Abstract
Purpose: We have previously reported that PRDX2 plays an oncogenic role in colon cancer. In this study, the mRNA expression profile of PRDX2 in HCT116 cells was investigated. Furthermore, we selected Dynamin 3 (DNM3), which is up-regulated by siPRDX2, to investigate its expression pattern and functions in colon cancer.
Patients and methods: PRDX2 siRNA was transfected into HCT116 cells and the mRNA profile was tested by RNA-Sequencing. The expression of interest proteins was determined by Western blot. DNM3 expression in colon cancer tissues and para-carcinoma tissues was evaluated by Western blot and immunohistochemistry assays. Full-length cDNA of DNM3 was cloned into pcDNA3.1 and introduced into HCT116 and HT29 cells. Cell proliferation was tested by CCK-8 and colony formation assays. Cell invasion and migration were tested by transwell assays. Gelatin zymography was utilized for detection of MMP9 activity. Cell apoptosis was investigated with Annexin V/PI staining and flow cytometry and visualized with Hoechst/PI staining assay. All statistical analysis was performed with SPSS 17.0 software.
Results: PRDX2 knockdown led to 210 up-regulated genes and 16 down-regulated genes in HCT116 cells. We also found that DNM3 expression was up-regulated following PRDX2 silencing in HCT116 and HT29 cells. In colon cancer patients, DNM3 was down-regulated and showed a significant association with pathologic grading. DNM3 overexpression inhibited cell proliferation and induced apoptosis in HCT116 and HT29 cells. Cell migration and invasion were also down-regulated in DNM3 overexpressing colon cancer cells, which might be due to the inhibition of MMP9 proteolytic activities. After thorough investigation of the potential mechanism involved, we hypothesized that DNM3 overexpression induced activation of the mitochondrial apoptosis pathway and inhibition of the AKT pathway.
Conclusion: These data suggest that DNM3 is down-regulated in colon cancer, serving as a tumor suppressor. Our study provides new sights into the prognostic value and therapeutic application of DNM3 in colon cancer.
Keywords: mRNA expression profile, proliferation, invasion; migration, apoptosis, AKT
Introduction
Colon cancer is the third most common cancer worldwide and the leading cause of cancer-related deaths.1 Current treatment options for colon cancer include surgery, radiation therapy, chemotherapy, and immunotherapy. Despite advances in these therapies, the prognosis of colon cancer patients still remains poor due to local recurrence and distant metastasis.2,3 The 5-year survival rate is about 90% among patients with early-stage colon cancer, but this statistic reduces to 10% in metastasized cancer.4 An increasing amount of evidences has shown that the growth and metastasis of colon tumor involve numerous genetic and epigenetic changes, but the specific molecular mechanisms involved are still poorly understood.5 Therefore, exploring the pathogenesis of colon cancer will provide new strategies for diagnosis and targeted therapy.
PRDX2 belongs to the peroxiredoxin family of antioxidant enzymes, which are responsible for protection of cells from damage induced by reactive oxygen species.6 Although PRDX2 is ubiquitously expressed, it is up-regulated in breast cancer, cervical cancer and colorectal cancer.7–9 Previous studies have shown that PRDX2 is involved in numerous biological processes, such as relieving oxidative stress10 and regulating signaling pathways that control proliferation, differentiation and survival.11–13 As for tumorigenesis, PRDX2 is known to be dysregulated and to play important roles in multiple cancers, including colorectal cancer and cervical cancer.14–16 However, the contributions of PRDX2 to cancer genomic expression remain largely unknown.
Dynamins (DNMs) are a family of guanylate triphosphatases (GTPases) that participate in vesicle budding and membrane fusion via GTP hydrolysis.17,18 Emerging evidence has shown that DNM3 is associated with tumor progression. Inokawa et al have revealed that methylation of DNM3, which is down-regulated by promoter methylation, predicts a poor prognosis for hepatocellular carcinoma (HCC) patients.19 Numerous studies have reported that DNM3 inhibits the growth and metastasis of HCC by up-regulating p53 expression or down-regulating MMP2.19,20 Similarly, DNM3 is found to play a tumor suppressive role in both colon and cervical cancer by regulating the activities of the MMP family.21,22 However, our knowledge regarding the importance of DNM3 in tumor progression remains insufficient, and most knowledge has been obtained via observation. The precise molecular mechanism is not yet clearly understood.
In this study, we investigated the mRNA expression profile of PRDX2 in colon cancer. Next, DNM3 was identified as a downstream protein of PRDX2 for further analysis. We then investigated the expression of DNM3 and its correlation with the prognosis of colon cancer patients. Function analysis was also performed to determine the influence of DNM3 on the proliferation and metastasis of colon cancer cells. Finally, we sought to elucidate the molecular mechanism of DNM3 by focusing on mitochondrial apoptosis and AKT signaling pathways.
Methods
Tissue specimens
A total of 64 cases of colon cancer tissues and 57 cases of para-carcinoma tissues were collected from Shandong Provincial Hospital affiliated to Shandong University for immunohistochemistry assay. An additional 10 cases of recently diagnosed colon cancer specimens and their matched para-carcinoma tissues were also collected from Shandong Provincial Hospital affiliated to Shandong University and stored at −80°C for Western blot assay. The study was approved by the Medical Ethics Review Committee of the Shandong Provincial Hospital affiliated to Shandong University. Written informed consent was obtained from all patients involved. This research study was performed in accordance with the Declaration of Helsinki involving human material.
Immunohistochemistry
Colon cancer tissues and para-carcinoma tissues were resected to at a thickness of 4 μm on SuperFrost slides (Braunschweig, Germany). The sections were then dewaxed, rehydrated and antigen-retrieved by using the microwave. Endogenous peroxidase activity was eliminated via 0.5% H2O2. After blocking for 1 hr at 25°C, the tissues were incubated with primary antibodies and then developed using 3,3ʹ-diaminobenzidine tetrahydrochloride developer. After counterstaining with hematoxylin, dehydration with gradient ethanol, and mounting with neutral resins, tissue sections were observed under a microscope (Leica Microsystems, Wetzlar, Germany).
Images were taken in 5 random fields under 40× magnification. Staining intensity was categorized as follows: “0” (no staining), “1” (weakly positive), “2” (moderately positive), and “3” (strongly positive). The percentage of positively stained cells was scored as: 0=0%, 1=1–25%, 2=26–50%, 3=51–75%, and 4=76–100%. Evaluations were performed by two independent pathologists. Total scores were determined by the sum of the staining intensity and percentage of positively stained cells. Low expression was identified when the total score was calculated as <6, otherwise they were defined as high expression.
Cell culture and transfection
Human colon cancer cell lines HT29, SW480, HCT116 and SW1116 were purchased from ATCC (Manassas, VA, USA). All cell lines were routinely cultured in DMEM (Invitrogen, Carlsbad, CA, USA) supplemented with 10% FBS(Gibco, Life Technologies, Carlsbad, CA, USA), 100 U/mL penicillin, and 100 mg/mLstreptomycin, at 37°C in 5% CO2. The siRNA sequences targeting PRDX2 were synthesized from Ribobio (Guangzhou, China) and transfected into HT29 and HCT116 cells using Lipofectamine 2000 (Invitrogen) to knock down the expression of PRDX2. Nonsense siRNA was used as the negative control (NC). The siRNA-PRDX2 sequence was: 5ˊ-CCTTCGCCAGATCACTGTTAA-3ˊ. The PRDX2 or DNM3 cDNA sequence was cloned into pcDNA3.1 vector to up-regulated its expression. Cells were cultured to 80% confluence in 6-well plates, then either pcDNA3.1 vector (NC) or pcDNA3.1-DNM3, pcDNA3.1-PRDX2 was transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. After transfection for 48 hrs, DNM3 protein expression was determined by Western blot.
Rna-sequencing analysis
Following 24 hrd of transfection with siRNA, HCT116 cells were collected, sequenced and analyzed by the Beijing Genomics Institute (BGI,Shenzhen, China). DEseq2, EBseq, NOIseq and PossionDis algorithms were used to detect differentially expressed genes.
Western blot
The total proteins content of the cells was extracted using RIPA lysis buffer (Shennengbocai, Shanghai, China) with the addition of 1% proteinase inhibitor. Protein concentrations were evaluated using the BCA method (Beyotime, Jiangsu, China). After 5 mins of heating at 95°C, cell proteins from each sample were separated by 10% SDS-PAGE at 20 μg per lane and then transferred onto polyvinylidene fluoride membranes. The proteins were blocked with 5% non-fat milk for 1 hr, then protein bands were sequentially incubated with primary antibodies overnight at 4°C. Next, the protein bands were with TBST (TBS buffer with 0.1% Tween 20) for three times and incubated with secondary antibodies for 1 hr at room temperature. The protein bands were washed with TBST an additional four times, then visualized with an Odyssey Scanning system (Li-Cor, Lincoln, NE, USA). The primary antibodies against PRDX2 (Cat# 60202-1-Ig, 1:1,000), DNM3 (Cat# 14737-1-AP, 1:1,000), GAPDH (Cat# 60004-1-Ig, 1:10,000), Bcl2 (Cat# 12789-1-AP, 1:1,000), Bax (Cat# 60267-1-Ig, 1:1,000), Caspase-3-P17 (Cat# 19677-1-AP, 1:10,000), AKT (Cat# 60203-2-Ig, 1:5,000), p-AKT (Cat# 66444-1-Ig, 1:2,000), p70S6K (Cat# 66638-1-Ig, 1:3,000) and Cyclin D1 (Cat# 60186-1-Ig, 1:10,000) were purchased from ProteinTech (Rosemont, IL, USA). All secondary antibodies were also purchased from ProteinTech.
Proliferation assays
CCK-8 assay: After transfection for 24 hrs, the cells were digested and made into a cell suspension at 3×104/mL. 100 µL of cell suspension was added into each well of a 96-well plate. The cells were routinely cultured and cell viability was tested every 24 hrs. 10 µLof CCK-8 reagent was added to each well before the test, and incubated for 1.5 hr in a 37°C incubator. The OD values were detected using a microplate reader (BioTek, VT, USA) at 450 nm, and a proliferation curve was drawn.
Colony formation: Colon cancer cells transfected with pcDNA3.1-DNM3 were digested and seeded into a 6-cm petri dish at a concentration of about 400 cells. After 14 days to allow for clone formation, the cells were dried in the air, fixed by 4% paraformaldehyde and stained by 0.1% crystal violet. The cell clones were imaged under a light microscope (Leica Microsystems) and counted.
Transwell assays
For cell invasion assay, HCT116 and HT29 cells transfected with pCDNA3.1-DNM3 for 24 hrs were seeded into Matrigel-coated transwell chambers (8 μm pore size;Costar, Corning, NY, USA) with a total of 2.5×104 cells in 500 μLof medium. The lower chambers received 500 μL of medium containing 10% FBS. The cells were incubated at 37°C for 24 hrs. Residual cells on the upper surface of transwell membranes were removed. The invaded cells on the lower surface were fixed with methanol, stained by 0.1% crystal violet and imaged using phase contrast microscopy. Cell number was counted in five random fields of view. The cell migration assay was performed in the same way, but without matrigel.
Gelatin zymography
After transfection for 24 hr, HCT116 and HT29 cells were seeded into a 6-well plate with a total of 1×105 cells per well with serum-free medium. Cells were normally cultured for 24 hrs. Next, cell supernatants were collected and slightly centrifuged to remove cell debris. Protein concentrations were tested using the BCA method. Sequentially, a total of 20 μg of proteins were separated on 10% SDS-PAGE with 1 mg/mL gelatin A (Sigma-Aldrich, St. Louis, MO, USA). After electrophoresis, the gels were stained with 0.1% Coomassie Brilliant Blue. They were then destained with 45% methanol and 10% (v/v) acetic acid until clear bands suggestive of gelatin digestion were present. The bands were scanned by Image Scanner (Amersham, USA) and analyzed by ImageQuant TL V2003 software.
Flow cytometry detection for apoptosis
The effects of pCDNA3.1-DNM3 on the apoptosis of HCT116 and HT29 cells were tested using an Annexin V-FITC/PI Apoptosis Detection Kit (Beijing Biosea Biotechnology, Beijing, China). Briefly, HCT116 and HT29 cells treated with pCDNA3.1-DNM3 or NC were collected by centrifugation (1000 rpm, 5 mins) and resuspended in the binding buffer. Then, Annexin V‑FITC and PI were added into the cell suspension and incubated for 15 mins at room temperature in the dark. Finally, the percentage of apoptotic cells was determined using a flow cytometer (BD, San Diego, CA, USA).
Hoechst/PI staining assay
To observe cell apoptosis visually, HCT116 and HT29 cells with overexpressed DNM3 were seeded into 6-well plates and stained with Hoechst solution (10 μg/mL; Beyotime, Shanghai, China) and PI solution (5 μg/mL;Beyotime) for 15 mins. Then, fluorescent microscopy was utilized for imaging of the cells. Approximately 200 cells from three random view fields were selected to calculate the percentage of PI-positive cells in each sample.
Statistical analysis
All statistical analysis was performed with SPSS 17.0 software (SPSS Inc., Chicago, IL, USA). The measurement data were expressed as mean ± SD, and the comparison between groups was performed using Student’s t test. P<0.05 was considered statistically significant.
Results
DNM3 is negatively regulated by PRDX2 in human colon cancer cells
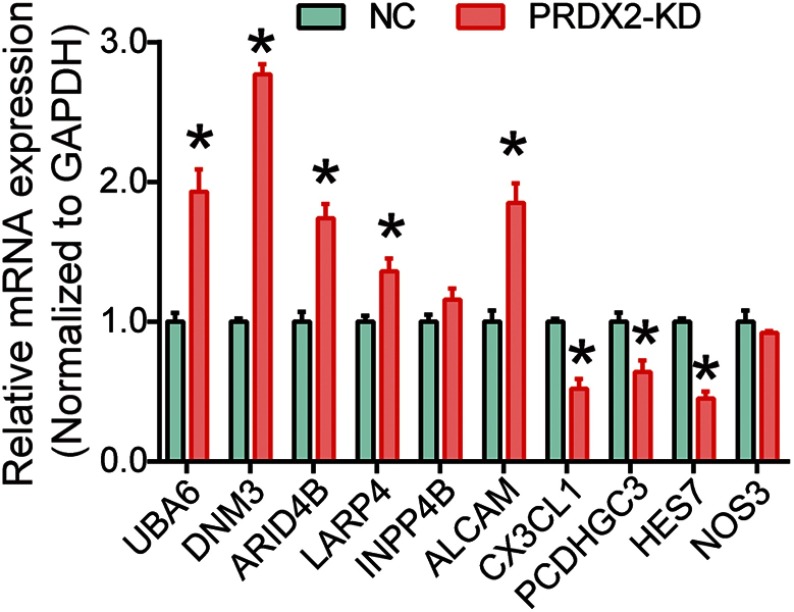
To investigate the mRNA expression profile of PRDX2 in human colon cancer, RNA-Seq by PossionDis was performed to detect significantly differentially expressed genes (|log2 fold change|>1 & Q value <0.001) in PRDX2 silenced HCT116 cells. The results indicated that PRDX2 knockdown led to 210 up-regulated genes and 16 down-regulated genes (Figure 1A–C). Ten differentially expressed genes were selected (Table S1) to be further validated by qRT-PCR analysis. We found that in the selected candidate genes, the expression of DNM3 was strongly regulated by silencing of PRDX2 (Figure S1). Therefore, DNM3 was selected for further experiments. Then, we investigated the protein expression of PRDX2 and DNM3 in colon cancer lines HT29, SW480, HCT116 and SW1116 (Figure 1D). Protein expression of PRDX2 was highest in HT29 cells, second highest in HCT116 cells and lowest in SW1116 cells (Figure 1D). Interestingly, DNM3 expression was lowest in HT29 cells and highest in SW1116 cells (Figure 1D). To further validate the correlation between PRDX2 and DNM3 protein expression, PRDX2 was knocked down in HCT116 and HT29 cells (with higher PRDX2 expression) and up-regulated in SW480 and SW1116 cells (with lower PRDX2 expression). As shown in Figure 1E, PRDX2 knockdown resulted into a significantly increased expression of DNM3 in both HCT116 and HT29 cells (P=0.001, P=0.0002) compared with NC cells, while PRDX2 overexpression significantly reduced the DNM3 expression in both SW480 and SW1116 cells compared with the NC group (P=0.0002, P=0.0002). These suggested that DNM3 might be a downstream effector of PRDX2.
Figure 1.
The mRNA expression profile of PRDX2 knockdown in HCT116 cells and DNM3 is a downstream protein of PRDX2. (A–C) PRDX2 siRNA was conducted and introduced into HCT116 cells. The mRNA expression profile was determined by RNA-Sequencing. The volcano plot (A) and heatmap (B) of different RNA expression profile, and the number of differently expressed genes (C). (D) The protein expression of PRDX2 and DNM3 was detected by Western blot in HT29, SW480, HCT116 and SW1116 cells. (E) The effects of PRDX2 knockdown or PRDX2 overexpression on DNM3 expression were tested by Western blot. The intensities of protein bands were quantified by Image J software. GAPDH was used as an internal control. *P<0.05. All experiments were performed in triplicate.
Abbreviations: DNM, Dynamin; DEGs, differently expressed genes; PRDX2-KD, cells transfected with siRNA-PRDX2 (PRDX2 knockdown); PRDX2-OV, cells transfected with pcDNA3.1-PRDX2 (PRDX2 overexpression); NC, negative control.
Table S1.
Expression level of candidate genes after PRDX2 knockdown
| Symbol | NC-expression | PRDX2-expression | log2FoldChange(PRDX2-KD/NC | P-value | Up/Down-Regulation(PRDX2-KD/NC) |
|---|---|---|---|---|---|
| UBA6 | 2.49 | 4.98 | 1 | 5.08E-13 | Up |
| DNM3 | 0.51 | 1.91 | 1.9050 | 2.33E-05 | Up |
| ARID4B | 1.59 | 3.2 | 1.0090 | 1.69E-08 | Up |
| LARP4 | 6.2 | 12.48 | 1.0093 | 3.74E-18 | Up |
| INPP4B | 0.77 | 1.55 | 1.0093 | 1.48E-07 | Up |
| ALCAM | 3.03 | 6.1 | 1.0095 | 7.44E-12 | Up |
| CX3CL1 | 0.44 | 0.01 | −5.4594 | 6.00E-05 | Down |
| PCDHGC3 | 1.8 | 0.59 | −1.6092 | 0.000105 | Down |
| HES7 | 2.73 | 0.92 | −1.5692 | 0.000149 | Down |
| NOS3 | 1.64 | 0.73 | −1.1677 | 0.000106 | Down |
Figure S1.
Expression of selected genes.
Notes: After PRDX2 was silenced, the mRNA expression of selected genes was examined using RT-PCR. *P<0.05. All experiments were performed in triplicate.
Abbreviation: NC, negative control.
DNM3 is down-regulated in colon cancer tissues and demonstrates an association with the pathological grading of patients
To explore the potential involvement of DNM3 in the progression of colon cancer, we assessed DNM3 expression using Western blot and IHC analysis. Results of Western blot analysis revealed a decrease in DNM3 expression in colon cancer tissues compared with paired para-carcinoma tissues (Figure 2A and B, P=0.0434). As indicated by IHC analysis, we observed glandular hyperplasia in cancer tissue, the cancer cell nucleus increased in volume and rounded, and the nucleolus was obvious, the chromatin was rough, and the cancerous gland infiltrated in the interstitial (Figure 2C). However, the glands in para-cancerous tissues were arranged regularly. The glandular epithelial cells were rod-shaped and located at the basal part. The nucleoli were not obvious and the chromatin was fine. And the local glandular epithelial cells proliferated in a stratified arrangement with low-grade intraepithelial neoplasia (Figure 2C). Moreover, the DNM3 protein expression was down-regulated in colon cancer tissues compared with para-carcinoma tissues (Figure 2C). Results of statistical analysis indicated that the low-expression rate of DNM3 in colon cancer tissues was 68.75% (44/64), which was significantly higher than that in para-carcinoma tissues (38.6%, 22/57, P=0.002, Table 1). Next, we explored whether DNM3 expression was associated with clinicopathological characteristics in colon cancer patients. The results shown in Table 2 demonstrate that DNM3 expression was significantly associated with the pathological grading of colon cancer patients (P=0.025). However, no significant correlation was found between DNM3 expression and gender, age or tumor diameter.
Figure 2.
Dynamin 3 (DNM3) was down-regulated in colon cancer. (A) The expression of DNM3 in colon cancer and paired para-carcinoma tissues was investigated by Western blot. (B) Quantitative analysis of Western blot results. (C) Immunohistochemistry analysis was performed to detect the expression of DNM3 protein. The intensities of protein bands were quantified by Image J software. GAPDH was used as an internal control. *P<0.05.
Table 1.
DNM3 expression in colon cancer tissues compared with para-carcinoma tissue
| Group | n | DNM3 expression | P | |
|---|---|---|---|---|
| Low (n%) | High (n%) | |||
| Colon cancer | 64 | 44 (68.75) | 20 (31.25) | 0.002* |
| Para-carcinoma | 57 | 22 (38.6) | 35 (61.4) | |
Note: *P<0.05.
Abbreviation: DNM, Dynamin.
Table 2.
DNM3 expression is associated with the clinicopathological parameters in colon cancer
| Clinicopathological parameters | n | DNM3 Low (n%) | DNM3 High (n%) | P |
|---|---|---|---|---|
| Gender | ||||
| Male | 36 | 21 (58.3) | 15 (41.7) | 0.077 |
| Female | 28 | 23 (82.1) | 5 (17.9) | |
| Age (years) | ||||
| ≤60 | 25 | 16 (64.0) | 9 (36.0) | 0.959 |
| >60 | 39 | 27 (69.2) | 12 (30.8) | |
| Tumor diameter (cm) | ||||
| ≤5 | 37 | 28 (75.7) | 9 (24.3) | 0.260 |
| >5 | 27 | 16 (59.3) | 11 (40.7) | |
| Pathological grading | ||||
| I–II | 52 | 32 (61.5) | 20 (38.5) | 0.025* |
| II–III | 12 | 12 (100.0) | 0 (0.0) |
Note: *P<0.05.
Abbreviation: DNM, Dynamin.
Overexpression of DNM3 inhibits the proliferation of HCT116 and HT29 cells
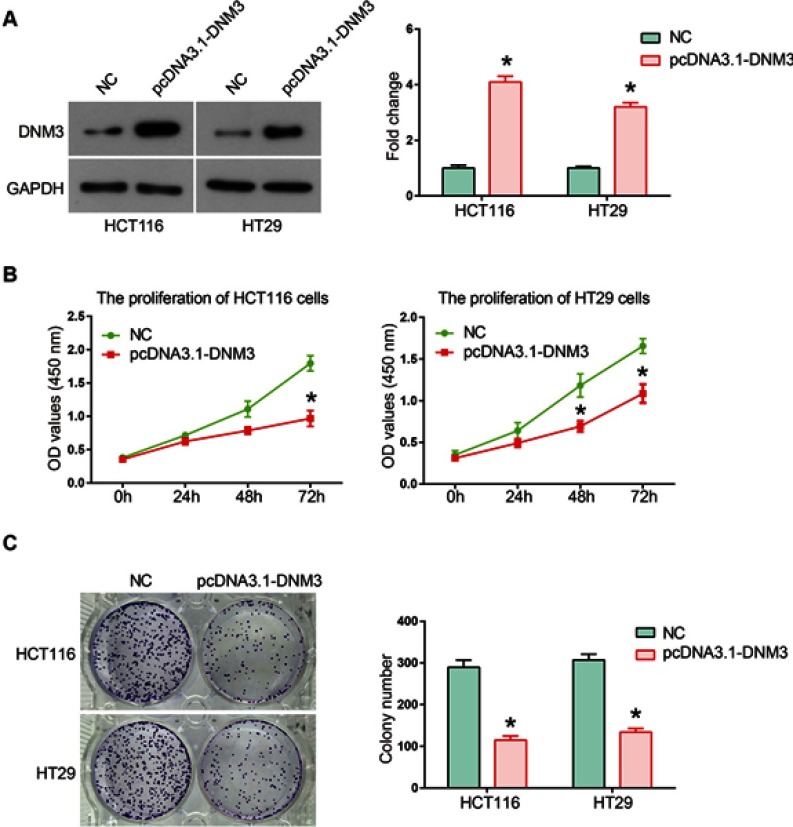
To investigate the influence of DNM3 on tumor cell proliferation, we overexpressed DNM3 in HCT116 and HT29 cells by using pcDNA 3.1 as a vector. Western blot results showed that DNM32 expression was overexpressed by 4-fold in HCT116 cells and 3-fold in HT29 cells by plasmid transfection (Figure 3A, both P=0.0001). Cell proliferation was tested using CCK-8 assay, which suggested that overexpression of DNM3 significantly decreased the living HCT116 and HT29 cells compared with the NC group after transfection of 72 hrs (Figure 3B, P=0.016, P=0.0014). Colony formation assay also showed less clones in DNM3 overexpressing HCT116 and HT29 cells compared with the NC group (Figure 3C, both P=0.0001). Taken together, these data suggest a negative role of DNM3 in the proliferation of HCT116 and HT29 cells.
Figure 3.
Dynamin 3 (DNM3) overexpression decreases cell viability in colon cancer cells. (A) DNM3 cDNA was cloned into pcDNA3.1 plasmid and introduced into HCT116 and HT29 cells, and the empty pcDNA3.1 plasmid was used as negative control (NC). The overexpression of DNM3 was validated by Western blot. The intensities of protein bands were quantified by Image J software. GAPDH was used as an internal control. (B) The effects of DNM3 overexpression on cell proliferation of HCT116 and HT29 cells were investigated by CKK-8 assay. (C) The cologenic ability was tested in DNM3 overexpressing HCT116 and HT29 cells. *P<0.05. All experiments were performed in triplicate.
Overexpression of DNM3 inhibits the invasion and migration of HCT116 and HT29 cells by regulating MMP9 activity
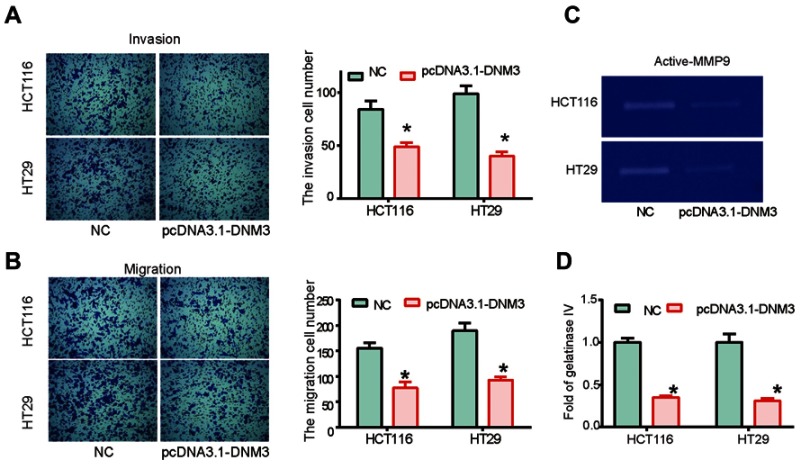
Cell invasion and migration are important features of tumor cells, which frequently initiate tumor metastasis in vivo.23 In order to determine whether DNM3 influenced the invasion and migration of tumor cells, transwell assays were performed. Cell invasion results indicated that DNM3 overexpression resulted in a significantly decreased invasion number in HCT116 and HT29 cells (Figure 4A, P=0.0022, P=0.003). Similarly, cell migration was also inhibited in HCT116 and HT29 cells overexpressing DNM3 (Figure 4B, P=0.0011, P=0.006). Due to the important role of MMP9 in degradation of extracellular matrix and promotion of cell invasion, MMP9 activity was examined in DNM3 overexpressing cells. Gelatin zymography analysis indicated that DNM3 overexpression resulted in a significant decrease in the activity of secreted MMP9 in HCT116 and HT29 cells (Figure 4C and D, both P=0.001).
Figure 4.
Dynamin 3 (DNM3) overexpression inhibits the migration and invasion of colon cancer cells. Cell invasion (A) and migration (B) were investigated by transwell assays in DNM3 overexpressing HCT116 and HT29 cells. (C) The proteolytic activities of secreted MMP9 were investigated by gelatin zymography assay. (D) Quantitative analysis of gelatin zymography assay results. *P<0.05. All experiments were performed in triplicate.
Abbreviation: NC, negative control.
Overexpression of DNM3 promotes apoptosis in HCT116 and HT29 cells via the mitochondrial apoptosis pathway
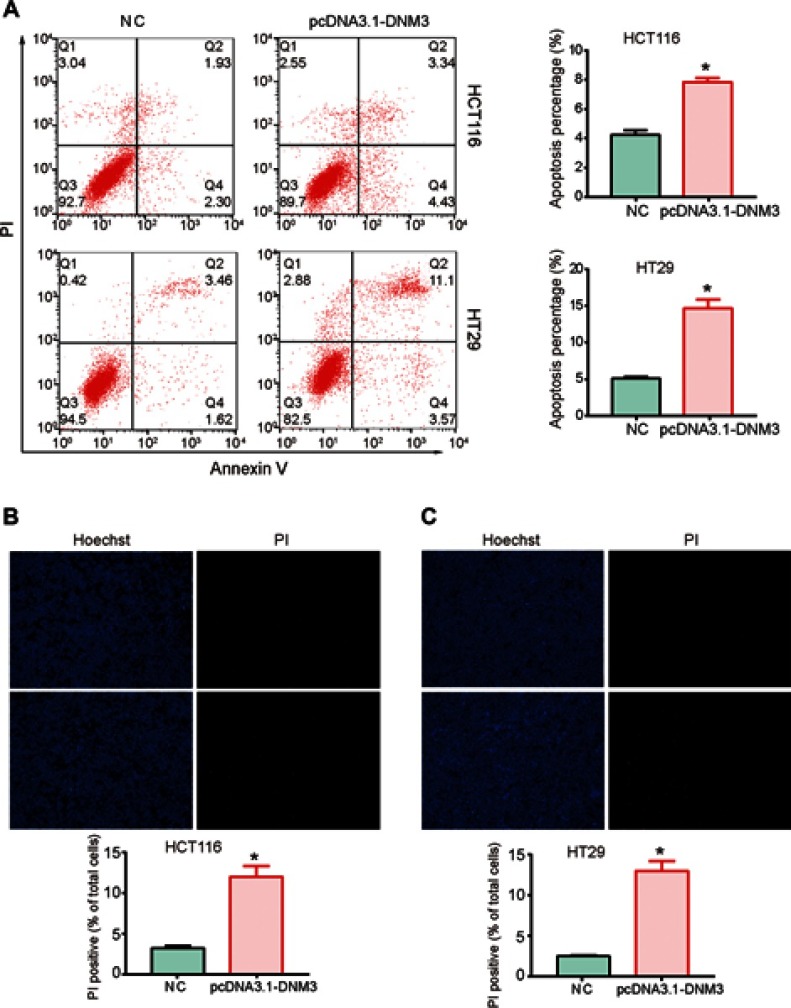
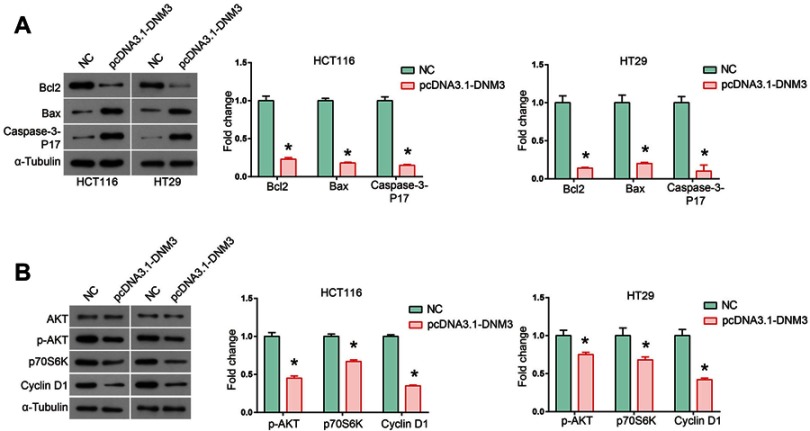
After observing significant inhibition of the proliferation and metastasis induced by overexpression of DNM3, we further investigated whether cell apoptosis contributed to this effect. As shown in Figure 5A, flow cytometry results indicated that overexpression of DNM3 resulted in a significant increase in the percentage of apoptotic cells in both HCT116 (P=0.001) and HT29 (P=0.002) cells. The pro-apoptotic effects of DNM3 overexpression were further visualized by a Hoechst/PI staining assay. Photomicrographs of double-fluorescent staining with PI (red) and Hoechst 33342 (blue) indicated that DNM3 overexpression significantly increased the uptake of PI in HCT116 and HT29 cells (Figure 5B and C, P=0.003, P=0.001). Next, we investigated the pro-apoptotic mechanism of DNM3 by focusing on the mitochondrial apoptotic pathway. As shown in Figure 6A, Western blot analysis revealed that overexpression of DNM3 led to decreased expression of anti-apoptotic protein Bcl2 (P=0.002, P=0.001) and increased expression of pro-apoptotic protein Bax (P=0.0001, P=0.008) and Caspase-3-P17 (P=0.0122, P=0.0001) in both HCT116 and HT29 cells. These results indicate that the mitochondrial apoptotic pathway was activated.
Figure 5.
Dynamin 3 (DNM3) overexpression induces apoptosis in colon cancer cells. (A) Cell apoptosis was investigated by Annexin V/PI staining and flow cytometry. (B) and (C) Photomicrographs of double-fluorescent staining with PI (red) and Hoechst 33342 (blue) in HCT116 (B) and HT29 (C) cells. PI-positive cells were counted in approximately 200 cells from three random microscopic fields for each sample and are expressed as percentages of the total cells. *P<0.05. All experiments were performed in triplicate.
Figure 6.
Dynamin 3 (DNM3) overexpression leads to activation of mitochondria apoptosis pathway and down-regulation of AKT signaling pathway. (A) The expression of mitochondria apoptosis pathway members Bcl2, Bax and Caspase-3-P17 was investigated by Western blot. (B) The expression of AKT signaling pathway members AKT, p-AKT, p70S6K and Cyclin D1 was investigated by Western blot. The intensities of protein bands were quantified by Image J software. GAPDH was used as an internal control. *P<0.05. All experiments were performed in triplicate. Supplementary data.
Abbreviation: NC, negative control.
Overexpression of DNM3 induces the inactivation of AKT signaling pathway
The AKT signaling pathway was investigated in DNM3 overexpressing HCT116 and HT29 cells. Activation of the AKT pathway corresponds with an elevation of phosphorylated AKT. Furthermore, phosphorylated AKT is involved in the regulation of numerous downstream effectors important in cellular growth, such as p70S6 kinase (p70S6K). This results in enhanced translation of a subset of genes that are required for protein synthesis, cell growth, inhibition of cell apoptosis, and acceleration of cell proliferation, which finally lead to tumorigenesis.24 We found that overexpression of DNM3 led to a significant decrease in phosphorylated AKT (P=0.007, P=0.0143) and its down-stream effectors p70S6K (P=0.001, P=0.0068) and Cyclin D1 (P=0.001, P=0.006) in both HCT116 and HT29 cells, suggesting that the AKT signaling pathway was inactivated (Figure 6B).
Discussion
PRDX2 is an important member of the ROS scavenging system that exerts a pro-tumor role in colon cancer. For example, it has been found that silencing of PRDX2 disrupts the drug resistance of colon cancer cells to 5-FU by inhibiting the PI3K/AKT pathway.16 Rong Wang et al have reported that PRDX2 is essential for maintaining the stem cell-like phenotype of cancer cells through activation of the Hedgehog signaling pathway in colon cancer.25 Our previous study also suggests that PRDX2 down-regulation inhibits colorectal cancer growth via the Wnt/β-catenin signaling pathway.26 Therefore, we hypothesize that PRDX2 may induce significant changes in the genomic expression of colon cancer. Herein, we found that PRDX2 silencing in HCT116 cells resulted in 210 up-regulated genes and 16 down-regulated genes, which demonstrated an important function of PRDX2 in genomic regulation and provided insight on potential therapeutic targets for colon cancer.
For further analysis, we selected DNM3, which was up-regulated by PRDX2 silencing in RNA-Seq. DNM3 is a GTPase involved in endocytosis that possesses mechanochemical properties of tabulating and severing membranes. In this study, we found that PRDX2 siRNA promoted the protein expression of DNM3 in HCT116 and HT29 cells, and PRDX2 overexpression decreased the expression of DNM3 in SW480 and SW1116 cells. Furthermore, Western blot and IHC analysis suggested that the expression of DNM3 was significantly decreased in colon cancer tissues compared with para-carcinoma tissues, which was consistent with the results of HCC tissues.27 The correlation of DNM3 with the clinicopathological characteristics of colon cancer patients suggested that down-regulation of DNM3 predicts a high pathological grading of colon cancer, while no significant correlation was found amongst patient gender, age or tumor diameter. Inokawa et al have reported that HCC patients with reduced expression of DNM3 in tumor tissues exhibited a worse prognosis compared to patients with high expression.19 These data suggest that DNM3 may serve as a clinical prognostic biomarker for HCC and colon cancer patients.
In this study, function analysis suggested that DNM3 overexpression inhibited the proliferation, migration and invasion of HCT116 and HT29 cells. Moreover, up-regulation of DNM3 also inhibited the MMP9 activity. These results were consistent with the previous studies that DNM3 plays a tumor suppressive role in tumor progression, including HCC and cervical cancer.19,20 Mechanism investigation indicated that DNM3 overexpression induced cell apoptosis and activated the mitochondrial apoptosis pathway in colon cancer cells, including decreased Bcl2 and increased Bax and Caspase-3-P17. The mitochondrial apoptotic pathway, which is one of many pathways involved in regulation of cell death, stands for the canonical intrinsic death pathway.28 Bax is a pro-apoptotic member of the Bcl-2 family that is involved in promoting mitochondrial outer membrane permeabilization (MOMP) and triggering the release of cytochrome C to the cytoplasm to activate caspase cascade.29 Bcl2 functions as an anti-apoptotic protein by counteracting pro-apoptotic effectors, such as Bax.29 Therefore, these results suggest that DNM3 promotes cell apoptosis in a mitochondrial apoptotic pathway in colon cancer. Finally, we identified that overexpression of DNM3 down-regulated the Akt signaling pathway by decreasing the expression of p-AKT, p70S6K and Cyclin D1. The AKT signaling pathway is a key pathway in the regulation of multiple biological processes, including cell proliferation, survival, and migration of tumors.24 Combined with current evidence that PRDX2 knockdown promotes DNM3 expression and suppresses the PI3K/AKT pathway, we speculate that DNM3 mediates the activation effect of PRDX2 on AKT. However, this signaling pathway requires further analysis.
In this study, we concluded that DNM3 was negatively regulated by PRDX2. Moreover, DNM3 functions as a tumor suppressor in the progression of colon cancer, up-regulation of DNM3 could inhibit the proliferation and invasion of colon cancer cells, which the AKT signaling pathway may be involved in. Therefore, our study suggests that PRDX2 may be a potential therapeutic target for colon cancer, and that the expression of DNM3 can be up-regulated by the silencing of PRDX2.
Acknowledgments
This work was funded by National Natural Science Foundation of China (No. 81502058), Science and Technology Development Program of Shandong Province (No. 2017WS099) and China Postdoctoral Science Foundation (No. 2016M592197).
Ethics approval and informed consent
The study was approved by the Medical Ethics Review Committee of the Shandong Provincial Hospital affiliated to Shandong University.
Consent for publication
Informed consent was given by all patients involved. Research has been performed in accordance with the Declaration of Helsinki involving human material.
Supplementary materials
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387 [DOI] [PubMed] [Google Scholar]
- 2.Maeda H, Kashiwabara K, Aoyama T, et al. Hazard rate of tumor recurrence over time in patients with colon cancer: implications for postoperative surveillance from three Japanese foundation for multi-disciplinary treatment of cancer (JFMC) clinical trials. J Cancer. 2017;8(19):4057–4064. doi: 10.7150/jca.21365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Veenstra CM, Krauss JC. Emerging systemic therapies for colorectal cancer. Clin Colon Rectal Surg. 2018;31(3):179–191. doi: 10.1055/s-0037-1602238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vecchione L, Gambino V, Raaijmakers J, et al. A vulnerability of a subset of colon cancers with potential clinical utility. Cell. 2016;165(2):317–330. doi: 10.1016/j.cell.2016.02.059 [DOI] [PubMed] [Google Scholar]
- 5.Yu F, Zhou C, Zeng H, Liu Y, Li S. BMI1 activates WNT signaling in colon cancer by negatively regulating the WNT antagonist IDAX. Biochem Biophys Res Commun. 2018;496(2):468–474. doi: 10.1016/j.bbrc.2018.01.063 [DOI] [PubMed] [Google Scholar]
- 6.Brown JD, Day AM, Taylor SR, Tomalin LE, Morgan BA, Veal EA. A peroxiredoxin promotes H2O2 signaling and oxidative stress resistance by oxidizing a thioredoxin family protein. Cell Rep. 2013;5(5):1425–1435. doi: 10.1016/j.celrep.2013.10.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim K, Yu M, Han S, et al. Expression of human peroxiredoxin isoforms in response to cervical carcinogenesis. Oncol Rep. 2009;21(6):1391–1396. [PubMed] [Google Scholar]
- 8.Stresing V, Baltziskueta E, Rubio N, et al. Peroxiredoxin 2 specifically regulates the oxidative and metabolic stress response of human metastatic breast cancer cells in lungs. Oncogene. 2013;32(6):724–735. doi: 10.1038/onc.2012.93 [DOI] [PubMed] [Google Scholar]
- 9.Lu W, Fu Z, Wang H, Feng J, Wei J, Guo J. Peroxiredoxin 2 is upregulated in colorectal cancer and contributes to colorectal cancer cells’ survival by protecting cells from oxidative stress. Mol Cell Biochem. 2014;387(1–2):261–270. doi: 10.1007/s11010-013-1891-4 [DOI] [PubMed] [Google Scholar]
- 10.Yim MB, Chae HZ, Rhee SG, Chock PB, Stadtman ER. On the protective mechanism of the thiol-specific antioxidant enzyme against the oxidative damage of biomacromolecules. J Biol Chem. 1994;269(3):1621–1626. [PubMed] [Google Scholar]
- 11.Fang J, Nakamura T, Cho D-H, Gu Z, Lipton SA. S-nitrosylation of peroxiredoxin 2 promotes oxidative stress-induced neuronal cell death in Parkinson’s disease. Proc Natl Acad Sci U S A. 2007;104(47):18742–18747. doi: 10.1073/pnas.0705904104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Immenschuh S, Baumgart-Vogt E. Peroxiredoxins, oxidative stress, and cell proliferation. Antioxid Redox Signal. 2005;7(5–6):768–777. doi: 10.1089/ars.2005.7.768 [DOI] [PubMed] [Google Scholar]
- 13.Kim H, Lee TH, Park ES, et al. Role of peroxiredoxins in regulating intracellular hydrogen peroxide and hydrogen peroxide-induced apoptosis in thyroid cells. J Biol Chem. 2000;275(24):18266–18270. doi: 10.1074/jbc.275.24.18266 [DOI] [PubMed] [Google Scholar]
- 14.Kontostathi G, Zoidakis J, Makridakis M, et al. Cervical cancer cell line secretome highlights the roles of transforming growth factor-beta-induced protein Ig-h3, peroxiredoxin-2, and NRF2 on cervical carcinogenesis. Biomed Res Int. 2017;2017:1–15. doi: 10.1155/2017/4180703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang S, Fu Z, Wei J, Guo J, Liu M, Du K. Peroxiredoxin 2 is involved in vasculogenic mimicry formation by targeting VEGFR2 activation in colorectal cancer. Med Oncol. 2015;32(1):414. [DOI] [PubMed] [Google Scholar]
- 16.Xu J, Zhang S, Wang R, Wu X, Zeng L, Fu Z. Knockdown of PRDX2 sensitizes colon cancer cells to 5-FU by suppressing the PI3K/AKT signaling pathway. Biosci Rep. 2017;37(3):BSR20160447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perrais D, Merrifield CJ. Dynamics of endocytic vesicle creation. Dev Cell. 2005;9(5):581–592. doi: 10.1016/j.devcel.2005.10.002 [DOI] [PubMed] [Google Scholar]
- 18.Ochoa GC, Slepnev VI, Neff L, et al. A functional link between dynamin and the actin cytoskeleton at podosomes. J Cell Biol. 2000;150(2):377–389. doi: 10.1083/jcb.150.2.377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inokawa Y, Nomoto S, Hishida M, et al. Dynamin 3: a new candidate tumor suppressor gene in hepatocellular carcinoma detected by triple combination array analysis. Onco Targets Ther. 2013;6:1417–1424. doi: 10.2147/OTT.S51913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Z, Chen C, Guo W, Zheng S, Sun Z, Geng X. DNM3 attenuates hepatocellular carcinoma growth by activating P53. Med Sci Monit. 2016;22:197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang L, Lang Q-L, Liang W-M, et al. Construction of a recombinant eukaryotic expression vector containing DNM3 gene and its expression in colon cancer cells. Onco Targets Ther. 2018;11:6665–6671. doi: 10.2147/OTT.S176388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee Y-Y, Do I-G, Park YA, et al. Low dynamin 2 expression is associated with tumor invasion and metastasis in invasive squamous cell carcinoma of cervix. Cancer Biol Ther. 2010;10(4):329–335. doi: 10.4161/cbt.10.4.12275 [DOI] [PubMed] [Google Scholar]
- 23.Thaper D, Vahid S, Nip KM, et al. Targeting lyn regulates snail family shuttling and inhibits metastasis. Oncogene. 2017;36(28):3964–3975. doi: 10.1038/onc.2017.5 [DOI] [PubMed] [Google Scholar]
- 24.Martini M, De Santis MC, Braccini L, Gulluni F, Hirsch E. PI3K/AKT signaling pathway and cancer: an updated review. Ann Med. 2014;46(6):372–383. doi: 10.3109/07853890.2014.912836 [DOI] [PubMed] [Google Scholar]
- 25.Wang R, Wei J, Zhang S, et al. Peroxiredoxin 2 is essential for maintaining cancer stem cell-like phenotype through activation of Hedgehog signaling pathway in colon cancer. Oncotarget. 2016;7(52):86816–86828. doi: 10.18632/oncotarget.13559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu W, Fu Z, Wang H, Feng J, Wei J, Guo J. Peroxiredoxin 2 knockdown by RNA interference inhibits the growth of colorectal cancer cells by downregulating Wnt/beta-catenin signaling (vol 343, pg 190, 2014). Cancer Lett. 2014;350(1–2):69–69. doi: 10.1016/j.canlet.2014.02.020 [DOI] [PubMed] [Google Scholar]
- 27.Zhang H-J, Yuan G-L, Liang Q-L, et al. Progress of dynamin 3 in tumors. Int J Clin Exp Med. 2017;10(11):15060–15063. [Google Scholar]
- 28.Li Y, Zhang D, Yu K, et al. CMPD1 inhibited human gastric cancer cell proliferation by inducing apoptosis and G2/M cell cycle arrest. Biol Res. 2018;51:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Green DR, Llambi F. Cell death signaling. Cold Spring Harb Perspect Biol. 2015;7(12):a006080. doi: 10.1101/cshperspect.a006080 [DOI] [PMC free article] [PubMed] [Google Scholar]