Abstract
Approximately 30% patients with stage III colon cancer (CC) develop local recurrence and/or distant metastasis, even if postoperative adjuvant chemotherapy with oxaliplatin plus 5-fluorouracil and leucovorin (5-FU/LV) has been completed. In the present study, molecular analysis was performed to identify molecular markers of tumor recurrence in patients with stage III CC receiving oxaliplatin-based adjuvant chemotherapy. The FACOS study was conducted as a phase II study to evaluate the safety and efficacy of oxaliplatin-based treatment for stage III CC patients. Of the 132 CC patients enrolled in the present study, gene expression analysis using a microarray was conducted in 51 patients. Of these 51 patients, 6 developed recurrence within 5 years. The topmost 5% genes that showed differential expressions between cases that developed/did not develop recurrence were selected, and a set of predictive molecular markers for recurrence was identified. Of the 34,694 genes in the microarray, 1,734 genes were extracted as topmost 5% genes showing differential expressions between cases with and without recurrence. Among these, 10 genes, includingADH1A, ADH1C, CA12, CHP2, HMGCS2, SNAR-A1, TPI1, MS4A12, PLA2G10 and PTPRO, were identified as markers that could clearly divide patients with and without recurrence. Although several prediction models of tumor recurrence have been reported for CC, the set of 10 genes that the present study identified may be useful to predict the risk of recurrence in stage III CC patients receiving oxaliplatin-based adjuvant chemotherapy. Based on these results, high-risk patients with CC should be carefully observed to detect tumor recurrence during the follow-up period.
Keywords: colon cancer, oxaliplatin, molecular marker
Introduction
Previous studies (1–3) have demonstrated that oxaliplatin (Ox)-based adjuvant chemotherapy is superior to 5-furuolouracil (5-FU)-based adjuvant chemotherapy for stage III colorectal cancer (CRC) patients, in terms of extending the disease-free survival (DFS). For patients with stage III CRC, recent clinical guidelines, including the National Comprehensive Cancer Network (NCCN) (4) and European Society for Medical Oncology (ESMO) (5), recommend Ox-based postoperative adjuvant chemotherapy after successful surgical resection to prevent recurrence or metastasis; however, 30% of the patients still develop recurrence, even if postoperative adjuvant chemotherapy has been successfully completed.
In Japan, oral 5-FU regimens have been used for stage III CRC patients after curative resection (6). We conducted the FACOS study to verify the efficacy and safety of Ox+5-FU/LV (FOLFOX therapy) and Ox+capecitabine (XELOX therapy) as postoperative adjuvant chemotherapy for Japanese patients with stage III colon cancer (CC) (7,8). Even in stage III CC patients in whom Ox-based adjuvant chemotherapy was successfully completed, a subset of patients still developed tumor recurrence, regardless of clinicopathological factors. Although there are several reports (9–11) of prognostic factors, including the KRAS or BRAF mutation status and proficient or deficient mismatch repair genes in stage III CC patients receiving Ox-based adjuvant chemotherapy, only a few reports (12,13) have demonstrated prognostic molecular markers at the mRNA expression level to predict recurrence in stage III CC patients receiving Ox-based adjuvant chemotherapy. Validation studies of the expressions of 12 genes in Oncotype DX have been reported in stage II and III CC patients receiving adjuvant FOLFOX treatment (12). The recurrence score, calculated based on the gene expression levels (14), was positively correlated with the disease stage. The SUNRISE Study (15) also reported results consistent with previous reports (12), even in stage II and III CC Japanese patients who did not receive any adjuvant chemotherapy. Another report (13) showed that classification based on the gene expression profiles is useful to predict the prognosis in stage III CRC patients receiving adjuvant FOLFOX therapy. The DFS and overall survival (OS) associated with these classifications were similar to those associated with classifications based on the consensus molecular subtypes (CMS) (16). While these classifications are available to predict poor prognosis in CRC patients, these molecular signatures are not specific for detecting tumor recurrence in stage III CC patients. Furthermore, the efficacy of Ox-based adjuvant chemotherapy following curative resection for stage III CRC patients is not included in the molecular signature. Therefore, we investigated the predictive molecular markers of tumor recurrence specifically in stage III CC patients receiving adjuvant FOLFOX or XELOX therapy.
In the present study, we performed microarray-based gene expression profiling for detecting tumor recurrence in a half of the patients that were registered for the FACOS study. Microarray-based gene expression profiling has been established as a method to identify CRC patients with relapse (17–19). In a previous study (17), 58 genes were found to be upregulated and 160 genes were found to be downregulated in Dukes' C CRC patients with a poor prognosis, as compared to the expression levels in those with a good prognosis, even though the status of KRAS and TP53 mutations failed to predict tumor recurrence. It would be convenient to identify patients with a high risk of tumor recurrence using the expression profiles of molecular markers, especially a small number of genes, in cancer tissues derived from resected specimens. In the present study, we identified a set of molecular markers in patients with a high risk of tumor recurrence among stage III CC patients receiving FOLFOX or XELOX treatment.
Patients and methods
Ethical considerations
The present study was conducted with the approval of the local Ethics Committee of Saitama Medical Center. From every patient registered for this study, informed consent for registration was obtained.
Patients and tissue samples
The study was a phase II clinical study to investigate the efficacy and safety of FOLFOX and XELOX therapy as postoperative adjuvant chemotherapy for Japanese patients with stage III CC. Among the 132 CC patients enrolled (7,8), gene expression analyses using a microarray was conducted in 51 patients. The regimens of mFOLFOX6 and XELOX are described in our previous report (7,8): Briefly, the mFOLFOX6 regimen comprises intravenous infusions of oxaliplatin (85 mg/m2) and LV (200 mg/m2) for 2 h, followed by rapid intravenous bolus infusion of 5-FU (400 mg/m2) for 5 min, and continuous intravenous infusion of 5-FU (2,400 mg/m2) for 46 h. This regimen is repeated every 2 weeks for 12 cycles. The XELOX regimen comprises intravenous infusion of oxaliplatin (130 mg/m2 over 2 h) on day 1 and oral administration of capecitabine (1,000 mg/m2 twice daily) from the evening of day 1 to the morning of day 15. This regimen is repeated every 3 weeks for 8 cycles.
Of 51 patients in whom we analyzed the gene expression profiles, tumor recurrence was observed in 6 patients within 5 years. These patients were categorized into the recurrence group and remaining 45 patients were categorized into the non-recurrence group for this study.
The cancer and/or normal tissues taken from resected specimens were immediately frozen in liquid nitrogen and stored at −80°C until RNA extraction.
Microarray analysis
Before extraction of the total RNAs, each tissue sample was minced and homogenized in TRIzol reagents (Thermo Fisher Scientific, Inc., Waltham, MA, USA) on ice. The total RNA extraction procedure using TRIzol was performed in accordance with the manufacturer's instructions. The quality of all of the total RNAs was estimated using BioAnalyzer and the RNA600 nano kit (Agilent Technologies, Inc., Santa Clara, CA, USA). After DNase I treatment to remove contaminating DNA from each sample, the total RNAs were purified using phenol/chloroform/isoamyl alcohol (25:24:1, v/v) and isopropyl alcohol. The quality and quantity of the RNAs were finally confirmed with the BioAnalyzer and NanoDrop spectrometer (Thermo Fisher Scientific, Inc.), respectively.
For the microarray analysis, the extracted RNAs were amplified and labeled with Cy3-streptavidin (Amersham Biosciences, Buckinghamshire, UK), using the TargetAmp nano labeling kit for Illumina Expression BeadChip (Epicentre Biotechnologies, Madison, WI, USA). The human HT-12 v4 Expression BeadChip kit (Illumina Inc., San Diego, CA, USA) was used to hybridize the labeled samples and washed, in accordance with the manufacturer's instructions. Scanning and data measurements were performed using BeadsStation 500GXDW and GenomeStudio software (Illumina), in accordance with the manufacturer's standard protocol.
The microarray data were analyzed using the lumi (2.18.0) (20) and limma (3.22.7) (21) packages in R/Bioconductor (version 3.1.3 (22) and 3.0 (23), respectively). Gene annotations and related information were acquired from NCBI Entrez Gene (https://www.ncbi.nlm.nih.gov/gene/), UCSC Genome Browser (https://genome.ucsc.edu/), and Ensembl (https://genome.ucsc.edu), based on human genome GRCh38/hg38. Construction of scatter plots, hierarchical clustering, and heat maps was also performed with R/Bioconductor. Takeru for Sequencer IV (NABE International Corp, Tsukuba, Japan) was used for all the calculation of the microarray analysis and other relevant analyses. All expression data are represented by their logarithmically transformed (base; 2) values.
Statistical analysis
Associations between categorical variables were evaluated by the χ2 test, Fisher's exact test or Mann-Whitney U test. P<0.05 was considered to indicate a statistically significant difference. A survival analysis was conducted using the Kaplan-Meier method, and the log-rank test was used to determine the significance of differences between the survival curves. The period of OS was calculated from the time of surgery to the date of death from any cause, and was censored at the time of the last visit to our hospital or March 2018, whichever came first. All statistical analyses were performed using the SPSS v.11.0 software (SPSS Inc., Chicago, IL, USA).
Results
Comparison of the clinicopathological characteristics between the recurrence and non-recurrence groups
Comparison of the clinicopathological characteristics between the recurrence and non-recurrence groups is shown in Table I. Deeper invasion was significantly associated with recurrence (p=0.0075). There was no significant difference between the recurrence and non-recurrence groups in terms of the age, gender, PS, treatment regimen, primary tumor site, histology, lymph node metastasis, stage classification, or presence/absence of lymphatic/venous invasion.
Table I.
Patient characteristics in recurrent and non-recurrent patients with stage III colon cancer.
| Characteristics | Recurrent group (n=6) | Non-recurrent group (n=45) | P-value |
|---|---|---|---|
| Agea | 68.5 (55–75) | 67 (34–75) | 0.44 |
| Sex (male:female) | 5:1 | 33:12 | 0.98 |
| PS (0:1)b | 6:0 | 42:3 | 0.51 |
| Treatment | 0.17 | ||
| mFOLFOX6 | 1 | 25 | |
| XELOX | 5 | 20 | |
| Primary tumor site | 0.8 | ||
| Cecum | 1 | 1 | (C,S,T vs. D,S,R) |
| Ascending colon | 0 | 8 | |
| Transverse colon | 0 | 5 | |
| Descending colon | 0 | 3 | |
| Sigmoid colon | 3 | 15 | |
| Rectosigmoid | 2 | 13 | |
| Depth of invasionc | 0.0075 | ||
| T1 | 0 | 1 | (T1-3 vs. T4a,b) |
| T2 | 0 | 2 | |
| T3 | 0 | 27 | |
| T4a | 4 | 14 | |
| T4b | 2 | 1 | |
| Type of histology | 0.6 | ||
| tub1 | 0 | 0 | |
| tub2 | 6 | 37 | |
| por | 0 | 5 | |
| muc | 0 | 3 | |
| Lymph node metastasisc | 0.3 | ||
| N1 | 3 | 32 | |
| N2 | 3 | 13 | |
| Stagec | 0.15 | ||
| IIIA | 0 | 3 | |
| IIIB | 2 | 32 | |
| IIIC | 4 | 10 | |
| No. of lymph node dissectiona | 19.5 (9–28) | 20 (5–67) | 0.35 |
| Lymphatic invasion | 5 | 38 | 0.6 |
| Venous invasion | 6 | 38 | 0.68 |
| Preoperative complications | |||
| Perforation | 1 | 0 | |
| Colon obstruction | 0 | 1 | 0.55 |
| Lymph node dissectiond | |||
| D2 | 0 | 4 | |
| D3 | 6 | 41 | 0.45 |
Median (range)
American society of anesthesiologists physical status
According to the 7th TNM Classification
According to the Japaneses Classification of Colorectal Carcinoma.
Candidate genes for prediction of tumor recurrence
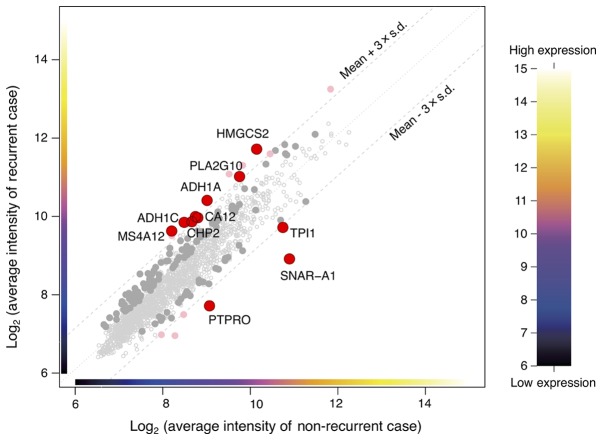
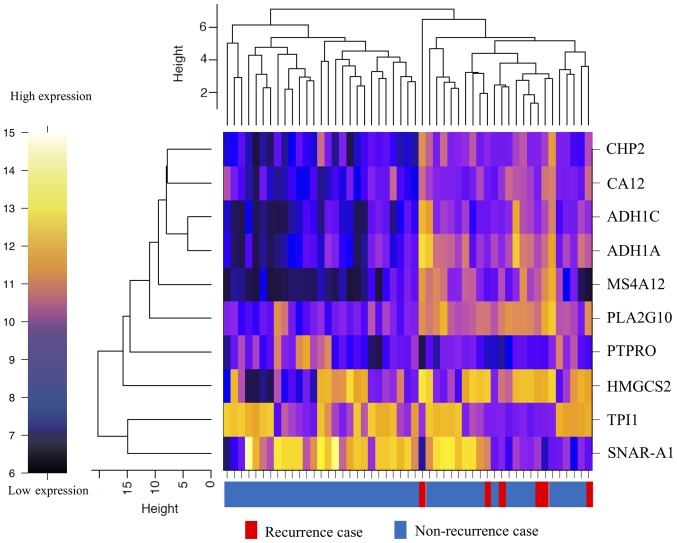
From the microarray (which contained a total of 34,694 genes), initially, the analysis dataset of 1,734 genes was selected as a collection of genes showing the top 5% values of the coefficient of variation among the 51 samples. Then, the differentially expressed genes (10 genes) between the two groups were detected using two criteria to filter the dataset. One of the criteria was a P-value of the difference of less than 0.05, and the other was a difference in the expression levels of the genes in dataset by at least threefold of the standard deviation (±) from the mean (Fig. 1). The heat map showed that determination of the expressions of 10 genes (alcohol dehydrogenase 1A (ADH1A), alcohol dehydrogenase 1C (ADH1C), carbonic anhydrase XII (CA12), calcineurin-like EF-hand protein 2 (CHP2), mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase (HMGCS2), small NF90 (ILF3)-associated RNA A1 (SNAR-A1), triosephosphate isomerase 1 (TPI1), membrane-spanning 4-domain A12 (MS4A12), phospholipase A2 group X (PLA2G10), protein tyrosine phosphatase, receptor type O (PTPRO)) allowed classification of the specimens into the recurrence and non-recurrence groups (Fig. 2). The expression level of each of these genes was significantly different between the recurrence and non-recurrence groups (Table II); while the expression levels of 7 of the genes (ADH1A, ADH1C, CA12, CHP2, HMGCS2, MS4A12, and PLA2G10) were significantly higher in the recurrence group, those of 3 of the remaining genes (SNAR-A1, TPI1, and PTPRO) were significantly lower in the recurrence group as compared to the non-recurrence group (Table II).
Figure 1.
Expression plot of the 1,734 genes that were selected as the genes showing the top 5% values of the coefficient of variation between the groups with and without recurrence. The red circles show the differentially expressed genes (10 genes) in the dataset by over at least threefold of the standard deviation (±) from the mean.
Figure 2.
Heat map of the hierarchical clustering analysis of the differentially expressed genes.
Table II.
The differental expression of 10 genes for prediction of tumor recurrence.
| Gene symbol | Log2 ratio (RG/NRG) | P-value | Gene name | Accession no. |
|---|---|---|---|---|
| ADH1A | 1.412 | 0.026 | Alcohol dehydrogenase 1A | NM_000667 |
| ADH1C | 1.377 | 0.015 | Alcohol dehydrogenase 1C | NM_000669 |
| CA12 | 1.182 | 0.009 | Carbonic anhydrase XII | NM_001218 |
| CHP2 | 1.217 | 0.011 | Calcineurin-like EF-hand protein 2 | NM_022097 |
| HMGCS2 | 1.589 | 0.029 | Mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase 2 | NM_005518, NM_001166107 |
| SNAR-A1 | −1.962 | 0.009 | Small ILF3/NF90-associated RNA A1 (non-coding RNA) | BU536065, NR_004435 |
| TPI1 | −1.008 | 0.043 | Triosephosphate isomerase 1 | XM_001725700, NM_000365 |
| MS4A12 | 1.441 | 0.036 | Membrane spanning 4-domains A12 | NM_017716 |
| PLA2G10 | 1.282 | 0.005 | Phospholipase A2 group X | NM_003561 |
| PTPRO | −1.335 | 0.007 | Protein tyrosine phosphatase, receptor type O | NM_030667, NM_002848 |
Discussion
In the present study, conducting using the database of the FACOS study, we found novel predictive molecular markers for recurrence in stage III CC patients receiving postoperative adjuvant oxaliplatin-based chemotherapy (8). The recurrence group could be definitively identified according to the expression levels of 10 genes, including ADH1A, ADH1C, CA12, CHP2, HMGCS2, SNAR-A1, TPI1, MS4A12, PLA2G10, and PTPRO; high expression levels of 7 genes (ADH1A, ADH1C, CA12, CHP2, HMGCS2, MS4A12, and PLA2G10) and low expression levels of 3 genes (SNAR-A1, TPI1, and PTPRO) were associated with tumor recurrence. In the FACOS study, tumor recurrence developed in 27 (20.5%) of the 132 CC patients. In this study, 6 patients (11.8%) with tumor recurrence included for the mRNA expression analysis in 51 CC patients. Of the 6 patients with relapse, 2 patients were stage IIIB [among the 34 stage IIIB patients (5.9%)] and the remaining 4 patients were stage IIIC [among the 14 stage IIIC patients (28.6%)]. In regard to the depth of invasion, all the 6 patients with tumor recurrence had T4 disease [among the 21 CC patients with T4 disease (28.6%)]. Although tumor recurrence was statistically associated with deep invasion, it may be difficult to predict recurrence strictly based on the clinicopathological characteristics alone, since patients with recurrence constituted only a small part of the clinicopathological categories. Along with deep invasion as one of the predictive clinicopathological characteristics, our 10-gene signature is expected to be useful to predict patients with tumor recurrence in stage III CC patients receiving Ox-based adjuvant chemotherapy.
Previous studied have identified KRAS or BRAF mutation is one of the prognostic biomarkers in stage III CC patients receiving adjuvant FOLFOX therapy (9,11,24). Defective mismatch repair gene (dMMR) is also reported as an independent prognostic factor in stage III CC patients receiving adjuvant FOLFOX therapy (9–11,24); in particular, right-sided colon patients with dMMR had good outcomes (11,24). Consequently, patients with both proficient MMR (pMMR)-tumors and KRAS or BRAF mutations are at risk for poor outcomes. These evidences may be relevant to clinical practice, since they are consistent with the results of randomized clinical trials, including the N0470 (9,11), conducted in stage III CC patients. However, the KRAS and/or BRAF mutation status alone selects approximately a half of stage III CC patients since these mutation types account for 40 and 10% of CC patients, respectively. Moreover, the frequency of CC patients with dMMR is 15% at the most, while that of CC patients with proficient MMR (pMMR) accounts for the remaining 85%. The range of prediction for patients with tumor recurrence is still wide. In this study, 3 of the patients with recurrence were found to have wild-type KRAS, when they were examined as having KRAS after the detection of tumor recurrence. Therefore, the KRAS and/or BRAF status alone is not sufficient to identify patients having the potential for tumor recurrence.
A 12-gene CC recurrence score in the Oncotype DX colon cancer assay has been advocated for the prediction of tumor recurrence in patients with stage II and III CC (12,15,25). The 12 genes include 7 recurrence genes (FAP, INHBA, BGN, Ki-67, C-MYC, MYBL2, GADD45B) and 5 reference genes (ATP5E, GPX1, PGK1, UBB, VDAC2), and the recurrence score is calculated using reference-normalized expression measurements (25). This 12-gene CC recurrence score has been reported to be correlated with the risk of tumor recurrence in a large number of stage II and III CC patients (12,15). However, this score does not bear a direct relationship to the use of Ox-based adjuvant chemotherapy. We analyzed the expression profile of the 12 genes in our samples (data not shown), and the 6 patients with tumor recurrence could not be classified into same cluster. Although we did not calculate the recurrence score according to the formula (25), we consider that the molecular set of the 10 genes that we identified in the present study is more reliable to classify patients with tumor recurrence.
Recently, CC has been classified under 4 CMSs according to the genetic changes, including in MMR, RAS, BRAF, methylation pattern, and the gene related to the WNT signaling pathway (16). Of these 4 subtypes, CMS4 tumor (mesenchymal type), which is characterized by stromal infiltration, activation of transforming growth factor-beta (TGF-β), and angiogenesis, and is associated with a worse relapse-free survival and OS. CMS1 tumor (microsatellite instability immune type), which shows microsatellite instability, hypermethylation, and immune activation, is associated with worse survival after relapse. Kwon et al (13), found molecular subtypes in stage III CRC patients given FOLFOX adjuvant chemotherapy. These subtypes were similar to the CMS classification in terms of the relapse-free survival and OS. Although 10 up-regulated genes were identified in each subtype, the 10 genes that were identified in this study did not include those genes. Classification according to the CMS and other molecular subtypes are promising classifications for prediction of the prognosis in CRC patients. However, we have tried to identify patients with tumor recurrence in stage III CC patients according to expression profiling of a small number of specific genes.
None of the 10 genes identified in this study, including ADH1A, ADH1C, CA12, CHP2, HMGCS2, SNAR-A1, TPI1, MS4A12, PLA2G10 and PTPRO, have been documented in previous reports (17–19) in which microarray-based gene expression profiling has been performed for prediction of tumor recurrence in stage II/III CRC patients not receiving Ox-based adjuvant chemotherapy. Among the 10 genes, overexpression of CA12 and HMGCS2 seem to be the most likely to be associated with chemoresistance and poor prognosis in patients with CRC. CA12 is one of the carbonic anhydrase isoforms which catalyzes reversible hydration of CO2 to bicarbonate for maintenance of pH homeostasis in the human body (26). A previous report (27) demonstrated that CA12 expression was correlated with the expression of P-glycoprotein, resulting in the acquisition of chemoresistance in CRC cancer cells. In regard to the clinical significance of CA12 expression, increased intensity of immunohistochemical staining for CA12 was reported to be significantly associated with poor survival in CRC patients (26). HMGCS2 is the rate-limiting enzyme that catalyzes acetyl-CoA to ketone bodies (28). HMGCS2 expression has been reported to be enhanced in the tumor tissue in rectal cancer patients administered preoperative chemoradiotherapy, and HMGCS2 overexpression to be associated with a poor disease-free survival, local recurrence-free survival, and metastasis-free survival (29). Furthermore, a recent report (28) has indicated that HMGCS2 enhances cellular invasion and metastasis in CRC. Both CA12 and HMGCS2 are considered to be potential targets for cancer treatment. Of the remaining 8 genes, MS4A12 (30) and PTPRO (31) have been documented to be involved in the epidermal growth factor signaling pathway in CRC. TPI1 has been detected as an auto-antibody in the sera of CRC patients (32). No associations between the expressions of other genes and the clinical outcomes in CRC patients have been reported. Although tumor recurrence was statistically associated with deep invasion in this clinical information, the genes associated with deep invasion may be missing. In this study, we found a novel 10 gene-expression signature. Since the biological functions of several of these genes have been established, our 10 gene-expression signature may be a promising biomarker for the prediction of tumor recurrence in stage III CC patients receiving Ox-based adjuvant chemotherapy.
The main limitations of this study were the small study population and the fact that there were only six recurrence events. The cut-off value of the 10 genes could not be determined using a realtime PCR method due to insufficient samples. However, we would still like to emphasize the potential usefulness of determining the 10-gene expression status in Japanese patients with stage III CC receiving Ox-based adjuvant chemotherapy for prediction of the risk of tumor recurrence. We will plan to perform prospective study to confirm the relationship between the expression levels of these 10 genes and tumor recurrence in stage III CC patients.
In summary, determination of the expression levels of a novel set of 10 genes that we found in the present study may be a useful means to predict the risk of recurrence in stage III CC patients receiving an Ox-based adjuvant chemotherapy, although several prediction models of recurrence have reported for CC. Based on the presence of the 10 gene-expression signature, high-risk CC patients should be carefully observed to detect tumor recurrence during the follow-up period.
Acknowledgements
Not applicable.
Funding
The present study was conducted in collaboration with Yakult Honsha Co., Ltd., and was sponsored by Yakult Honsha Co., Ltd. (Tokyo, Japan). (grant no. Kyodo23021). The involvement of our collaborators had no impact or bearing on study design or analysis.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
KKu and YN drafted the manuscript. YN, YM, HE and YO performed microarray experiments and contributed to the bioinformatics analysis. MY, KI, CK, KKo, MK, KT and TM provided the tissue samples and clinical information, and made substantial contributions to conception, analysis and interpretation of data. KKu, YO and HI conceived and designed the study, and edited the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was conducted with the approval of the local Ethics Committee of Saitama Medical Center (approval no. 305-IV). Patients were required to provide written informed consent prior to enrollment.
Patient consent for publication
All the patients have written informed consent for the publication of any associated data.
Competing interest
The authors declare that they have no competing interests.
References
- 1.Kuebler JP, Wieand HS, O'Connell MJ, Smith RE, Colangelo LH, Yothers G, Petrelli NJ, Findlay MP, Seay TE, Atkins JN, et al. Oxaliplatin combined with weekly bolus fluorouracil and leucovorin as surgical adjuvant chemotherapy for stage II and III colon cancer: Results from NSABP C-07. J Clin Oncol. 2007;25:2198–2204. doi: 10.1200/JCO.2006.08.2974. [DOI] [PubMed] [Google Scholar]
- 2.André T, Boni C, Navarro M, Tabernero J, Hickish T, Topham C, Bonetti A, Clingan P, Bridgewater J, Rivera F, de Gramont A. Improved overall survival with oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment in stage II or III colon cancer in the MOSAIC trial. J Clin Oncol. 2009;27:3109–3116. doi: 10.1200/JCO.2008.20.6771. [DOI] [PubMed] [Google Scholar]
- 3.Schmoll HJ, Tabernero J, Maroun J, de Braud F, Price T, Van Cutsem E, Hill M, Hoersch S, Rittweger K, Haller DG. Capecitabine plus oxaliplatin compared with fluorouracil/folinic acid as adjuvant therapy for stage III colon cancer: Final results of the NO16968 randomized controlled phase III trial. J Clin Oncol. 2015;33:3733–3740. doi: 10.1200/JCO.2015.60.9107. [DOI] [PubMed] [Google Scholar]
- 4.NCCN Guidelines Version 2. 2018, corp-author. Colon cancer. National comprehensive cancer network. https://www.nccn.org/professionals/physician_gls/pdf/colon.pdf
- 5.Schmoll HJ, Van Cutsem E, Stein A, Valentini V, Glimelius B, Haustermans K, Nordlinger B, van de Velde CJ, Balmana J, Regula J, et al. ESMO consensus guidelines for management of patients with colon and rectal cancer. A personalized approach to clinical decision making. Ann Oncol. 2012;23:2479–2516. doi: 10.1093/annonc/mds236. [DOI] [PubMed] [Google Scholar]
- 6.Hamaguchi T, Shirao K, Moriya Y, Yoshida S, Kodaira S, Ohashi Y, NSAS-CC Group Final results of randomized trials by the national surgical adjuvant study of colorectal cancer (NSAS-CC) Cancer Chemother Pharmacol. 2011;67:587–596. doi: 10.1007/s00280-010-1358-1. [DOI] [PubMed] [Google Scholar]
- 7.Yoshimatsu K, Ishibashi K, Koda K, Yokomizo H, Oda N, Oshiro M, Kato H, Oya M, Nakajima H, Ooki S, et al. A Japanese multicenter phase II study of adjuvant chemotherapy with mFOLFOX6/CAPOX for stage III colon cancer treatment after D2/D3 lymphadenectomy. Surg Today. 2019;49:498–506. doi: 10.1007/s00595-019-1771-y. [DOI] [PubMed] [Google Scholar]
- 8.Kosugi C, Koda K, Ishibashi K, Yoshimatsu K, Tanaka S, Kato R, Kato H, Oya M, Narushima K, Mori M, et al. Safety of mFOLFOX6/XELOX as adjuvant chemotherapy after curative resection of stage III colon cancer: Phase II clinical study (The FACOS study) Int J Colorectal Dis. 2018;33:809–817. doi: 10.1007/s00384-018-2979-9. [DOI] [PubMed] [Google Scholar]
- 9.Sinicrope FA, Shi Q, Smyrk TC, Thibodeau SN, Dienstmann R, Guinney J, Bot BM, Tejpar S, Delorenzi M, Goldberg RM, et al. Molecular markers identify subtypes of stage III colon cancer associated with patient outcomes. Gastroenterology. 2015;148:88–99. doi: 10.1053/j.gastro.2014.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zaanan A, Fléjou JF, Emile JF, Des GG, Cuilliere-Dartigues P, Malka D, Lecaille C, Validire P, Louvet C, Rougier P, et al. Defective mismatch repair status as a prognostic biomarker of disease-free survival in stage III colon cancer patients treated with adjuvant FOLFOX chemotherapy. Clin Cancer Res. 2011;17:7470–7478. doi: 10.1158/1078-0432.CCR-11-1048. [DOI] [PubMed] [Google Scholar]
- 11.Sinicrope FA, Mahoney MR, Smyrk TC, Thibodeau SN, Warren RS, Bertagnolli MM, Nelson GD, Goldberg RM, Sargent DJ, Alberts SR. Prognostic impact of deficient DNA mismatch repair in patients with stage III colon cancer from a randomized trial of FOLFOX-based adjuvant chemotherapy. J Clin Oncol. 2013;31:3664–3672. doi: 10.1200/JCO.2013.48.9591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yothers G, O'Connell MJ, Lee M, Lopatin M, Clark-Langone KM, Millward C, Paik S, Sharif S, Shak S, Wolmark N. Validation of the 12-gene colon cancer recurrence score in NSABP C-07 as a predictor of recurrence in patients with stage II and III colon cancer treated with fluorouracil and leucovorin (FU/LV) and FU/LV plus oxaliplatin. J Clin Oncol. 2013;31:4512–4519. doi: 10.1200/JCO.2012.47.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwon Y, Park M, Jang M, Yun S, Kim WK, Kim S, Paik S, Lee HJ, Hong S, Kim TI, et al. Prognosis of stage III colorectal carcinomas with FOLFOX adjuvant chemotherapy can be predicted by molecular subtype. Oncotarget. 2017;8:39367–39381. doi: 10.18632/oncotarget.17023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gray RG, Quirke P, Handley K, Lopatin M, Magill L, Baehner FL, Beaumont C, Clark-Langone KM, Yoshizawa CN, Lee M, et al. Validation study of a quantitative multigene reverse transcriptase-polymerase chain reaction assay for assessment of recurrence risk in patients with stage II colon cancer. J Clin Oncol. 2011;29:4611–4619. doi: 10.1200/JCO.2010.32.8732. [DOI] [PubMed] [Google Scholar]
- 15.Yamanaka T, Oki E, Yamazaki K, Yamaguchi K, Muro K, Uetake H, Sato T, Nishina T, Ikeda M, Kato T, et al. 12-Gene recurrence score assay stratifies the recurrence risk in stage II/III colon cancer with surgery alone: The SUNRISE study. J Clin Oncol. 2016;34:2906–2913. doi: 10.1200/JCO.2016.67.0414. [DOI] [PubMed] [Google Scholar]
- 16.Guinney J, Dienstmann R, Wang X, de Reyniès A, Schlicker A, Soneson C, Marisa L, Roepman P, Nyamundanda G, Angelino P, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arango D, Laiho P, Kokko A, Alhopuro P, Sammalkorpi H, Salovaara R, Nicorici D, Hautaniemi S, Alazzouzi H, Mecklin JP, et al. Gene-expression profiling predicts recurrence in Dukes' C colorectal cancer. Gastroenterology. 2005;129:874–884. doi: 10.1053/j.gastro.2005.06.066. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Jatkoe T, Zhang Y, Mutch MG, Talantov D, Jiang J, McLeod HL, Atkins D. Gene expression profiles and molecular markers to predict recurrence of Dukes' B colon cancer. J Clin Oncol. 2004;22:1564–1571. doi: 10.1200/JCO.2004.08.186. [DOI] [PubMed] [Google Scholar]
- 19.Bandrés E, Malumbres R, Cubedo E, Honorato B, Zarate R, Labarga A, Gabisu U, Sola JJ, García-Foncillas J. A gene signature of 8 genes could identify the risk of recurrence and progression in Dukes' B colon cancer patients. Oncol Rep. 2007;17:1089–1094. [PubMed] [Google Scholar]
- 20.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du P, Kibbe WA, Lin SM. lumi: A pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 22.Team RC. R Foundation for statistical computing; Vienna, Austria: 2015. R: A language and environment for statistical computing. [Google Scholar]
- 23.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sinicrope FA, Shi Q, Allegra CJ, Smyrk TC, Thibodeau SN, Goldberg RM, Meyers JP, Pogue-Geile KL, Yothers G, Sargent DJ, Alberts SR. Association of DNA mismatch repair and mutations in BRAF and KRAS with survival after recurrence in stage III colon cancers: A secondary analysis of 2 randomized clinical trials. JAMA Oncol. 2017;3:472–480. doi: 10.1001/jamaoncol.2016.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.You YN, Rustin RB, Sullivan JD. Oncotype DX(®) colon cancer assay for prediction of recurrence risk in patients with stage II and III colon cancer: A review of the evidence. Surg Oncol. 2015;24:61–66. doi: 10.1016/j.suronc.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 26.Viikilä P, Kivelä AJ, Mustonen H, Koskensalo S, Waheed A, Sly WS, Pastorek J, Pastorekova S, Parkkila S, Haglund C. Carbonic anhydrase enzymes II, VII, IX and XII in colorectal carcinomas. World J Gastroenterol. 2016;22:8168–8177. doi: 10.3748/wjg.v22.i36.8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kopecka J, Campia I, Jacobs A, Frei AP, Ghigo D, Wollscheid B, Riganti C. Carbonic anhydrase XII is a new therapeutic target to overcome chemoresistance in cancer cells. Oncotarget. 2015;6:6776–6793. doi: 10.18632/oncotarget.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen SW, Chou CT, Chang CC, Li YJ, Chen ST, Lin IC, Kok SH, Cheng SJ, Lee JJ, Wu TS, et al. HMGCS2 enhances invasion and metastasis via direct interaction with PPARα to activate Src signaling in colorectal cancer and oral cancer. Oncotarget. 2017;8:22460–22476. doi: 10.18632/oncotarget.13006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee YE, He HL, Shiue YL, Lee SW, Lin LC, Wu TF, Chang IW, Lee HH, Li CF. The prognostic impact of lipid biosynthesis-associated markers, HSD17B2 and HMGCS2, in rectal cancer treated with neoadjuvant concurrent chemoradiotherapy. Tumour Biol. 2015;36:7675–7683. doi: 10.1007/s13277-015-3503-2. [DOI] [PubMed] [Google Scholar]
- 30.Koslowski M, Sahin U, Dhaene K, Huber C, Türeci O. MS4A12 is a colon-selective store-operated calcium channel promoting malignant cell processes. Cancer Res. 2008;68:3458–3466. doi: 10.1158/0008-5472.CAN-07-5768. [DOI] [PubMed] [Google Scholar]
- 31.Asbagh LA, Vazquez I, Vecchione L, Budinska E, De Vriendt V, Baietti MF, Steklov M, Jacobs B, Hoe N, Singh S. The tyrosine phosphatase PTPRO sensitizes colon cancer cells to anti-EGFR therapy through activation of SRC-mediated EGFR signaling. Oncotarget. 2014;5:10070–10083. doi: 10.18632/oncotarget.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kobayashi S, Hiwasa T, Arasawa T, Kagaya A, Ishii S, Shimada H, Ito M, Suzuki M, Kano M, Rahmutulla B, et al. Identification of specific and common diagnostic antibody markers for gastrointestinal cancers by SEREX screening using testis cDNA phage library. Oncotarget. 2018;9:18559–18569. doi: 10.18632/oncotarget.24963. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.