Abstract
Despite the clinical requirement for early diagnosis, the early events in lung cancer and their mechanisms are not fully understood. Pituitary tumor transforming gene 1 binding factor (PTTG1IP) is a tumor-associated gene; however, to the best of our knowledge, its association with lung cancer has not been reported. The present study analyzed PTTG1IP expression in early-stage non-small cell lung cancer (NSCLC) samples and investigated its epigenetic regulatory mechanisms. The results revealed that the mRNA level of PTTG1IP in NSCLC tissues was significantly downregulated by 43% compared with that in adjacent tissues. In addition, overexpression of this gene significantly inhibited cell proliferation. According to data from The Cancer Genome Atlas, a significant negative correlation was identified between the PTTG1IP gene methylation level and expression level in lung adenocarcinoma and lung squamous cell carcinoma cases. Reduced representation bisulfite sequencing (RRBS) analysis of six paired early-stage NSCLC tissue samples indicated that the CpG island shore of the PTTG1IP promoter is hypermethylated in lung cancer tissues, which was further validated in 12 paired early-stage NSCLC samples via bisulfite amplicon sequencing. Following treatment with 5-aza-2′-deoxycytidine to reduce DNA methylation in the promoter region, the PTTG1IP mRNA level increased, indicating that the PTTG1IP promoter DNA methylation level negatively regulates PTTG1IP transcription. In conclusion, in early-stage NSCLC, the PTTG1IP gene is regulated by DNA methylation in its promoter region, which may participate in the development and progression of lung cancer.
Keywords: PTTG1IP, lung cancer, DNA methylation, expression, promoter
Introduction
Lung cancer, a complex disease involving both epigenetic and genetic changes, is the leading cause of cancer-associated mortality worldwide (1,2). Lung cancer has had a high incidence rate and a poor 5-year survival rate of <19% in the United States between 2006 and 2012 (2). One cause of the high mortality rate is the lack of specific early detection methods and the majority of patients are diagnosed with middle- or late-stage disease (3). Therefore, early detection and treatment strategies for lung cancer are urgently required.
Several imaging and cytology-based strategies have been utilized for early lung cancer detection. However, none have been demonstrated to completely reduce lung cancer mortality (3–5). Previous studies have reported that aberrant epigenetic changes are one of the most frequent cancer-associated events and are regarded as important mechanisms in carcinogenesis (6). Investigation of the associated molecular mechanisms can be exploited to diagnose early-stage lung cancer (3–5). Furthermore, methylation profiles may be potential biomarkers for early cancer diagnosis and they have been demonstrated to exhibit good prognostic value (4,7–9). Previously, accumulating evidence has confirmed that tumor tissues can be characterized by hypermethylation at promoter-associated CpG islands (CGIs) or global hypomethylation of the genome compared with normal tissues (9–11). Furthermore, certain studies have suggested that methylation of DNA CpG sites is an epigenetic regulator of gene expression that usually results in gene silencing (12,13). Hao et al (12) reported that methylation patterns can predict prognosis and survival, and identified an association between differential methylation of CpG sites and the expression of cancer-associated genes. Their findings demonstrate the utility of methylation biomarkers for cancer molecular characterization, diagnosis and prognosis determination. Therefore, a number of specific tumor targets can be developed for use as DNA methylation-based biomarkers (4,7,9).
At present, numerous useful cancer biomarkers have been identified. Pituitary tumor transforming gene 1 binding factor (PTTG1IP; also termed PBF) is a ubiquitously expressed proto-oncogene. PTTG1IP was first identified through its ability to bind to human securin, also termed pituitary tumor transforming gene (PTTG) (14,15). Thus far, PTTG1IP has been reported to be highly expressed in thyroid, breast, colorectal, and liver cancer (16–19). However, to the best of our knowledge, its expression levels in lung cancer have not been reported. The present study investigated PTTG1IP expression in early non-small cell lung cancer (NSCLC) and examined the correlation between the PTTG1IP promoter region methylation level and the gene expression level.
Materials and methods
Tissue samples
In total, 18 pairs of early-stage (stage I or II) NSCLC tissues and adjacent tissues were obtained from the South Hospital of Renji Hospital Shanghai Jiao Tong University School of Medicine (Shanghai, China) between January 2014 and March 2015 (Table I). A total of 12 male and 6 female patients aged between 45 and 75 years were included in the present study. During excision surgery, 50 mg fresh cancer tissue and 50 mg adjacent normal tissue (<2 cm from cancer margin) was obtained from each patient. Tissue samples were immediately frozen in liquid nitrogen following resection and stored at −80°C until use. All included samples were histologically confirmed primary NSCLC and pathological stage I or II according to the Tumor-Node-Metastasis staging system (20). Written informed consent was obtained from all patients and the study was approved by the Ethics Committee of South Hospital of Renji Hospital Shanghai Jiao Tong University School of Medicine.
Table I.
Basic information of the paired lung cancer tissue and adjacent tissue samples.
| Sample no. | Sex | Diagnosis | Staged |
|---|---|---|---|
| Pair 1c | Female | Lung adenocarcinoma | II |
| Pair 2a,c | Male | Lung squamous cell carcinoma | II |
| Pair 3a,c | Female | Lung adenocarcinoma | II |
| Pair 4a,c | Male | Lung squamous cell carcinoma | II |
| Pair 5a,c | Male | Lung squamous cell carcinoma | II |
| Pair 6a,c | Female | Lung adenocarcinoma | I |
| Pair 7a,c | Male | Lung adenocarcinoma | II |
| Pair 8a,c | Female | Lung adenocarcinoma | II |
| Pair 9a,c | Male | Lung adenocarcinoma | II |
| Pair 10a,c | Male | Lung adenocarcinoma | II |
| Pair 11a,c | Male | Lung adenocarcinoma | II |
| Pair 12c | Male | Lung adenocarcinoma | I |
| Pair 13b | Male | Lung adenocarcinoma | I |
| Pair 14b | Male | Lung adenocarcinoma | II |
| Pair 15b | Male | Lung squamous cell carcinoma | II |
| Pair 16b | Female | Lung adenocarcinoma | I |
| Pair 17b | Male | Lung adenocarcinoma | I |
| Pair 18b | Female | Lung adenocarcinoma | II |
Analyzed by reverse transcription-quantitative polymerase chain reaction.
Analyzed by reduced representation bisulfite sequencing.
Analyzed by bisulfite amplicon sequencing.
According to the Tumor-Node-Metastasis staging system (20).
Cell culture and treatments
A549 cells of the human lung adenocarcinoma cell line were cultured in Roswell Park Memorial Institute-1640 medium (Thermo Fisher Scientific, Inc., Waltham, MA, USA) with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and 1% penicillin/streptomycin. MRC5 cells of the human embryonic lung fibroblast cell line were cultured in minimum essential medium (Thermo Fisher Scientific, Inc., Waltham, MA, USA) with 10% FBS and 1% penicillin/streptomycin. Cell cultures were incubated at 37°C in a humidified atmosphere with 5% CO2. Following A549 cell culture to ~90% confluence, 1 µM 5-aza-2′-deoxycytidine (5-aza-dC, Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was added to the culture medium. DMSO (Sigma-Aldrich; Merck KGaA) treatment was used as a control. The cells were harvested following 48-h treatment, and total RNA and genomic DNA were obtained according to standard protocols. In brief, TRIzol® reagent (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was used for RNA extraction. Precipitated RNA was washed to remove impurities, and then resuspended for use in downstream applications. DNA extractions were performed using the High Pure PCR Template Preparation kit (Roche Diagnostics, Basel, Switzerland). Cells were lysed with Lysis Buffer and Proteinase K, and released DNA was bound on a glass fiber filter and washed prior to elution.
Plasmid construction and cell transfection
To generate a vector expressing myc-PTTG1IP, the corresponding sequences were subcloned into a pcDNA3.1 vector (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) that was subsequently termed pcDNA3.1/3Xmyc-PTTG1IP. Cells were plated the day prior to transfection and cultured to 70% confluence. Transfection was performed with Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) with 1 µg plasmid/well, according to the manufacturer's protocol. Cells transfected with empty pcDNA3.1 vector was used as a control. The medium was replaced with new culture medium 6 h post-transfection. Cells were harvested 48 h following transfection and then prepared for subsequent assays.
Cell proliferation assay
Cell proliferation was assessed by Cell Counting Kit-8 (Dojindo Molecular Technologies, Inc., Kumamoto, Japan) assays. Cells were seeded at 1,000 cells/well into 96-well plates with 100 µl culture medium. Subsequently, 10 µl CCK-8 solution was added to the cells at every 24 h for 5 days and the cells were incubated for 2 h at 37°C. The reaction product was quantified according to the manufacturer's protocol by measuring the absorbance at 450 nm.
RNA and DNA extraction
Total RNA was extracted from cultured cells and tissue samples using TRIzol® reagent (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), according to the manufacturer's protocol. Genomic DNA was extracted from cultured cells and tissue samples using a High Pure PCR Template Preparation kit (Roche Diagnostics, Basel, Switzerland), according to the manufacturer's protocol.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
RT was performed with a mix of oligo dT primer and random primers for mRNA using a PrimeScript RT Reagent kit (Takara Biotechnology Co., Ltd., Dalian, China). qPCR was performed using a CFX96 Real-Time PCR detector (Bio-Rad Laboratories, Inc., Hercules, CA, USA) and SYBR Premix Ex Taq™ (Takara Biotechnology Co., Ltd., Dalian, China). The thermocycling conditions were: 95°C for 2 min; 40 cycles of 95°C for 20 sec, 60°C for 20 sec and 72°C for 20 sec. The comparative 2−ΔΔCq method was used to calculate fold changes (21). GAPDH was used as an endogenous reference. The primers used for qPCR were as follows: PTTG1IP forward, 5′-GTCTGGACTACCCAGTTACAAGC-3′ and reverse, 5′-CGCCTCAAAGTTCACCCAA-3′; GAPDH forward, 5′-GGAGTCCACTGGCGTCTTC-3′ and reverse, 5′-GCTGATGATCTTGAGGCTGTTG-3′. The experiment was performed in triplicate.
Reduced representation bisulfite sequencing (RRBS)
Genomic DNA was used to perform RRBS. RRBS library construction was performed as described previously (22). The library was sequenced on a next-generation sequencing (NGS) HiSeq platform (Illumina, Inc., San Diego, CA, USA). The sequencing data were aligned to a reference genome (UCSC hg19) using Bismark (a flexible aligner and methylation caller for Bisulfite-Seq applications) with default parameters. The methylation level of each cytosine was calculated using the R package MethylKit (version 1.0.0; http://code.google.com/p/methylkit), which is a comprehensive R package for analysis of genome-wide DNA methylation profiles (23).
Bisulfite amplicon sequencing
The DNA methylation level of the PTTG1IP promoter was analyzed in cells or tissue samples via bisulfite amplification followed by NGS. A total of 500 ng DNA was bisulfite treated using an EZ DNA Methylation Gold-kit (Zymo Research Corp., Irvine, CA, USA). The bisulfite-converted DNA was used to amplify the candidate fragment with a Takara EX Taq Hot Start Version kit (Takara Biotechnology Co., Ltd., Dalian, China). The PCR products were loaded on a 1.5% agarose gel for analysis and recovered for library construction and NGS using a MiSeq platform (Illumina, Inc., San Diego, CA, USA). The DNA methylation level of candidate fragments was determined by analyzing the NGS data. The primers for amplification were as follows: PTTG1IP forward, 5′-GTATTGTTGAAGGGTGTAGAGATG-3′ and PTTG1IP reverse, 5′-CCACCCACCAAAACTTAATAATTA-3′.
Statistical analysis
The statistical significance of mean values in a two-sample comparison was determined with Student's t-test. P<0.05 was considered to indicate a statistically significant difference. Data are presented as the mean ± standard error. The lung adenocarcinoma and lung squamous cell carcinoma data sets from The Cancer Genome Atlas (TCGA) were used to further validate the relationship between promoter methylation and gene expression of PTTG1IP. Gene expression data (RNASeq) and DNA methylation data (Illumina methylation beadchip HM450 K) from 456 lung adenocarcinoma samples and 370 squamous cell carcinoma samples were downloaded from TCGA database on the cBioportal website (www.cbioportal.org). Spearman's non-parametric correlation test was performed to evaluate the correlation between gene methylation and expression using R software (version 3.3.2; http://www.R-project.org) (24).
Results
Decreased PTTG1IP expression in early-stage non-small cell lung cancer
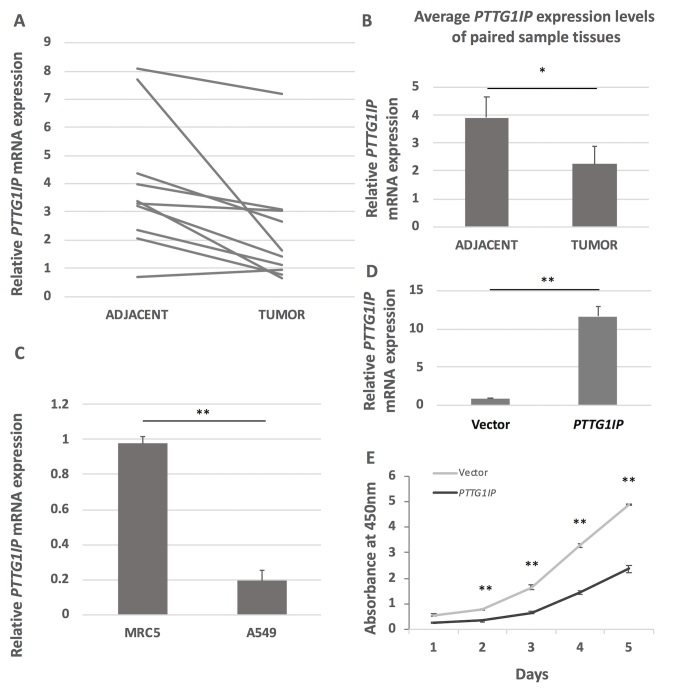
Although PTTG1IP has been reported to be abnormally expressed in a variety of tumor types (17–19,25), to the best of our knowledge, its association with lung cancer remains to be reported. The present study analyzed PTTG1IP expression in 10 paired early-stage NSCLC tissue samples (Table I). The RT-qPCR results revealed that PTTG1IP expression was decreased in all the cancer tissues except sample pair 10 (Fig. 1A). The mean mRNA level in the lung cancer tissues was significantly lower compared with that in the adjacent tissues (Fig. 1B). PTTG1IP expression was reduced by 80.1% in the lung cancer cell line A549 compared with the normal lung cell line MRC5 (Fig. 1C). To evaluate whether the expression level of PTTG1IP is associated with the proliferation capacity of lung cancer cells, PTTG1IP was overexpressed in A549 cells. The expression level of PTTG1IP was ~11 times higher in cells transfected with pcDNA3.1/3Xmyc-PTTG1IP compared with those transfected with empty pcDNA3.1 vector 2 days after transfection (Fig. 1D). A cell proliferation assay revealed that the proliferation of pcDNA3.1/3Xmyc-PTTG1IP-transfected cells was significantly inhibited compared with the control cells. By day five, the number of transfected cells was <50% of that in the control group (Fig. 1E).
Figure 1.
PTTG1IP expression is decreased in lung cancer and increased expression in a cancer cell line decreases cell proliferation. (A) The relative PTTG1IP mRNA expression levels in paired early-stage lung cancer tissue samples. (B) The mean PTTG1IP expression levels in paired tissue samples. P<0.05. (C) The relative PTTG1IP mRNA levels in normal lung cells (MRC5) and lung cancer cells (A549). **P<0.01. (D) PTTG1IP overexpression was achieved in A549 cells. (E) The proliferation of PTTG1IP-overxpressing cells and control cells was determined by Cell Counting Kit-8 assay at 24 h intervals over 5 days. Overexpression of PTTG1IP significantly decreased cell proliferation. *P<0.05, **P<0.01 vs. PTTG1IP. Data are presented as the mean ± standard error (n=3). PTTG1IP, pituitary tumor transforming gene 1 binding factor.
DNA methylation analysis of the PTTG1IP promoter
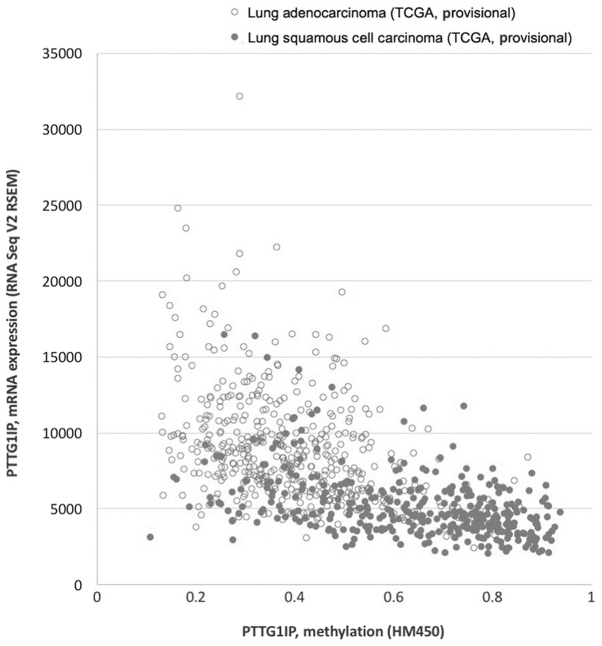
To investigate the regulatory mechanism driving the decreased expression of PTTG1IP in lung cancer, the present study first downloaded RNAseq data and DNA methylation chip 450k data of lung adenocarcinoma and lung squamous cell carcinoma from The Cancer Genome Atlas (TCGA) database on the cBioportal website (www.cbioportal.org/). Correlation analysis revealed a significant negative correlation between the PTTG1IP gene methylation level and mRNA level in both lung adenocarcinoma and lung squamous cell carcinoma, with Spearman correlation coefficients of −0.415 and −0.457, respectively (Fig. 2).
Figure 2.
Expression level of PTTG1IP is correlated with the DNA methylation level in lung adenocarcinoma and lung squamous cell carcinoma. The expression level is presented as reads per kilobase of exon model per million mapped reads from RNAseq data in TCGA database. The DNA methylation level in each sample is presented as the mean methylation level of all CpG sites within the PTTG1IP gene body based on HumanMethylation450 BeadChip (HM450) data in TCGA database. TCGA, The Cancer Genome Atlas; PTTG1IP, pituitary tumor transforming gene 1 binding factor.
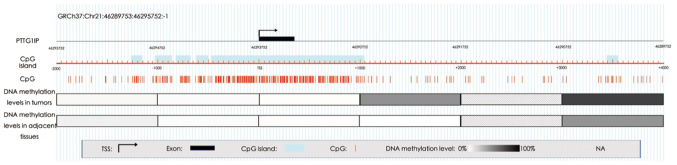
To further determine the association between PTTG1IP promoter methylation and gene expression, an RRBS study was conducted with six pairs of early-stage NSCLC tissue samples (Table I). As presented in Fig. 3, a plurality of CGIs were distributed among the 2,000 bp upstream and the 2,000 bp downstream of the PTTG1IP transcription start site (TSS). However, regional DNA methylation analysis demonstrated that the region from 2,000 bp upstream to 1,000 bp downstream of the TSS was hypomethylated both in tumor tissues and adjacent tissues, although CpG loci were very concentrated in this region. However, a difference was identified in the CGI shore region of 1,000-2,000 bp downstream of the TSS between lung cancer tissues and adjacent tissues, with an mean DNA methylation difference of 50%. In the region 5,000-6,000 bp downstream of the TSS, DNA was hypermethylated and the methylation level in cancer tissues was higher compared with that in adjacent tissues. Therefore, hypermethylation of the CGI shore region within the PTTG1IP gene promoter might be associated with its low expression.
Figure 3.
DNA hypermethylation is identified in the PTTG1IP promoter region. The location on chromosome 21, the distribution of the CpG islands and CpG sites in the PTTG1IP promoter are listed in the top half of the figure. The DNA methylation levels in tumors and adjacent tissues based on reduced representation bisulfite sequencing analysis are presented in grayscale. Darker colors indicate higher levels of methylation. PTTG1IP, pituitary tumor transforming gene 1 binding factor; TSS, transcription start site.
DNA methylation level of the CGI shore region within the PTTG1IP gene promoter is associated with PTTG1IP expression
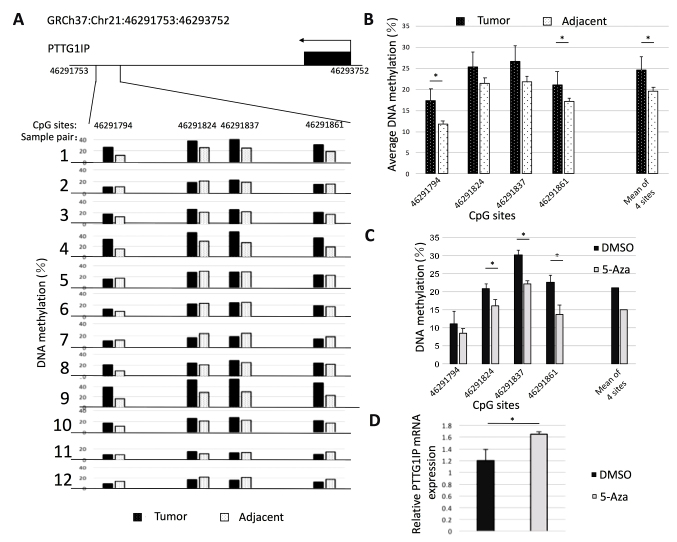
Subsequently, the methylation level of a fragment composed of four CG sites in the CGI shore region within the PTTG1IP promoter was measured in 12 pairs of early-stage NSCLC samples using bisulfite amplicon sequencing (Fig. 4A). Hypermethylation was identified in >50% of the cancer tissues in the sample pairs. As presented in Fig. 4B, the mean methylation level of the four CG loci in tumor tissues was higher compared with that in adjacent cancerous tissues. The mean methylation level of the four CG loci was 22.6 and 18.0%, respectively, and the difference was significant. The trend of these results was consistent with that observed in the RRBS analysis. To verify the association between DNA methylation and gene expression in this region, A549 cells were treated with 5-aza-2′-deoxycytidine (1 µM) to reduce DNA methylation levels. Following 48 h of treatment, a significant decrease in methylation of the three CG sites (except for site 46291794) was observed (Fig. 4C). The mean methylation level of the fragment was reduced from 21 to 15%. Furthermore, RT-qPCR revealed that PTTG1IP gene expression was significantly increased following treatment with 5-aza-2′-deoxycytidine (Fig. 4D). These results suggest that hypermethylation in the CGI shore within the PTTG1IP promoter is essential for silencing of PTTG1IP.
Figure 4.
Validation of the hypermethylation in the PTTG1IP promoter region and its association with gene expression. (A) The methylation level of CpG sites in the PTTG1IP promoter in paired tumor samples based on bisulfite amplicon sequencing. (B) The mean methylation level of CpG sites in the PTTG1IP promoter in paired tumor samples. (C) The methylation level of CpG sites in the PTTG1IP promoter and (D) the expression level of PTTG1IP in A549 cells following treatment with 1 µm 5-Aza. Data are presented as the mean ± standard error *P<0.05. PTTG1IP, pituitary tumor transforming gene 1 binding factor; 5-Aza, 5-aza-2′-deoxycytidine.
Discussion
The present study reported a negative correlation between PTTG1IP gene expression and the methylation level of its promoter region in lung cancer. In addition, it was identified that PTTG1IP was highly methylated in the early stage of lung cancer and exhibited a low expression level. Cytological experiments indicated that PTTG1IP overexpression may inhibit lung cancer cell proliferation. The present study provides a possible new mechanism for lung cancer development and a potential novel marker for early diagnosis of lung cancer.
The National Lung Screening Trial demonstrated a 20% reduction in lung cancer mortality using low-dose computed tomography (CT) screening (19). This survival benefit comes at the cost of testing numerous indeterminate pulmonary nodules, with an overall false-positive rate of 96.4% (26,27). One possible way to improve CT screening specificity is to use cancer-specific biomarkers from sputum and plasma. Previous studies have examined DNA methylation as a biomarker of cancer risk; however, the current low sensitivity and/or specificity of lung cancer screening is not sufficient (28–31). Epigenetic biomarkers, particularly DNA methylation, have become one of the most promising options for improving cancer diagnosis and have several advantages compared with other markers, including gene expression or genetic markers (32).
One surprising finding in cancer biology that has emerged from TCGA sequencing projects is the wide diversity of mutations that promote cancer development (33). DNA methylation changes are covalent modifications that are very stable and usually occur early in carcinogenesis. In addition, DNA methylation can be detected by a variety of sensitive and low-cost techniques, even in samples with low tumor cell purity (32). This epigenetic modification can also be detected in different biological fluids and is one of the most promising noninvasive cancer detection tools (32).
Previously, different epigenetic candidates have been proposed but have not yet reached clinical requirements, which is predominantly due to the fact that the majority of studies are based on a single candidate gene (34–38). For example, methylated CDKN2A, commonly referred to as p16, was an early focus in the search for diagnostic biomarkers in lung cancer plasma; however, although earlier studies identified CDKN2A promoter methylation in the plasma of patients with lung cancer (39–42), subsequent studies have described low sensitivity and specificity of this method (32,43,44). Methylated plasma CDKN2A may be used to detect lung cancer; however, it is more likely to be used as one part of a biomarker panel rather than as a single gene diagnostic marker. Other candidate genes include adenomatous polyposis coli (45,46), ras association domain family 1A gene (34,43,44,46,47), retinoic acid receptor β (43,44,46,48) and cadherin 13 (43,44,46); however, the sensitivity of these genes is generally low. The diagnostic firm Theracode identified short stature homeobox protein 2 as a potential biomarker (49); however, only 60% sensitivity (95% confidence interval, 53–67%) and 90% specificity (95% confidence interval, 84–94%) were identified (49). A multigene panel is a viable solution to the sensitivity and specificity concerns; however, more candidate genes need to be identified. Another consideration is that if early diagnosis of lung cancer requires a panel approach to assess plasma circulating tumor DNA, a panel with tumor type specificity is required, which requires a single gene methylation change in the panel or a combination of gene methylation changes indicating lung cancer. The present study demonstrated that PTTG1IP may be a new and specific gene that is aberrantly methylated in lung cancer.
PTTG1IP, also termed PBF, was originally reported to bind and promote the nuclear translocation of PTTG1 (50). PTTG1 is a marker of invasive colorectal cancer (51) and is a key signature gene associated with tumor metastasis (52). The functional interaction between PTTG1 and p53 has been demonstrated in transformed cells (53,54).
A number of studies have suggested that the subcellular localization of PTTG1IP and PTTG1 is crucial for progression of mitosis through the metaphase-anaphase transition (14,15,18). PTTG1IP promotes PTTG1 activation by promoting transfer of PTTG1 from the cytoplasm to the nucleus, thereby allowing the interaction between separase and PTTG1 (50). In addition to its role in metaphase/anaphase transition, PTTG1IP is also involved in transactivation of fibroblast growth factor 2 (50) and regulation of the human symporter in thyroid cells through its interaction with PTTG1 (55). However, to date, the full functionality of PTTG1IP has not been revealed.
PTTG1IP overexpression has been previously observed in certain types of malignancy, including thyroid (25), breast (53) and colorectal (52) cancer. However, to the best of our knowledge, PTTG1IP expression in other cancer types, including lung cancer, has not been reported. Expression data for all genes in lung adenocarcinoma, breast cancer, colorectal cancer, kidney cancer, melanoma, liver cancer and ovarian cancer (GSE1007, GSE20347, GSE32323, GSE6344, GSE3189, GSE14520 and GSE14407) were downloaded from the Gene Expression Omnibus database in NCBI. The ID_REF for PTTG1IP is 200677_at. The results of the analysis demonstrated that expression changes were not consistent among the tumor types, suggesting that PTTG1IP may perform different roles in different tumors (data not shown). Furthermore, it was revealed that the expression of PTTG1IP was regulated by the DNA methylation level. Further investigation demonstrated that DNA methylation at the shore of the CGI in the promoter region was negatively associated with PTTG1IP expression. More importantly, this region was hypermethylated in early-stage NSCLC. An appropriate gene methylation marker for early diagnosis of lung cancer may be a lung cancer-specific hypermethylated DNA site. Therefore, the unique performance of PTTG1IP in early-stage NSCLC suggests it can be used as an early biomarker for lung cancer diagnosis. Of course, prior to application in the clinic, further investigations are required to verify whether hypermethylation of the PTTG1IP promoter can be detected in body fluids, including sputum and plasma, from patients with early-stage NSCLC.
In conclusion, to the best of our knowledge, the present study investigated the expression of PTTG1IP in early-stage lung cancer for the first time. Low expression and promoter hypermethylation were identified. Furthermore, a negative correlation between PTTG1IP expression and methylation levels was revealed. These findings indicate that the methylation level of the PTTG1IP promoter region may be a candidate biomarker for early diagnosis of lung cancer.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- CT
computed tomography
- NGS
next-generation sequencing
- PTTG1IP
pituitary tumor transforming gene 1 binding factor
- RRBS
reduced representation bisulfite sequencing
- TCGA
The Cancer Genome Atlas
- TSS
transcription start site
Funding
The present study was supported by The National Natural Science Foundation of China (grant no. 81872103 and no. 81372768), Natural Science Foundation of Minhang District (grant no. 2015MHZ069), Training Program of Renji Hospital (grant no. 2017PYQA09) and the Science and Technology Climbing Fund of SIPPR (grant nos. PD2017-2 and PD2017-4).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
XT and HJ provided the samples. XT, SZ and HG performed the experiments. WH, MX and QW analyzed the data. XT and QW wrote the manuscript. XN and HJ designed and supervised the study and wrote the manuscript.
Ethics approval and consent to participate
All experimental protocols were approved by the Ethics Committee of South Hospital of Renji Hospital Shanghai Jiao Tong University School of Medicine (Shanghai, China). Written informed consent was obtained from each patient prior to participation.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 3.Anglim PP, Alonzo TA, Laird-Offringa IA. DNA methylation-based biomarkers for early detection of non-small cell lung cancer: An update. Mol Cancer. 2008;7:81. doi: 10.1186/1476-4598-7-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo S, Yan F, Xu J, Bao Y, Zhu J, Wang X, Wu J, Li Y, Pu W, Liu Y, et al. Identification and validation of the methylation biomarkers of non-small cell lung cancer (NSCLC) Clin Epigenetics. 2015;7:3. doi: 10.1186/s13148-014-0035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sardi AH, Islam S. Early lung cancer detection, mucosal, and alveolar imaging. Curr Opin Pulm Med. 2016;22:271–280. doi: 10.1097/MCP.0000000000000270. [DOI] [PubMed] [Google Scholar]
- 6.Yang IV, Schwartz DA. Epigenetic control of gene expression in the lung. Am J Respir Crit Care Med. 2011;183:1295–1301. doi: 10.1164/rccm.201010-1579PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belinsky SA. Gene-promoter hypermethylation as a biomarker in lung cancer. Nat Rev Cancer. 2004;4:707–717. doi: 10.1038/nrc1432. [DOI] [PubMed] [Google Scholar]
- 8.Ooki A, Maleki Z, Tsay JJ, Goparaju C, Brait M, Turaga N, Nam HS, Rom WN, Pass HI, Sidransky D, et al. A panel of novel detection and prognostic methylated DNA markers in primary non-small cell lung cancer and serum DNA. Clin Cancer Res. 2017;23:7141–7152. doi: 10.1158/1078-0432.CCR-17-1222. [DOI] [PubMed] [Google Scholar]
- 9.Walter K, Holcomb T, Januario T, Yauch RL, Du P, Bourgon R, Seshagiri S, Amler LC, Hampton GM, S Shames D. Discovery and development of DNA methylation-based biomarkers for lung cancer. Epigenomics. 2014;6:59–72. doi: 10.2217/epi.13.81. [DOI] [PubMed] [Google Scholar]
- 10.Bedford MT, van Helden PD. Hypomethylation of DNA in pathological conditions of the human prostate. Cancer Res. 1987;47:5274–5276. [PubMed] [Google Scholar]
- 11.Portela A, Liz J, Nogales V, Setién F, Villanueva A, Esteller M. DNA methylation determines nucleosome occupancy in the 5′-CpG islands of tumor suppressor genes. Oncogene. 2013;32:5421–5428. doi: 10.1038/onc.2013.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hao X, Luo H, Krawczyk M, Wei W, Wang W, Wang J, Flagg K, Hou J, Zhang H, Yi S, et al. DNA methylation markers for diagnosis and prognosis of common cancers. Proc Natl Acad Sci USA. 2017;114:7414–7419. doi: 10.1073/pnas.1703577114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 14.Read ML, Seed RI, Fong JC, Modasia B, Ryan GA, Watkins RJ, Gagliano T, Smith VE, Stratford AL, Kwan PK, et al. The PTTG1-binding factor (PBF/PTTG1IP) regulates p53 activity in thyroid cells. Endocrinology. 2014;155:1222–1234. doi: 10.1210/en.2013-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imruetaicharoenchoke W, Fletcher A, Lu W, Watkins RJ, Modasia B, Poole VL, Nieto HR, Thompson RJ, Boelaert K, Read ML, et al. Functional consequences of the first reported mutations of the proto-oncogene PTTG1IP/PBF. Endocr Relat Cancer. 2017;24:459–474. doi: 10.1530/ERC-16-0340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stratford AL, Boelaert K, Tannahill LA, Kim DS, Warfield A, Eggo MC, Gittoes NJ, Young LS, Franklyn JA, McCabe CJ. Pituitary tumor transforming gene binding factor: A novel transforming gene in thyroid tumorigenesis. J Clin Endocrinol Metab. 2005;90:4341–4349. doi: 10.1210/jc.2005-0523. [DOI] [PubMed] [Google Scholar]
- 17.Watkins RJ, Read ML, Smith VE, Sharma N, Reynolds GM, Buckley L, Doig C, Campbell MJ, Lewy G, Eggo MC, et al. Pituitary tumor transforming gene binding factor: A new gene in breast cancer. Cancer Res. 2010;70:3739–3749. doi: 10.1158/0008-5472.CAN-09-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Read ML, Seed RI, Modasia B, Kwan PP, Sharma N, Smith VE, Watkins RJ, Bansal S, Gagliano T, Stratford AL, et al. The proto-oncogene PBF binds p53 and is associated with prognostic features in colorectal cancer. Mol Carcinog. 2016;55:15–26. doi: 10.1002/mc.22254. [DOI] [PubMed] [Google Scholar]
- 19.Li C, Wang Y, Wang S, Wu B, Hao J, Fan H, Ju Y, Ding Y, Chen L, Chu X, et al. Hepatitis B virus mRNA-mediated miR-122 inhibition upregulates PTTG1-binding protein, which promotes hepatocellular carcinoma tumor growth and cell invasion. J Virol. 2013;87:2193–2205. doi: 10.1128/JVI.02831-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edge SB, Compton CC. The American Joint Committee on Cancer: The 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Gu H, Bock C, Boyle P, Gnirke A, Meissner A. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat Protoc. 2011;6:468–481. doi: 10.1038/nprot.2010.190. [DOI] [PubMed] [Google Scholar]
- 23.Akalin A, Kormaksson M, Li S, Garrett-Bakelman FE, Figueroa ME, Melnick A, Mason CE. methylKit: A comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012;13:R87. doi: 10.1186/gb-2012-13-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.R Core Team, corp-author. computing. R Foundation for Statistical Computing; Vienna, Austria: 2013. R: A language and environment for statistical. [Google Scholar]
- 25.Hsueh C, Lin JD, Chang YS, Hsueh S, Chao TC, Yu JS, Jung SM, Tseng NM, Sun JH, Kuo SY, Ueng SH. Prognostic significance of pituitary tumour-transforming gene-binding factor (PBF) expression in papillary thyroid carcinoma. Clin Endocrinol (Oxf) 2013;78:303–309. doi: 10.1111/cen.12007. [DOI] [PubMed] [Google Scholar]
- 26.National Lung Screening Trial Research Team. Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, Fagerstrom RM, Gareen IF, Gatsonis C, Marcus PM, Sicks JD. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011;365:395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tammemägi MC, Katki HA, Hocking WG, Church TR, Caporaso N, Kvale PA, Chaturvedi AK, Silvestri GA, Riley TL, Commins J, Berg CD. Selection criteria for lung-cancer screening. N Engl J Med. 2013;368:728–736. doi: 10.1056/NEJMoa1211776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brock MV, Hooker CM, Ota-Machida E, Han Y, Guo M, Ames S, Glöckner S, Piantadosi S, Gabrielson E, Pridham G, et al. DNA methylation markers and early recurrence in stage I lung cancer. N Engl J Med. 2008;358:1118–1128. doi: 10.1056/NEJMoa0706550. [DOI] [PubMed] [Google Scholar]
- 29.Leng S, Do K, Yingling CM, Picchi MA, Wolf HJ, Kennedy TC, Feser WJ, Baron AE, Franklin WA, Brock MV, et al. Defining a gene promoter methylation signature in sputum for lung cancer risk assessment. Clin Cancer Res. 2012;18:3387–3395. doi: 10.1158/1078-0432.CCR-11-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandoval J, Mendez-Gonzalez J, Nadal E, Chen G, Carmona FJ, Sayols S, Moran S, Heyn H, Vizoso M, Gomez A, et al. A prognostic DNA methylation signature for stage I non-small-cell lung cancer. J Clin Oncol. 2013;31:4140–4147. doi: 10.1200/JCO.2012.48.5516. [DOI] [PubMed] [Google Scholar]
- 31.Yang X, Dai W, Kwong DL, Szeto CY, Wong EH, Ng WT, Lee AW, Ngan RK, Yau CC, Tung SY, et al. Epigenetic markers for noninvasive early detection of nasopharyngeal carcinoma by methylation-sensitive high resolution melting. Int J Cancer. 2015;136:E127–E135. doi: 10.1002/ijc.29192. [DOI] [PubMed] [Google Scholar]
- 32.Heyn H, Esteller M. DNA methylation profiling in the clinic: Applications and challenges. Nat Rev Genet. 2012;13:679–692. doi: 10.1038/nrg3270. [DOI] [PubMed] [Google Scholar]
- 33.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Belinsky SA, Klinge DM, Dekker JD, Smith MW, Bocklage TJ, Gilliland FD, Crowell RE, Karp DD, Stidley CA, Picchi MA. Gene promoter methylation in plasma and sputum increases with lung cancer risk. Clin Cancer Res. 2005;11:6505–6511. doi: 10.1158/1078-0432.CCR-05-0625. [DOI] [PubMed] [Google Scholar]
- 35.Topaloglu O, Hoque MO, Tokumaru Y, Lee J, Ratovitski E, Sidransky D, Moon CS. Detection of promoter hypermethylation of multiple genes in the tumor and bronchoalveolar lavage of patients with lung cancer. Clin Cancer Res. 2004;10:2284–2288. doi: 10.1158/1078-0432.CCR-1111-3. [DOI] [PubMed] [Google Scholar]
- 36.Geng J, Sun J, Lin Q, Gu J, Zhao Y, Zhang H, Feng X, He Y, Wang W, Zhou X, Yu J. Methylation status of NEUROG2 and NID2 improves the diagnosis of stage I NSCLC. Oncol Lett. 2012;3:901–906. doi: 10.3892/ol.2012.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt B, Liebenberg V, Dietrich D, Schlegel T, Kneip C, Seegebarth A, Flemming N, Seemann S, Distler J, Lewin J, et al. SHOX2 DNA methylation is a biomarker for the diagnosis of lung cancer based on bronchial aspirates. BMC Cancer. 2010;10:600. doi: 10.1186/1471-2407-10-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nikolaidis G, Raji OY, Markopoulou S, Gosney JR, Bryan J, Warburton C, Walshaw M, Sheard J, Field JK, Liloglou T. DNA methylation biomarkers offer improved diagnostic efficiency in lung cancer. Cancer Res. 2012;72:5692–5701. doi: 10.1158/0008-5472.CAN-12-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bearzatto A, Conte D, Frattini M, Zaffaroni N, Andriani F, Balestra D, Tavecchio L, Daidone MG, Sozzi G. p16(INK4A) Hypermethylation detected by fluorescent methylation-specific PCR in plasmas from non-small cell lung cancer. Clin Cancer Res. 2002;8:3782–3787. [PubMed] [Google Scholar]
- 40.Kurakawa E, Shimamoto T, Utsumi K, Hirano T, Kato H, Ohyashiki K. Hypermethylation of p16(INK4a) and p15(INK4b) genes in non-small cell lung cancer. Int J Oncol. 2001;19:277–281. [PubMed] [Google Scholar]
- 41.An Q, Liu Y, Gao Y, Huang J, Fong X, Li L, Zhang D, Cheng S. Detection of p16 hypermethylation in circulating plasma DNA of non-small cell lung cancer patients. Cancer Lett. 2002;188:109–114. doi: 10.1016/S0304-3835(02)00496-2. [DOI] [PubMed] [Google Scholar]
- 42.Ng CS, Zhang J, Wan S, Lee TW, Arifi AA, Mok T, Lo DY, Yim AP. Tumor p16M is a possible marker of advanced stage in non-small cell lung cancer. J Surg Oncol. 2002;79:101–106. doi: 10.1002/jso.10046. [DOI] [PubMed] [Google Scholar]
- 43.Hsu HS, Chen TP, Hung CH, Wen CK, Lin RK, Lee HC, Wang YC. Characterization of a multiple epigenetic marker panel for lung cancer detection and risk assessment in plasma. Cancer. 2007;110:2019–2026. doi: 10.1002/cncr.23001. [DOI] [PubMed] [Google Scholar]
- 44.Wang YC, Hsu HS, Chen TP, Chen JT. Molecular diagnostic markers for lung cancer in sputum and plasma. Ann N Y Acad Sci 1075. 2006:179–184. doi: 10.1196/annals.1368.024. [DOI] [PubMed] [Google Scholar]
- 45.Usadel H, Brabender J, Danenberg KD, Jerónimo C, Harden S, Engles J, Danenberg PV, Yang S, Sidransky D. Quantitative adenomatous polyposis coli promoter methylation analysis in tumor tissue, serum and plasma DNA of patients with lung cancer. Cancer Res. 2002;62:371–375. [PubMed] [Google Scholar]
- 46.Rykova EY, Skvortsova TE, Laktionov PP, Tamkovich SN, Bryzgunova OE, Starikov AV, Kuznetsova NP, Kolomiets SA, Sevostianova NV, Vlassov VV. Investigation of tumor-derived extracellular DNA in blood of cancer patients by methylation-specific PCR. Nucleosides Nucleotides Nucleic Acids. 2004;23:855–859. doi: 10.1081/NCN-200026031. [DOI] [PubMed] [Google Scholar]
- 47.Ponomaryova AA, Rykova EY, Cherdyntseva NV, Skvortsova TE, Dobrodeev AY, Zav'yalov AA, Tuzikov SA, Vlassov VV, Laktionov PP. RARβ2 gene methylation level in the circulating DNA from blood of patients with lung cancer. Eur J Cancer Prev. 2011;20:453–455. doi: 10.1097/CEJ.0b013e3283498eb4. [DOI] [PubMed] [Google Scholar]
- 48.Ponomaryova AA, Rykova EY, Cherdyntseva NV, Skvortsova TE, Dobrodeev AY, Zav'yalov AA, Bryzgalov LO, Tuzikov SA, Vlassov VV, Laktionov PP. Potentialities of aberrantly methylated circulating DNA for diagnostics and post-treatment follow-up of lung cancer patients. Lung Cancer. 2013;81:397–403. doi: 10.1016/j.lungcan.2013.05.016. [DOI] [PubMed] [Google Scholar]
- 49.Kneip C, Schmidt B, Seegebarth A, Weickmann S, Fleischhacker M, Liebenberg V, Field JK, Dietrich D. SHOX2 DNA methylation is a biomarker for the diagnosis of lung cancer in plasma. J Thorac Oncol. 2011;6:1632–1638. doi: 10.1097/JTO.0b013e318220ef9a. [DOI] [PubMed] [Google Scholar]
- 50.Chien W, Pei L. A novel binding factor facilitates nuclear translocation and transcriptional activation function of the pituitary tumor-transforming gene product. J Biol Chem. 2000;275:19422–19427. doi: 10.1074/jbc.M910105199. [DOI] [PubMed] [Google Scholar]
- 51.Heaney AP, Singson R, McCabe CJ, Nelson V, Nakashima M, Melmed S. Expression of pituitary-tumour transforming gene in colorectal tumours. Lancet. 2000;355:716–719. doi: 10.1016/S0140-6736(99)10238-1. [DOI] [PubMed] [Google Scholar]
- 52.Carvalho L, Yu J, Schwartsmann G, McLeod HL, Fleshman JW. RNA expression of the molecular signature genes for metastasis in colorectal cancer. Oncol Rep. 2011;25:1321–1327. doi: 10.3892/or.2011.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bernal JA, Luna R, Espina A, Lázaro I, Ramos-Morales F, Romero F, Arias C, Silva A, Tortolero M, Pintor-Toro JA. Human securin interacts with p53 and modulates p53-mediated transcriptional activity and apoptosis. Nat Genet. 2002;32:306–311. doi: 10.1038/ng997. [DOI] [PubMed] [Google Scholar]
- 54.Kim DS, Fong J, Read ML, McCabe CJ. The emerging role of pituitary tumour transforming gene (PTTG) in endocrine tumourigenesis. Mol Cell Endocrinol. 2007;278:1–6. doi: 10.1016/j.mce.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 55.Boelaert K, Smith VE, Stratford AL, Kogai T, Tannahill LA, Watkinson JC, Eggo MC, Franklyn JA, McCabe CJ. PTTG and PBF repress the human sodium iodide symporter. Oncogene. 2007;26:4344–4356. doi: 10.1038/sj.onc.1210221. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.