Abstract
The expression of microRNA (miR)-100 in non-small cell lung cancer (NSCLC) and its association with clinicopathological features and poor prognosis were investigated. A total of 283 patients with NSCLC were enrolled in The First Hospital of Jiaxing from February 2013 to April 2015. Total RNA was extracted from cancer tissues and corresponding adjacent normal tissues. The expression of miR-100 was detected by RT-qPCR. Association between the expression level of miR-100 with clinicopathological features and prognosis of NSCLC were analyzed. The expression level of miR-100 in NSCLC tissues was lower than that in the normal tissues (P<0.05). According to the median expression level of miR-100 in cancer tissue, patients were divided into the high expression and low expression groups. Cross-tabulation analysis showed that the expression level of miR-100 was significantly associated with patients' age, TNM stage, metastasis and histological type (P<0.05), but not with sex (P>0.05). The proportion of patients with low miR-100 expression was higher in patients who died than in those who survived (P<0.05). Univariate prognostic analysis showed that miR-100 expression, age, TNM staging, and metastasis may be risk factors for poor prognosis in patients with NSCLC. Cox multivariate regression analysis showed that the downregulated miR-100 expression, advanced TNM stage, and metastasis were independent risk factors for poor prognosis of NSCLC. The relatively low expression level of miR-100 in NSCLC is associated with poor prognosis of patients. Therefore, miR-100 shows potential as a prognostic marker for NSCLC.
Keywords: miR-100, pathological features, prognosis, non-small cell lung cancer, correlation
Introduction
Lung cancer develops from the respiratory system. Incidence and mortality rate of lung cancer ranks first among all malignancies in the world. With the aggregated environmental pollution, increased number of smokers and growth of aging population, incidence of lung cancer shows an increasing trend (1,2). Lung cancer affects 1.6 million individuals every year, and this number is still increasing. As the most common type of lung cancer, non-small cell lung cancer (NSCLC) accounts for 80–85% of all the cases (3,4).
Radiotherapy and chemotherapy are conventional treatments for NSCLC. However, chemotherapy has been proved to be ineffective in improving the survival of patients (5,6). In recent years, studies on patients with stage II and IIIa NSCLC have shown that postoperative radiotherapy significantly shortens patients' survival time. Therefore, identification of novel therapeutic targets is urgently needed (7,8). miRNAs are widely expressed in eukaryotic cell organisms and regulate cell proliferation, differentiation, and apoptosis. Abnormal changes in miRNA biosynthesis are involved in a variety of pathophysiological processes (9,10). Chen et al (11) reported that the downregulation of miR-100 is closely related to the progression and prognosis of hepatocellular carcinoma. Xu et al (12) found that miR-100 is downregulated in human bladder epithelial carcinoma and that the elevated miR-100 expression in bladder cancer cells inhibits cell proliferation and migration.
However, the involvement of miR-100 in lung cancer still has not been reported. In this study, the expression of miR-100 in 283 patients with NSCLC was investigated and the association with patients' clinicopathological features was investigated to study the effects of miR-100 on patient prognosis.
Patients and methods
Subjects
A total of 283 patients with NSCLC were selected from February 2013 to April 2015 in The First Hospital of Jiaxing (Jiaxing, China). The patients included 128 males and 155 females, with a mean age of 56.32±11.03 years. All patients were pathologically diagnosed as NSCLC, including 176 patients with invasive adenocarcinoma, 70 patients with squamous cell carcinoma and 37 large cell carcinoma. None of the patients had previous history of a tumor. Patients with organ dysfunction such as liver or kidney, abnormal bleeding or abnormal coagulation function were excluded. All the patients have complete clinical and follow-up data. Patients who received treatment, patients with large tumors, patients with other pulmonary or chest wall disease, and patients who died of other diseases were excluded. This study was approved by the Ethics Committee of The First Hospital of Jiaxing. All patients or their families signed an informed consent.
Extraction of total RNA
Total RNA was extracted from cancer and normal adjacent tissues using TRIzol reagent (Shanghai Mingjing Biotechnology Co., Ltd.) according to the manufacturers instructions. A micro-ultraviolet spectrophotometer MD1000 (Thmorgan Biotechnology Co., Ltd.) was used to measure the concentration and analyze the purity of RNA samples, and 3% agarose gel electrophoresis (Jingke Chemical Technology Co., Ltd.) was used to analyze the integrity of RNA.
Total RNA was subjected to reverse transcription (45°C for 45 min and 95°C for 5 min) to synthesize cDNA, followed by preparation of PCR reaction system using fluorescence quantitative SYBR-Green PCR kit (cat. no. 4364344; Thermo Fisher Scientific, Inc.). PCR reaction conditions were: 95°C for 10 min, followed by 95°C for 10 sec, 60°C for 20 sec, 72°C for 10 sec, and 72°C for 5 min. U6 was used as an endogenous control, and each experiment was performed 3 times. Data were analyzed by the 2−ΔΔCq method (13). Primers were synthesized by Suzhou Yaxun Biotechnology Co., Ltd. The primer sequences are shown in Table I.
Table I.
Primer sequences.
| Variables | Forward primers | Reverse primers |
|---|---|---|
| miR-100 | 5′-CGACGAGGC | 5′-CCATCGATG |
| GTTGCCTGCACC-3′ | GAATCTTTAAC-3 | |
| U6 | 5′-CGCTTCGGC | 5′-TTCACGAAT |
| AGCACATATAC-3′ | TTGCGTGTCAT-3′ |
miR-100, microRNA-100.
Observation indicators
Association between the expression of miR-100 and clinicopathological features of NSCLC were explored. All patients were followed up for a maximum of 60 months. The association between miR-100 expression and survival was analyzed.
Statistical analysis
SPSS19.0 [AsiaAnalytics (formerly SPSS China)] was used for all statistical analyses. Enumeration data were expressed as rate and compared by χ2 test. Measurement data were expressed as mean ± standard deviation and normal distribution was tested by K-S test. Normal distribution measurement data were compared by t-test, and non-normal distribution data were compared by Chi-square test. Association between the miR-100 expression and clinicopathological features of patients were analyzed by cross-tabulation analysis. Univariate analysis was performed using Kaplan-Meier analysis and the log-rank test. Multivariate analysis was performed using the Cox model for multivariate analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
General information
Among the 283 NSCLC patients, there were 176 invasive adenocarcinoma, 70 squamous cell carcinoma, and 37 large cell carcinoma. According to TNM staging, 113 patients were in stage I, 71 in stage II, 88 in stage III, and 11 in stage IV. Follow-up time was no more than 60 months, and median was 31 months, and mean follow-up time was 25.32±14.33 months. During follow-up, 155 patients died and lymph node metastasis occurred in 150 patients (Table II).
Table II.
General information.
| Variables | No. (%) |
|---|---|
| Sex [n (%)] | |
| Male | 128 (45.23) |
| Female | 155 (54.77) |
| Age | 56.32±11.03 |
| Histological type [n (%)] | |
| Invasive adenocarcinoma | 176 (62.19) |
| Squamous cell carcinoma | 70 (24.73) |
| Large cell carcinoma | 37 (13.07) |
| TNM stage [n (%)] | |
| I | 113 (39.93) |
| II | 71 (25.09) |
| III | 88 (31.10) |
| IV | 11 (3.89) |
| Follow-up time (month) | |
| Median follow-up time | 31 |
| Mean follow-up time | 25.32±14.33 |
| Follow-up results [n (%)] | |
| Survival | |
| Metastasis | 15 (5.30) |
| Non-metastasis | 113 (39.93) |
| Death | |
| Metastasis | 135 (47.70) |
| Non-metastasis | 20 (7.07) |
TNM, tumor-node-metastasis.
Expression of miR-100 in cancer tissues and adjacent healthy tissues of NSCLC patients
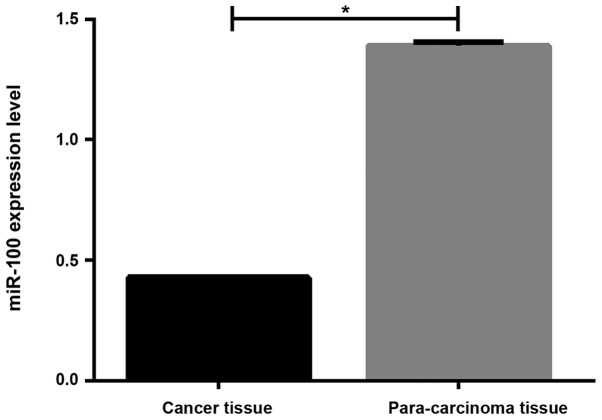
RT-qPCT results showed that the miR-100 expression level was significantly lower in tumor tissues than in adjacent tissues (0.429±0.004 vs. 1.292±0.013, P<0.05, Fig. 1).
Figure 1.
Comparison of miR-100 expression levels in cancer and adjacent normal tissues in patients with NSCLC. RT-qPCR results showed that miR-100 expression level was significantly lower in cancer tissues than in adjacent tissues (0.429±0.004 vs. 1.292±0.013, P<0.05). *P<0.05; NSCLC, non-small cell lung cancer; miR-100, microRNA-100.
Association between miR-100 expression and clinicopathological features
According to the median expression level of miR-100 in cancer tissue (0.413), patients were divided into the high expression and low expression groups. Results of cross-tabulation analysis showed that the low expression level of miR-100 was associated with the patients' age, TNM stage, metastasis, histological type (P<0.05), but not sex (P>0.05). The proportion of patients with low miR-100 expression was higher in patients who died than in those who survived (P<0.05; Table III).
Table III.
Association between miR-100 expression and clinicopathological features.
| Variables | Low expression | High expression | χ2 value | P-value |
|---|---|---|---|---|
| Cases | 170 | 113 | ||
| Sex | 1.134 | 0.233 | ||
| Male | 75 | 53 | ||
| Female | 95 | 60 | ||
| Age | 2.050 | 0.042 | ||
| <56.32 years | 71 | 59 | ||
| ≥56.32 years | 99 | 54 | ||
| TNM stage | 2.039 | 0.041 | ||
| I+II | 101 | 83 | ||
| III+IV | 69 | 30 | ||
| Metastasis | 8.138 | 0.001 | ||
| Yes | 145 | 5 | ||
| No | 25 | 108 | ||
| Histological type | 2.012 | 0.045 | ||
| Invasive adenocarcinoma (n=176) | 113 | 63 | ||
| Squamous cell carcinoma (n=70) | 36 | 34 | ||
| Large cell carcinoma (n=37) | 21 | 16 | ||
| Follow-up results | 2.349 | 0.019 | ||
| Survival | 61 | 67 | ||
| Death | 109 | 46 |
miR-100, microRNA-100; TNM, tumor-node-metastasis.
Univariate and multivariate analysis of association between miR-100 expression and prognosis using Kaplan-Meier analysis and the log-rank test
Cox single factor regression analysis was performed on 283 patients. Univariate prognostic analysis showed that the miR-100 expression level, age, TNM staging, and metastasis may be risk factors for poor prognosis in patients with NSCLC. Cox multivariate regression analysis showed that the low miR-100 expression levels, advanced TNM stages and tumor metastasis are independent risk factors for poor prognosis of NSCLC (Tables IV and V).
Table IV.
Univariate and multivariate analysis of association between miR-100 expression and prognosis.
| Univariate analysis | ||
|---|---|---|
| Variables | HR (95% CI) | P-value |
| miRNA-100 (low vs. high) | 1.648 (1.122–2.896) | 0.010 |
| Sex (male vs. female) | 1.012 (0.797–1.620) | 0.559 |
| Age (<56.32 vs. ≥56.32 years) | 1.132 (1.028–1.715) | 0.014 |
| TNM stages (I and II vs. III and IV) | 2.821 (1.346–2.857) | 0.013 |
| Metastasis (yes vs. no) | 3.053 (1.282–7.323) | 0.011 |
| Histological type (ACC vs. SCC) | 0.921 (0.831–1.525) | 0.325 |
miR-100, microRNA-100; TNM, tumor-node-metastasis; ACC, invasive adenocarcinoma; SCC, squamous cell carcinoma.
Table V.
Multivariate analysis of association between miR-100 expression and prognosis.
| Multivariate analysis | ||
|---|---|---|
| Variables | HR (95% CI) | P-value |
| miRNA-100 (low vs. high) | 1.328 (1.022–2.416) | 0.006 |
| TNM stages (I and II vs. III and IV) | 2.231 (1.357–2.547) | 0.005 |
| Metastasis (yes vs. no) | 3.053 (1.282–7.323) | 0.011 |
miR-100, microRNA-100; TNM, tumor-node-metastasis.
Discussion
Occurrence and development of NSCLC, including adenocarcinoma, squamous cell carcinoma, and large cell carcinoma, are closely related to internal gene expression and external environment (14). miR-100 is located on human chromosome 11q24.1 position, and its nucleic acid sequence is highly conserved, and may be closely related to the growth and development of human body (15). Studies have shown that miR-100 is closely associated with the occurrence, development, invasion and metastasis of colorectal, endometrial and breast cancers (16–18). In this study, we explored the expression and clinical significance of miR-100 in patients with NSCLC with an expectation of providing a new target for clinical treatment and prognosis assessment of this disease.
Results of this study showed that the expression level of miR-100 was significantly lower in cancer tissues of NSCLC patients than in normal adjacent tissues. Therefore, we hypothesized that miR-100 may act as a tumor suppressor gene in this disease. Huang et al (19) found that the miR-100 expression was low in cancerous tissues of patients with pancreatic cancer. Leite et al (20) also showed that miR-100 functions as a tumor suppressor gene in patients with prostate cancer. In this study, we analyzed the association between the expression level of miR-100 and clinicopathological features of patients with NSCLC. We found that the low expression level of miR-100 was associated to the patients' age, TNM stage, metastasis, and histological type. The involvement of miR-100 in NSCLC still has not been well studied. Chen et al (11) found that the low expression of miR-100 was closely associated with clinical grade, lymph node metastasis, and TNM stages of hepatocellular carcinoma. Consistent with this study, we also found that advanced tumor stages and tumor metastasis were associated with the low expression level of miR-100. However, Chen et al (11) reported no significant association between the low expression of miR-100 and the age of the patients, which may be explained by the number of subjects included in this study. We also analyzed the association between miR-100 expression and patients' 5-year follow-up results. We found that the proportion of patients with low miR-100 expression was higher in patients who died than in those who survived. Therefore, we hypothesize that the low expression level of miR-100 is closely related to the patients' age, TNM staging, metastasis, histological type, and affects the prognosis of patients.
We analyzed the expression level of miR-100 and prognosis of patients. Analysis of the results showed that the low expression of miR-100, advanced TNM stage and metastasis are independent risk factors for NSCLC. A meta-analysis carried out by Chen et al (21) showed that expression of miR-100 is associated with the survival of cancer patients and may be a clinical prognostic factor for cancer. Cao et al (22) also found that the low expression of miR-100 may be a prognostic risk factor for bladder cancer. Similar results were found in this study, indicating that expression level of miR-100 may serve as a predictor of prognosis of NSCLC. Our study provided new insights for diagnosis and treatment of NSCLC. However, only cancer tissues and adjacent healthy tissues were used in this study and the clinical value of serum miR-100 was not investigated. In addition, the sample size of this study is small. Therefore, further studies with a larger number of samples are needed to confirm the conclusions in this study.
In summary, the expression level of miR-100 is relatively lower in cancer tissues than in normal adjacent tissues in NSCLC patients. The low expression level of miR-100 is closely associated with poor prognosis of patients. Therefore, miR-100 shows potential as a prognostic marker for NSCLC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
XM drafted the manuscript and performed PCR. JZ and HM extracted total RNA. YY was responsible for statistical analysis. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of The First Hospital of Jiaxing (Jiaxing, China). Signed informed consents were obtained from the patients and/or guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lortet-Tieulent J, Soerjomataram I, Ferlay J, Rutherford M, Weiderpass E, Bray F. International trends in lung cancer incidence by histological subtype: adenocarcinoma stabilizing in men but still increasing in women. Lung Cancer. 2014;84:13–22. doi: 10.1016/j.lungcan.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 3.Markou A, Sourvinou I, Vorkas PA, Yousef GM, Lianidou E. Clinical evaluation of microRNA expression profiling in non small cell lung cancer. Lung Cancer. 2013;81:388–396. doi: 10.1016/j.lungcan.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 4.Yang JJ, Chen HJ, Yan HH, Zhang XC, Zhou Q, Su J, Wang Z, Xu CR, Huang YS, Wang BC, et al. Clinical modes of EGFR tyrosine kinase inhibitor failure and subsequent management in advanced non-small cell lung cancer. Lung Cancer. 2013;79:33–39. doi: 10.1016/j.lungcan.2012.09.016. [DOI] [PubMed] [Google Scholar]
- 5.Brahmer J, Reckamp KL, Baas P, Crinò L, Eberhardt WE, Poddubskaya E, Antonia S, Pluzanski A, Vokes EE, Holgado E, et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med. 2015;373:123–135. doi: 10.1056/NEJMoa1504627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herbst RS, Baas P, Kim DW, Felip E, Pérez-Gracia JL, Han JY, Molina J, Kim JH, Arvis CD, Ahn MJ, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet. 2016;387:1540–1550. doi: 10.1016/S0140-6736(15)01281-7. [DOI] [PubMed] [Google Scholar]
- 7.Shaw AT, Kim DW, Nakagawa K, Seto T, Crinó L, Ahn MJ, De Pas T, Besse B, Solomon BJ, Blackhall F, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med. 2013;368:2385–2394. doi: 10.1056/NEJMoa1214886. [DOI] [PubMed] [Google Scholar]
- 8.Molina R, Filella X, Augé JM, Fuentes R, Bover I, Rifa J, Moreno V, Canals E, Viñolas N, Marquez A, et al. Tumor markers (CEA, CA 125, CYFRA 21-1, SCC and NSE) in patients with non-small cell lung cancer as an aid in histological diagnosis and prognosis. Comparison with the main clinical and pathological prognostic factors. Tumour Biol. 2003;24:209–218. doi: 10.1159/000074432. [DOI] [PubMed] [Google Scholar]
- 9.Okamura K, Takayama K, Izumi M, Harada T, Furuyama K, Nakanishi Y. Diagnostic value of CEA and CYFRA 21-1 tumor markers in primary lung cancer. Lung Cancer. 2013;80:45–49. doi: 10.1016/j.lungcan.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 10.Köttgen A, Albrecht E, Teumer A, Vitart V, Krumsiek J, Hundertmark C, Pistis G, Ruggiero D, O'Seaghdha CM, Haller T, et al. LifeLines Cohort Study; CARDIoGRAM Consortium; DIAGRAM Consortium; ICBP Consortium; MAGIC Consortium Genome-wide association analyses identify 18 new loci associated with serum urate concentrations. Nat Genet. 2013;45:145–154. doi: 10.1038/ng.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen P, Zhao X, Ma L. Downregulation of microRNA-100 correlates with tumor progression and poor prognosis in hepatocellular carcinoma. Mol Cell Biochem. 2013;383:49–58. doi: 10.1007/s11010-013-1753-0. [DOI] [PubMed] [Google Scholar]
- 12.Xu C, Zeng Q, Xu W, Jiao L, Chen Y, Zhang Z, Wu C, Jin T, Pan A, Wei R, et al. miRNA-100 inhibits human bladder urothelial carcinogenesis by directly targeting mTOR. Mol Cancer Ther. 2013;12:207–219. doi: 10.1158/1535-7163.MCT-12-0273. [DOI] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Ashworth A, Rodrigues G, Boldt G, Palma D. Is there an oligometastatic state in non-small cell lung cancer? A systematic review of the literature. Lung Cancer. 2013;82:197–203. doi: 10.1016/j.lungcan.2013.07.026. [DOI] [PubMed] [Google Scholar]
- 15.Fu HL, Wu DP, Wang XF, Wang JG, Jiao F, Song LL, Xie H, Wen XY, Shan HS, Du YX, et al. Altered miRNA expression is associated with differentiation, invasion, and metastasis of esophageal squamous cell carcinoma (ESCC) in patients from Huaian, China. Cell Biochem Biophys. 2013;67:657–668. doi: 10.1007/s12013-013-9554-3. [DOI] [PubMed] [Google Scholar]
- 16.Yang XD, Xu XH, Zhang SY, Wu Y, Xing CG, Ru G, Xu HT, Cao JP. Role of miR-100 in the radioresistance of colorectal cancer cells. Am J Cancer Res. 2015;5:545–559. [PMC free article] [PubMed] [Google Scholar]
- 17.Qin C, Huang RY, Wang ZX. Potential role of miR-100 in cancer diagnosis, prognosis, and therapy. Tumour Biol. 2015;36:1403–1409. doi: 10.1007/s13277-015-3267-8. [DOI] [PubMed] [Google Scholar]
- 18.Gong Y, He T, Yang L, Yang G, Chen Y, Zhang X. The role of miR-100 in regulating apoptosis of breast cancer cells. Sci Rep. 2015;5:11650. doi: 10.1038/srep11650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang JS, Egger ME, Grizzle WE, McNally LR. MicroRNA-100 regulates IGF1-receptor expression in metastatic pancreatic cancer cells. Biotech Histochem. 2013;88:397–402. doi: 10.3109/10520295.2012.762460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leite KR, Morais DR, Reis ST, Viana N, Moura C, Florez MG, Silva IA, Dip N, Srougi M. MicroRNA 100: a context dependent miRNA in prostate cancer. Clinics (São Paulo) 2013;68:797–802. doi: 10.6061/clinics/2013(06)12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J, Zheng B, Wang C, Chen Y, Du C, Zhao G, Zhou Y, Shi Y. Prognostic role of microRNA-100 in various carcinomas: evidence from six studies. Tumour Biol. 2014;35:3067–3071. doi: 10.1007/s13277-013-1398-3. [DOI] [PubMed] [Google Scholar]
- 22.Cao YH, Zhang HH, Xu HF, Duan YJ, Li Q, Huang B. Prognostic role of microRNA-100 in patients with bladder cancer. Genet Mol Res. 2015;14:15948–15954. doi: 10.4238/2015.December.7.6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.