistant GC.
Figure 4.
SUZ12 may be associated with miRNA-362-5p in cisplatin-resistant GC cells. (A) Putative target sites of miR-362-5p at nucleotide positions 113–139, 449–472, 932–954 and 1196–1222 in the 3′-UTR of SUZ12 mRNA. (B) Reverse transcription-quantitative PCR analysis demonstrated that SUZ12 mRNA expression level was significantly higher in the SGC7901/DDP cells compared with the SGC7901 cells. (C) Western blotting revealed that the SUZ12 protein level was higher in the SGC7901/DDP cells compared with the SGC7901 cells; β-actin was used as a loading control. n=3; **P<0.01. 3′-UTR, 3′untranslated region; miR-362-5p, microRNA-362-5p; SUZ12, suppressor of zeste 12 protein.
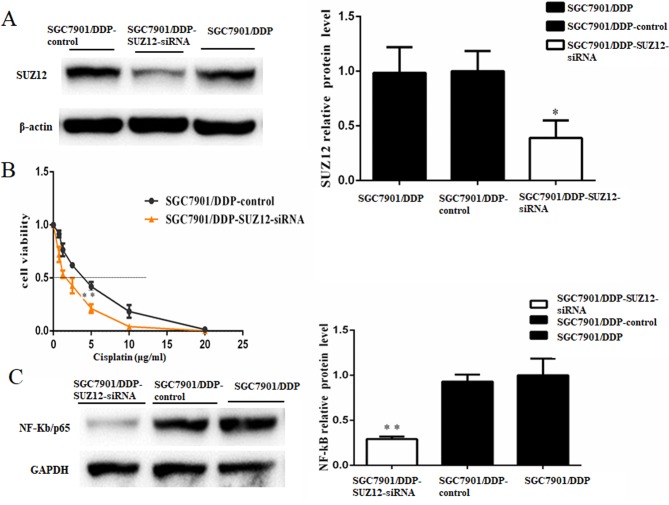
To study the role of SUZ12, si-SUZ12 was used to knock down SUZ12 expression in SGC7901/DDP cells, and the protein expression level of SUZ12 was significantly suppressed (Fig. 5A). MTT assays confirmed that knockdown of SUZ12 in SGC7901/DDP cells treated with cisplatin for 48 h significantly decreased cell survival status (IC50, 2.569±0.479 µg/ml in SGC7901/DDP-SUZ12-siRNA cells vs. 5.097±0.629 µg/ml in SGC7901/DDP cells; P<0.01; Fig. 5B). Therefore, downregulation of SUZ12 enhanced the cisplatin sensitivity of SGC7901/DDP cells. SUZ12 has been demonstrated to act as a positive regulator of NF-κB signaling (20). In the present study, western blot analysis revealed that the NF-κB/p65 protein levels were significantly decreased following SUZ12-knockdown compared with the control group (Fig. 5C).
Figure 5.
SUZ12 acts as a promoter of cisplatin resistance in GC cells. (A) Western blotting demonstrated that the protein expression level of SUZ12 was significantly reduced by si-SUZ12 transfection; β-actin was used as a loading control. (B) Viability of cells treated with cisplatin for 48 h. MTT assays were conducted on SGC7901/DDP cells treated with various concentrations of cisplatin (0, 0.75, 1.25, 2.5, 5, 10 or 20 µg/ml). The IC50 for cisplatin was estimated based on the cell viability. The IC50 for cisplatin of the SGC7901/DDP cells transfected with SUZ12-siRNA was notably lower compared with the control cells (P<0.01). (C) Western blotting demonstrated that NF-κB/p65 protein level was decreased in SGC7901/DDP cells transfected with SUZ12-siRNA compared with control cells; GADPH was used as a loading control. n=3; *P<0.05; **P<0.01; IC50, median inhibitory concentration; miR-362-5p, microRNA-362-5p; NC, negative control; siRNA, short interfering RNA; SUZ12, suppressor of zeste 12 protein.
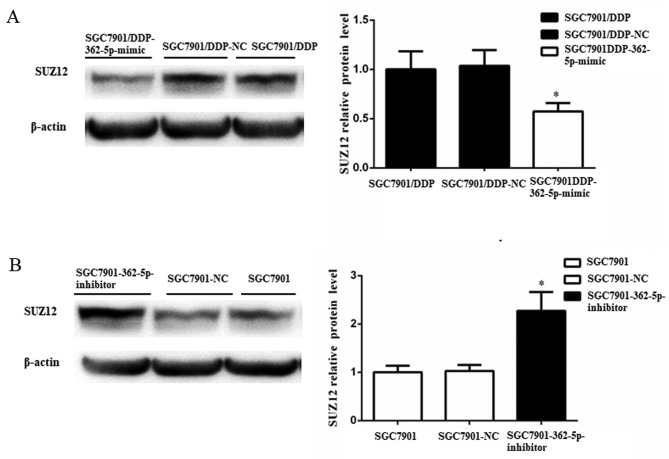
Subsequently, the changes in the expression of SUZ12 protein were further evaluated by western blot analysis after the modulation of miR-362-5p expression. SUZ12 protein expression was significantly reduced in SGC7901/DDP cells transfected with miR-362-5p mimic compared with those transfected with its negative control (Fig. 6A). Additionally, elevated expression of SUZ12 protein was detected following the downregulation of miR-362-5p in SGC7901 cells (Fig. 6B). Taken together, the results of the present study demonstrated that miR-362-5p may modulate cisplatin resistance by targeting SUZ12.
Figure 6.
Negative regulation of SUZ12 protein expression by miR-362-5p. (A) The SUZ12 protein level in SGC7901/DDP cells transfected with miR-362-5p mimic was decreased compared with NC-transfected and untransfected cells. (B) The SUZ12 protein expression level was elevated in SGC7901 cells transfected with miR-362-5p inhibitor compared with control cells. n=3; *P<0.05; miR-362-5p, microRNA-362-5p; NC, negative control; SUZ12, suppressor of zeste 12 protein.
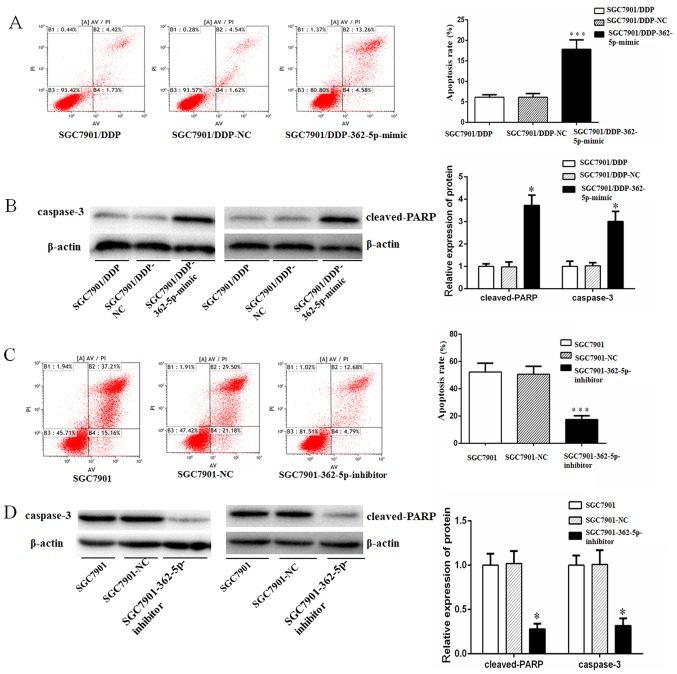
miR-362-5p sensitizes GC cells to cisplatin-induced apoptosis
A previous study has demonstrated that the development of chemotherapy resistance in a variety of cancer types is associated with decreased susceptibility to drug-induced apoptosis (21). Therefore, it was hypothesized that miR-362-5p may also serve a role in the cisplatin resistance of GC cells through the modulation of apoptosis. To test this hypothesis, cisplatin-induced apoptosis in SGC7901 and SGC7901/DDP cells transfected with miR-362-5p negative control, mimic or inhibitor, as well as untransfected cells, was quantified by flow cytometry. Additionally, the protein expression of apoptotic markers caspase-3 and cleaved PARP was determined by western blotting. The activation of caspase-3 and its downstream target PARP mediate apoptosis by chromatin condensation and DNA fragmentation (22). Following 48-h treatment with cisplatin at a final concentration of 0.8 µg/ml, the SGC7901/DDP cells overexpression miR-362-5p exhibited significantly higher apoptotic rates and expression levels of caspase-3 and cleaved-PARP compared with the control cells (Fig. 7A and B). Furthermore, following 48-h treatment with cisplatin at a final concentration of 0.3 µg/ml, the SGC7901 cells transfected with miR-362-5p inhibitor displayed significantly lower cisplatin-induced apoptosis, and expression levels of caspase-3 and cleaved-PARP compared with the control cells (Fig. 7C and D). Taken together, the results of the present study demonstrated that miR-362-5p may modulate the cisplatin resistance of human GC in part by sensitizing cells to cisplatin-induced apoptosis.
Figure 7.
miR-362-5p sensitizes gastric carcinoma cells to cisplatin-induced apoptosis. (A) Flow cytometry and (B) western blot analysis results demonstrated an increase in apoptotic rate (%) and the expression of apoptotic markers (caspase-3 and cleaved-PARP), respectively, following 48-h cisplatin treatment (final concentration of 0.8 µg/ml) in SGC7901/DDP cells transfected with miR-362-5p mimic compared with control cells. (C) Flow cytometry and (D) western blotting results indicated significant decreases in apoptotic rate and the expression of apoptotic markers (caspase-3 and cleaved-PARP), respectively, after 48-h cisplatin treatment (final concentration 0.3 µg/ml) in SGC7901 cells treated with miR-362-5p inhibitor compared with control cells. Columns indicate the mean of three independent experiments. *P<0.05; ***P<0.001. AV, annexin V-FITC; miR-362-5p, microRNA-362-5p; NC, negative control; PARP, poly-[ADP-ribose] polymerase; PI, propidium iodide.
Discussion
GC is one of the most common types of malignancy worldwide (1). To date, surgery and chemotherapy have been the major methods of GC treatment (23). For post-operative patients with GC or patients with unresectable GC, chemotherapy may increase survival rates (3). Cisplatin-based chemotherapy, as the most common chemotherapy regimen for GC, induces tumor cell death through DNA damage (24). However, its therapeutic effectiveness is limited, as demonstrated by the occurrence of cisplatin resistance, which is the primary cause of disease recurrence and metastasis, and which results in GC treatment failure (4). Numerous studies (25–27) have indicated various mechanisms of drug resistance; for instance, mutations of target genes, increases in drug efflux, enhancement of DNA repair activity or dysfunction of pro-apoptotic proteins, the mechanisms of cancer cell drug resistance are still not clearly understood. Therefore, the identification of novel molecular mechanisms of cisplatin resistance is urgently needed to improve the survival status of patients with GC.
miRNAs have been demonstrated to regulate the progression of various types of cancer (28,29). It has been extensively reported that miRNAs can modulate cisplatin resistance in tumor cells. For instance, it has been reported that miR-130a targeting copper transporter protein 1 (30) and miR-1284 targeting high mobility group box 1 (31) can regulate cisplatin resistance in cervical cancer. Furthermore, miR-144-3p targeting nuclear factor erythroid 2-related factor 2 (32) and microRNA-133b targeting epidermal growth factor receptor (33) also regulate lung cancer resistance to cisplatin. To explore novel molecular mechanisms of cisplatin resistance, the differential miRNA expression profiles among GC cells (cisplatin-resistant SGC7901/DDP cells vs. cisplatin-sensitive SGC7901 cells) were determined by miRNA microarray analysis. A total of 48 miRNAs, including miR-362-5p, were identified as significantly differentially expressed (by >2-fold). miR-362-5p, which was identified by the miRNA microarray screen and confirmed by RT-qPCR analysis, was notably downregulated in human GC SGC7901/DDP cells compared with SGC7901 cells. Therefore, it was hypothesized that miR-362-5p may be involved in the cisplatin resistance of GC cells. Though miR-362-5p has been reported to contribute to the regulation of cancer processes (11,12), its role in SGC7901/DDP cells is not fully understood. Results from the present study demonstrated the regulation of miR-362-5p in GC SGC7901 and SGC7901/DDP cells. The results also revealed that upregulation of miR-362-5p in GC cells increased cisplatin sensitivity and cisplatin-induced apoptosis, whereas downregulation of miR-362-5p in GC cells attenuated cisplatin sensitivity and cisplatin-induced apoptosis. These results revealed that miR-362-5p may act as a novel effector molecule that serves crucial roles in the regulation of the cisplatin resistance of GC cells.
Generally, miRNAs bind to a target mRNA at the 3′-UTR to promote target mRNA degradation or block target mRNA translation (6,7). Bioinformatics analysis predicted that SUZ12 may be a target of miR-362-5p. SUZ12 is a carcinogenic factor and a catalyst of migration and invasion in various cancer types (16–18). The expression of SUZ12 is significantly increased in GC tissues, and increased SUZ12 expression promotes GC cell proliferation and metastasis (16). Furthermore, SUZ12 is involved in long non-coding RNA (lncRNA)-mediated promotion of cancer progression (34). However, whether SUZ12 is involved in the cisplatin resistance of GC cells is still unknown. To verify whether the cisplatin resistance induced by miR-362-5p was mediated through SUZ12, further experiments are required. The results of the present study demonstrated that the SUZ12 expression level in SGC7901/DDP cells was markedly higher compared with expression in SGC7901 cells and that knockdown of SUZ12 enhanced the cisplatin sensitivity of SGC7901/DDP cells. These results implied that SUZ12 may act as a promoter of cisplatin resistance in GC. Notably, a previous study has confirmed that SUZ12 acts as positive regulator for NF-κB signaling (20). The NF-κB signaling pathway is involved in anti-apoptosis, inflammation and immunity. The present study demonstrated that the NF-κB/p65 protein levels were significantly decreased following knockdown of SUZ12. Therefore, SUZ12 may inhibit the cisplatin-induced apoptosis of GC cells by increasing the expression of NF-κB/p65. Notably, SUZ12 protein expression was increased in SGC7901 cells transfected with miR-362-5p inhibitor. By contrast, the expression levels of SUZ12 protein were reduced in SGC7901/DDP cells with upregulated miR-362-5p expression. Therefore, SUZ12 may be a target of miR-362-5p in GC cells.
In summary, the results of the present study revealed that miR-362-5p may sensitize SGC7901/DDP cells to cisplatin by regulating the SUZ12/NF-κB/p65 pathway. miR-362-5p may be a potential therapeutic target in combination with anti-GC chemotherapies. One potential adjunct therapy may be treatment with mimics that reinforce the expression and function of miR-362-5p. However, miR-362-5p may also non-specifically bind to other functional mRNAs to cause adverse side effects. In addition, it should be noted that experiments were conducted in only one type of GC cells. Further investigation on miR-362-5p function is therefore required.
Acknowledgements
Not applicable.
Funding
The present study was funded by The Anhui Provincial Science and Technology Plan Project (grant no. 1604b0602027).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
XW, MeG, YA and KG performed the experiments and wrote the manuscript. KG made substantial contributions to conception and design of the manuscript. XW, MeG and YA were responsible for the design of the experiments. MiG, WL, YZ, XX, QZ and HW analyzed the experimental data. KG assisted with the statistical analysis. KG critically revised the manuscript and provided final approval, and made substantial contributions to conception and design of the study. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Petrillo A, Pompella L, Tirino G, Pappalardo A, Laterza MM, Caterino M, Orditura M, Ciardiello F, Lieto E, Galizia G, et al. Perioperative treatment in resectable gastric cancer: Current perspectives and future directions. Cancers (Basel) 2019;11(pii):E399. doi: 10.3390/cancers11030399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marin JJ, Al-Abdulla R, Lozano E, Briz O, Bujanda L, Banales JM, Macias RI. Mechanisms of resistance to chemotherapy in gastric cancer. Anticancer Agents Med Chem. 2016;16:318–334. doi: 10.2174/1871520615666150803125121. [DOI] [PubMed] [Google Scholar]
- 4.Orditura M, Galizia G, Sforza V, Gambardella V, Fabozzi A, Laterza MM, Andreozzi F, Ventriglia J, Savastano B, Mabilia A, et al. Treatment of gastric cancer. World J Gastroenterol. 2014;20:1635–1649. doi: 10.3748/wjg.v20.i7.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaufmann SH, Earnshaw WC. Induction of apoptosis by cancer chemotherapy. Exp Cell Res. 2000;256:42–49. doi: 10.1006/excr.2000.4838. [DOI] [PubMed] [Google Scholar]
- 6.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Li C, Zou J, Zheng G, Chu J. MiR-30a decreases multidrug resistance (MDR) of gastric cancer cells. Med Sci Monit. 2016;0:0. doi: 10.12659/MSM.898415. [DOI] [PubMed] [Google Scholar]
- 9.Bao J, Xu Y, Wang Q, Zhang J, Li Z, Li D, Li J. miR-101 alleviates chemoresistance of gastric cancer cells by targeting ANXA2. Biomed Pharmacother. 2017;92:1030–1037. doi: 10.1016/j.biopha.2017.06.011. [DOI] [PubMed] [Google Scholar]
- 10.Ying G, Wu R, Xia M, Fei X, He QE, Zha C, Wu F. Identification of eight key miRNAs associated with renal cell carcinoma: A meta-analysis. Oncol Lett. 2018;16:5847–5855. doi: 10.3892/ol.2018.9384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ni F, Zhao H, Cui H, Wu Z, Chen L, Hu Z, Guo C, Liu Y, Chen Z, Wang X, et al. MicroRNA-362-5p promotes tumor growth and metastasis by targeting CYLD in hepatocellular carcinoma. Cancer Lett. 2015;356:809–818. doi: 10.1016/j.canlet.2014.10.041. [DOI] [PubMed] [Google Scholar]
- 12.Ni F, Gui Z, Guo Q, Hu Z, Wang X, Chen D, Wang S. Downregulation of miR-362-5p inhibits proliferation, migration and invasion of human breast cancer MCF7 cells. Oncol Lett. 2016;11:1155–1160. doi: 10.3892/ol.2015.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 14.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He LJ, Cai MY, Xu GL, Li JJ, Weng ZJ, Xu DZ, Luo GY, Zhu SL, Xie D. Prognostic significance of overexpression of EZH2 and H3k27me3 proteins in gastric cancer. Asian Pac J Cancer Prev. 2012;13:3173–3178. doi: 10.7314/APJCP.2012.13.7.3173. [DOI] [PubMed] [Google Scholar]
- 16.Xia R, Jin FY, Lu K, Wan L, Xie M, Xu TP, De W, Wang ZX. SUZ12 promotes gastric cancer cell proliferation and metastasis by regulating KLF2 and E-cadherin. Tumour Biol. 2015;36:5341–5351. doi: 10.1007/s13277-015-3195-7. [DOI] [PubMed] [Google Scholar]
- 17.Li H, Cai Q, Wu H, Vathipadiekal V, Dobbin ZC, Li T, Hua X, Landen CN, Birrer MJ, Sánchez-Beato M, Zhang R. SUZ12 promotes human epithelial ovarian cancer by suppressing apoptosis via silencing HRK. Mol Cancer Res. 2012;10:1462–1472. doi: 10.1158/1541-7786.MCR-12-0335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu C, Shi X, Wang L, Wu Y, Jin F, Bai C, Song Y. SUZ12 is involved in progression of non-small cell lung cancer by promoting cell proliferation and metastasis. Tumour Biol. 2014;35:6073–6082. doi: 10.1007/s13277-014-1804-5. [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Su SK, Li CY, Lei PJ, Wang X, Zhao QY, Cai Y, Wang Z, Li L, Wu M. The EZH1-SUZ12 complex positively regulates the transcription of NF-kB target genes through interaction with UXT. J Cell Sci. 2016;129:2343–2353. doi: 10.1242/jcs.185546. [DOI] [PubMed] [Google Scholar]
- 21.Zhu W, Shan X, Wang T, Shu Y, Liu P. miR-181b modulates multidrug resistance by targeting BCL2 in human cancer cell lines. Int J Cancer. 2010;127:2520–2529. doi: 10.1002/ijc.25260. [DOI] [PubMed] [Google Scholar]
- 22.Elmore S. Apoptosis: A review of programmed cell death. Toxicol Pathol. 2007;35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petrelli F, Zaniboni A, Ghidini A, Ghidini M, Turati L, Pizzo C, Ratti M, Libertini M, Tomasello G. Timing of adjuvant chemotherapy and survival in colorectal, gastric, and pancreatic cancer. A systematic review and meta-analysis. Cancers (Basel) 2019;11(pii):E550. doi: 10.3390/cancers11040550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu W, Chen Q, Wang Q, Sun Y, Wang S, Li A, Xu S, Røe OD, Wang M, Zhang R, et al. JWA reverses cisplatin resistance via the CK2-XRCC1 pathway in human gastric cancer cells. Cell Death Dis. 2014;5:e1551. doi: 10.1038/cddis.2014.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galluzzi L, Senovilla L, Vitale I, Michels J, Martins I, Kepp O, Castedo M, Kroemer G. Molecular mechanisms of cisplatin resistance. Oncogene. 2012;31:1869–1883. doi: 10.1038/onc.2011.384. [DOI] [PubMed] [Google Scholar]
- 26.Ghosh S. Cisplatin: The first metal based anticancer drug. Bioorg Chem. 2019;88:102925. doi: 10.1016/j.bioorg.2019.102925. [DOI] [PubMed] [Google Scholar]
- 27.Rocha CRR, Silva MM, Quinet A, Cabral-Neto JB, Menck CFM. DNA repair pathways and cisplatin resistance: An intimate relationship. Clinics (Sao Paulo) 2018;73(Suppl 1):e478s. doi: 10.6061/clinics/2018/e478s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin Y, Gu Q, Sun Z, Sheng B, Qi C, Liu B, Fu T, Liu C, Zhang Y. Upregulation of miR-3607 promotes lung adenocarcinoma proliferation by suppressing APC expression. Biomed Pharmacother. 2017;95:497–503. doi: 10.1016/j.biopha.2017.08.052. [DOI] [PubMed] [Google Scholar]
- 29.Herr I, Sähr H, Zhao Z, Yin L, Omlor G, Lehner B, Fellenberg J. MiR-127 and miR-376a act as tumor suppressors by in vivo targeting of COA1 and PDIA6 in giant cell tumor of bone. Cancer Lett. 2017;409:49–55. doi: 10.1016/j.canlet.2017.08.029. [DOI] [PubMed] [Google Scholar]
- 30.Feng C, Ma F, Hu C, Ma JA, Wang J, Zhang Y, Wu F, Hou T, Jiang S, Wang Y, Feng Y. SOX9/miR-130a/CTR1 axis modulates DDP-resistance of cervical cancer cell. Cell Cycle. 2018;17:448–458. doi: 10.1080/15384101.2017.1395533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen J, Li G. MiR-1284 enhances sensitivity of cervical cancer cells to cisplatin via downregulating HMGB1. Biomed Pharmacother. 2018;107:997–1003. doi: 10.1016/j.biopha.2018.08.059. [DOI] [PubMed] [Google Scholar]
- 32.Yin Y, Liu H, Xu J, Shi D, Zhai L, Liu B, Wang L, Liu G, Qin J. miR-144-3p regulates the resistance of lung cancer to cisplatin by targeting Nrf2. Oncol Rep. 2018;40:3479–3488. doi: 10.3892/or.2018.6772. [DOI] [PubMed] [Google Scholar]
- 33.Li B, Ding CM, Li YX, Peng JC, Geng N, Qin WW. Over-regulation of microRNA-133b inhibits cell proliferation of cisplatin-induced non-small cell lung cancer cells through PI3K/Akt and JAK2/STAT3 signaling pathway by targeting EGFR. Oncol Rep. 2018;39:1227–1234. doi: 10.3892/or.2018.6215. [DOI] [PubMed] [Google Scholar]
- 34.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.