Abstract
Esophageal squamous cell carcinoma (ESCC) is one of the most common types of cancer worldwide. However, operative diagnostic and prognostic systems for ESCC remain to be established. To improve assessment of the prognosis for patients with ESCC, the present study developed an online consensus survival tool for ESCC, termed OSescc. OSescc was built using 264 ESCC cases with gene expression data and relevant clinical information obtained from the Gene Expression Omnibus and The Cancer Genome Atlas databases. Kaplan-Meier survival plots with hazard ratios and P-values were generated by OSescc to predict the association between potential biomarkers and relapse free survival and overall survival. In addition, the current study integrated a function by which one could assess the prognosis based on an individual probe or the mean value of multiple probes for each gene, which helped improve the evaluation of the validity and reliability of the potential prognosis biomarkers. OSescc can be accessed at bioinfo.henu.edu.cn/DBList.jsp.
Keywords: survival, tool, esophageal squamous cell carcinoma, prognosis, database
Introduction
Esophageal cancer (EC) is the tenth most common cause of cancer-associated mortality in United States in 2018 (1), and consists of two histological types; esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (2). ESCC has been the more common histological type of EC for centuries, particularly in Eastern Asia, and accounted for ~90% of all cases of EC worldwide, in 2012 (3). In addition, ESCC was the fourth most fatal type of cancer in China, in 2015 (4). A common treatment method for ESCC is esophagectomy, followed by radiation or neoadjuvant chemotherapy for the residual tumor tissues (5–7). However, these treatment regimens have failed to substantially improve the survival for patients with ESCC over the last few decades (8). Despite the rapid and successful identification of prognostic biomarkers by genome-wide gene expression profiling in numerous types of cancer, including breast cancer (9–11), lung adenocarcinoma (12), non-small cell lung cancer (13) and rectal adenocarcinoma (14), only a small number of biomarkers for ESCC have been identified, including cortactin (15) and ribonuclease 3 (RNASEN; also known as drosha ribonuclease III) (16).
The intra- and inter-patient molecular heterogeneity of cancer has been repeatedly reported (17–25), and makes it difficult to identify and establish universal prognostic biomarkers. To assist in the development of novel prognostic biomarkers, a number of prognostic databases have been established for breast, ovarian, lung and gastric cancer, and numerous new prognostic molecular markers have been identified (26–29). However, to the best of our knowledge, there is no relevant prognostic database for ESCC.
To facilitate the validation of prognostic biomarkers of ESCC and the evaluation of their sensitivity in independent studies, and to help clinicians and scientists improve their understanding of the clinically relevant gene expression data, the present study developed an online consensus survival tool for ESCC, termed OSecc, a web-based interactive survival analysis tool for ESCC. Using OSescc enables individuals to assess overall survival (OS) and relapse free survival (RFS) based on the expression of given genes or probes. OSescc is accessible at bioinfo.henu.edu.cn/DBList.jsp.
Materials and methods
Data acquisition
Datasets were obtained from Gene Expression Omnibus (GEO; ncbi.nlm.nih.gov/geo/) and The Cancer Genome Atlas (TCGA; cancergenome.nih.gov) by searching with the keyword ‘esophageal carcinoma’, followed by manually checking for ‘more than 20 ESCC cases with clinical survival data and gene expression profiling data’.
Development of OSescc
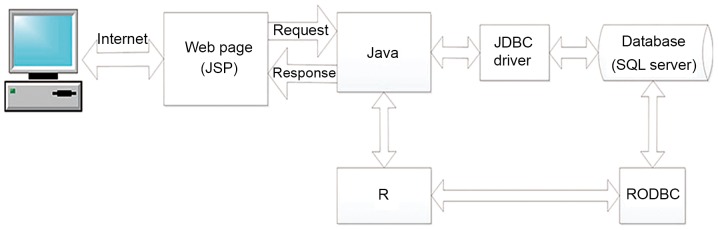
The OSescc server is hosted in a Windows Tomcat server. Server-side scripts are developed in Java v8 (https://www.oracle.com/technetwork/java/javaee/overview/index.html), which control the request of analysis and the return of the results. The database system is managed by a Microsoft Standard Query Language (SQL) server 2008 (https://www.microsoft.com/de-de/download/details.aspx?id=22985), which stores the gene expression and clinical data. The RODBC package of R serves as a connecting layer between the R v3.5.2 (https://www.r-project.org/) and the SQL server database. The Java Database Connectivity driver serves as a connecting layer between the Java and the SQL Server database. The R package ‘survival’ was used to generate Kaplan-Meier (KM) survival curves and calculate the hazard ratio (HR) and log-rank P-value. The central server for OSescc can be accessed at bioinfo.henu.edu.cn/DBList.jsp. A system architecture diagram is presented in Fig. 1. Screenshots of the database interface and the result retrieval are presented in Fig. 2.
Figure 1.
Diagram of the system architecture. JSP, Java Server Pages; JDBC, Java Database Connectivity; SQL, Standard Query Language; RODBC, R Open Database Connectivity.
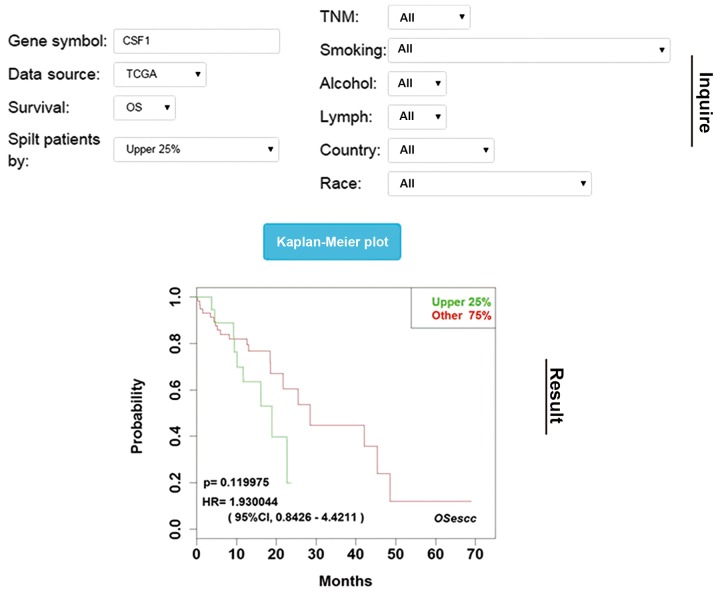
Figure 2.
Screenshot of the database interface. HR, hazard ratio; CI, confidence interval; OSecc, online consensus survival tool for esophageal squamous cell carcinoma; TCGA, The Cancer Genome Atlas; CSF1, colony stimulating factor 1; OS, overall survival.
Results
Two datasets that met the criteria were obtained from GEO (GSE53625) (30) and TCGA (31) database (Table I). These datasets were composed of a total of 264 unique ESCC cases with comprehensive clinical information, including smoking history, alcohol history, TNM stage (32), lymph node status, country and ethnicity.
Table I.
Clinical characteristics of the patients with esophageal squamous cell carcinoma collected in OSescc.
| Characteristic | GSE53625 | TCGA |
|---|---|---|
| Cohort size, n | 179 | 85 |
| Alcohol drinker, n (%) | ||
| Yes | 106 (59.22) | 61 (71.76) |
| No | 73 (40.78) | 23 (27.06) |
| NA | 0 (0.00) | 1 (1.18) |
| Smoker, n (%) | ||
| Yes | 114 (63.69) | 56 (65.88) |
| No | 65 (36.31) | 27 (31.76) |
| NA | 0 (0.00) | 2 (2.36) |
| TNM stage, n (%) | ||
| I | 10 (5.59) | 1 (1.18) |
| II | 77 (43.01) | 24 (28.24) |
| III | 92 (51.40) | 13 (15.29) |
| IV | 0 (0.00) | 4 (4.70) |
| NA | 0 (0.00) | 43 (50.59) |
| Lymph node status, n (%) | ||
| Yes | 0 (0.00) | 20 (23.53) |
| No | 0 (0.00) | 34 (40.00) |
| NA | 179 (100.00) | 31 (36.47) |
| Ethnicity, n (%) | ||
| Caucasian | 0 (0.00) | 40 (47.06) |
| Asian | 179 (100.00) | 40 (47.06) |
| African-American | 0 (0.00) | 2 (2.35) |
| NA | 0 (0.00) | 3 (3.53) |
TCGA, The Cancer Genome Atlas; NA, not available; OSescc, online consensus survival tool for esophageal squamous cell carcinoma.
In OSescc, users could input the gene symbol into the ‘Gene symbol’ dialog window (as presented in Fig. 2) to assess the prognostic significance of interested genes/probes in ESCC. As a result, the KM plot would display the association between the inquired gene and the OS or RFS in which the samples can be stratified by different expression levels of the selected genes or probes depending on the user's choice of median, quartile or 30%. The results can be retrieved in a cohort-specific manner or a cohort-combined manner to increase the statistic power, and the HR and log-rank test P-value are generated. In addition, the population can be co-stratified by alcohol, smoking, TNM stage, lymph node status, country and ethnicity prior to running the analysis to investigate the subclass specific prognostic power.
Clinicians may also be interested in the association between survival and the expression of an individual prognosis-associated probe or transcript variant. Therefore, the present study developed a function by which OSescc can generate a KM plot not only in a gene-specific manner, with the mean value expression of multiple probes for a given gene, but also in a probe-specific manner, with the expression of a single probe for a given gene, on the GSE53625 dataset. This particular function enables individuals to compare the outcomes from a single probe or from multiple probes, which in turn improves the evaluation of the validity and reliability of the potential prognosis biomarkers. By doing so, the current study was able to further analyze four groups of genes.
The first group contained 79 genes; each gene in this group possessed multiple probes and all probes predicted the prognosis with statistical significance towards the same trend of either good or bad prognosis. Therefore, genes in this group could serve as the prognostic biomarkers with high degrees of prediction power.
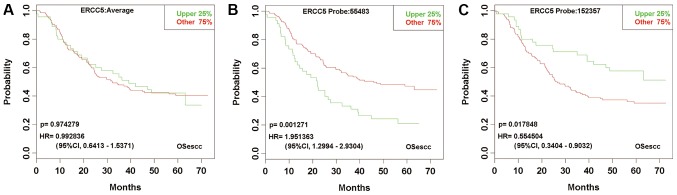
The second group contained only two genes. Probes of either gene contradicted with each other with regard to the predicted prognosis and distinct HR. For example, the ERCC excision repair 5 (ERCC5) gene had two probes (probe nos. 55483 and 152357) in GSE53625. A high expression of probe no. 55483 predicted a significantly worse overall survival rate (HR, 1.9514, P=0.0013), whereas a high expression of probe no. 152375 indicated a significantly improved overall survival rate (HR, 0.5545; P=0.0178). The contradiction between the two probes for the ERCC5 gene resulted in an insignificant prognostic prediction and decreased the sensitivity of outcome prediction using the mean value expression detected by the two probes (Fig. 3).
Figure 3.
Kaplan-Meier plots for the expression of each ERCC5 probe and the mean value expression. A high expression of probe no. 55483 predicted a significantly worse overall survival rate, whereas a high expression of probe no. 152375 indicated a significantly improved overall survival rate. (A) The mean value expression of ERCC5 detected by the two probes. (B) ERCC5 probe no. 55483. (C) ERCC5 probe no. 152357. Esophageal squamous cell carcinoma cases, n=179. ERCC5, ERCC excision repair 5; HR, hazard ratio; CI, confidence interval; OSecc, online consensus survival tool for esophageal squamous cell carcinoma.
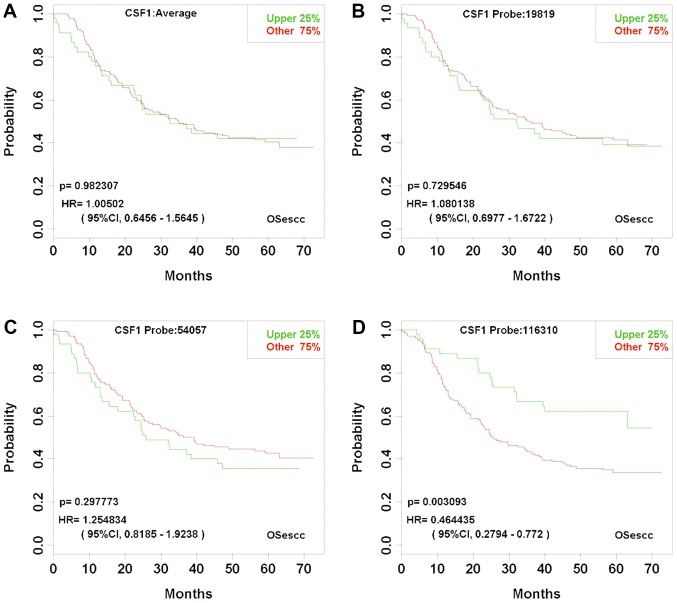
The third group included 532 genes, each of which had multiple probes. Notably, for each given gene, although ≥1 probe could offer a statistically significant prognosis prediction, the prediction based on the mean value expression of all probes was not significant. An example is the colony stimulating factor 1 gene, which was detected using three probes (probe nos. 19819, 54057 and 116310), with P-values of 0.7295, 0.2978 or 0.0031, respectively. Whereas probe no. 116310 appeared to be a promising prognosis predictor with P=0.0031, the P-value of log-rank test for the mean value expression of the three probes was 0.9823, which indicates the prognosis prediction was insignificant (Fig. 4).
Figure 4.
Kaplan-Meier plots for the expression of each CSF1 probe and the mean value expression of three probes. Probe no. 116310 demonstrated a statistically significant prognosis prediction; however, the other probes and the mean value expression of all probes were not significant. (A) The mean value expression detected using three probes. (B) Probe no. 19819. (C) Probe no. 54057. (D) Probe no. 116310. Esophageal squamous cell carcinoma cases, n=179. CSF1, colony stimulating factor 1; HR, hazard ratio; CI, confidence interval; OSecc, online consensus survival tool for esophageal squamous cell carcinoma.
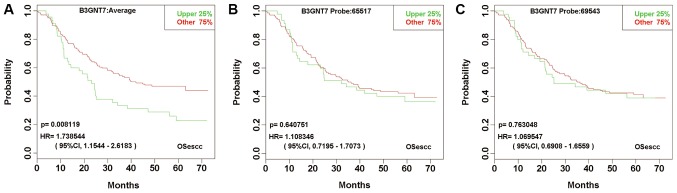
The last group included 135 genes, all with multiple probes. Notably, for each gene, although none of the probes predicted the prognosis with a statistically significant power, the prognosis prediction based on the mean value of all probes was significant. In other words, for genes in this group, the prognosis prediction in a gene-specific manner, but not in a probe-specific manner, was statistically significant. For example, the β-1,3-N-acetylglucosaminyltransferase 7 gene has two probes (probe nos. 65517 and 69543). The prognostic significance of each probe was 0.64075 and 0.76305, respectively. However, the P-value of log-rank test for the mean value expression detected by the two probes was 0.0081 (Fig. 5).
Figure 5.
Kaplan-Meier plots for the expression of each B3GNT7 probe and the mean value expression. Neither probe nos. 65517 or 69543 for B3GNT7 predicted the prognosis with a statistically significant power; however, the prognosis prediction based on the mean value of all probes was significant. (A) The mean value expression of B3GNT7 detected using two probes. (B) B3GNT7 probe no. 65517. (C) B3GNT7 probe no. 69543. Esophageal squamous cell carcinoma cases, n=179. B3GNT7, β-1,3-N-acetylglucosaminyltransferase 7; HR, hazard ratio; CI, confidence interval; OSecc, online consensus survival tool for esophageal squamous cell carcinoma.
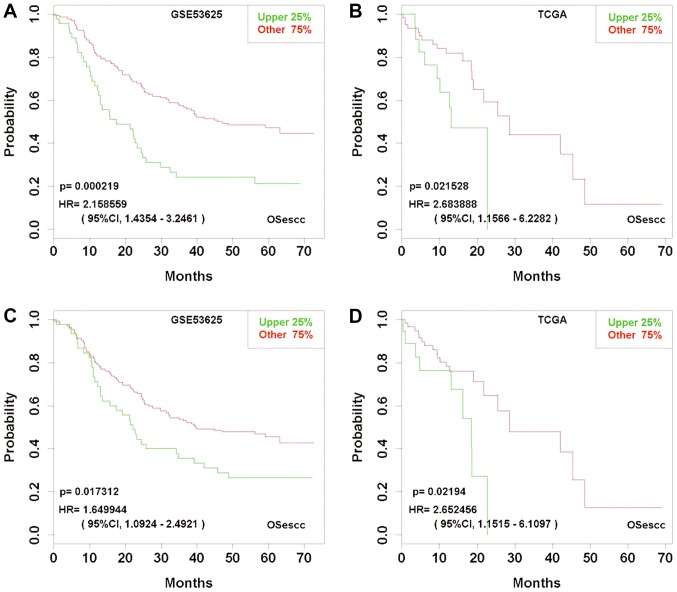
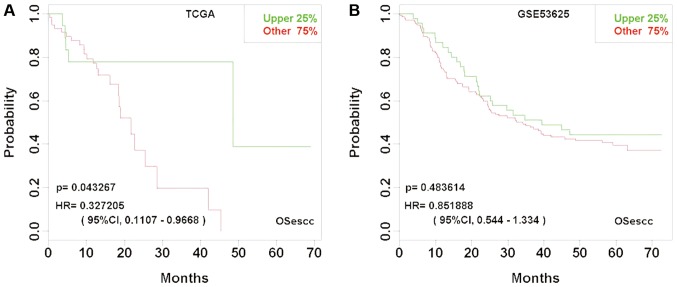
Previous studies have reported several potential biomarkers for ESCC prognosis, including ABL proto-oncogene 1, non-receptor tyrosine kinase (ABL1) (33), transcription elongation factor A protein like 1 (TCEAL1) (34), tropomyosin 2 (35), cystatin C (36), REV3 like, DNA directed polymerase ζ catalytic subunit (37) and urokinase-type plasminogen activator (38). In OSescc, as expected, ABL1 and TCEAL1 demonstrated significant prognostic prediction in the GEO and TCGA datasets (Fig. 6). Furthermore, OSescc could detect the variations of a prognostic biomarker among different cohorts. For example, RNASEN, which has previously been reported as a biomarker of worse prognosis in patients with ESCC (15), was also demonstrated to be associated with worse prognosis by OSescc in the TCGA dataset; however, the prognosis analysis based on the GSE53625 dataset was not statistically significant (Fig. 7).
Figure 6.
Kaplan-Meier plots for two previously published esophageal squamous cell carcinoma biomarkers cross-validated between TCGA and GSE53625 datasets. (A) Prognostic significance of gene ABL1 in GSE53625. (B) Prognostic significance of gene ABL1 in TCGA. (C) Prognostic significance of gene TCEAL1 in GSE53625. (D) Prognostic significance of gene TCEAL1 in TCGA. ESCC cases in TCGA, n=85 and in GSE53625, n=179. TCGA, The Cancer Genome Atlas; ABL1, ABL proto-oncogene 1, non-receptor tyrosine kinase; TCEAL1, transcription elongation factor A protein like 1; HR, hazard ratio; CI, confidence interval; OSecc, online consensus survival tool for esophageal squamous cell carcinoma; ESCC, esophageal squamous cell carcinoma.
Figure 7.
Kaplan-Meier plots for gene RNASEN reveal different prognostic significance between TCGA and GSE53625 datasets. (A) Prognostic significance of RNASEN in TCGA. (B) Prognostic significance of RNASEN in GSE53625. ESCC cases in TCGA, n=85 and in GSE53625, n=179. RNASEN, ribonuclease 3; TCGA, The Cancer Genome Atlas; HR, hazard ratio; CI, confidence interval; OSecc, online consensus survival tool for esophageal squamous cell carcinoma; ESCC, esophageal squamous cell carcinoma.
Discussion
Discovering prognostic biomarkers is important for translational cancer research. The present study analyzed two large gene expression profiling datasets of patients with ESCC with comprehensive clinical follow-up information from GEO and TCGA, and developed OSescc, an online free tool, to assess the prognostic power of potential prognostic biomarkers.
The advantage of OSescc is its convenience to assess and validate potential prognostic biomarkers for ESCC. It can assist clinicians and biologists to determine the prognostic power of given genes in an easy and interactive way. The disadvantage of current study is the sample size; only two available cohorts were implemented in OSescc. This is one reason why the OSescc database was generated to collect all available ESCC clinical follow-up information and gene expression profiling data to facilitate the prognosis analysis for other researchers. When novel ESCC clinical and profiling data becomes available, they will be add to the OSescc database to improve its power and reliability. OSescc was developed to assist researchers and clinicians to investigate the prognostic value of interested genes specific to patients with ESCC. So far, the clinical and gene expression data collected from GEO and TCGA only contained patients with ESCC. In addition to OSescc, we are working on developing online survival tools for other types of cancer and these prognostic tools for other types of cancer should be publically accessible in the future.
Furthermore, the prognosis of patients with ESCC had been demonstrated to be highly dependent on biological factors of the patient, including immune function, nutrition, alcohol drinking and smoking status (39,40); however, the aim of current study was to evaluate the association between gene and prognosis. Nevertheless, certain factors, including TNM stage, smoking, alcohol, lymph node status, country and ethnicity, were also implemented in OSescc to facilitate users to limit their prognostic analysis in particular groups of ESCC. In summary, OSescc may become a guidance tool for selecting suitable prognostic markers for ESCC.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- OSescc
online consensus survival tool for esophageal squamous cell carcinoma
- TCGA
The Cancer Genome Atlas
- GEO
Gene Expression Omnibus
- OS
overall survival
- RFS
relapse free survival
- ESCC
esophageal squamous cell carcinoma
Funding
This work was supported by the National Natural Science Foundation of China (grant no. 81602362), the Program for Science and Technology Development in Henan Province (grant no. 162102310391), the Program for Young Key Teacher of Henan Province (grant no. 2016GGJS-214), Supporting Grants of Henan University (grant nos. 2015YBZR048 and B2015151), the Program for Innovative Talents of Science and Technology in Henan Province (grant no. 18HASTIT048) and the Yellow River Scholar Program (grant no. H2016012).
Availability of data and materials
The datasets used and analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
QW, FW, WZ, YT, XL and XG conceived and designed the study, designed and implemented the software and drafted the manuscript. QW, FW, LX, WZ, YT, JL, JX, YL, XZ, YW, XL and XG acquired and analyzed the data, and designed and implemented the database. QW, FW, LX, WZ, YT, YW, XL and XZ critically revised the manuscript for intellectual content.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Arnold M, Soerjomataram I, Ferlay J, Forman D. Global incidence of oesophageal cancer by histological subtype in 2012. Gut. 2015;64:381–387. doi: 10.1136/gutjnl-2014-308124. [DOI] [PubMed] [Google Scholar]
- 4.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 5.Medical Research Council Oesophageal Cancer Working Group Surgical resection with or without preoperative chemotherapy in oesophageal cancer: A randomised controlled trial. Lancet. 2002;359:1727–1733. doi: 10.1016/S0140-6736(02)08651-8. [DOI] [PubMed] [Google Scholar]
- 6.Shapiro J, van Lanschot JJB, Hulshof MCCM, van Hagen P, van Berge Henegouwen MI, Wijnhoven BPL, van Laarhoven HWM, Nieuwenhuijzen GAP, Hospers GAP, Bonenkamp JJ, et al. Neoadjuvant chemoradiotherapy plus surgery versus surgery alone for oesophageal or junctional cancer (CROSS): Long-term results of a randomised controlled trial. Lancet Oncol. 2015;16:1090–1098. doi: 10.1016/S1470-2045(15)00040-6. [DOI] [PubMed] [Google Scholar]
- 7.van Hagen P, Hulshof MC, van Lanschot JJ, Steyerberg EW, van Berge Henegouwen MI, Wijnhoven BP, Richel DJ, Nieuwenhuijzen GA, Hospers GA, Bonenkamp JJ, et al. Preoperative chemoradiotherapy for esophageal or junctional cancer. N Engl J Med. 2012;366:2074–2084. doi: 10.1056/NEJMoa1112088. [DOI] [PubMed] [Google Scholar]
- 8.Vellayappan BA, Soon YY, Ku GY, Leong CN, Lu JJ, Tey JC. Chemoradiotherapy versus chemoradiotherapy plus surgery for esophageal cancer. Cochrane Database Syst Rev. 2017;8:CD010511. doi: 10.1002/14651858.CD010511.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 10.van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 11.Chang JC, Wooten EC, Tsimelzon A, Hilsenbeck SG, Gutierrez MC, Elledge R, Mohsin S, Osborne CK, Chamness GC, Allred DC, O'Connell P. Gene expression profiling for the prediction of therapeutic response to docetaxel in patients with breast cancer. Lancet. 2003;362:362–369. doi: 10.1016/S0140-6736(03)14023-8. [DOI] [PubMed] [Google Scholar]
- 12.Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, Misek DE, Lin L, Chen G, Gharib TG, Thomas DG, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 13.Lu Y, Lemon W, Liu PY, Yi Y, Morrison C, Yang P, Sun Z, Szoke J, Gerald WL, Watson M, et al. A gene expression signature predicts survival of patients with stage I non-small cell lung cancer. PLoS Med. 2006;3:e467. doi: 10.1371/journal.pmed.0030467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghadimi BM, Grade M, Difilippantonio MJ, Varma S, Simon R, Montagna C, Füzesi L, Langer C, Becker H, Liersch T, et al. Effectiveness of gene expression profiling for response prediction of rectal adenocarcinomas to preoperative chemoradiotherapy. J Clin Oncol. 2005;23:1826–1838. doi: 10.1200/JCO.2005.00.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu P, Qiao J, He W, Wang J, Jia Y, Sun Y, Tang S, Fu L, Qin Y. Genome-wide gene expression profile analyses identify CTTN as a potential prognostic marker in esophageal cancer. PLoS One. 2014;9:e88918. doi: 10.1371/journal.pone.0088918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sugito N, Ishiguro H, Kuwabara Y, Kimura M, Mitsui A, Kurehara H, Ando T, Mori R, Takashima N, Ogawa R, Fujii Y. RNASEN regulates cell proliferation and affects survival in esophageal cancer patients. Clin Cancer Res. 2006;12:7322–7328. doi: 10.1158/1078-0432.CCR-06-0515. [DOI] [PubMed] [Google Scholar]
- 17.Cancer Genome Atlas Research Network, corp-author. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ceccarelli M, Barthel FP, Malta TM, Sabedot TS, Salama SR, Murray BA, Morozova O, Newton Y, Radenbaugh A, Pagnotta SM, et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164:550–563. doi: 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi W, Porten S, Kim S, Willis D, Plimack ER, Hoffman-Censits J, Roth B, Cheng T, Tran M, Lee IL, et al. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell. 2014;25:152–65. doi: 10.1016/j.ccr.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Damrauer JS, Hoadley KA, Chism DD, Fan C, Tiganelli CJ, Wobker SE, Yeh JJ, Milowsky MI, Iyer G, Parker JS, Kim WY. Intrinsic subtypes of high-grade bladder cancer reflect the hallmarks of breast cancer biology. Proc Natl Acad Sci USA. 2014;111:3110–3115. doi: 10.1073/pnas.1318376111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo X, Jo VY, Mills AM, Zhu SX, Lee CH, Espinosa I, Nucci MR, Varma S, Forgó E, Hastie T, et al. Clinically relevant molecular subtypes in leiomyosarcoma. Clin Cancer Res. 2015;21:3501–3511. doi: 10.1158/1078-0432.CCR-14-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lapointe J, Li C, Higgins JP, Van De Rijn M, Bair E, Montgomery K, Ferrari M, Egevad L, Rayford W, Bergerheim U, et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci USA. 2004;101:811–816. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J, Dang H, Wang XW. The significance of intertumor and intratumor heterogeneity in liver cancer. Exp Mol Med. 2018;50:e416. doi: 10.1038/emm.2017.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee WC, Diao L, Wang J, Zhang J, Roarty EB, Varghese S, Chow CW, Fujimoto J, Behrens C, Cascone T, et al. Multiregion gene expression profiling reveals heterogeneity in molecular subtypes and immunotherapy response signatures in lung cancer. Mod Pathol. 2018;31:947–955. doi: 10.1038/s41379-018-0029-3. [DOI] [PubMed] [Google Scholar]
- 25.Rouzier R, Perou CM, Symmans WF, Ibrahim N, Cristofanilli M, Anderson K, Hess KR, Stec J, Ayers M, Wagner P, et al. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin Cancer Res. 2005;11:5678–1885. doi: 10.1158/1078-0432.CCR-04-2421. [DOI] [PubMed] [Google Scholar]
- 26.Szász AM, Lánczky A, Nagy Á, Förster S, Hark K, Green JE, Boussioutas A, Busuttil R, Szabó A, Győrffy B. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7:49322–49333. doi: 10.18632/oncotarget.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Györffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 28.Győrffy B, Lánczky A, Szállási Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr Relat Cancer. 2012;19:197–208. doi: 10.1530/ERC-11-0329. [DOI] [PubMed] [Google Scholar]
- 29.Staff PO. Correction: Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2014;9:e111842. doi: 10.1371/journal.pone.0111842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li J, Chen Z, Tian L, Zhou C, He MY, Gao Y, Wang S, Zhou F, Shi S, Feng X, et al. LncRNA profile study reveals a three-lncRNA signature associated with the survival of patients with oesophageal squamous cell carcinoma. Gut. 2014;63:1700–1710. doi: 10.1136/gutjnl-2013-305806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cancer Genome Atlas Research Network, Analysis Working Group: Asan University; BC Cancer Agency; Brigham and Women's Hospital; Broad Institute; Brown University; Case Western Reserve University; Dana-Farber Cancer Institute; Duke University; Greater Poland Cancer Centre, et al, corp-author. Integrated genomic characterization of oesophageal carcinoma. Nature. 2017;541:169–175. doi: 10.1038/nature20805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edge SB, Compton CC. The American Joint Committee on Cancer: The 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 33.Zhang J, Zhou Y, Wu YJ, Li MJ, Wang RJ, Huang SQ, Gao RR, Ma L, Shi HJ, Zhang J. Hyper-methylated miR-203 dysregulates ABL1 and contributes to the nickel-induced tumorigenesis. Toxicol Lett. 2013;223:42–51. doi: 10.1016/j.toxlet.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 34.Makino H, Tajiri T, Miyashita M, Sasajima K, Anbazhagan R, Johnston J, Gabrielson E. Differential expression of TCEAL1 in esophageal cancers by custom cDNA microarray analysis. Dis Esophagus. 2005;18:37–40. doi: 10.1111/j.1442-2050.2005.00432.x. [DOI] [PubMed] [Google Scholar]
- 35.Zhang J, Wang K, Zhang J, Liu SS, Dai L, Zhang JY. Using proteomic approach to identify tumor-associated proteins as biomarkers in human esophageal squamous cell carcinoma. J Proteome Res. 2011;10:2863–2872. doi: 10.1021/pr200141c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan Y, Fan Q, Wang L, Zhou Y, Li J, Zhou K. LncRNA Snhg1, a non-degradable sponge for miR-338, promotes expression of proto-oncogene CST3 in primary esophageal cancer cells. Oncotarget. 2017;8:35750–35760. doi: 10.18632/oncotarget.16189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu X, Zou S, Zhou J, Zhu H, Zhang S, Shang Z, Ding WQ, Wu J, Chen Y. REV3L, the catalytic subunit of DNA polymerase ζ, is involved in the progression and chemoresistance of esophageal squamous cell carcinoma. Oncol Rep. 2016;35:1664–1670. doi: 10.3892/or.2016.4549. [DOI] [PubMed] [Google Scholar]
- 38.Xu H, Zhang H, Feng C. Over-expression of PLAU regulates the radiosensitivity of esophageal cancer cell. Int J Clin Exp Pathol. 2016;9:10020–10027. [Google Scholar]
- 39.Clavier JB, Antoni D, Atlani D, Ben Abdelghani M, Schumacher C, Dufour P, Kurtz JE, Noel G. Baseline nutritional status is prognostic factor after definitive radiochemotherapy for esophageal cancer. Dis Esophagus. 2014;27:560–567. doi: 10.1111/j.1442-2050.2012.01441.x. [DOI] [PubMed] [Google Scholar]
- 40.Wu IC, Wu CC, Lu CY, Hsu WH, Wu MC, Lee JY, Chou SH, Lee JM, Chou YP, Wu DC, Wu MT. Substance use (alcohol, areca nut and cigarette) is associated with poor prognosis of esophageal squamous cell carcinoma. PLoS One. 2013;8:e55834. doi: 10.1371/journal.pone.0055834. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed during the present study are available from the corresponding author on reasonable request.