Abstract
Tumor associated macrophages (TAMs) are a major type of inflammatory cell in a tumor microenvironment. Previous evidence has suggested that TAMs promote tumorigenesis, growth, invasion and metastasis, thereby affecting tumor metabolism. The mechanisms through which they affect the invasion and metastasis of lung cancer cells remain unclear. The present study investigated the effects and molecular mechanisms of TAMs on the proliferation, invasion and migration of lung adenocarcinoma A549 cells. Human mononuclear leukemia THP-1 cells were induced into TAMs. The morphological changes that occurred during the induction of the THP-1 cells were examined with a light microscope. Successful TAM formation was confirmed via flow cytometry. Proliferation, invasion and migration of the lung adenocarcinoma A549 cells were detected by EDU proliferation, scratch wound and Transwell invasion and migration assays, respectively. The expression levels of key proteins involved in the PI3K/AKT signaling pathway were detected by western blot analysis. It was identified that treatment with interleukin (IL)-4, IL-13 and Phorbol-12-myristate-13-acetate successfully induced THP-1 to form TAMs. The indirect coculture model of TAMs was established by Transwell chamber detection, and the proliferation, invasion and migration ability of lung adenocarcinoma A549 cells were enhanced. Western blot analysis indicated that the expression levels of phosphorylated (p)-PI3K and p-AKT proteins were significantly upregulated in the TAMs coculture group compared with that of the blank control group. In summary, the present study demonstrated that TAMs may promote the proliferation, invasion and migration of lung adenocarcinoma A549 cells in vitro, perhaps through the activation of the PI3K/AKT signaling pathway.
Keywords: lung neoplasms, tumor-associated macrophages, cell proliferation, invasion, migration
Introduction
Lung cancer is the most common type of malignant tumor, accounting for 11.6% of total global cancer incidence and 18.4% of cancer-associated mortalities (1). As lung cancer has no specific symptoms and signs, patients usually present with advanced stage disease at primary diagnosis. Despite the availability of comprehensive treatments, including active surgery and radiotherapy, a large percentage of patients inevitably succumb to the disease due to recurrence and distant metastasis. A study of risk factors associated with lung cancer and a comprehensive understanding of its pathogenesis is vital in the prevention and treatment of this disease.
The tumor microenvironment is the environment surrounding a tumor that may promote immune escape and induce tumor resistance (2–4). Macrophages are an important component of the tumor microenvironment. They may be divided into two primary groups, termed M1 and M2, which may be identified by their cell surface markers and functional phenotypes (5). Tumor-associated macrophages (TAMs) are recruited to tumors as monocytes from the blood, and as they escape across the vasculature of the tumor, they differentiate into macrophages. Immature monocytes have good plasticity and may therefore be induced by interleukin (IL)-4 and IL-13 to form M2 macrophages, which have similar functions to TAMs (6,7). In general, increased levels of M1-like infiltrates in the tumor are correlated with an improved prognosis, exhibiting an antitumoral effect, whereas increased levels of M2-like TAM infiltrate are correlated with poor prognoses, demonstrating pro-tumoral effects (8,9). TAMs are abundant in all the stages of tumor progression (4). In early phases, macrophages recognize the malignant cells and present their antigens to the effectors of the immune system. In later stage disease, TAMs serve a role in tumor progression by stimulating tumor growth, angiogenesis, metastasis and immunosuppression (10). By producing growth factors, they stimulate carcinoma cell proliferation (11). They are also able to produce proteolytic enzymes that digest the extracellular matrix to assist with tumor cell dissemination. Finally, they provide a supportive niche for metastatic tumor cells at distant sites (12).
Previous studies have indicated that TAMs may be a diagnostic and prognostic indicator for malignant pleural effusion (13,14), thereby suggesting a close association with lung cancer invasion and metastasis. Stromal macrophages expressing macrophage scavenger receptor 1 are associated with tumor aggressiveness in lung adenocarcinoma (15). A previous study hypothesized that the developmental origin of TAMs dictates their relative distribution, function and response to cancer therapies in lung tumors (16). Furthermore, the distribution patterns of M1 and M2 macrophages in tumor islets and stroma were demonstrated to be associated with the prognosis of non-small cell lung cancer (NSCLC); high levels of M1 macrophage infiltration in the tumor islets and low levels of total tumor-infiltrating M2 macrophages were associated with improved survival of patients with NSCLC (17). However, the mechanism of its effect on lung cancer invasion and metastasis remains unclear. The aim of the present study was to investigate the effects of TAMs on the proliferation, invasion and migration of lung adenocarcinoma A549 cells, and to explore its molecular mechanism.
Materials and methods
Cell culture and treatment
A549 and THP-1 human mononuclear leukemia cell lines were provided by the Jiangxi Provincial Key Laboratory of Molecular Medicine. A549 cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Biological Industries Ltd.) supplemented with 10% fetal bovine serum (FBS; Biological Industries Ltd.) and maintained in a humidified atmosphere of 5% CO2 and 95% air at 37°C. THP-1 cells were cultured in RPMI-1640 medium supplemented with 10% FBS and 0.05 mM β-mercaptoethanol in a humidified atmosphere of 5% CO2 and 95% air at 37°C.
Induced differentiation of THP-1 cells
The THP-1 cells were cultured and differentiation was induced (β-mercaptoethanol was removed during induction, and the serum was inactivated). Initially, the cells were cultured in RPMI-1640 medium supplemented with 320 nM phorbol 12-myristate 13-acetate (PMA, ab120297). After 48 h, the cells adapted from suspension to adherent growth (cultured M0 macrophages). Thereafter, the original medium was discarded and changed to RPMI-1640 medium supplemented with 20 ng/ml IL-4 and IL-13 for 72 h. The majority of the cells protruded from pseudopods, and were identified as TAMs. Following repeated washing with aseptic PBS, subsequent co-cultivation was possible. The cytokine recombinant human IL-4 (200–04)/IL-13 (200–13) was purchased from PeproTech, Inc.
ELISA assay
Following normal culture of THP-1 and TAMs for 48 h, cell culture supernatant was centrifuged for 20 min at 1,000 × g and at a temperature of 2–8°C. Cell debris and impurities were removed and supernatant was collected. Samples were stored at −20°C to avoid repeated freezing and thawing. These assays were performed according to the manufacturer's instructions, using the Invitrogen ELISA Kit (cat. no., KHC0101; Thermo Fisher Scientific, Inc.) for human IL-10. The standard curve assay range was 7.8–500 pg/ml, sensitivity <1 pg/ml.
Flow cytometry
M0 and TAMs were digested with trypsin without phenol red. THP-1 cells grown normally without treatment were used as normal controls. They were resuspended following low-speed (600 × g) centrifugation at room temperature for 5 min. A total of 1×106 cells were counted in each flow tube, then washed 3 times with sterile PBS. Cluster of differentiation (CD) 14 (cat. no., 555399; dilution 1:5) and CD163 (cat. no., 556018; 1:5) flow and isotype control antibodies (BD Biosciences) were then added and incubated for 45 min in the dark at room temperature. Following washing, they were resuspended in 500 µl PBS and examined using a flow cytometer. The results were analyzed using BD FACSDiva 6.1 software (BD Biosciences).
Transwell chamber indirect coculture and experimental grouping
Using a 0.4 µm Transwell coculture system (Sigma-Aldrich; Merck KGaA), THP-1 cells and TAMs were indirectly cocultured with A549 cells, respectively. A549 cells in the logarithmic growth phase were plated in 6- or 24-well plates (lower chamber) and untreated THP-1 cells or induced TAMs were placed in the Transwell chamber (upper chamber). The experimental groups were as follows: Blank control (A549 cells were cultured in 1640-RPMI medium containing 1% FBS); THP-1 cell coculture (A549 cells cocultured with THP-1 cells); and TAMs coculture (A549 cells cocultured with TAMs).
EDU proliferation experiment
Each group contained 3 duplicate wells. A549 cells were seeded at a density of 1.0×104/600 µl in DMEM medium and placed in a 24-well plate. The normal culture was at 70–80% confluence. The old medium was removed via a Transwell chamber membrane (0.4 µM). THP-1 cells and TAMs cells were seeded at a density of 1×105/200 µl according to the aforementioned experimental coculture groups. The Transwell membrane was removed after 24 h and suspended in DMEM medium containing 50 µM EDU reagent A (Guangzhou RiboBio Co., Ltd.) for 2 h, following removal of the lower chamber culture medium. Cells were fixed with 4% formaldehyde for 15 min at room temperature and treated with 0.5% Triton ×100 for 20 min at room temperature to permeabilize cells. After being washed with PBS three times, cells were incubated with 1X Apollo reaction cocktail (200 µl/well) for 30 min. DNA was stained with 10 µg/ml of Hoechst 33342 stain (200 µl/well) for 20 min and visualized with fluorescence microscopy. Images were captured with a fluorescence microscope (magnification, ×100). Five groups of confluent cells were randomly selected from each sample image. EdU-positive cells were counted in the fluorescent image, and the relative positive ratio was calculated as the number of red positive cells as a percentage of blue DAPI-stained cells from the average of the five group values.
Transwell invasion and migration assay
A549 cells cocultured with THP-1 cells or TAMs for 48 h were trypsinized, centrifuged at 600 × g for 5 min at room temperature and resuspended in serum-free DMEM medium, and cell density was adjusted to 1×105/ml. The cell suspension (200 µl) was added to the upper chamber, which was precoated with 50 µg/ml Matrigel gel at 37°C for 30 min, and 600 µl DMEM supplemented with 10% FBS was added to the lower chamber. After 24 h, the Transwell membrane (8 µm) was removed and fixed with 100% methanol at room temperature for 30 min. The cells were wiped with a cotton swab to avoid penetration of the membranes of the upper chamber, and then stained with 0.1% crystal violet at room temperature for 20 min. A light microscope (magnification, ×100) was used to capture images and count the number of transmembrane cells and examine the invasive ability of A549 cells. The protocols for the Transwell migration and invasion experiments were identical, with the exception of the use of Matrigel gel in the invasion experiments only.
Scratch wound assay
A549 cells were seeded in 6-well plates. At a confluence of 80–90%, a sterile 200 µl pipette tip was used to scratch cells. Following 2 washing steps with sterile PBS, fresh DMEM high glucose medium containing 1% FBS was added. In the upper chambers of Transwell (membrane pore size, 0.4 µm), THP-1 or TAM cells (1×105 cells/well) were plated in 500 µl RPMI-1640 medium supplemented with 1% FBS, and incubated at 37°C for 2 h. Transwells were then placed into the 6-well plates containing A549 cells which had been previously prepared. Images were captured under an light microscope (magnification, ×100) at 0 and 24 h, respectively, and the wound area was measured.
Western blot analysis
Following treatment, A549 cells were harvested and lysed using radioimmunoprecipitation assay lysis buffer (Thermo Fisher Scientific, Inc.) supplemented with PMSF (1 mM). The whole cell lysates were centrifuged at 12,000 × g for 15 min at 4°C, and protein content in the supernatant was determined by the bicinchoninic acid protein assay kit (Applygen Technologies, Inc.). Proteins samples (30 µg per lane) were separated by 10% SDS-PAGE and transferred to PVDF membranes at 4°C for 90 min. After blocking with 5% non-fat milk at room temperature for 60 min, membranes were incubated with the following primary antibodies overnight at 4°C: Anti-PI3K p85α (cat. no., ab191606; 1:1,000); anti-phosphorylated (p)-PI3K p85α (Y607; cat. no., ab182651; 1:1,000); anti-AKT1/2/3 (cat. no., ab179463; 1:10,000); anti-p-AKT1 (Ser473; cat. no., ab81283; 1:1,000); and anti-GAPDH (cat. no., ab181602; 1:10,000). All primary antibodies were purchased from Abcam. The membranes were then probed with IgG-horseradish peroxidase-conjugated goat anti-rabbit (cat. no., ab2057; Abcam; 1:10,000) and goat anti-mouse (cat. no., ab6789; Abcam; 1:10,000) secondary antibodies for 1 h at room temperature. Finally, the proteins were detected using ECL Enhanced Chemiluminescent kit (Thermo Fisher Scientific, Inc.). Densitometric analysis was performed using ImageJ 1.44p software (National Institutes of Health).
Statistical analysis
Experiments were performed in triplicate. Data are presented as the mean ± standard deviation. Statistical analysis was performed using GraphPad Prism 6 software (GraphPad Software, Inc.). Data were evaluated using one-way ANOVA with Tukey's post hoc test or unpaired two-tailed Student's t-test. P<0.05 was considered to indicate a statistically significant difference.
Results
Formation of TAMs
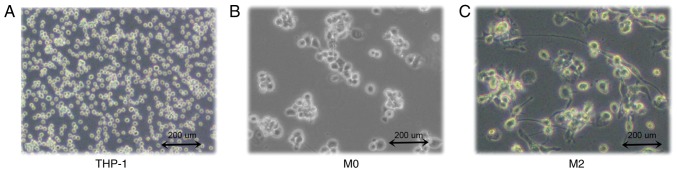
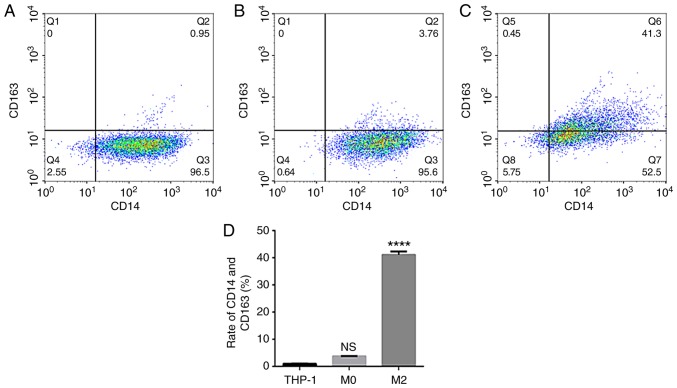
THP-1 human mononuclear leukemia cells were successfully induced to form TAMs (Fig. 1A). After 48 h of PMA stimulation, the cells adhered to the wall of the culture plate but no longer proliferated. Cell became large, with round or elliptical morphologies, and there was a pseudopod extension (Fig. 1B). After 72 h of stimulation with IL-4 and IL-13, the majority of the cells were spindle-shaped, protruding from the pseudopod (Fig. 1C). Then, ELISA was used to detect the expression of IL-10 in THP-1 and TAM culture supernatants. The results indicated that IL-10 expression levels in the TAM culture supernatant were significantly increased compared with the THP-1 supernatant (P<0.0001; Fig. 2). The flow cytometry data suggested that the expression rate of uninduced THP-1 CD14+/CD163+ cells was 0.96±0.09% (Fig. 3A). The expression rate of CD14+/CD163+ cells after 48 h of PMA stimulation was 3.77±0.10% (Fig. 3B). The expression rate of CD14+/CD163+ cells following stimulation with IL-4 and IL-13 was 41.10±1.21% (Fig. 3C), and the successful induction of CD14+/CD163+ cells was demonstrated in the TAMs culture group (Fig. 3D). The rate of expression was significantly increased compared with that of uninduced THP-1 cells, and the difference was statistically significant (P<0.0001).
Figure 1.
Differentiation of THP-1 human monocytic leukemia cells into TAMs. (A) THP-1 human monocytic leukemia cells. (B) Stage I induced undifferentiated macrophages (M0). (C) Stage II induced TAMs. Images are at magnification, ×100. TAMs, tumor associated macrophages.
Figure 2.
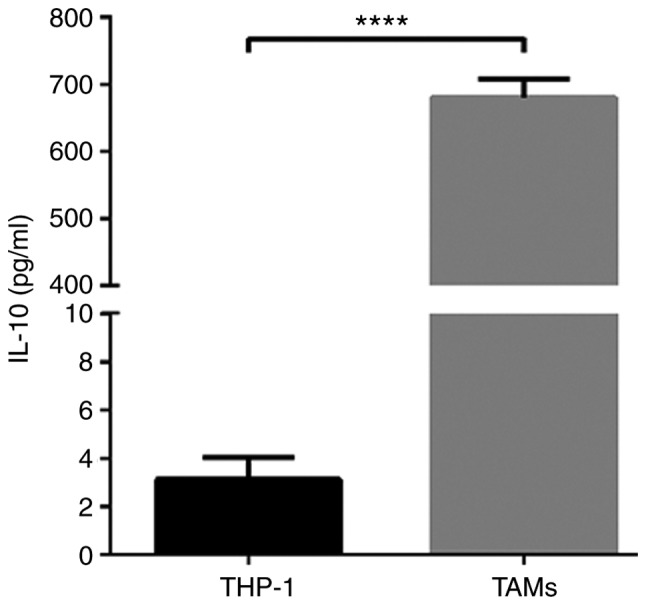
ELISA detection of IL-10 expression in THP-1 cells and TAM culture supernatants. *P<0.0001 vs. control. TAMs, tumor associated macrophages; IL-10, interleukin 10.
Figure 3.
Identification of TAMs by flow cytometry. The expression of CD14 and CD163 in (A) THP-1 cells, (B) M0 macrophages and (C) TAMs was examined. (D) Quantification of the flow cytometry data. ****P<0.0001 vs. control. TAMs, tumor associated macrophages; CD, cluster of differentiation; NS, not significant.
TAMs affect the biological activity of A549 cells
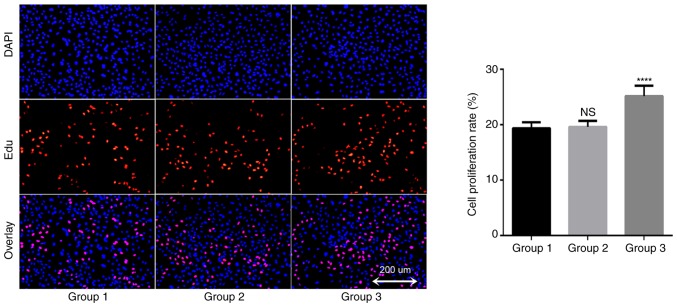
In order to understand the potential role of TAMs in the growth of A549 cells, an EDU proliferation kit was used to detect changes in the proliferation rate of lung cancer A549 cells. The results demonstrated that the proliferation rate of the blank control group was 19.4±1.1% after 24 h of A549 cell culture; the proliferation rate of the THP-1 coculture group was 19.6±1.1%, and the proliferation rate of TAMs coculture group was 25.2±1.9% (Fig. 4). There was no significant difference between the THP-1 coculture and blank control groups (P=0.945), but the proliferation rate of the TAMs coculture group was significantly increased compared with that of the blank control group. These data suggested that TAMs in coculture system may promote the proliferation of lung adenocarcinoma A549 cells.
Figure 4.
TAMs may promote proliferation of A549 cells. DAPI- and EDU-positive staining indicates the total cells and proliferating cells, respectively (magnification, ×100). Group 1 represents normal control group; group 2 represents THP-1 coculture group; and group 3 represents TAMs coculture group. The EDU proliferation assay test indicated that the proliferation rate of the TAMs coculture group was significantly increased compared with that of the blank control group. ****P<0.0001 vs. control. TAMs, tumor associated macrophages.
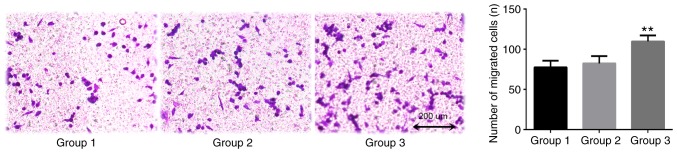
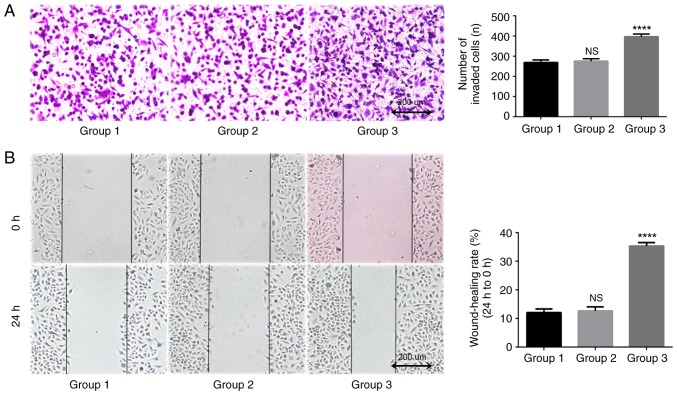
TAMs in the coculture system may promote the invasion of lung adenocarcinoma A549 cells, as indicated by data from the Transwell assay conducted in the presents study (Fig. 5). After 24 h coculture, the number of transmembrane cells in the blank control group was 77.3±8.3. The number of transmembrane cells in the THP-1 coculture group (group 2) was 82.3±9.1. The difference between these groups was not statistically significant (P=0.753). The number of transmembrane cells in the TAMs coculture group was 109.7±7.5, which was significantly increased compared with that of the blank control group (P=0.007). This confirmed that TAMs promoted the invasive ability of A549 cells.
Figure 5.
TAMs promote the invasion of A549 cells. Group 1 represents normal control group; group 2 represents THP-1 coculture group; and group 3 represents TAMs coculture group. The Transwell invasion assay demonstrated that, compared with the control group, TAMs significantly promoted the invasion of A549 cells. Magnification, ×100. **P<0.01 vs. control. TAMs, tumor associated macrophages.
TAMs in coculture system may promote the migration of lung adenocarcinoma A549 cells. In the present study, a Transwell assay was used to detect longitudinal migration ability of A549 cells. The results indicated that after 24 h of coculture, the number of transmembrane cells in the blank control group was 269.0±12.5, and the number in the THP-1 coculture group was 275.3±12.1. The difference between these two groups was statistically significant (P=0.816; Fig. 6A). The number of transmembrane cells in the TAMs coculture group was 396.7±13.0, which was significantly increased compared with that in the blank control group and the THP-1 coculture group (P<0.0001). A scratch wound assay was used to detect lateral migration ability of A549 cells. The results indicated that the differences between the TAMs coculture group compared with the blank control group was 12.0±1.3%, and between the THP-1 coculture group and the blank control group was 12.6±1.4% (Fig. 6B). The migration rate of A549 cells after 24 h (35.3±1.2%) was significantly increased (P<0.0001). These data suggested that TAMs may improve the migration ability of A549 cells in the coculture system.
Figure 6.
TAMs promote the migration of A549 cells. Group 1 represents normal control group; group 2 represents THP-1 coculture group; and group 3 represents TAMs coculture group. (A) Images from the Transwell migration assay. Magnification, ×100. Compared with the control group, TAMs significantly promoted the longitudinal migration of A549 cells. ****P<0.0001 vs. control. (B) Images from the scratch wound assay. Magnification, ×100. Compared with the control group, TAMs significantly promoted the lateral migration of A549 cells. ****P<0.0001 vs. control. TAMs, tumor associated macrophages; NS, not significant.
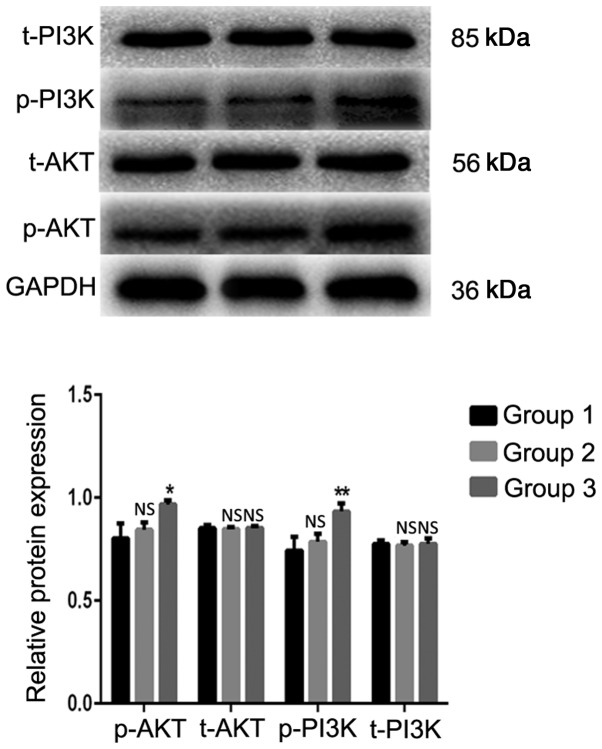
PI3K/AKT signaling pathway is activated in A549 cells by indirect coculture with TAMs
The western blot analysis results indicated that the expression levels of the p-AKT and p-PI3K proteins in the TAMs coculture group were significantly increased compared with those in the blank control group (Fig. 7). The differences were statistically significant (P=0.014 and P=0.008, respectively). The expression levels of the total (t)-AKT and t-PI3K proteins were not significantly different from those in the blank control group (P>0.05). These results demonstrated that TAMs may affect the biological activity of lung adenocarcinoma A549 cells by activating the PI3K/AKT signaling pathway in coculture systems.
Figure 7.
TAMs participate in regulating the biological activity of lung adenocarcinoma A549 cells via the PI3K/AKT signaling pathway. Group 1 represents normal control group; group 2 represents THP-1 coculture group; and group 3 represents TAMs coculture group. *P<0.05, **P<0.01 vs. control. TAMs, tumor associated macrophages; p, phosphorylated; t, total; NS, not significant.
Discussion
The tumor microenvironment is the internal environment on which tumor cells depend for their survival and development. In previous years, the interaction between immune cells and other cells or factors in the tumor microenvironment has been demonstrated. Immature monocytes in the blood are recruited by various cytokines in the tumor microenvironment and induced to undergo a phenotypic and functional transformation, thereby forming TAMs. Furthermore, chemical factors including chemokines, growth factors, angiogenic factors and matrix proteases are produced to act on tumor cells in order to promote tumor growth, invasion and metastasis. It is hypothesized that immature monocytes may be induced into different functional macrophages by different cytokines, including M1 macrophages induced by granulocyte- macrophage-colony-stimulating factor/lipopolysaccharide to promote inflammation and tumor suppressor activity via NF-κB and p-signal transducer and activator of transcription 1 pathways, and induced M2 macrophages by IL-4 and IL-13 in order to promote tumor growth (6,7). As TAMs have a similar function to M2 macrophages, M2 macrophages are usually induced in vitro to replace TAMs in research models (18). The present study also used specific concentrations of PMA, IL-4 and IL-13 to stimulate the differentiation of THP-1 cells into M2 macrophages as opposed to TAMs, and used ELISA to detect the expression of IL-10 in THP-1 cells and TAM culture supernatants. In addition, flow cytometry was used to detect the expression ratio of CD14+/CD163+ cells to verify that the model was successfully established. The experimental results indicated that following induction, the growth characteristics of THP-1 cells changed from suspension to adherence, and their morphology changed from round to fusiform with no proliferative activity. The flow cytometry data suggested that the expression ratio of CD14+/CD163+ pattern in the induced cells was significantly increased, thereby confirming that the in vitro study model of TAMs was successfully established in the present study.
Previous studies have demonstrated that TAMs may affect the efficacy of epidermal growth factor receptor-tyrosine kinase inhibitors (EGFR-TKI) targeted therapy in NSCLC, and its pathological score may be used as an effective prognostic biomarker (19–21). However, at a cellular level, there is no relevant basic research to confirm that TAMs directly affect the the ability of TKIs to induce cell death in NSCLC cells. Additional studies are vital to explore the potential involvement of TAMs in mediating NSCLC TKI-targeted drug resistance and TKI resistance reversal via immunotherapy. In the present study, a Transwell coculture method was used to simulate a tumor microenvironment, and the effects of cytokines secreted by TAMs on the biological activity of lung adenocarcinoma A549 cells were explored. The molecular mechanism was initially examined on a preliminary basis. In the present study, an indirect coculture model of TAMs and lung adenocarcinoma A549 cells was successfully established and an EDU proliferation assay kit was used to detect the proliferation rate of A549 cells. The results demonstrated that TAMs significantly promoted the proliferation and clonality of A549 cells. Through Transwell invasion, migration and scratch wound assays, it was also identified that TAMs also significantly promoted the invasion and migration of A549 cells.
The PI3K/AKT cell signal transduction pathway a signaling pathways closely associated with tumorigenesis and development. In lung cancer, it not only promotes proliferation, invasion and migration of tumor cells through cascade phosphorylation of a series of proteins, but it may also affect the efficacy of EGFR-targeted therapy for non-small NSCLC (22,23). Therefore, the present study aimed to verify whether TAMs affect the biological activity of lung cancer cells by activating the PI3K/AKT signaling pathway. In the present study, the expression levels of t-PI3K, t-AKT and corresponding phosphorylated p-AKT and p-PI3K proteins were detected by western blot analysis. The results suggested that TAMs may significantly promote the expression of phosphorylated PI3K and AKT proteins. In other words, cytokines secreted by TAMs may activate the PI3K/AKT signaling pathway in order to promote the proliferation, invasion and migration of A549 cells. Whether TAMs are involved in the TKI resistance observed in lung cancer cells exhibiting EGFR-activating mutations would be an interesting topic of future study.
The data from the present study is inconclusive in certain aspects, for example: TAMs in the microenvironment may affect tumor cells through paracrine effects and direct contact, but the present study only established an indirect coculture environment. The proportion of TAMs in the in vitro TAMs study model was high, but it was not pure. Although the present study established the THP-1 coculture group to exclude the effects of undifferentiated or other different types of macrophages, purification of TAMs by specific antibody-modified magnetic beads would produce more convincing results. In conclusion, TAMs promoted the proliferation, invasion and migration of lung adenocarcinoma A549 cells, and this may be associated with the activation of the PI3K/AKT signaling pathway.
Acknowledgements
Not applicable.
Funding
The present study was supported by Key Projects of External Scientific and Technological Cooperation of Jiangxi Province Science and Technology Department (grant no. 20181BBH80005) and the Natural Science Foundation of Jiangxi Province (grant no. 20161BAB205265).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Author's contributions
JX designed the experiment and revised the manuscript. SY and YD performed the experiments and were major contributors in writing the manuscript. LP, MY, ZL and LN collected and collated the relevant literature and data and processed the experimental data and images. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for approval
Not applicable.
Competing interest
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Kerkar SP, Restifo NP. Cellular constituents of immune escape within the tumor microenvironment. Cancer Res. 2012;72:3125–3130. doi: 10.1158/0008-5472.CAN-11-4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ostrand-Rosenberg S. Tolerance and immune suppression in the tumor microenvironment. Cell Immunol. 2016;299:23–29. doi: 10.1016/j.cellimm.2015.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ruffell B, Coussens LM. Macrophages and therapeutic resistance in cancer. Cancer Cell. 2015;27:462–472. doi: 10.1016/j.ccell.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prenen H, Mazzone M. Tumor-associated macrophages: A short compendium. Cell Mol Life Sci. 2019;76:1447–1458. doi: 10.1007/s00018-018-2997-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goswami KK, Ghosh T, Ghosh S, Sarkar M, Bose A, Baral R. Tumor promoting role of anti-tumor macrophages in tumor microenvironment. Cell Immunol. 2017;316:1–10. doi: 10.1016/j.cellimm.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 7.Aras S, Zaidi MR. TAMeless traitors: Macrophages in cancer progression and metastasis. Br J Cancer. 2017;117:1583–1591. doi: 10.1038/bjc.2017.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141:39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elinav E, Nowarski R, Thaiss CA, Hu B, Jin C, Flavell RA. Inflammation-induced cancer: Crosstalk between tumours, immune cells and microorganisms. Nat Rev Cancer. 2013;13:759–771. doi: 10.1038/nrc3611. [DOI] [PubMed] [Google Scholar]
- 10.Mittal D, Gubin MM, Schreiber RD, Smyth MJ. New insights into cancer immunoediting and its three component phases-elimination, equilibrium and escape. Curr Opin Immunol. 2014;27:16–25. doi: 10.1016/j.coi.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 12.Mantovani A, Allavena P. The interaction of anticancer therapies with tumor-associated macrophages. J Exp Med. 2015;212:435–445. doi: 10.1084/jem.20150295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang F, Yang L, Gao Q, Huang L, Wang L, Wang J, Wang S, Zhang B, Zhang Y. CD163+CD14+ macrophages, a potential immune biomarker for malignant pleural effusion. Cancer Immunol Immunother. 2015;64:965–976. doi: 10.1007/s00262-015-1701-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang L, Wang F, Wang L, Huang L, Wang J, Zhang B, Zhang Y. CD163+ tumor-associated macrophage is a prognostic biomarker and is associated with therapeutic effect on malignant pleural effusion of lung cancer patients. Oncotarget. 2015;6:10592–10603. doi: 10.18632/oncotarget.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohtaki Y, Ishii G, Nagai K, Ashimine S, Kuwata T, Hishida T, Nishimura M, Yoshida J, Takeyoshi I, Ochiai A. Stromal macrophage expressing CD204 is associated with tumor aggressiveness in lung adenocarcinoma. J Thorac Oncol. 2010;5:1507–1515. doi: 10.1097/JTO.0b013e3181eba692. [DOI] [PubMed] [Google Scholar]
- 16.Loyher PL, Hamon P, Laviron M, Meghraoui-Kheddar A, Goncalves E, Deng Z, Torstensson S, Bercovici N, Baudesson de Chanville C, et al. Macrophages of distinct origins contribute to tumor development in the lung. J Exp Med. 2018;215:2536–2553. doi: 10.1084/jem.20180534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackute J, Zemaitis M, Pranys D, Sitkauskiene B, Miliauskas S, Vaitkiene S, Sakalauskas R. Distribution of M1 and M2 macrophages in tumor islets and stroma in relation to prognosis of non-small cell lung cancer. BMC Immunol. 2018;19:3. doi: 10.1186/s12865-018-0241-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tjiu JW, Chen JS, Shun CT, Lin SJ, Liao YH, Chu CY, Tsai TF, Chiu HC, Dai YS, Inoue H, et al. Tumor-associated macrophage-induced invasion and angiogenesis of human basal cell carcinoma cells by cyclooxygenase-2 induction. J Invest Dermatol. 2009;129:1016–1025. doi: 10.1038/jid.2008.310. [DOI] [PubMed] [Google Scholar]
- 19.Chung FT, Lee KY, Wang CW, Heh CC, Chan YF, Chen HW, Kuo CH, Feng PH, Lin TY, Wang CH, et al. Tumor-associated macrophages correlate with response to epidermal growth factor receptor-tyrosine kinase inhibitors in advanced non-small cell lung cancer. Int J Cancer. 2012;131:E227–E235. doi: 10.1002/ijc.27403. [DOI] [PubMed] [Google Scholar]
- 20.Zhang B, Zhang Y, Zhao J, Wang Z, Wu T, Ou W, Wang J, Yang B, Zhao Y, Rao Z, Gao J. M2-polarized macrophages contribute to the decreased sensitivity of EGFR-TKIs treatment in patients with advanced lung adenocarcinoma. Med Oncol. 2014;31:127. doi: 10.1007/s12032-014-0127-0. [DOI] [PubMed] [Google Scholar]
- 21.Peng H, Chen B, Huang W, Tang Y, Jiang Y, Zhang W, Huang Y. Reprogramming tumor-associated macrophages to reverse EGFRT790M resistance by dual-targeting codelivery of gefitinib/vorinostat. Nano Lett. 2017;17:7684–7690. doi: 10.1021/acs.nanolett.7b03756. [DOI] [PubMed] [Google Scholar]
- 22.Gadgeel SM, Wozniak A. Preclinical rationale for PI3K/Akt/mTOR pathway inhibitors as therapy for epidermal growth factor receptor inhibitor-resistant non-small-cell lung cancer. Clin Lung Cancer. 2013;14:322–332. doi: 10.1016/j.cllc.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 23.Pérez-Ramírez C, Cañadas-Garre M, Molina MÁ, Faus-Dáder MJ, Calleja-Hernández MÁ. PTEN and PI3K/AKT in non-small-cell lung cancer. Pharmacogenomics. 2015;16:1843–1862. doi: 10.2217/pgs.15.122. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.