Abstract
Esophageal squamous cell carcinoma (ESCC) is one of the six most commonly diagnosed tumor types in the Chinese population. Gene expression profiles help to predict the prognosis of patients with ESCC. Disease recurrence as the survival endpoint has been analyzed in the majority of previous studies; therefore, the aim of the present study was to construct a robust gene signature in order to determine the overall survival (OS) of patients with ESCC. The gene expression and clinical data of patients with ESCC were downloaded from The Cancer Genome Atlas (TCGA) database. Of the selected data (172 samples from surviving patients), 72 samples were randomly selected as modeling data, and verification was conducted using the entire dataset. Data from the Gene Expression Omnibus was analyzed simultaneously, and a venn diagram was constructed to determine the intersection between these two sets of results; a total of 97 genes were found to be associated with OS. Kyoto Encyclopedia of Genes and Genomes analysis demonstrated that these genes were primarily associated with specific pathways (Homo sapiens), including DNA replication, protein processing in endoplasmic reticulum and influenza A. A five-gene signature was identified with a robust likelihood-based survival modeling approach. Using regression coefficient modeling, a prognostic model consisting of the C-X-C motif chemokine ligand 8, DNA damage inducible transcript 3, RAB27A, member RAS oncogene family, replication factor C subunit 2 and elongation factor for RNA polymerase II 2 genes was constructed and validated. Based on these results, patients were subdivided into high and low-risk groups. Compared with the high-risk group, the OS time of patients in the low-risk group was significantly increased. Furthermore, it was determined that the five genes were all differentially expressed in ESCC tissues compared with normal tissues, indicating the potential role of these genes in ESCC initiation and progression. In another independent cohort, this five-gene signature was further confirmed and was considered as an independent prognostic biomarker for OS prediction in patients with ESCC. In conclusion, the OS of patients with ESCC may be predicted using this five-gene signature, which may be useful in identifying patients with high-risk ESCC.
Keywords: esophageal squamous cell carcinoma, gene signature, overall survival
Introduction
Esophageal squamous cell carcinoma (ESCC) is the eighth most common type of cancer globally. A total of 456,000 new cases were diagnosed during 2014, in addition to ~400,000 deaths worldwide (1); however, the incidence of ESCC in different countries varies considerably, with approximately half of the newly diagnosed cases occurring in China (1). Early-stage ESCC frequently exhibits a number of non-specific symptoms, which the majority of patients do not act on; therefore due to late-stage diagnosis, patient outcome is often poor, with a 5-year survival rate of 13–18%. Although the diagnosis and treatment of ESCC have improved, the overall mortality and annual incidence rates continue to increase (1,2).
ESCC is a heterogeneous disease that involves multiple complex pathways. The Wnt signaling (3), epidermal growth factor receptor (4), T cell factor (5) and tissue factor (6) pathways are commonly dysregulated during the initiation, progression and metastasis of ESCC; therefore, in order to devise individualized diagnosis and treatment approaches, the abnormalities in these molecular pathways require further investigation. Extensive research has focused on the identification of molecular markers for ESCC; however, only limited success has been achieved when researching gene mutations or single proteins (7). Gene expression profiles are an effective means to determine tumor type and to evaluate patient prognosis (8). Using the systemic immune-inflammation index (9), the expression of cancer/testis antigens (10) and determining progression markers (11) in ESCC has been demonstrated to effectively improve diagnosis and treatment. However, due to the heterogeneity of ESCC it is difficult to apply all of these prognostic signatures at once.
In the majority of studies, relapse-free or disease-free survival have been used to effectively identify molecular markers or to determine the prognosis of patients; however, overall survival (OS) is considered to be the most reliable indicator of treatment success. Therefore, the aim of the present study was to determine whether a robust gene signature was an effective means for predicting the OS of patients with ESCC. Using univariate survival analysis, 1,319 genes associated with OS in patients with ESCC were selected from The Cancer Genome Atlas (TCGA), and a five-gene signature was developed using a robust likelihood-based survival modeling approach. Furthermore, the five-gene signature was used to generate a prognostic model using BRB-Array Tools; the prognostic value of this five-gene signature was validated in another independent cohort. The results demonstrated that the five-gene signature was able to identify patients at high-risk of developing ESCC.
Materials and methods
Data sources
Data from the frozen tumor specimens (n=161) and adjacent non-tumor tissues (n=11) of 172 patients with ESCC were retrieved from TCGA (https://cancergenome.nih.gov/; date of access, September 15, 2017). A total of 72 samples were randomly selected as modeling data (70 tumor and 2 normal samples), and the data were validated using the entire TCGA dataset. The GSE20347 dataset (including 17 tumor and 17 matched samples) was downloaded from the Gene Expression Omnibus (GEO) database on September 15, 2017 (12).
Gene expression microarray and data analysis
Statistical analysis of the microarray data was performed using R software (13). The data were normalized and transformed in order to calculate expression values using the MA function in the ‘affy’ library. The signal values of all of the genes were subsequently transformed to log base 2, and quantile normalization was conducted to determine the tantamount distributions for the probe signal intensities.
Identification of gene signatures
A robust likelihood-based survival modeling approach (14) was used to select the gene signatures. Analysis was performed using the ‘rbsurv’ package in R. The calculation algorithm was as follows: i) The samples were divided into the training set with n*(1-p) random samples, and the validation set with n*p samples, where p=1/3. A gene was fitted to the training set and the parameter estimate of this gene was calculated. Log likelihood was evaluated with the parameter estimate and the validation set, and the evaluation of each gene was repeated; ii) the aforementioned procedure was repeated 10 times in order to calculate 10 log likelihoods for each gene. The best gene, g (1), was selected based on the largest mean log likelihood; iii) the next best gene was identified by evaluating every two-gene model. Subsequently, an ideal gene-model was selected based on the largest mean log likelihood; and iv) the forward gene selection procedure was continued, resulting in a series of models. The Akaike information criterion (AIC) was then calculated for all candidate models, and the optimal model (with the smallest AIC) was selected.
Unsupervised hierarchical clustering and Kaplan-Meier analysis
Unsupervised hierarchical clustering analysis (15) was performed in R using the ‘hclust’ function with Euclidean distance. Kaplan-Meier curves were plotted for two distinct groups of patients using the ‘survfit’ function, and the P-values (log-rank test) were calculated using the ‘survdiff’ function (16).
Development and validation of the five-gene survival risk score system
In order to calculate the regression coefficient for each gene (based on the 72 training samples from TCGA), the five genes were evaluated using BRB-Array Tools v4.6.0 Beta 2 (http://linus.nci.nih.gov/BRB-ArrayTools.html) with the survival risk group prediction tool (17); the survival risk score is the sum of the product of the expression level of a gene and the corresponding regression coefficient. Patients were then divided into high- and low-risk groups based on the 50th percentile, and the leave-one-out cross-validation method was used to ascertain robustness. Using the RMA function in R, the gene expression data with OS information (n=100; TCGA) were used as the validation dataset.
The survival risk scores of an independent patient cohort downloaded from the GEO (GSE20347; including 17 tumor and 17 normal samples) were calculated using the coefficient derived from the training dataset, and receiver operating characteristic (ROC) curve analyses were conducted for survival prediction. Subsequently, the sensitivity and specificity of the gene signature were evaluated to estimate the discriminatory power of the prognostic gene expression signatures. The area under the curve (AUC) was calculated and a bootstrap method was used to calculate the associated 95% confidence internal. The optimal cut-off value (obtained based on the ROC curves) was used to divide the patients into high- and low-risk groups, and Kaplan-Meier curves were plotted for both groups of patients using the ‘survfit’ function in R.
Enrichment analysis of kyoto encyclopedia of genes and genomes (KEGG) pathways
Genes associated with the patient OS were evaluated using the Cox proportional hazard regression model. The genes were subjected to KEGG pathway enrichment analysis using the Gene Set Analysis Toolkit V2 (18,19), were a hypergeometric statistical test and BH multiple adjustment test were employed. All human genes were used as references, and the top eight pathways were considered to be significantly enriched.
Statistical analysis
In order to ensure the integrity and comparability of the analyzed datasets, the GEO gene chip data were normalized and background-corrected using the gcrma package to eliminate systematic errors between the chips (20). The R package limma (21) was used to identify the control group and differentially expressed genes between the treated samples, and those with log2 (fold change) >1 and P<0.05 were screened.
The verification data consisted of the entire 172 samples from TCGA. The adjacent normal (n=11) and ESCC tissues (n=161) are unpaired groups, and Mann-Whitney U tests were performed to compare gene expression between these groups. All statistical analyses were performed using GraphPad Prism 5.0 (GraphPad Software, Inc.,) and P<0.05 (two-tailed) was considered to indicate a statistically significant difference.
Results
The Cox proportional hazard regression model and forward selection method were used to identify a five-gene signature to predict the OS of patients with ESCC. The datasets were collected from TCGA and the GEO, and the performance of the five-gene signature was subsequently evaluated.
Identification of genes associated with the OS of patients with ESCC
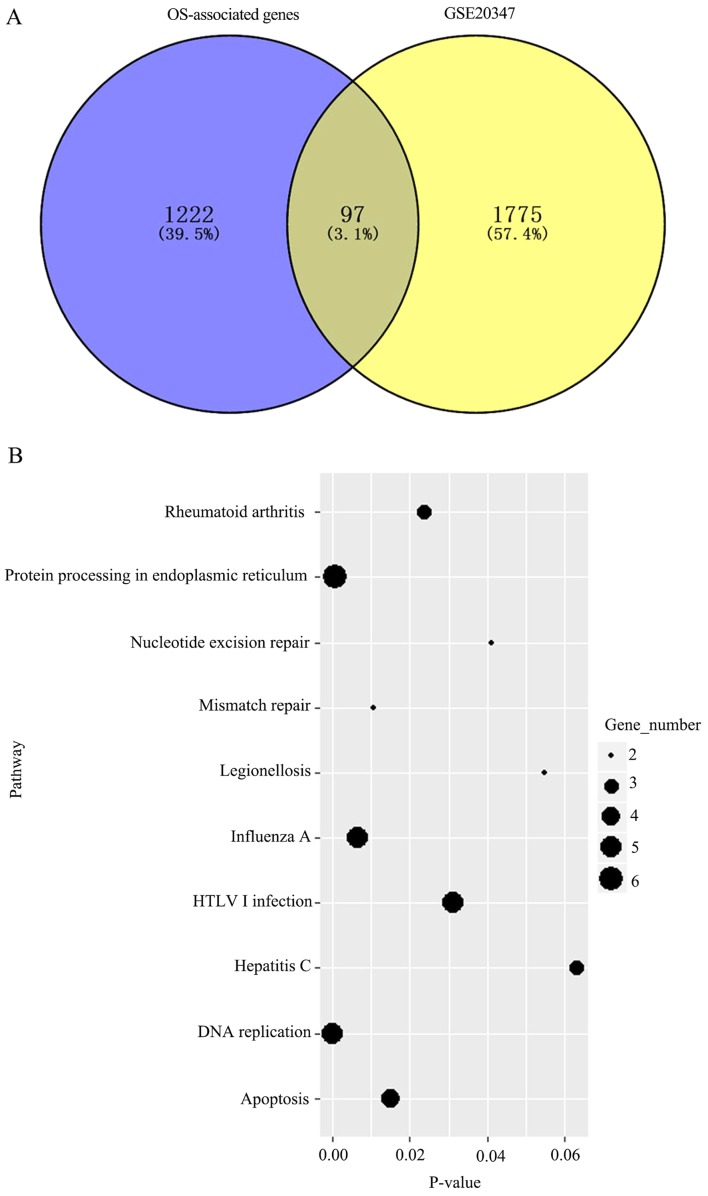
Univariate survival analysis was initially performed with the Cox proportional hazard regression model. The analysis was based on 72 samples with observed survival time and censoring status data, and the threshold was set to P<0.05. A total of 1,222 genes were determined to be associated with OS. Additionally, the GSE20347 dataset (GEO) was analyzed and 1,775 differentially expressed genes between tumor and normal tissues were determined; a total of 97 intersecting genes were identified between these two sets of results (Fig. 1A). KEGG pathway enrichment analysis was subsequently performed for these 97 genes to determine key pathways associated with OS. The genes were enriched in eight metabolic pathways (Fig. 1B), including DNA replication, protein processing in endoplasmic reticulum and influenza A, which were the most significant.
Figure 1.
(A) Venn diagram of survival-associated gene analysis in TCGA and differentially expressed genes in the GEO database. OS- and survival-associated genes based on 72 samples from TCGA. Differentially expressed genes in the GSE20347 database from the GEO database. (B) Enrichment of KEGG pathway analysis for 97 OS-associated genes. The top 8 pathways are shown. TCGA, The Cancer Genome Atlas; GEO, Gene expression Omnibus; OS, overall survival; KEGG, Kyoto Encyclopedia of Genes and Genomes; HTLV1, Human T lymphotropic virus type 1.
Screening of the five-gene signature
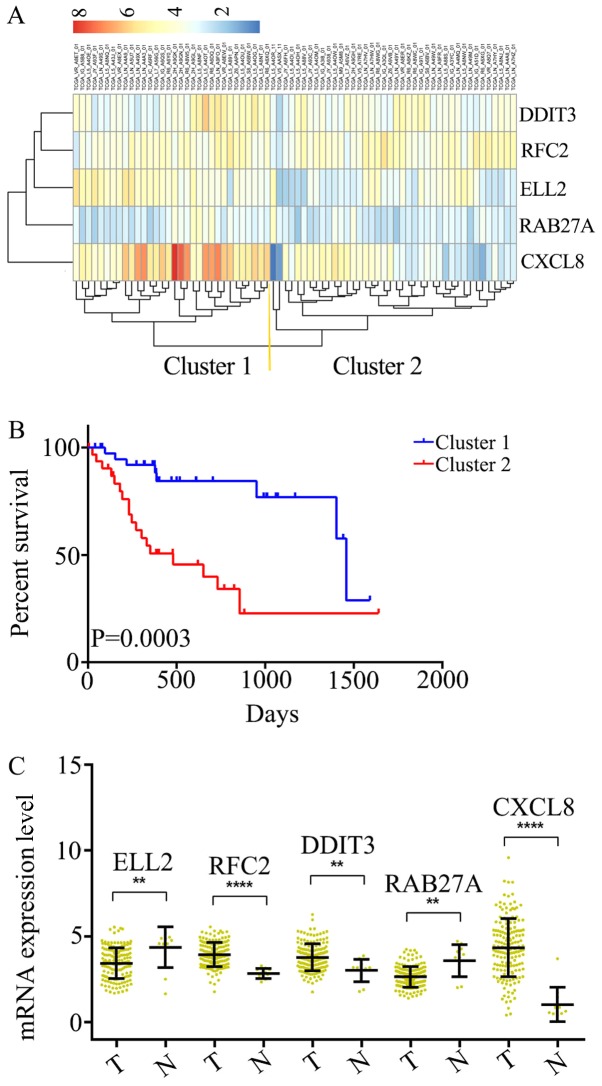
Subsequently, the optimal survival-associated signature genes were screened based on the Cox proportional hazard regression model. In view of the large degree of variability in the data, the cross-validation technique was used to divide the samples into training and validation sets. Forward selection was employed in order to generate a series of gene models, and the minimal AIC was used to select the most suitable model (Table I). Ultimately, five genes were selected for the signature: C-X-C motif chemokine ligand 8 (CXCL8), DNA damage inducible transcript 3 (DDIT3), RAB27A, replication factor C subunit 2 (RFC2) and elongation factor for RNA polymerase II 2 (ELL2). This five-gene signature was able to optimally predict the OS of patients with ESCC. Subsequently, unsupervised hierarchical clustering analysis was performed and the patient population was classified into Cluster 1 and 2 (Fig. 2A). The patients in Cluster 1 were compared with those in Cluster 2, and the mortality risk was revealed to be significantly reduced in patients in Cluster 1 (P=0.0003; Fig. 2B). Thus, the OS of patients with ESCC was able to be predicted using the five-gene signature.
Table I.
Survival-associated gene signature screening using forward selection.
| Gene | nLogLik | AIC |
|---|---|---|
| 0 | 93.1 | 186.21 |
| CXCL8a | 89.9 | 181.79 |
| DDIT3a | 86.31 | 176.62 |
| RAB27Aa | 84.72 | 175.43 |
| RFC2a | 81.95 | 171.9 |
| ELL2a | 76.79 | 163.57 |
| SAC3D1 | 76.34 | 164.67 |
| LMNB2 | 75.11 | 164.21 |
| PTDSS1 | 75.1 | 166.19 |
| ADORA2B | 74.16 | 166.32 |
| CDC45 | 73.71 | 167.43 |
| RPA1 | 73.09 | 168.18 |
| CAPN1 | 72.97 | 169.93 |
| ASF1B | 72.43 | 170.87 |
| CAPNS1 | 70.53 | 169.07 |
| SREBF1 | 70.35 | 170.7 |
Genes selected for the five-gene signature. CXCL8, C-X-C motif chemokine ligand 8; DDIT3, DNA damage inducible transcript 3; RFC2, replication factor C subunit 2; ELL2, elongation factor for RNA polymerase II 2; SAC3D1, SAC3 domain containing 1; LMNB2, lamin B2; PTDSS1, phosphatidylserine synthase 1; ADORA2B, adenosine A2b receptor; CDC45, cell division cycle 45; RPA1, replication protein A1; CAPN1, calpain 1; ASF1B, anti-silencing function 1B histone chaperone; SREBF1, sterol regulatory element binding transcription factor 1; AIC, Akaike information criterion.
Figure 2.
Identification of an optimal gene signature for overall survival prediction. (A) Unsupervised hierarchical clustering analysis was performed with the five-gene signature, which divided patients into two clusters, namely Cluster 1 and 2. (B) Kaplan–Meier curves for patients in different clusters. (C) The mRNA expression of CXCL8, DDIT3, RAB27A, RFC2 andELL2 in 161 ESCC tissues and 11 adjacent normal tissues in TCGA cohort. Data is represented by mean ± SD and P-values were obtained by Mann–Whitney U test. (**P<0.01, ****P<0.0001). T, tumor tissue; N, normal tissue; CXCL8, C-X-C motif chemokine ligand 8; DDIT3, DNA damage inducible transcript 3; RFC2, replication factor C subunit 2; ELL2, elongation factor for RNA polymerase II 2.
Differential expression of the five-gene signature in ESCC and adjacent normal tissues
The mRNA expression levels in ESCC (n=161) and adjacent normal tissues (n=11) were analyzed among all of the 172 samples from TCGA. It was determined that RFC2, DDIT3 and CXCL8 were significantly overexpressed at the mRNA level in ESCC tissues, compared with the adjacent normal tissues (Fig. 2C). Additionally, ELL2 and RAB27A were found to be downregulated in ESCC tissues (Fig. 2C). Thus, these five genes were considered to serve key roles as either tumor suppressors or oncogenes in ESCC.
Construction of a survival risk score system based on the five-gene signature
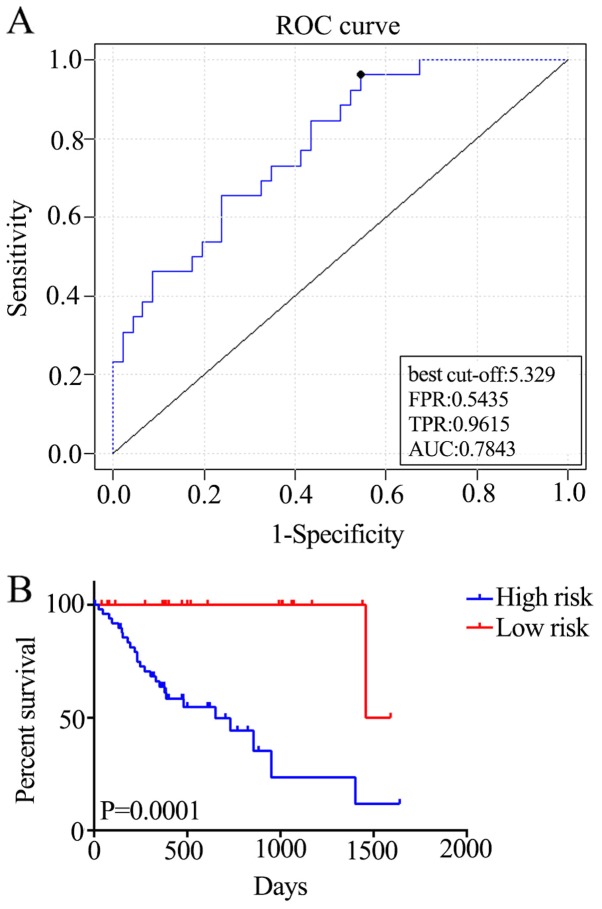
In order to generate the regression coefficient of each gene, the five genes were assessed using BRB-Array Tools. Subsequently, a survival risk scoring system was constructed based on 72 training samples. The survival risk score was calculated as follows: Risk score=(844*ELL2)-(0.614*RFC2) + (0.88*DDIT3) + (0.625*RAB27A) + (0.135*CXCL8); a higher survival risk score equates to higher mortality risk in patients with ESCC. The total AUC of the respective cross-validated time-dependent ROC curves was 0.7843 (Fig. 3A), which confirmed the prediction accuracy of this model. Subsequently, the 72 patients with ESCC were categorized into high-(n=50) and low-risk (n=22) groups, and the OS of the high-risk group was significantly reduced compared with that of the low-risk group [hazard ratio (HR)=16.37; P=0.0001; Fig. 3B].
Figure 3.
Construction of the survival risk score system based on the five-gene signature. (A) The cross-validated time-dependent ROC curve was generated for survival predictions with an AUC of 0.7843. (B) Patients were divided into high- and low-risk groups by cross-validated Kaplan–Meier curve. ROC, receiver-operating curve; AUC, area under curve; FPR, false positive rate; TPR, true positive rate.
External validation of the five-gene signature
In order to assess the robustness and effectiveness of the five-gene signature, the remaining samples (n=100) were used as the validation dataset. ROC curve analyses demonstrated that the five-gene signature was able to accurately predict the OS of patients with ESCC (AUC=0.6225; Fig. 4A). Furthermore, the patients were divided into two risk groups, based on the best cut-off risk scores, 126 (73.3%) patients in the low-risk group and 46 (26.7%) in the high-risk group of the validation dataset. Kaplan-Meier analysis demonstrated significant differences in OS between the two groups (P=0.0291; Fig. 4B). Additionally, the five-gene signature predicted the OS of patients with ESCC using univariate analysis (Table II). Therefore, the five-gene signature is an effective method for predicting the OS of patients with ESCC as an independent prognostic factor.
Figure 4.
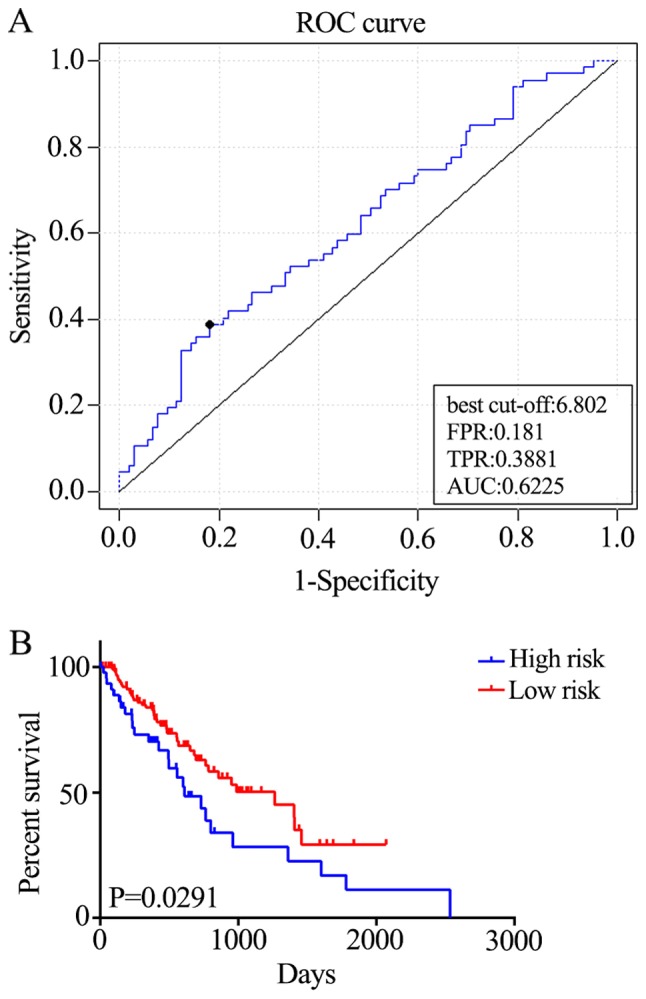
Performance of the five-gene signature in predicting the overall survival of patients with esophageal squamous cell carcinoma. (A) The cross-validated time-dependent ROC curve was generated for survival predictions with an AUC of 0.6225. (B) Patients were divided into high- and low-risk groups by cross-validated Kaplan–Meier curve. ROC, receiver-operating curve; AUC, area under curve; FPR, false positive rate; TPR, true positive rate.
Table II.
Univariate Cox regression analysis of potential prognostic factors for patients with esophageal squamous cell carcinoma.
| Overall survival | |||
|---|---|---|---|
| Characteristics | Patients, n | HR (95% CI) | P-value |
| Sex | 0.0411 | ||
| Female | 26 | 1 | |
| Male | 146 | 2.486 (1.032–3.728) | |
| Age (years) | 0.7015 | ||
| ≥60 | 88 | 1 | |
| <60 | 84 | 1.097 (0.6811–1.775) | |
| Stage (TNM) | <0.0001 | ||
| I–II | 92 | 1 | |
| III–IV | 59 | 2.889 (2.048–6.533) | |
| Risk | 0.0239 | ||
| Low | 126 | 1 | |
| High | 46 | 1.69 (1.083–3.173) | |
HR, hazard ratio; CI, confidence interval; TNM, Tumor-Node-Metastasis.
The association between the risk score and the survival status of patients at different tumor stages was also investigated. Stage I and II tumors were associated with a significant increase in 5-year mortality rate compared with stage III and IV tumors (HR=2.889; P<0.0001; Fig. 5A). Generally, there is a critical time period for the treatment of stage III and IV ESCC; therefore, it is crucial to evaluate the prognosis of the patients within this period. Patients in the high-risk group exhibited reduced survival compared with those in the low-risk group (HR=2.196; P=0.0287; Fig. 5B). Therefore, the five-gene signature may be able to predict the prognosis of patients with stage III and IV ESCC.
Figure 5.
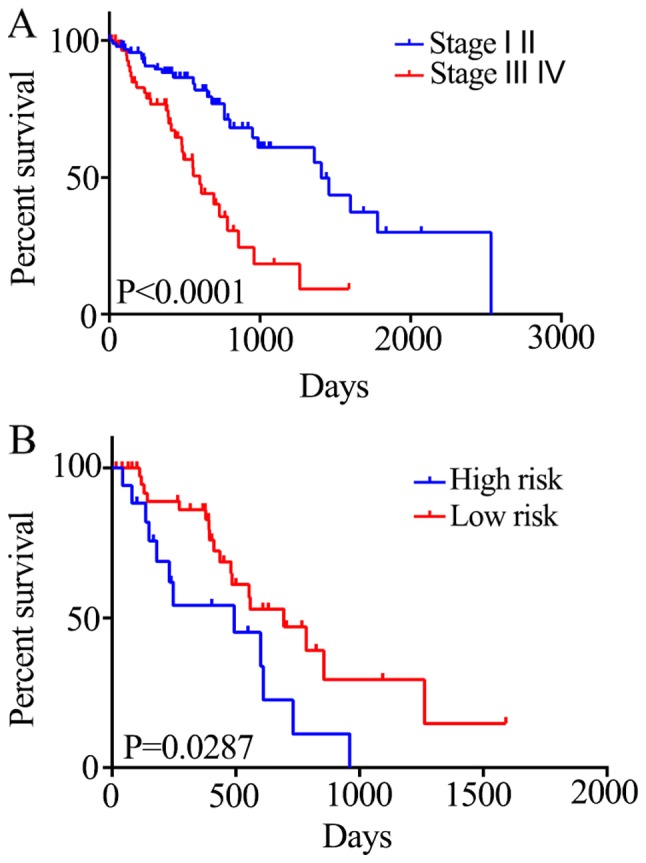
Association between the five-gene signature and overall survival in patients with stage II and III esophageal squamous cell carcinoma. (A) For 172 ESCC patients from TCGA, stage I and II tumors are associated with a significantly increased 5-year mortality rate compared with stage III and IV tumors (HR=2.889; P<0.0001). (B) Stage III and IV tumors were divided into high and low risk group based on their survival risk scores. Patients in the high-risk group exhibited reduced survival compared with patients in the low-risk group (HR=2.196;P=0.0287).
Discussion
In the present study, a robust five-gene signature to predict the prognosis of patients with ESCC was constructed and validated. However, additional confounding factors should also be considered, including the number of negative lymph nodes (22), metabolic tumor volume, total lesion glycolysis, regional lymph node metastasis and concurrent chemotherapy (23). Furthermore, the included data were limited to two databases, thus the impact of these confounding factors could not be fully elucidated. Nonetheless, the five-gene signature highlighted the possibility of predicting the OS of patients with ESCC. Additionally, high-risk patients who may require targeted treatment interventions may be identified with the five-gene signature.
Frozen tumor specimens and adjacent non-tumor tissue of patients with ESCC were retrieved from TCGA, and a total of 72 samples were randomly selected as modeling data (70 tumor and 2 normal samples); 1,222 OS-associated genes were identified. Furthermore, data from the GEO identified 1,775 genes associated with OS. A venn diagram was constructed to intersect the two sets of results, identifying a total of 97 genes associated with OS. A number of genes were found to have prognostic value for patients with ESCC, including FOS (24), Adenosine Deaminase RNA Specific (25,26), Karyopherin Subunit α 2 (27) and Translocator protein (28). These genes were enriched in a number of signaling pathways, including DNA replication, protein processing in endoplasmic reticulum and influenza, as demonstrated using KEGG analysis.
Activation of the Wnt signaling pathway frequently (and critically) occurs during the development of ESCC, therefore presents a therapeutic target for the disease (29). Additionally, the ubiquitin-proteasome system is vital for apoptosis regulation and cell growth, and may also represent a potential molecular target for the treatment and prevention of cancer (30). In the present study, the highlighted genes were condensed and the most suitable five-gene signature (CXCL8, DDIT3, RAB27A, RFC2 and ELL2) was selected for prognosis prediction. According to the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/gene/), none of these genes were associated with cancer research. However, when ESCC tissues were compared with adjacent normal tissues, these five genes were differentially expressed, demonstrating their potential to promote or suppress the development of cancer. The prognostic signature was also validated using 172 samples from TCGA, though elucidation of the true function of these genes requires further investigation.
In the present study, the OS of patients with ESCC was predicted using the five-gene signature. In future studies, comparison of high-risk patients with the entirety if the selected cohort may indicate common responses to different therapy options. Currently, treatment and prognosis of patients with ESCC are primarily determined using the American Joint Committee on Cancer Tumor-Node-Metastasis (TNM) staging system; however, the TNM system is limited in clinical practice, and may not result in an accurate prognosis. Currently, increased attention is focused on patients with stage III and IV ESCC, as patients who received adjuvant chemotherapy were indicated to have had improved outcomes (31–33). Additionally, a number of clinical laboratory tests (including the detection of tumor markers in the serum) have been developed to predict the prognosis of patients with ESCC. These tests have been evaluated in numerous validation cohorts, and are able to assist in the application of personalized chemotherapy programs (34,35). In the present study, the OS of patients with stage III and IV ESCC was predicted using the five-gene signature, tough a larger sample population may have been more beneficial to validate the prognostic value of the five-gene signature in patients with stage III and IV ESCC; further studies will be conducted to demonstrate whether the five-gene signature can help to determine the benefit of adjuvant chemotherapy for patients with stage III and IV ESCC.
In conclusion, the five-gene signature may represent a novel biomarker for determining the prognosis of patients with ESCC, and may also indicate potential therapeutic targets for treating the disease.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Sichuan Science and Technology Program (grant no. 2019JDRC0076).
Availability of data and materials
All the data generated and analyzed in the present study are available from the corresponding author on reasonable request.
Authors' contributions
WH contributed to the study design, data acquisition and analysis, and drafted the manuscript. QY was involved in analyzing the data and drafting the manuscript. LF was involved in GEO data and TCGA data acquisition. YH contributed to the study design and revised it critically for important intellectual content. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.McGuire S. World Cancer Report 2014. Geneva, Switzerland: World Health Organization, International Agency for Research on Cancer, WHO Press, 2015. Adv Nutr. 2016;7:418–419. doi: 10.3945/an.116.012211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349:2241–2252. doi: 10.1056/NEJMra035010. [DOI] [PubMed] [Google Scholar]
- 3.Luo J, Wang W, Tang Y, Zhou D, Gao Y, Zhang Q, Zhou X, Zhu H, Xing L, Yu J. mRNA and methylation profiling of radioresistant esophageal cancer cells: The involvement of Sall2 in acquired aggressive phenotypes. J Cancer. 2017;8:646–656. doi: 10.7150/jca.15652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang W, Pang Q, Yan C, Wang Q, Yang J, Yu S, Liu X, Yuan Z, Wang P, Xiao Z. Induction of PD-L1 expression by epidermal growth factor receptor-mediated signaling in esophageal squamous cell carcinoma. Onco Targets Ther. 2017;10:763–771. doi: 10.2147/OTT.S118982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang W, Yan S, Liu M, Zhang G, Yang S, He S, Bai J, Quan L, Zhu H, Dong Y, Xu N. Beta-catenin/TCF pathway plays a vital role in selenium induced-growth inhibition and apoptosis in esophageal squamous cell carcinoma (ESCC) cells. Cancer Lett. 2010;296:113–122. doi: 10.1016/j.canlet.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 6.Xiang J, Zang W, Che J, Chen K, Hang J. Regulation network analysis in the esophageal squamous cell carcinoma. Eur Rev Med Pharmacol Sci. 2012;16:2051–2056. [PubMed] [Google Scholar]
- 7.Ashktorab H, Kupfer SS, Brim H, Carethers JM. Racial disparity in gastrointestinal cancer risk. Gastroenterology. 2017;153:910–923. doi: 10.1053/j.gastro.2017.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quackenbush J. Microarray analysis and tumor classification. N Engl J Med. 2006;354:2463–2472. doi: 10.1056/NEJMra042342. [DOI] [PubMed] [Google Scholar]
- 9.Wang L, Wang C, Wang J, Huang X, Cheng Y. A novel systemic immune-inflammation index predicts survival and quality of life of patients after curative resection for esophageal squamous cell carcinoma. J Cancer Res Clin Oncol. 2017;143:2077–2086. doi: 10.1007/s00432-017-2451-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salmaninejad A, Zamani MR, Pourvahedi M, Golchehre Z, Hosseini Bereshneh A, Rezaei N. Cancer/testis antigens: Expression, regulation, tumor invasion, and use in immunotherapy of cancers. Immunol Invest. 2016;45:619–640. doi: 10.1080/08820139.2016.1197241. [DOI] [PubMed] [Google Scholar]
- 11.Lin ZW, Gu J, Liu RH, Liu XM, Xu FK, Zhao GY, Lu CL, Ge D. Genome-wide screening and co-expression network analysis identify recurrence-specific biomarkers of esophageal squamous cell carcinoma. Tumour Biol. 2014;35:10959–10968. doi: 10.1007/s13277-014-2388-9. [DOI] [PubMed] [Google Scholar]
- 12.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Edgar R. NCBI GEO: Mining tens of millions of expression profiles-database and tools update. Nucleic Acids Res 35 (Database Issue) 2007:D760–D765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.James G, Witten D, Hastie T, Tibshirani R. Springer-Verlag New York; 2013. An introduction to statistical learning with applications in R. [Google Scholar]
- 14.Cho HJ, Ami Y, Kim S, Kang J, Seung-Mo H, Kang Robust likelihood-based survival modeling with microarray data. J Statist Software. 2008;29:1–16. [Google Scholar]
- 15.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Therneau T. A package for survival analysis in S. R package version 2.37–7. 2014 [Google Scholar]
- 17.Simon R, Lam A, Li MC, Ngan M, Menenzes S, Zhao Y. Analysis of gene expression data using BRB-ArrayTools. Cancer Inform. 2007;3:11–17. doi: 10.1177/117693510700300022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J, Vasaikar S, Shi Z, Greer M, Zhang B. WebGestalt 2017: A more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res 45 (W1) 2017:W130–W137. doi: 10.1093/nar/gkx356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu C, Irizarry R, MacDonald J, Gentry J. gcrma: Background adjustment using sequence information. 2005 [Google Scholar]
- 21.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu H, Liu C, Xu M, Guo M, Xu S, Xie M. Prognostic value of the number of negative lymph nodes in esophageal carcinoma without lymphatic metastasis. Thorac Cancer. 2018;9:1129–1135. doi: 10.1111/1759-7714.12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yildirim BA, Torun N, Guler OC, Onal C. Prognostic value of metabolic tumor volume and total lesion glycolysis in esophageal carcinoma patients treated with definitive chemoradiotherapy. Nucl Med Commun. 2018;39:553–563. doi: 10.1097/MNM.0000000000000837. [DOI] [PubMed] [Google Scholar]
- 24.Li JC, Zhao YH, Wang XY, Yang Y, Pan DL, Qiu ZD, Su Y, Pan JJ. Clinical significance of the expression of EGFR signaling pathway-related proteins in esophageal squamous cell carcinoma. Tumour Biol. 2014;35:651–657. doi: 10.1007/s13277-013-1089-0. [DOI] [PubMed] [Google Scholar]
- 25.Qin YR, Qiao JJ, Chan TH, Zhu YH, Li FF, Liu H, Fei J, Li Y, Guan XY, Chen L. Adenosine-to-inosine RNA editing mediated by ADARs in esophageal squamous cell carcinoma. Cancer Res. 2014;74:840–851. doi: 10.1158/0008-5472.CAN-13-2545. [DOI] [PubMed] [Google Scholar]
- 26.Zhang J, Chen Z, Tang Z, Huang J, Hu X, He J. RNA editing is induced by type I interferon in esophageal squamous cell carcinoma. Tumour Biol. 2017;39:1010428317708546. doi: 10.1177/1010428317708546. [DOI] [PubMed] [Google Scholar]
- 27.Sakai M, Sohda M, Miyazaki T, Suzuki S, Sano A, Tanaka N, Inose T, Nakajima M, Kato H, Kuwano H. Significance of karyopherin-{alpha} 2 (KPNA2) expression in esophageal squamous cell carcinoma. Anticancer Res. 2010;30:851–856. [PubMed] [Google Scholar]
- 28.Yuan Y, Xue L, Fan F. Screening of differentially expressed genes related to esophageal squamous cell carcinoma and functional analysis with DNA microarrays. Int J Oncol. 2014;44:1163–1170. doi: 10.3892/ijo.2014.2262. [DOI] [PubMed] [Google Scholar]
- 29.Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer. 2013;13:11–26. doi: 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
- 30.McBride WH, Iwamoto KS, Syljuasen R, Pervan M, Pajonk F. The role of the ubiquitin/proteasome system in cellular responses to radiation. Oncogene. 2003;22:5755–5773. doi: 10.1038/sj.onc.1206676. [DOI] [PubMed] [Google Scholar]
- 31.Wang BY, Lin PY, Wu SC, Chen HS, Hsu PK, Shih CS, Liu CY, Liu CC, Chen YL. Comparison of pathologic stage in patients receiving esophagectomy with and without preoperative chemoradiation therapy for esophageal SCC. J Natl Compr Canc Netw. 2014;12:1697–1705. doi: 10.6004/jnccn.2014.0171. [DOI] [PubMed] [Google Scholar]
- 32.Li L, Zhao L, Lin B, Su H, Su M, Xie D, Jin X, Xie C. Adjuvant Therapeutic modalities following three-field lymph node dissection for stage II/III esophageal squamous cell carcinoma. J Cancer. 2017;8:2051–2059. doi: 10.7150/jca.18981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang L, Li W, Lyu X, Song Y, Mao Y, Wang S, Huang J. Adjuvant chemotherapy with paclitaxel and cisplatin in lymph node-positive thoracic esophageal squamous cell carcinoma. Chin J Cancer Res. 2017;29:149–155. doi: 10.21147/j.issn.1000-9604.2017.02.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tong Q, Wang XL, Li SB, Yang GL, Jin S, Gao ZY, Liu XB. Combined detection of IL-6 and IL-8 is beneficial to the diagnosis of early stage esophageal squamous cell cancer: A preliminary study based on the screening of serum markers using protein chips. Onco Targets Ther. 2018;11:5777–5787. doi: 10.2147/OTT.S171242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kunizaki M, Hamasaki K, Wakata K, Tobinaga S, Sumida Y, Hidaka S, Yasutake T, Miyazaki T, Matsumoto K, Yamasaki T, et al. Clinical value of serum p53 antibody in the diagnosis and prognosis of esophageal squamous cell carcinoma. Anticancer Res. 2018;38:1807–1813. doi: 10.21873/anticanres.12419. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the data generated and analyzed in the present study are available from the corresponding author on reasonable request.