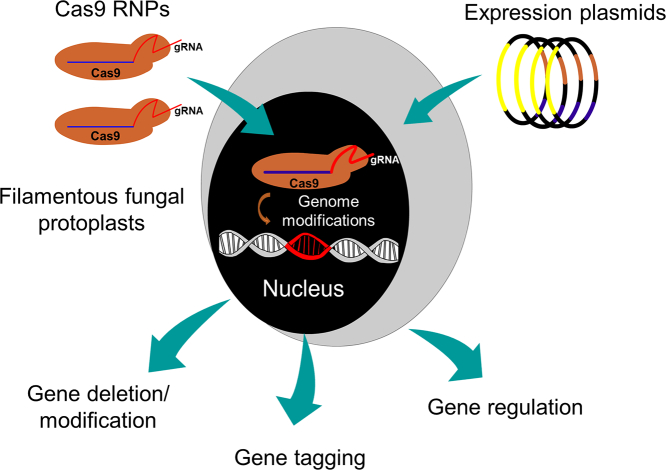
Abstract
Widely distributed in various environmental niches, filamentous fungi play an important role in industry, drug development, and plant/animal health. Manipulation of the genome and the coding sequences are essential for a better understanding of the function of genes and their regulation, but traditional genetic approaches in some filamentous fungi are either inefficient or nonfunctional. The rapid development and wide implementation of CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats /(CRISPR)-associated protein-9 nuclease) technology for various model and non-model organisms has provided the initial framework to adapt this gene editing technology for filamentous fungi. In this review, an overview of the CRISPR/Cas9 tools and strategies that have been developed for different filamentous fungi is presented, including integration of the CAS9 gene into the genome, transient expression of Cas9/sgRNA, the AMA1-based plasmid approach, and the Cas9 RNP method. The various applications of CRISPR/Cas9 technology in filamentous fungi that have been implemented are explored, with particular emphasis on gene disruption/deletion and precise genome modification through gene tagging and alteration in gene regulation. Potential challenges that are confronted when developing a CRISPR/Cas9 system for filamentous fungi are also discussed such as the nuclear localization sequence for the CAS9 gene, potential off-target effects, and highly efficient transformation methods. Overcoming these obstacles may further facilitate wide application of this technology. As a simple, economical, and powerful tool, CRISPR/Cas9 systems have the potential for future implementation into many molecular aspects of filamentous fungi.
Keywords: Filamentous fungus, Cas9, CRISPR, Gene deletion, Secondary metabolite, Transformation, Protoplast
Graphical Abstract
1. Introduction
Filamentous fungi significantly impact food security, crop production, drug development, and animal/human health. Several important discoveries in molecular biology were derived from studies using filamentous fungi. For instance, Beadle and Tatum used the model filamentous fungus Neurospora crassa in their early research in cell biology and proposed the well-known “one gene, one enzyme” hypothesis [1]. To this day, N. crassa remains a model system and is used in various areas of research including investigation on circadian rhythms, gene silencing, fungal development, and cell polarity [2]. Importantly, discoveries of fungal secondary metabolites, including antibiotics and statins, have contributed to saving the lives of millions of people, and enabled the development of several medical procedures and treatments. Penicillin, produced by Penicillium chrysogenum, was discovered for its ability to inhibit gram-positive bacteria and has led to clinically used derivatives that serve as broad-spectrum antibiotics [3].
Although filamentous fungi have contributed greatly to science, many filamentous fungi are pathogens that cause a reduction in crop production and human/animal health problems. Magnaporthe oryzae is the causal agent of rice blast disease, the most important fungal rice disease worldwide, and results in lesions on all parts of the plants such as leaves, panicles, and seeds [4]. Fusarium oxysporum causes the well-known Fusarium wilt disease on a wide range of agriculturally important host plants [5]. Typical symptoms include vascular browning of the xylem tissue, leaf yellowing, and wilting. Along with Fusarium solani, F. oxysporum is responsible for the majority of the cases of fusariosis in immunocompromisied patients resulting in a high mortality [6,7]. Clinical infections caused by fungi within the genus Aspergillus are associated with 30%~90% mortality [8,9]. Some filamentous fungi produce mycotoxins that contaminate the food supply or affect plant or animal health. Some of the best known examples of mycotoxins include the carcinogenic aflatoxins synthesized by several Aspergillus species [10], and deoxynivalenol, a type B trichothecene produced predominantly by Fusarium graminearum, which may function by inhibiting protein synthesis [11].
The economic importance of filamentous fungi, has led to more focus on molecular studies on these microbes, but limitations in reverse genetic tools including gene deletion/editing, RNAi, protein expression, and gene tagging procedures have in some instances remained a challenge. While most filamentous fungi are haploid, those that are diploid or multinuclieated pose an additional complication for investigating the function of a gene, since all alleles need to be mutated to determine the contribution of the gene of interest to a particular phenotype. A popular method for gene deletion/disruption in fungi is based on homologous recombination (HR), where selective markers conferring resistance to antifungal compounds, usually to hygromycin or neomycin, are used to replace or integrate within the target locus. While some fungi such as Saccharomyces cerevisiae, N. crassa, and Aspergillus spp. may have a relatively high HR efficiency, gene disruption can be difficult for many non-model fungi due to a low HR efficiency. The development of the simple, rapid, powerful, and economical gene manipulation tool known as CRISPR/Cas9 technology opened a new era for genome editing. This technology has been widely used for genome editing of model and non-model higher organisms [12]. This technique and its rapid application to fungi has shown obvious advantages in gene manipulation when compared to previous methods since the CRISPR/Cas9 system induces double strand breaks in host cell DNA, enhancing the efficiency of gene manipulation and integration.
2. The Working Mechanism of the CRISPR/Cas9 System
CRISPR (clustered regularly interspaced short palindromic repeats) is a family of DNA repeats found in ~40% of the sequenced bacterial genomes and ~90% of the archaeal genomes [13]. Cas9, a DNA endonuclease, forms a simple bacterial immune system with a crRNA/tracrRNA hybrid to recognize, bind, and ultimately cleave foreign DNA [14]. The crRNA/tracrRNA hybrid was later engineered into a single guide RNA (sgRNA) which facilitated experimental design [14].
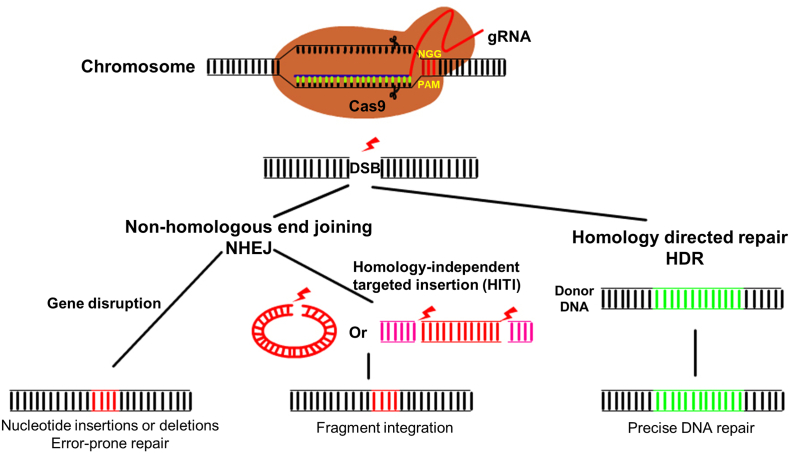
The recognition of a DNA target region depends on a protospacer adjacent motif (PAM) which is located at the 3′-terminus of a 20 bp target sequence. Once the CRISPR/Cas9 complex recognizes the target DNA sequence, it will generate a double strand break (DSB) at the DNA target locus between the final third and fourth nucleotides which will activate the host DNA repair process. Two cellular DNA repair mechanisms, non-homologous end joining (NHEJ) and homologous recombination (HR), play essential roles in precise genome editing and gene manipulation (Fig. 1). NHEJ has been regarded as an error-prone repair mechanism that generates either short insertions or deletions of nucleotides in close proximity to the DSB site(s) [15]. If these short insertions or deletions exist in a gene coding region, or within a portion of the promoter involved in recruiting proteins involved in transcription, the function of the endogenous gene will be disrupted. Consequently, this procedure is widely used for generating gene mutations. However, an increasing amount of evidence has indicated that NHEJ is not always error-prone [16]. Recently, a homology independent targeted integration (HITI) strategy was devised that allows fragments to be integrated into the genome by NHEJ repair. Interestingly, the integration can occur in both dividing and non-dividing cells with a high probability of perfect DNA repair at the sgRNA cleavage site [[17], [18], [19]]. In HR, the second DSB repair pathway, the mechanism relies on a DNA template, allowing the foreign DNA fragment(s) to be precisely integrated into the host genome. However, HR-mediated DSB repair only occurs in dividing cells, limiting the application of this method to a broad range of uses.
Fig. 1.
Overview of CRISPR/Cas9 gene editing system and DNA repair mechanisms. When complexed with a sgRNA, Cas9 will cleave double stranded DNA at a specific site directed by the target sequence and PAM of the sgRNA. Once the DNA is cleaved, two DNA repair pathways, NHEJ and HDR, occur and may alter the genome of the cell. In NHEJ, insertions or deletions of nucleotides may occur at the cleavage site; alternatively if multiple DSBs are generated, repair can occur between different fragments. During HDR homologous recombination occurs at the loci adjacent to the DBS using a DNA template.
3. Various Strategies for Developing CRISPR/Cas9 Tools in Filamentous Fungi
Various versions of CRISPR/Cas9 tools have been developed for fungi and each of these approaches may be the preferred method for a particular fungus (Table 1). The yeast S. cerevisiae was the first fungus for which CRISPR/Cas9 was applied to modify the genome [20], and the ability of this fungus to stably maintain plasmids facilitated the development of tools for CRISPR/Cas9. Usually these plasmids contain the CAS9 gene under constitutive expression [20,21], and when coupled with transient sgRNA cassette transformation, generate DSBs thereby increasing homologous recombination rates [20]. Previously, gene manipulation in the clinically relevant yeast Candida albicans was tedious as the fungus is a diploid; however development of CRISPR/Cas9 methods has provided an efficient way to generate homozygous mutations and facilitated genome engineering in this organism [[21], [22], [23]].
Table 1.
Filamentous fungi in which CRISPR gene editing technology has been utilized.
| Strategies | Filamentous fungus | Genome editing |
Reference | |
|---|---|---|---|---|
| NHEJ | HDRI | |||
| CAS9 integration into fungal genome for expression | Beauveria bassiana | + | [85] | |
| Blastomyces dermatitidis | + | [86] | ||
| Coprinopsis cinerea | + | [87] | ||
| Cordyceps militaris | + | [88] | ||
| Fusarium graminearum | + | + | [60] | |
| Ganoderma spp. | + | [89] | ||
| Leptosphaeria maculans | + | [59] | ||
| Myceliophthora spp. | + | + | [90] | |
| Nodulisporium sp. | + | [91] | ||
| Rhizopus delemar | + | [92] | ||
| Sporisorium scitamineum | + | [93] | ||
| Trichoderma reesei | + | + | [24] | |
| Ustilaginoidea virens | + | [94] | ||
| CAS9 transient expression | Fusarium fujikuroi | + | + | [69] |
| Neurospora crassa | + | [26] | ||
| Sclerotinia sclerotiorum | + | [34] | ||
| Ustilago tricophora | + | + | [95] | |
| Self-replicating plasmids (eg. AMA1-based) | Alternaria alternata | + | [58] | |
| Aspergillus aculeatus | + | [37] | ||
| Aspergillus brasiliensis | + | |||
| Aspergillus carbonarius | + | |||
| Aspergillus luchuensis | + | |||
| Aspergillus nidulans | + | |||
| Aspergillus niger | + | |||
| Aspergillus oryzae | + | + | [96] | |
| Penicillium chrysogenum | + | [38] | ||
| Talaromyces atroroseus | + | [36] | ||
| Ustilago maydis | + | [97] | ||
| Cas9 RNPs | Aspergillus fumigatus | + | [43] | |
| Fusarium oxysporum | + | + | [44] | |
| Magnaporthe oryzae | + | + | [46] | |
| Mucor circinelloides | + | + | [48] | |
| Penicillium chrysogenum | + | [38] | ||
| Penicillium decumbens | + | [98] | ||
| Trichoderma reesei | + | [99] | ||
| Other | Shiraia bambusicola | + | [100] | |
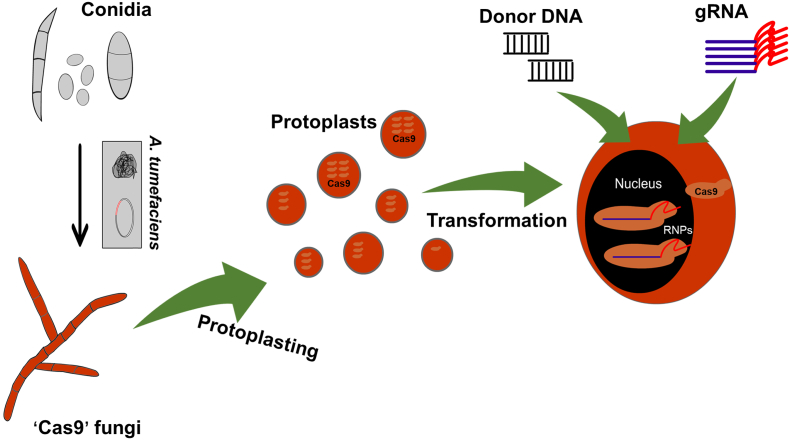
Following these initial CRISPR/Cas9 experiments with yeasts, the gene editing tool was adapted for both model and non-model filamentous fungi (Table 1; Fig. 2). A CRISPR/Cas9 system was first established in the filamentous fungus Trichoderma reesei using a codon-optimized CAS9 gene that was introduced into the genome using Agrobacterium tumefaciens-mediated transformation [24]. When the expression of CAS9 was under the control of either a constitutive or inducible promoter, there was no obvious alteration in the function of other proteins as observed by fungal morphology and various other phenotypes. The successful integration of Cas9 into the fungal genome generated a strain (a ‘Cas9’ strain) that requires only the sgRNA for genome editing through protoplast transformation, providing a method to rapidly target multiple genes simultaneously [24].
Fig. 2.
Scheme of generating a ‘Cas9’ strain for gene editing. A fungus is transformed through protoplasts or Agrobacterium tumefaciens-mediated transformation with the CAS9 gene under control of either an inducible or constitutive promoter generating a ‘Cas9’ strain. Once the CAS9 gene has been stably integrated into the fungal genome, subsequent transformations allow various other loci of interest to be targeted by sgRNAs.
The random integration of the CAS9 gene into the host genome may induce undesired mutations, and obtaining a ‘Cas9’ fungal strain may require screening several transformants or optimizing the codon usage of the CAS9 gene to a different codon preference for each species. Another caveat is the constitutive expression of CAS9 and/or its long-time retention in host cells which may generate off-target effects [25]. To overcome these limitations, transient expression systems of the CAS9 gene and sgRNA in filamentous fungi have been utilized (Fig. 3). A transient expression CRISPR/Cas9 system was developed in the model filamentous fungus, N. crassa [26], where the CAS9 gene was engineered with the trpC constitutive promoter from A. nidulans, and the Small Nucleolar RNA 52 (SNR52) promoter from S. cerevisiae was used to drive sgRNA expression. The editing efficiency gradually increased with an increasing number of copies of CAS9 and the quantity of sgRNA plasmids. The replacement of the endogenous promoter of the clr-2 gene with the β-tubulin promoter and the insertion of a codon optimized firefly luciferase cassette suggested that this strategy was efficient and could be applied to many aspects in filamentous fungi [26].
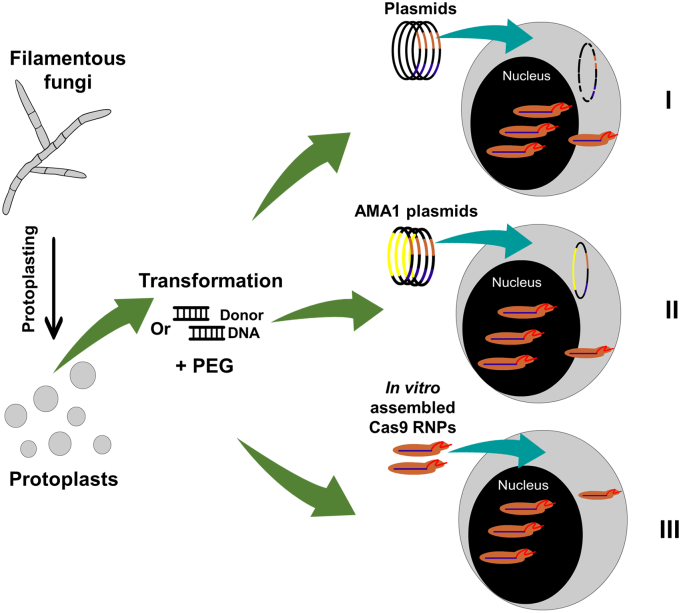
Fig. 3.
CRISPR/Cas9 systems for filamentous fungi that do not integrate CAS9. Initially, the fungal cell wall is degraded generating protoplasts followed by introduction of transient self-replicating plasmids or Cas9 RNPs. I) Plasmid(s) encoding the sgRNA and CAS9 are introduced into the protoplast and both of these components are expressed which combine to generate the DSB dictated by the sgRNA. The plasmids are rapidly degraded within the fungal cell. II) Self-replicating plasmids encoding the sgRNA and CAS9, and can be maintained for longer periods to time within the cell creating a higher probability of expression and Cas9 gene editing. III) Cas9 RNP transformation, where the purified Cas9 protein and the sgRNA are directly transformed into the fungal protoplasts. The Cas9 RNP is rapidly degraded during natural protein turnover.
It is not uncommon for fungi to have multiple nuclei within a single cell, have undergone whole or partial genome duplication [[27], [28], [29], [30]], or harbor supernumerary chromosomes [[31], [32], [33]], thus multiple genetic loci may need to be targeted within a single fungal cell. A CRISPR/Cas9 gene editing tool for the multinucleate filamentous phytopathogenic fungus Sclerotinia sclerotiorum has been reported [34]. CAS9 and sgRNAs were able to be transiently expressed in the protoplasts and function properly; however, plasmid fragments of various lengths were integrated into the Cas9/sgRNA cleavage site by the NHEJ repair pathway, generating mutants with insertions of the plasmid [34]. Using this method, the Ssoah1 gene, which is involved in the accumulation of oxalic acid, was disrupted and the ΔSsoah1 strain was unable to acidify the growth medium [34].
Although filamentous fungi cannot stably maintain most plasmids, several chromosomal replicators, including AMA1 and MATE elements, have provided a possible mechanism for extrachromosomal maintenance of plasmids in filamentous fungi [35]. AMA1 plasmids can significantly enhance transformation efficiency, have a low probability of being inserted into the fungal genome during transformation, and are able to be easily lost after transformation without selection, allowing the dominant selectable marker(s) to be reused for additional transformation procedures. Based on these advantages, several CRISPR/Cas9 systems utilize AMA1 plasmids for filamentous fungi [[36], [37], [38]]. In one of the first studies using this method, Nødvig et al. successfully mutated the yA gene in A. nidulans, resulting in a change of color of conidia from yellow to green [37]. In this study, the expression of the sgRNA depended on two key components, an intrinsic hammerhead ribozyme and a hepatitis delta virus ribozyme. Both can be self-cleaved and allow the precise initiation of the sgRNA [37].
All of these CRISPR/Cas9 systems in filamentous fungi require functional host transcription and translation systems for the expression of the foreign CAS9 gene and sgRNA transcription. Appropriate promoters must be chosen for both CAS9 and sgRNA expression, requiring optimization of promoters in different filamentous fungi. In addition, the synthesis of sgRNA is controlled by the RNA polymerase III (Pol III) promoters like the U6 promoter and requires precise transcription initiation [20,21]. A considerable amount of time and effort is necessary for optimizing a plasmid-based Cas9/sgRNA delivery method. Consequently, more recent attempts have used a Cas9 RNP-based delivery method for genome editing in different species, including Arabidopsis, fruit fly, zebrafish, and human cell lines [[39], [40], [41], [42]]. Cas9 RNPs consist of the purified Cas9 protein and an in vitro transcribed sgRNA, and both components are assembled in vitro into a complex and then directly transfected into the host cells. Compared to plasmid-based delivery, the Cas9 RNP-based delivery has several advantages including: 1) the expression of the CAS9 gene and transcription of the sgRNA does not rely on host machinery, and therefore the precise transcription initiation of the sgRNA is not a concern; 2) the assembled Cas9 RNPs have immediate cleavage activity and can be directly delivered into the host cells to target the specified DNA locus without having to express CAS9 or transcribe the sgRNA; 3) the cleavage efficiency of the different sgRNAs can be evaluated in vitro, allowing the optimal sgRNA(s) to be chosen for further experiments; 4) Cas9 RNPs will be depleted over a short timeframe from the host cells by protein degradation pathways, thereby reducing the possibility of Cas9 off-target activity.
Cas9 RNP-based delivery methods have been developed for filamentous fungi primarily using the polyethylene glycol (PEG)-mediated protoplast transformation technique to introduce the Cas9 RNPs to the recipient protoplasts. A simple and universal CRISPR/Cas9 system based on in vitro-assembled Cas9 RNPs has been reported in A. fumigatus [43], in which two strains, the WT and a ΔakuB strain (reduced in frequency of NHEJ), were used in a study of the CRISPR/Cas9-mediated HR repair mechanism to disrupt pksP, a gene responsible for the green color of the conidia. The hygromycin resistance cassette flanked by two microhomology sequences (35–50 bp) at both ends of the cassette were able to serve as both the selective marker and the donor template for insertion into the Cas9 RNP cleavage site(s). With the ΔakuB laboratory strain, up to 97% of the colonies showed the desired phenotype regardless of whether a 35 bp or 50 bp homologous sequence was used, while the efficiency was 40–74% in the WT, where the length of microhomology directly influenced the integration efficiency [43].
In another example, a stable Cas9 RNP transformation method was developed for the phytopathogenic filamentous fungus F. oxysporum. The nuclear localization sequence (NLS) from the histone H2B gene of F. oxysporum was fused with the Cas9 coding sequence and once purified, was transferred into fungal protoplasts for CRISPR-mediated gene editing. Mutations in two classical marker genes for fungal biology, URA5 and URA3, were generated during development of the system as a proof of concept via PEG-mediated transformation. When a homology directed repair strategy was utilized to integrate a dominant selectable marker into the FoBIK1 gene, encoding the key polyketide synthase responsible for the synthesis of the pigmented secondary metabolite bikaverin, the maximum efficiency was ~50% [44]. Based on this method, several endogenous genes were tagged at the native gene locus using the sGFP and mCherry fluorescent markers [45]. Importantly, as the NLS of this Cas9 protein was derived from the well conserved sequence of the histone H2B, this system may be widely applicable to the other members within the genus Fusarium and perhaps other fungi.
A Cas9 RNP-mediated co-editing and counter selection system has also been developed for the rice blast fungus, M. oryzae [46]. In this system, only a 30 bp homologous sequence was necessary to promote HR-directed repair flanking the Cas9 RNP cleavage site when generating an ALB1 mutant unable to produce melanin [46]. However, when the NHEJ strategy was attempted to mutate the ALB1 gene without addition of a donor template during the transformation, the frequency of generating the desired transformant was reduced, although mutants were able to be identified due to the easily distinguished phenotype. While these results suggest this strategy is less suitable for gene disruption, it was used to generate a single nucleotide substitution in the M. oryzae SDI1 gene, conferring resistance to the fungicide carboxin [46]. A strategy to edit two loci simultaneously was attempted, but the efficiency for this strategy was only ~1% and one genetic locus must have an obvious mutant phenotype enabling selection. Cas9 RNP-mediated transformation methods have been reported in other filamentous fungi including P. chrysogenum and Mucor circinelloides [47,48]. Collectively these studies indicate that Cas9 RNP-mediated transformation offers a simple and reliable method that could be adapted for use in other filamentous fungi and could be implemented for other CRISPR/Cas9-based molecular techniques.
4. Applications of CRISPR/Cas9 Tools for Filamentous Fungal Genome Engineering
The emergence of CRISPR/Cas9 tools has sparked a revolution in genome engineering for various organisms. The most exciting application of CRISPR/Cas9 technology focuses on precise gene editing, offering vast possibilities to modify organisms as desired (Fig. 4). In addition, derivatives of CRISPR/Cas9 enzymes can be utilized for several activities including base editing, DNA nicking, and epigenetic modification [49,50]. One of these variants that has found a wide range of uses is dCas9, which has a mutation in the endonuclease domain of Cas9 resulting in a loss of DNA cleavage activity. Transcriptional repression by dCas9 is achieved by targeting the transcriptional initiation or elongation regions thereby reducing the amount of mRNA generated through competitive inhibition. In human cells, this interference efficiency can be up to 90% [51]. Conversely, CRISPR/Cas9-based transcriptional activation (CRISPRa) is considered a powerful tool for overexpression of a target gene [52]. In this technique, either the transcriptional activation domain or the entire protein is fused with dCas9, facilitating the recruitment and assembly of the RNA transcriptional machinery to the target site specified by the sgRNA, ultimately driving an increase in gene expression. CRISPRa has the potential for widespread use in investigating fungal gene expression, especially concerning the expression of genes involved in secondary metabolite biosynthesis. Filamentous fungi are well-known to produce an arsenal of natural products with biological activity, and many of the genes responsible for the biosynthesis of these secondary metabolites are clustered together and are not transcribed under standard culturing conditions [53,54]. Identification of a suitable environment for activating the expression of these secondary metabolite biosynthetic clusters can be time-consuming and tedious, although some success has been achieved with a few activation strategies including the manipulation of global or specific regulators [55]. However, the specific secondary metabolite(s) produced by a large number of cryptic/silent secondary metabolite gene clusters remains unknown, and development of CRISPR/Cas9 technologies can serve as a valuable tool to elucidate the compounds produced by these clusters (Fig. 4). Although most of the currently developed CRISPR/Cas9 systems have been utilized primarily for functional characterization of genes in filamentous fungi, some other applications, including investigation of the subcellular localization of proteins and gene regulation, have been recently reported in filamentous fungi [45,56].
Fig. 4.
Application of CRISPR/Cas9 technology in filamentous fungi. While there are many potential applications of CRISPR/Cas9 technology for fungi, some of the most common that are currently employed are gene deletion/disruption and generation of gene fusions.
4.1. CRISPR/Cas9-Mediated Gene Disruption/Deletions
The initial establishment of CRISPR/Cas9 tools in filamentous fungi was mostly focused on generating gene disruptions using NHEJ and HR strategies (Fig. 1, Fig. 4). The plethora of suitable target genes that provide an easily observable phenotype played an important role in the rapid development of CRISPR/Cas9 tools. Usually the system involved targeting either biosynthetic genes responsible for pigment production and/or resistance-related genes since they have obvious phenotypes once disrupted. Some of the pigment biosynthetic genes that have been targeted include the yA gene (green/yellow) of A. nidulans, the albA gene (black/white) of A.niger [37], the pksP gene (green/white) of A. fumigatus [43,57], the pks17 gene (green/white) of P. chrysogenum [38], the pksA gene (black/white) of A. alternata [58], the FoBIK1 gene (purple/white) of F. oxysporum [44], and the ALB1 (black/white) and RSY1 genes (black/orange) of M. oryzae [46]. While the resistance-related genes that have been used in the development of CRISPR systems include orthologs of the uracil biosynthetic genes URA3 and/or URA5 which confer resistance to 5-fluoroorotic acid (5-FOA) when disrupted [37,44,48], the osmosensor histidine kinase Os1 in Leptosphaeria maculans and F. graminearum conferring resistance to the phenylpyrrole and dicarboximide classes of fungicides [59,60], and the SDI1 gene of M. oryzae conferring fungicide resistance to carboxin [46]. These studies indicated that most of the gene disruptions by NHEJ contained either short insertions or deletions of nucleotides at the Cas9 cleavage site, resulting in frameshift mutations. Gene disruptions by HR have been based on the integration of a dominant selectable marker, usually conferring resistance to an antifungal compound, at the Cas9/sgRNA cleavage site(s) disrupting the coding sequence of the gene of interest.
4.2. Generation of Gene Fusions Based on CRISPR/Cas9 Technology
Investigation of the protein subcellular localization using fluorescent markers plays an important role in characterizing the compartmentalization and intracellular movement of a protein. A gene can be tagged at the N- or C-terminus, but N-terminus tagging is usually a more complex process in order to avoid disruption of upstream regulatory elements. The precision provided by CRISPR/Cas9 genome editing has accelerated development in this area, and enabled fluorescent markers to be precisely fused to endogenous genes at their native locus (Fig. 4) [45,46,56,57]. Fluorescent marker tagging mainly depends on two strategies, homology-dependent and homology-independent, and currently most gene tagging using CRISPR/Cas9 technology in filamentous fungi is homology-dependent relying on HR-mediated DNA repair for integration of the fluorescent marker into the fungal genome. For example, in A. fumigatus, a GFP tag flanked with only 39 bp of homologous sequence was correctly integrated at the N-terminus of the cnaA gene, encoding the catalytic subunit of calcineurin, without the introduction of a selective marker [57]. A CRISPR-mediated co-editing strategy allowed editing at the SEP6 gene locus while simultaneously tagging the SEP5 gene with GFP at the N-terminus in M. oryzae, allowing visualization of GFP-Sep5 at the septin ring of the appressorium [46]. CRISPR/Cas9-mediated gene tagging can be also accomplished by a HITI strategy requiring the ligation of two separate DNA fragments/chromosomes at the Cas9 cleavage site [18]. Based on conservation in the DNA repair mechanism, this strategy has been used in F. oxysporum to fuse two endogenous genes with fluorescent markers [45]. This strategy enabled a Cas9 cleaved plasmid containing the fluorescent marker sequence to be ligated into the F. oxysporum genome at the Cas9 DNA cleavage site, tagging FoChs5 with 3 × sGFP at the C-terminus and FoSso2 with mCherry at the N-terminus [45]. This method has also been used to tag the scytalone dehydratase encoding SDH gene in M. oryzae at the C-terminus with GFP where a single CRISPR target site within the gene coding region facilitated the crossover event [56].
4.3. Investigation of Gene Regulation in Fungi via CRISPR/Cas9 Technology
Another important application of CRISPR/Cas9 technology has concerned the investigation of gene regulation in filamentous fungi. A large number of fungal genes are only expressed under specific conditions and include secondary metabolite biosynthetic genes, host-specific virulence factors, and some key regulatory genes. Expression profiles of many genes are dependent on environmental factors including light, salt, temperature, humidity, pH, and other stressors [61,62]. Use of CRISPR/Cas9-based genome editing has facilitated the exploration of gene expression profiles (Fig. 4). In N. crassa, the protein levels of histone H2B were used as an in vivo indicator for the amount of DNA in cells, and CRISPR/Cas9 was used to tag the endogenous histone H2B gene at the C-terminus with a codon-optimized firefly luciferase (H2B::LUC) [63]. The bioluminescence of the transformant strain displayed a circadian rhythm, indicating DNA synthesis in N. crassa was under control of the proteins that govern the circadian rhythm [63]. A similar strategy could be applied to study the expression of silent secondary metabolite gene clusters, where a molecular reporter, such as lacZ or fluorescent genes, can be fused to the key biosynthetic gene in the cluster enabling the expression of the gene/cluster of interest to be easily detected under different environmental conditions.
5. Challenges for Developing CRISPR/Cas9 Tools in Filamentous Fungi
Although a large number of filamentous fungal genomes have been sequenced and have contributed immensely to the understanding of fungal diversity and evolution, the reverse genetic techniques necessary to functionally characterize the genes of interest can be improved upon for most filamentous fungi. The emergence of CRISPR/Cas9-mediated genome editing has spurred a new era for functional genomics, and the rapid adoption of this technology for editing filamentous fungal genomes foreshadows the enormous potential for genome manipulation. Due to the diverse nature of the fungal kingdom, it can be difficult to develop a universal method for gene editing in filamentous fungi. Consequently, several CRISPR/Cas9 systems have been developed and optimized specifically for individual filamentous fungi or a specific genus, and each system may have different advantages and disadvantages. During the process of developing a CRISPR/Cas9 system for a particular fungus, several challenges may be encountered and can range from determining the optimal expression method for the CAS9 gene including the choice of promoter and the identification of a suitable NLS that allows it to be translocated to the nucleus, to the assessment and mitigation of undesired off-target cleavage by Cas9. Further exploration of these common challenges encountered during development of CRISPR/Cas9 systems is warranted as it could facilitate the development and use of Cas9 tools in other filamentous fungi.
5.1. Expression of the CAS9 Gene in Filamentous Fungi
The expression of CAS9 can be under the control of either a constitutive or an inducible promoter. Increasing evidence has shown that high levels of Cas9 protein may have a toxic effect to host cells. The influence of Cas protein levels on toxicity was investigated in the yeast S. cerevisiae, where the expression of CAS9 was placed under the control of a galactose inducible promoter [20]. When both the sgRNA and CAS9 were highly expressed there was a reduction in cell viability when compared to control yeast cells, indicating possible toxicity due to the high levels of Cas9 DNA cleavage activity [20]. These observations are further supported by similar findings in other organisms. Evidence of Cas9 toxicity was observed in the green alga Chlamydomonas reinhardtii when CAS9 was constitutively expressed [64], and in vivo expressed Cas9 toxicity has also been shown in the cyanobacterium Synechococcus elongatus [65], the parasite Trichomonas vaginalis [66], and in human cells [67].
Studies have indicated that the codon usage of the CAS9 gene may have an influence on Cas9 toxicity, while constitutive in vivo CAS9 expression might be suitable for some filamentous fungi. A codon-optimized CAS9 gene under the control of the amyB promoter was constitutively expressed in A. oryzae without causing detrimental effects in growth [68]. Similarly, a codon-optimized CAS9 fused with the GFP coding sequence under either a constitutive or an inducible promoter was integrated into the T. reesei genome via Agrobacterium-mediated transformation and the resulting transformants displayed strong nuclear fluorescence and appeared to have normal morphology and growth [24]. Conversely, Foster et al. was unable to obtain transformants that stably expressed the CAS9 gene in M. oryzae despite using different variants of CAS9 and several promoters [46]. Transformation with these plasmids and sgRNA generated a fewer number of transformants, suggesting constitutive CAS9 expression could be toxic in M. oryzae, and gene editing was ultimately accomplished by transient introduction of purified Cas9 protein with in vitro synthesized sgRNAs [46]. All of these results indicate that optimizing CAS9 expression in filamentous fungi can be a complex process.
5.2. Selection of the NLS for Cas9
Cas9 must enter the host nucleus to carry out cleavage of the DNA. The diversity within fungi complicate the identification of a universal NLS that is suitable for all fungi, and therefore the NLS sequence of Cas9 may need to be optimized for efficient nuclear entry in filamentous fungi. The selection of an appropriate NLS may be based on previous assessment of an NLS sequence or alternatively choosing a NLS of a native nuclear protein. The NLS from the endogenous gene H2B of F. oxysporum was identified by multiple sequence alignment and was fused to the N-terminus of the CAS9 gene. This NLS had protein solubility suitable for protein purification and did not significantly influence Cas9 cleavage activity [44]. A comparison of NLS sequences fused to Cas9 indicated the NLS can influence gene editing efficiency. Three different NLSs, SV40, H2BNLS, and VEVNLS, were fused to Cas9 and the gene editing efficiency assessed in F. fujikuroi, a close relative of F. oxysporum, and of the three variants, the Cas9 containing the endogenous NLS from histone H2B had a significantly higher gene editing efficiency [69].
5.3. Off-Target Activity
An important concern using CRISPR/Cas9 technology is the potential for off-target effects, where Cas9 causes an undesired DSB at an unintended chromosomal locus. Although the cleavage specificity of Cas9 is controlled by the 20 nt target sequence of the sgRNA and the presence of the PAM element, potential off-target cleavage is able to occur at loci with a high percentage of nucleotide identity to the sgRNA sequence [70,71]. In addition, different sgRNA structures can also influence the frequency of an off-target event occurring [71]. The off-target effect can limit the wide application of CRISPR/Cas9 technology in various organisms, and several approaches may increase CRISPR specificity and thereby reduce the likelihood of an off-target effect. The most critical parameter to limit off-target effects is the design of the sgRNA. The “seed” sequence of the sgRNA-guiding sequence, 10–12 bp adjacent to the PAM element, dictates Cas9 cleavage specificity [14], while the PAM element is necessary to permit unwinding and formation of secondary structures for DNA cleavage [14]. sgRNAs that contain over 17 identical nucleotides to another locus of the genome, are likely to have an unintended off-target cleavage at this secondary locus [71]. Additionally, truncated sgRNAs, containing 17–18 nt of target sequence, had higher specificity than the full length [72]. Taken together, designing a unique sgRNA, especially at the “seed” region, that does not share high DNA identity with other genetic loci greatly limits the probability of off-target cleavage with Cas9. Another factor that can influence the likelihood of an off-target event is the method of transformation, where plasmid-mediated transfection is responsible for more off-target events than Cas9 RNP-mediated transfection [73,74]. Newer modified versions of Cas9, such as “enhanced specificity” Cas9 [75] and high fidelity Cas9-HF1 [76], are available and may reduce the chances of off-target mutations occurring during transformation.
The probability of off-target mutations have been assessed in A. fumigatus. After transformation of the fungus with either a high concentration of Cas9 RNPs (1 μg/μL) or a low concentration (0.5 μg/μL), whole genome sequencing revealed there was no significant difference in the number of mutations in either one of the Cas9 RNP mediated transformations when compared to traditional transformation [77]. Additionally, there was no difference in the types of mutations that occurred between these groups, as the number of insertions and deletions were similar, and the locations of the mutations were predominantly located in intergenic regions [77]. In another study, off-target mutations were evaluated after CRISPR/Cas9-mediated transformation directed to TRI5 in F. graminearum [60]. A sgRNA targeting TRI5 shared 14 bp with two other loci within the genome, and sequencing of these loci after transformation revealed no mutations occurred at either location.
5.4. Transformation Methods With High Efficiency
Several genetic transformation methods are readily used for filamentous fungi including protoplast-mediated transformation, Agrobacterium-mediated transformation, and electroporation, but the transformation efficiency for these methods is usually low. The efficiency of protoplast-mediated transformation is in the order of one transformant generated per 1–10 million fungal protoplasts, although there is a very high degree of variability [78]. Because of this limitation, all established CRISPR/Cas9 tools in filamentous fungi have so far relied on the use of a selective marker or obvious phenotypic change to identify the desired transformant. Although techniques are available for removing these selective markers, these methods can be time-consuming and be experimentally challenging to conduct. Consequently, development of a highly efficient transformation method could greatly aid in the application CRISPR/Cas9 technology in filamentous fungi, and once transformation methods achieve a higher efficiency, gene editing could potentially be conducted without introducing a selective marker.
Several methods can potentially be used in filamentous fungi to increase transformation efficiencies. First, microinjection methods have been widely used in animal oomycetes and insect eggs with the advantage of directly delivering Cas9/sgRNA components into the host cells [79]. Although some filamentous fungi produce conidia with a single nucleus, the size of the conidia is usually small, making it difficult to adapt applications involving microinjection to filamentous fungi. In spite of difficulties, Toyoda et al. were able to successfully inject fluorescein-isothiocyanate-conjugated albumin into the cytoplasm of conidia from Erysiphe graminis f. sp. hordei [80], suggesting this technique may have potential for use in introduction of CRISPR/Cas9 molecules. Nano-delivery is another mechanism that has potential for transferring Cas9 RNPs and has been used to translocate genes, siRNA molecules, and proteins into host cells [81]. A nonviral delivery vehicle for the Cas9 RNP, termed CRISPR-Gold, has been used for gene editing in the brains of adult mice [82]. Once identified, suitable nanoparticles for use in filamentous fungi could be adapted for the delivery of exogenous genes or proteins into fungal cells. Alternatively, cell-penetrating peptides may have promise and have been shown to be able to enter into Candida spp. [83,84].
6. Conclusions
Filamentous fungi have played an integral role in agriculture, industry, drug development, and biodegradation. As genome sequencing techniques improve and the associated costs decrease, the gap between sequence information and functional assessment will widen. Although having been available for less than a decade, the precise genome editing provided by CRISPR/Cas9 systems facilitates genetic manipulation and has been already adapted for several filamentous fungi. CRISPR/Cas9 technology could greatly aid in the use of fungi as biological factories, in particular manipulation of secondary metabolite biosynthetic genes which may be useful in developing bioactive products or derivatives for medicine. In addition, this tool will be valuable in fungal biology and investigation of the molecular interactions between fungal pathogens and their hosts. Although this tool is primarily used for gene disruptions, further improvements to CRISPR/Cas9 systems will increase its utility for various applications in filamentous fungi in the future.
Acknowledgments
Acknowledgement
This publication was supported by the Alabama Agricultural Experiment Station and the Hatch program of the National Institute of Food and Agriculture, USDA.
Declaration of Competing Interest
In regards to the submitted manuscript entitled “Progress and challenges: development and implementation of CRISPR/Cas9 technology in filamentous fungi”, the authors declare there are no conflicts of interest.
References
- 1.Beadle G.W., Tatum E.L. Genetic control of biochemical reactions in Neurospora. Proc Natl Acad Sci USA. 1941;27:499–506. doi: 10.1073/pnas.27.11.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roche C.M., Loros J.J., McCluskey K., Glass N.L. Neurospora crassa : looking back and looking forward at a model microbe. Am J Bot. 2014;101:2022–2035. doi: 10.3732/ajb.1400377. [DOI] [PubMed] [Google Scholar]
- 3.Guzmán-Chávez F., Zwahlen R.D., Bovenberg R.A.L., Driessen A.J.M. Engineering of the filamentous fungus Penicillium chrysogenum as cell factory for natural products. Front Microbiol. 2018;9:2768. doi: 10.3389/fmicb.2018.02768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson R.A., Talbot N.J. Under pressure: investigating the biology of plant infection by Magnaporthe oryzae. Nat Rev Microbiol. 2009;7:185–195. doi: 10.1038/nrmicro2032. [DOI] [PubMed] [Google Scholar]
- 5.Michielse C.B., Rep M. Pathogen profile update: Fusarium oxysporum. Mol Plant Pathol. 2009;10:311–324. doi: 10.1111/j.1364-3703.2009.00538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coleman J.J. The Fusarium solani species complex: ubiquitous pathogens of agricultural importance. Mol Plant Pathol. 2016;17:146–158. doi: 10.1111/mpp.12289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muhammed M., Anagnostou T., Desalermos A., Kourkoumpetis T.K., Carneiro H.A., Glavis-Bloom J. Fusarium infection: report of 26 cases and review of 97 cases from the literature. Medicine (Baltimore) 2013;92:305–316. doi: 10.1097/MD.0000000000000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lass-Florl C. The changing face of epidemiology of invasive fungal disease in Europe. Mycoses. 2009;52:197–205. doi: 10.1111/j.1439-0507.2009.01691.x. [DOI] [PubMed] [Google Scholar]
- 9.Brown G.D., Denning D.W., Gow N.A., Levitz S.M., Netea M.G., White T.C. Hidden killers: human fungal infections. Sci Transl Med. 2012;4 doi: 10.1126/scitranslmed.3004404. (165rv13) [DOI] [PubMed] [Google Scholar]
- 10.Kumar P., Mahato D.K., Kamle M., Mohanta T.K., Kang S.G. Aflatoxins: a global concern for food safety, human health and their management. Front Microbiol. 2017;7:2170. doi: 10.3389/fmicb.2016.02170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goswami R.S., Kistler H.C. Heading for disaster: Fusarium graminearum on cereal crops. Mol Plant Pathol. 2004;5:515–525. doi: 10.1111/j.1364-3703.2004.00252.x. [DOI] [PubMed] [Google Scholar]
- 12.Komor A.C., Badran A.H., Liu D.R. CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell. 2017;168:20–36. doi: 10.1016/j.cell.2016.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marraffini L.A., Sontheimer E.J. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet. 2010;11:181–190. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodgers K., McVey M. Error-prone repair of DNA double-strand breaks. J Cell Physiol. 2016;231:15–24. doi: 10.1002/jcp.25053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bétermier M., Bertrand P., Lopez B.S. Is non-homologous end-joining really an inherently error-prone process? PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo T., Feng Y.L., Xiao J.J., Liu Q., Sun X.N., Xiang J.F. Harnessing accurate non-homologous end joining for efficient precise deletion in CRISPR/Cas9-mediated genome editing. Genome Biol. 2018;19:170. doi: 10.1186/s13059-018-1518-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzuki K., Tsunekawa Y., Hernandez-Benitez R., Wu J., Zhu J., Kim E.J. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature. 2016;540:144–149. doi: 10.1038/nature20565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimura Y., Hisano Y., Kawahara A., Higashijima S. Efficient generation of knock-in transgenic zebrafish carrying reporter/driver genes by CRISPR/Cas9-mediated genome engineering. Sci Rep UK. 2014;4:6545. doi: 10.1038/srep06545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DiCarlo J.E., Norville J.E., Mali P., Rios X., Aach J., Church G.M. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013;41:4336–4343. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vyas V.K., Barrasa M.I., Fink G.R. A Candida albicans CRISPR system permits genetic engineering of essential genes and gene families. Sci Adv. 2015;1 doi: 10.1126/sciadv.1500248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Min K., Ichikawa Y., Woolford C.A., Mitchell A.P. Candida albicans gene deletion with a transient CRISPR-Cas9 system. mSphere. 2016;1 doi: 10.1128/mSphere.00130-16. [e00130-16] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ng H., Dean N. Dramatic improvement of CRISPR/Cas9 editing in Candida albicans by increased single guide RNA expression. mSphere. 2017;2 doi: 10.1128/mSphere.00385-16. [e00385-16] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu R., Chen L., Jiang Y.P., Zhou Z.H., Zou G. Efficient genome editing in filamentous fungus Trichoderma reesei using the CRISPR/Cas9 system. Cell Discov. 2015;1:15007. doi: 10.1038/celldisc.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuscu C., Arslan S., Singh R., Thorpe J., Adli M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat Biotechnol. 2014;32:677–683. doi: 10.1038/nbt.2916. [DOI] [PubMed] [Google Scholar]
- 26.Matsu-Ura T., Baek M., Kwon J., Hong C. Efficient gene editing in Neurospora crassa with CRISPR technology. Fungal Biol Biotechnol. 2015;2:4. doi: 10.1186/s40694-015-0015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kellis M., Birren B.W., Lander E.S. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- 28.Wolfe K.H., Shields D.C. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- 29.Fedorova N.D., Khaldi N., Joardar V.S., Maiti R., Amedeo P., Anderson M.J. Genomic islands in the pathogenic filamentous fungus Aspergillus fumigatus. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma L.-J., Ibrahim A.S., Skory C., Grabherr M.G., Burger G., Butler M. Genomic analysis of the basal lineage fungus Rhizopus oryzae reveals a whole-genome duplication. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma L.-J., van der Does H.C., Borkovich K.A., Coleman J.J., Daboussi M.-J., Di Pietro A. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature. 2010;464:367–373. doi: 10.1038/nature08850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coleman J.J., Rounsley S.D., Rodriguez-Carres M., Kuo A., Wasmann C.C., Grimwood J. The genome of Nectria haematococca: contribution of supernumerary chromosomes to gene expansion. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goodwin S.B., Ben M'Barek S., Dhillon B., Wittenberg A.H.J., Crane C.F., Hane J.K. Finished genome of the fungal wheat pathogen Mycosphaerella graminicola reveals dispensome structure, chromosome plasticity, and stealth pathogenesis. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J., Zhang Y., Zhang Y., Yu P.L., Pan H., Rollins J.A. Introduction of large sequence inserts by CRISPR-Cas9 to create pathogenicity mutants in the multinucleate filamentous pathogen Sclerotinia sclerotiorum. mBio. 2018;9 doi: 10.1128/mBio.00567-18. [e00567-18] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aleksenko A., Clutterbuck A.J. Autonomous plasmid replication in Aspergillus nidulans: AMA1 and MATE elements. Fungal Genet Biol. 1997;21:373–387. doi: 10.1006/fgbi.1997.0980. [DOI] [PubMed] [Google Scholar]
- 36.Nielsen M.L., Isbrandt T., Rasmussen K.B., Thrane U., Hoof J.B., TO Larsen. Genes linked to production of secondary metabolites in Talaromyces atroroseus revealed using CRISPR-Cas9. PloS One. 2017;12 doi: 10.1371/journal.pone.0169712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nodvig C.S., Nielsen J.B., Kogle M.E., Mortensen U.H. A CRISPR-Cas9 system for genetic engineering of filamentous fungi. Plos One. 2015;10 doi: 10.1371/journal.pone.0133085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pohl C., Kiel J.A.K.W., Driessen A.J.M., Bovenberg R.A.L., Nygard Y. CRISPR/Cas9 based genome editing of Penicillium chrysogenum. ACS Synth Biol. 2016;5:754–764. doi: 10.1021/acssynbio.6b00082. [DOI] [PubMed] [Google Scholar]
- 39.Kim S., Kim D., Cho S.W., Kim J., Kim J.S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woo J.W., Kim J., Kwon S.I., Corvalan C., Cho S.W., Kim H. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat Biotechnol. 2015;33:1162–1164. doi: 10.1038/nbt.3389. [DOI] [PubMed] [Google Scholar]
- 41.Burger A., Lindsay H., Felker A., Hess C., Anders C., Chiavacci E. Maximizing mutagenesis with solubilized CRISPR-Cas9 ribonucleoprotein complexes. Development. 2016;143:2025–2037. doi: 10.1242/dev.134809. [DOI] [PubMed] [Google Scholar]
- 42.Bassett Andrew R., Tibbit C., Ponting Chris P., Liu J.-L. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep. 2013;4:220–228. doi: 10.1016/j.celrep.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Al Abdallah Q., Ge W., Fortwendel J.R. A simple and universal system for gene manipulation in Aspergillus fumigatus: in vitro-assembled Cas9-guide RNA ribonucleoproteins coupled with microhomology repair templates. mSphere. 2017;2 doi: 10.1128/mSphere.00446-17. (e00446-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Q., Cobine P.A., Coleman J.J. Efficient genome editing in Fusarium oxysporum based on CRISPR/Cas9 ribonucleoprotein complexes. Fungal Genet Biol. 2018;117:21–29. doi: 10.1016/j.fgb.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Q., Coleman J.J. CRISPR/Cas9-mediated endogenous gene tagging in Fusarium oxysporum. Fungal Genet Biol. 2019;126:17–24. doi: 10.1016/j.fgb.2019.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Foster A.J., Martin-Urdiroz M., Yan X., Wright H.S., Soanes D.M., Talbot N.J. CRISPR-Cas9 ribonucleoprotein-mediated co-editing and counterselection in the rice blast fungus. Sci Rep UK. 2018;8:14355. doi: 10.1038/s41598-018-32702-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pohl C., Mózsik L., Driessen A.J., Bovenberg R.A., Nygård Y.I. Synthetic biology. Springer; 2018. Genome editing in Penicillium chrysogenum using Cas9 ribonucleoprotein particles; pp. 213–232. [DOI] [PubMed] [Google Scholar]
- 48.Nagy G., Szebenyi C., Csernetics A., Vaz A.G., Toth E.J., Vagvolgyi C. Development of a plasmid free CRISPR-Cas9 system for the genetic modification of Mucor circinelloides. Sci Rep Uk. 2017;7:16800. doi: 10.1038/s41598-017-17118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eid A., Alshareef S., Mahfouz M.M. CRISPR base editors: genome editing without double-stranded breaks. Biochem J. 2018;475:1955–1964. doi: 10.1042/BCJ20170793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Terns M.P. CRISPR-based technologies: impact of RNA-targeting systems. Mol Cell. 2018;72:404–412. doi: 10.1016/j.molcel.2018.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gilbert Luke A., Horlbeck Max A., Adamson B., Villalta Jacqueline E., Chen Y., Whitehead Evan H. Genome-scale CRISPR-mediated control of gene repression and activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brakhage A.A., Schroeckh V. Fungal secondary metabolites – strategies to activate silent gene clusters. Fungal Genet Biol. 2011;48:15–22. doi: 10.1016/j.fgb.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 54.Keller N.P. Fungal secondary metabolism: regulation, function and drug discovery. Nat Rev Microbiol. 2019;17:167–180. doi: 10.1038/s41579-018-0121-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yin W., Keller N.P. Transcriptional regulatory elements in fungal secondary metabolism. J Microbiol. 2011;49:329–339. doi: 10.1007/s12275-011-1009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamato T., Handa A., Arazoe T., Kuroki M., Nozaka A., Kamakura T. Single crossover-mediated targeted nucleotide substitution and knock-in strategies with CRISPR/Cas9 system in the rice blast fungus. Sci Rep UK. 2019;9:7427. doi: 10.1038/s41598-019-43913-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang C., Meng X., Wei X., Lu L. Highly efficient CRISPR mutagenesis by microhomology-mediated end joining in Aspergillus fumigatus. Fungal Genet Biol. 2016;86:47–57. doi: 10.1016/j.fgb.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 58.Wenderoth M., Pinecker C., Voss B., Fischer R. Establishment of CRISPR/Cas9 in Alternaria alternata. Fungal Genet Biol. 2017;101:55–60. doi: 10.1016/j.fgb.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 59.Idnurm A., Urquhart A.S., Vummadi D.R., Chang S., Van de Wouw A.P., López-Ruiz F.J. Spontaneous and CRISPR/Cas9-induced mutation of the osmosensor histidine kinase of the canola pathogen Leptosphaeria maculans. Fungal Biol Biotechnol. 2017;4:12. doi: 10.1186/s40694-017-0043-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gardiner D.M., Kazan K. Selection is required for efficient Cas9-mediated genome editing in Fusarium graminearum. Fungal Biol. 2018;122:131–137. doi: 10.1016/j.funbio.2017.11.006. [DOI] [PubMed] [Google Scholar]
- 61.Selvig K., Alspaugh J.A. pH response pathways in fungi: adapting to host-derived and environmental signals. Mycobiology. 2011;39:249–256. doi: 10.5941/MYCO.2011.39.4.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tisch D., Schmoll M. Light regulation of metabolic pathways in fungi. Appl Microbiol Biotechnol. 2010;85:1259–1277. doi: 10.1007/s00253-009-2320-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu X., Dang Y.K., Matsu-ura T., He Y.B., He Q., Hong C.I. DNA replication is required for circadian clock function by regulating rhythmic nucleosome composition. Mol Cell. 2017;67:203–213. doi: 10.1016/j.molcel.2017.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jiang W.Z., Brueggeman A.J., Horken K.M., Plucinak T.M., Weeks D.P. Successful transient expression of Cas9 and single guide RNA genes in Chlamydomonas reinhardtii. Eukaryot Cell. 2014;13:1465–1469. doi: 10.1128/EC.00213-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wendt K.E., Ungerer J., Cobb R.E., Zhao H.M., Pakrasi H.B. CRISPR/Cas9 mediated targeted mutagenesis of the fast growing cyanobacterium Synechococcus elongatus UTEX 2973. Microb Cell Fact. 2016;15:115. doi: 10.1186/s12934-016-0514-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Janssen B.D., Chen Y.P., Molgora B.M., Wang S.E., Simoes-Barbosa A., Johnson P.J. CRISPR/Cas9-mediated gene modification and gene knock out in the human-infective parasite Trichomonas vaginalis. Sci Rep UK. 2018;8:270. doi: 10.1038/s41598-017-18442-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Morgens D.W., Wainberg M., Boyle E.A., Ursu O., Araya C.L., Tsui C.K. Genome-scale measurement of off-target activity using Cas9 toxicity in high-throughput screens. Nat Commun. 2017;8:15178. doi: 10.1038/ncomms15178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Katayama T., Tanaka Y., Okabe T., Nakamura H., Fujii W., Kitamoto K. Development of a genome editing technique using the CRISPR/Cas9 system in the industrial filamentous fungus Aspergillus oryzae. Biotechnol Lett. 2016;38:637–642. doi: 10.1007/s10529-015-2015-x. [DOI] [PubMed] [Google Scholar]
- 69.Shi T.Q., Gao J., Wang W.J., Wang K.F., Xu G.Q., Huang H. CRISPR/Cas9-based genome-editing in the filamentous fungus Fusarium fujikuroi and its application in strain engineering for gibberellic acid production. ACS Synth Biol. 2019;8:445–454. doi: 10.1021/acssynbio.8b00478. [DOI] [PubMed] [Google Scholar]
- 70.Pattanayak V., Lin S., Guilinger J.P., Ma E., Doudna J.A., Liu D.R. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol. 2013;31:839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fu Y.F., Sander J.D., Reyon D., Cascio V.M., Joung J.K. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lin S., Staahl B.T., Alla R.K., Doudna J.A. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife. 2014;3 doi: 10.7554/eLife.04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.DeWitt M.A., Corn J.E., Carroll D. Genome editing via delivery of Cas9 ribonucleoprotein. Methods. 2017;121-122:9–15. doi: 10.1016/j.ymeth.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Slaymaker I.M., Gao L.Y., Zetsche B., Scott D.A., Yan W.X., Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kleinstiver B.P., Pattanayak V., Prew M.S., Tsai S.Q., Nguyen N.T., Joung J.K. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Mol Ther. 2016;24 doi: 10.1038/nature16526. [S288-S] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Al Abdallah Q., Souza A.C.O., Martin-Vicente A., Ge W., Fortwendel J.R. Whole-genome sequencing reveals highly specific gene targeting by in vitro assembled Cas9-ribonucleoprotein complexes in Aspergillus fumigatus. Fungal Biol Biotechnol. 2018;5:11. doi: 10.1186/s40694-018-0057-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li D., Tang Y., Lin J., Cai W. Methods for genetic transformation of filamentous fungi. Microb Cell Fact. 2017;16:168. doi: 10.1186/s12934-017-0785-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim H., Kim J.S. A guide to genome engineering with programmable nucleases. Nat Rev Genet. 2014;15:321–334. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- 80.Toyoda H., Matsuda Y., Shoji R., Ouchi S. A microinjection technique for conidia of Erysiphe graminis f.sp. hordei. Phytopathology. 1987;77:815–818. [Google Scholar]
- 81.Singh R., Lillard J.W., Jr. Nanoparticle-based targeted drug delivery. Exp Mol Pathol. 2009;86:215–223. doi: 10.1016/j.yexmp.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee B., Lee K., Panda S., Gonzales-Rojas R., Chong A., Bugay V. Nanoparticle delivery of CRISPR into the brain rescues a mouse model of fragile X syndrome from exaggerated repetitive behaviours. Nat Biomed Eng. 2018;2:497–507. doi: 10.1038/s41551-018-0252-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gong Z.F., Karlsson A.J. Translocation of cell-penetrating peptides into Candida fungal pathogens. Protein Sci. 2017;26:1714–1725. doi: 10.1002/pro.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Farkhani S.M., Valizadeh A., Karami H., Mohammadi S., Sohrabi N., Badrzadeh F. Cell penetrating peptides: efficient vectors for delivery of nanoparticles, nanocarriers, therapeutic and diagnostic molecules. Peptides. 2014;57:78–94. doi: 10.1016/j.peptides.2014.04.015. [DOI] [PubMed] [Google Scholar]
- 85.Chen J., Lai Y., Wang L., Zhai S., Zou G., Zhou Z. CRISPR/Cas9-mediated efficient genome editing via blastospore-based transformation in entomopathogenic fungus Beauveria bassiana. Sci Rep UK. 2017;7:45763. doi: 10.1038/srep45763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kujoth G.C., Sullivan T.D., Merkhofer R., Lee T.-J., Wang H., Brandhorst T. CRISPR/Cas9-mediated gene disruption reveals the importance of zinc metabolism for fitness of the dimorphic fungal pathogen Blastomyces dermatitidis. mBio. 2018;9 doi: 10.1128/mBio.00412-18. [e00412-18] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sugano S.S., Suzuki H., Shimokita E., Chiba H., Noji S., Osakabe Y. Genome editing in the mushroom-forming basidiomycete Coprinopsis cinerea, optimized by a high-throughput transformation system. Sci Rep UK. 2017;7:1260. doi: 10.1038/s41598-017-00883-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen B.-X., Wei T., Ye Z.-W., Yun F., Kang L.-Z., Tang H.-B. Efficient CRISPR-Cas9 gene disruption system in edible-medicinal mushroom Cordyceps militaris. Front Microbiol. 2018;9:1157. doi: 10.3389/fmicb.2018.01157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Qin H., Xiao H., Zou G., Zhou Z., Zhong J.-J. CRISPR-Cas9 assisted gene disruption in the higher fungus Ganoderma species. Process Biochem. 2017;56:57–61. [Google Scholar]
- 90.Liu Q., Gao R., Li J., Lin L., Zhao J., Sun W. Development of a genome-editing CRISPR/Cas9 system in thermophilic fungal Myceliophthora species and its application to hyper-cellulase production strain engineering. Biotechnol Biofuels. 2017;10:1. doi: 10.1186/s13068-016-0693-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zheng Y.M., Lin F.L., Gao H., Zou G., Zhang J.W., Wang G.Q. Development of a versatile and conventional technique for gene disruption in filamentous fungi based on CRISPR-Cas9 technology. Sci Rep UK. 2017;7:9250. doi: 10.1038/s41598-017-10052-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bruni G.O., Zhong K., Lee S.C., Wang P. CRISPR-Cas9 induces point mutation in the mucormycosis fungus Rhizopus delemar. Fungal Genet Biol. 2019;124:1–7. doi: 10.1016/j.fgb.2018.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lu S., Shen X., Chen B. Development of an efficient vector system for gene knock-out and near in-cis gene complementation in the sugarcane smut fungus. Sci Rep UK. 2017;7:3113. doi: 10.1038/s41598-017-03233-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liang Y., Han Y., Wang C., Jiang C., Xu J.-R. Targeted deletion of the USTA and UvSLT2 genes efficiently in Ustilaginoidea virens with the CRISPR-Cas9 system. Front Plant Sci. 2018;9:699. doi: 10.3389/fpls.2018.00699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Huck S., Bock J., Girardello J., Gauert M., Pul Ü. Marker-free genome editing in Ustilago trichophora with the CRISPR-Cas9 technology. RNA Biol. 2019;16:397–403. doi: 10.1080/15476286.2018.1493329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Katayama T., Nakamura H., Zhang Y., Pascal A., Fujii W., Maruyama J.I. Forced recycling of an AMA1-based genome-editing plasmid allows for efficient multiple gene deletion/integration in the industrial filamentous fungus Aspergillus oryzae. Appl Environ Microbiol. 2019;85 doi: 10.1128/AEM.01896-18. [e01896-18] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schuster M., Schweizer G., Reissmann S., Kahmann R. Genome editing in Ustilago maydis using the CRISPR–Cas system. Fungal Genet Biol. 2016;89:3–9. doi: 10.1016/j.fgb.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 98.Grijseels S., Pohl C., Nielsen J.C., Wasil Z., Nygård Y., Nielsen J. Identification of the decumbenone biosynthetic gene cluster in Penicillium decumbens and the importance for production of calbistrin. Fungal Biol Biotechnol. 2018;5:18. doi: 10.1186/s40694-018-0063-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hao Z.Z., Su X.Y. Fast gene disruption in Trichoderma reesei using in vitro assembled Cas9/gRNA complex. BMC Biotechnol. 2019;19 doi: 10.1186/s12896-018-0498-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Deng H., Gao R., Liao X., Cai Y. Genome editing in Shiraia bambusicola using CRISPR-Cas9 system. J Biotechnol. 2017;259:228–234. doi: 10.1016/j.jbiotec.2017.06.1204. [DOI] [PubMed] [Google Scholar]