Abstract
microRNAs (miRs) serve critical roles in tumor progression. Low expression of miR-125a in gastric carcinoma (GC) may promote tumor development. In the present study, low expression of miR-125a was confirmed in cancer tissues using The Cancer Genome Atlas database. Additionally, the expression and clinical significance of miR-125a-5p was investigated using reverse transcription-quantitative PCR in 150 cases of GC. The results of the present study demonstrated that the level of miR-125a-5p expression was decreased in GC biopsies compared with that in matched adjacent normal tissues. Low expression of miR-125a-5p was associated with increased tumor diameter, high Ki67 expression and poor overall survival of patients with GC. Multivariate survival analysis demonstrated that low miR-125a-5p expression may be used as an independent prognostic factor for patients with GC. However, no effects on the cell viability in a Cell Counting kit-8 assay, and cell migration and invasion in Transwell assays were detected in response to treatment using miR-125a-5p mimics or inhibitors in vitro. Therefore, the results of the present study provide evidence that low expression of miR-125a-5p may be associated with a poor prognosis, suggesting its value as a tumor biomarker for patients with GC.
Keywords: microRNA-125a, gastric cancer, survival, The Cancer Genome Atlas
Introduction
Gastric cancer (GC) is the third leading cause of cancer-associated mortality worldwide (1). According to the National Central Cancer Registry of China, GC is the second most common cause of cancer-associated mortality in China, accounting for ~498,000 deaths in 2015 (2). The majority of patients with GC (>80%) are diagnosed at an advanced stage, therefore leading to 5-year survival rates of <25% (3).
MicroRNAs (miRNAs/miRs) are short non-coding RNA molecules of 18–25 nucleotides in length. They serve important roles in the initiation and progression of human cancer and maybe used as promising therapeutic targets (4) It has been reported that several miRNAs are involved in cancer progression and diagnosis, and are associated with patient survival or drug-resistance in GC (5–9). For example, circulating levels of miR-627, miR-629 and miR-652 may be used as promising classifiers for GC, and greatly enhance the feasibility of circulating miRNAs as non-invasive diagnostic markers (10). Downregulation of miR-491-5p is associated with larger tumor size, and promotes GC metastasis by regulating SNAIL family transcriptional repressor and fibroblast growth factor receptor 4 (11). A three-miRNA signature (miR-145-3p, miR-125b-5p and miR-99a-5p) is significantly associated with the survival of patients with GC (12). miR-17-5p may induce cisplatin-resistance of GC cells by modulating apoptosis via targeting p21 (13). Therefore, examining the expression profile of miRNAs in GC is important.
In the present study, expression of the miR-125a/let-7e/miR-99b cluster was analyzed using The Cancer Genome Atlas (TCGA) database; it was shown that miR-125a expression was decreased in GC tissues compared with in adjacent normal tissues. Additionally, the results showed that reduced miR-125a-5p expression was associated with high tumor diameter and poor survival of patients with GC. However, inhibitory effects of miR-125a-5p on cancer cell proliferation, migration and invasion were not identified in vitro.
Materials and methods
TCGA database
Stomach adenocarcinoma (STAD) miRNA expression data was downloaded. A total of 43 pairs of matched tumor and adjacent normal tissues were included within the TCGA-STAD dataset and were obtained using SPSS (version 18.0.0; SPSS, Inc.) software based on the sample IDs [criteria for each pair of matched samples: i) The first 13 characters of each sample ID were the same; and ii) the last two characters of each sample ID represented either primary tumor (code: 01) or solid tissue normal (code: 11)] from the TCGA database (https://cancergenome.nih.gov/). Then, the normalized expression data (level 3) of the miR-125a/let-7e/miR-99b cluster according to miRNA-seq calculated using the log2 algorithm (tumor/normal) in the selected samples were obtained, and the statistical significance between tumor and adjacent normal tissues was evaluated using a paired t-test.
Tissue specimens
A total of 150 GC tissue specimens and 97 matched adjacent normal tissues were obtained from patients (mean age, 58.96±11.76 years; age range, 32–82 years; 109 males and 41 females) that underwent surgery at Peking University Cancer Hospital and Institute (Beijing, China) between December 2004 and October 2008. The following inclusion criteria were applied: i) Stomach adenocarcinoma; ii) without preoperative radiation or chemotherapy; and iii) with records of overall survival (OS) information. The present study was approved by the Ethics Committee of Peking University Cancer Hospital and Institute (no. 2017KT79). Written informed consent was obtained from all participants.
Cell culture
The NCI-N87, NUGC-3, SGC-7901, AGS, MGC-803 and BGC-823 human GC cell lines were obtained from the Cell Center of Peking Union Medical University (Beijing, China). All cells were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 mg/ml streptomycin at 37°C with 5% CO2.
Cell transfection with miR-125a-5p mimics or inhibitors
miRNA mimics or inhibitors for miRabse accession no. MIMAT0000443 were synthesized by Guangzhou RiboBio Co., Ltd., and transiently transfected into 1×105 cells at a 50 nmol/l concentration using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol for 24 h prior to subsequent experimentation. The mimics sequence (5′-UCCCUGAGACCCUUUAACCUGUGA-3′), inhibitors sequence (5′-TCACAGGUUAAAGGGTCTCAGGGA-3′) and negative control sequence (NC; 5′-UUCUCCGAACGUGUCACGUU-3′) were all designed as chemically modified double strands.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was isolated from tissues and cells using miRNeasy Mini kits (Qiagen, Inc.) according to the manufacturer's protocols. Poly(A) tails were added to total RNA 3′-ends (14,15). Subsequently, RNA was reverse transcribed with an oligodT-adaptor primer (5′-GCGAGCACAGAATTAATACGACTCACTATAGGTTTTTTTTTTTTVN-3′) and Moloney Murine Leukemia Virus Reverse Transcriptase (Invitrogen; Thermo Fisher Scientific, Inc.) for 1 h at 37°C. The following primers were used: miR-125a-5p, forward 5′-TGAGACCCTTTAACCTGTGA-3′ and reverse 5′-GCGAGCACAGAATTAATACGAC-3′; U6, forward 5′-CTCGCTTCGGCAGCACA-3′and reverse 5′-AACGCTTCACGAATTTGCGT-3′. The expression levels of miRNA were evaluated using SYBR™ Select Master Mix (Thermo Fisher Scientific, Inc.) and an ABI7500 fast System (Applied Biosystems; Thermo Fisher Scientific, Inc.). The cycling conditions were as follows: 2 min at 50°C and 10 min at 95°C followed by 40 cycles of 95°C for 3 sec and 60°C for 30 sec. U6 was used as the reference gene. The expression level of miR-125a-5p was calculated using the 2−ΔΔCq method, whereby ΔCq=Cq (miR-125a-5p)-Cq (U6) (16,17).
Cell viability assay
A total of 5×103 cells/well were seeded into 96-well plates and cultured for 24, 48, 72 and 96 h. Cell growth was evaluated using a Cell Counting Kit-8 (CCK8; Dojindo Molecular Technologies, Inc.), according to the manufacturer's protocol. Absorbance was measured at a wavelength of 450 nm using a microplate reader (iMark; Bio-Rad Laboratories, Inc.).
Transwell migration and invasion assays
Cells (1×104) were seeded into the upper chambers of the Transwell plate, with or without Matrigel, in 100 µl RPMI-1640 medium containing 1% FBS. A total of 500 µl RPMI-1640 medium containing 10% FBS was placed in each lower chamber as a chemoattractant. After a 24-h incubation at 37°C, migrated or invaded cells were fixed with 4% formaldehyde at room temperature for 5 min and stained with 1% crystal violet for 5 min at room temperature. The number of migrated or invasive cells was counted using a light microscope (magnification, ×20) in five randomly-selected microscopic fields.
Statistical analysis
Data were analyzed using SPSS 13.0 software (SPSS, Inc.). The relevant data are expressed as the mean ± standard deviation from three independent experiments. Statistical significance was determined for two groups using a two-tailed Student's t-test, and for multiple comparisons using one-way ANOVA with a Bonferroni post hoc test. A Mann Whitney U test was used for the comparison of two independent groups as a nonparametric test on ranked data. A two-tailed χ2 test was used to evaluate the association between miR-125a-5p expression and clinicopathological factors. OS was analyzed using the Kaplan-Meier method and a log-rank test. The cut-off value for miR-125a-5p expression was based on the receiver-operator curve (ROC) corresponding to the maximum Youden index. A univariate Cox analysis was performed followed by a multivariate Cox regression analysis including only variables which were significant in the univariate Cox analysis (18,19). Pearson's correlation analysis was conducted to determine the correlation between the level of miR-125a-5p expression and the migration or invasion ability in GC cell lines. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of miR-125a in GC tissues
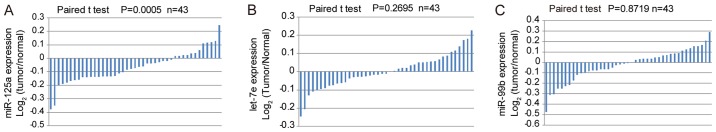
miR-125a/let-7e/miR-99b expression levels were analyzed in GC tissues based on data derived from the TCGA database. Even though the three miRNAs derive from the same precursor, only miR-125a exhibited low expression in GC tissues compared with adjacent normal tissues (Fig. 1A). There was no significant difference in the expression of let-7e or miR-99b between GC and matched adjacent normal tissues (Fig. 1B and C).
Figure 1.
Expression levels of miR-125a/let-7e/miR-99b cluster in patients with gastric carcinoma. miRNA expression data were downloaded from The Cancer Genome Atlas database. Expression levels of (A) miR-125a, (B) let-7e and (C) miR-99b were calculated using the log2 algorithm (tumor/normal). miR, microRNA.
Low expression of miR-125a-5p is associated with larger tumor diameter and poor survival of patients with GC
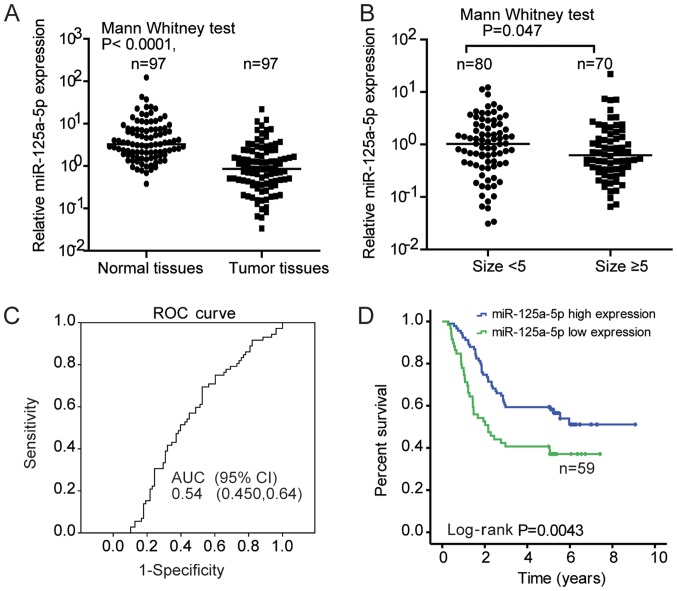
To further confirm the expression profile of miR-125a-5p, RT-qPCR was performed using 97 pairs of matched tumor and adjacent normal tissues. The results indicated significantly decreased expression levels of miR-125a-5p in GC tumor tissues compared with adjacent normal tissues (Mann Whitney U test; P<0.001; Fig. 2A). The expression level of miR-125a-5p was subsequently detected in 150 cases of patients with GC, and the association between miR-125a-5p expression and the clinicopathological parameters of patients with GC was evaluated. The average expression level of miR-125a-5p was decreased when the tumor diameter was ≥5 cm, compared with tumors of <5 cm diameter (1.491±2.929 vs. 1.778±2.278; P=0.0474; Fig. 2B). According to the cut-off value of the ROC curve corresponding to the maximum Youden index (Fig. 2C), patients were divided into high or low miR-125a expression groups. As shown in Table I, the number of patients with low miR-125a-5p expression was higher in if tumor size is ≥5 cm and Ki67 expression is high, indicating that low miR-125a-5p expression may be associated with tumor growth. However, there was no association between miR-125a-5p expression and other clinicopathological features, including sex, differentiation, TNM stage, lymph node metastasis, tumor embolus and distant metastasis (Table I). Nevertheless, the number of patients with low miR-125a-5p expression was higher in patients ≥60 years of age (Table I). The association between miR-125a-5p expression and aging requires further investigation.
Figure 2.
Association between reduced miR-125a-5p expression, tumor size and poor survival of patients with GC. (A) miR-125a-5p expression levels in GC vs. paired adjacent non-tumor tissue (n=97). (B) miR-125a-5p expression in tumor size <5 vs. >5 cm (n=80 vs. n=70; P=0.047). Lines in A and B indicate the median expression level in each group. (C) ROC curve analysis for miR-125a-5p expression in the 150 tumor tissue samples. (D) Overall survival analysis of miR-125a-5p expression in patients with GC using the Kaplan-Meier curve method. GC, gastric carcinoma; miR, microRNA; ROC, receiver-operator curve; AUC, area under curve; CI, confidence interval.
Table I.
Association between miR-125a-5p expression levels and clinical characteristics.
| microRNA-125a-5p | |||
|---|---|---|---|
| Characteristic | Low (%) | High (%) | P-value |
| Sex | 0.855 | ||
| Male | 55 (50.46) | 54 (49.54) | |
| Female | 20 (48.72) | 21 (51.22) | |
| Age (years) | 0.003 | ||
| <60 | 30 (38.46) | 48 (61.54) | |
| ≥60 | 45 (62.50) | 27 (37.50) | |
| Differentiation | 0.497 | ||
| Poor, undifferentiated | 62 (48.82) | 65 (51.18) | |
| Well, moderate | 13 (56.52) | 10 (43.48) | |
| Tumor size (cm) | 0.021 | ||
| <5 | 34 (42.50) | 46 (57.50) | |
| ≥5 | 43 (61.43) | 27 (38.57) | |
| TNM stage | 0.249 | ||
| I, II | 39 (45.88) | 46 (54.12) | |
| III, IV | 36 (55.38) | 29 (44.62) | |
| Lymph node metastasis | 0.402 | ||
| Negative (N0) | 16 (57.14) | 12 (42.86) | |
| Positive (N1, N2, N3) | 59 (48.36) | 63 (51.64) | |
| Tumor embolus | 0.570 | ||
| Negative | 32 (52.46) | 29 (47.54) | |
| Positive | 42 (47.73) | 46 (52.27) | |
| Metastasis | 0.206 | ||
| Negative | 50 (46.73) | 57 (53.27) | |
| Positive | 25 (58.14) | 18 (41.86) | |
| Ki67 | 0.032 | ||
| Negative | 0 (0.00) | 2 (100) | |
| <25% | 3 (23.08) | 10 (76.92) | |
| 25–50% | 9 (40.91) | 13 (59.09) | |
| >50% | 18 (66.67) | 9 (33.33) | |
The median expression value of miR-125a-5p in cancer tissues was used as the cut-off value to determine high and low groups.
Furthermore, the association between the expression level of miR-125a-5p and patient OS was assessed. Kaplan-Meier survival analysis revealed that patients with low miR-125a-5p expression exhibited a significantly shorter OS compared with those with high miR-125a-5p expression (Fig. 2D). Similarly, from the univariate analysis results of Cox proportional hazard model analysis, the higher tumor size, advanced TNM stage, and low miR-125a-5p expression are prognostic factors for survival (Table II). While using multivariate Cox's proportional hazard model analysis, it was identified that advanced TNM stage and low miR-125a-5p expression were independent risk factors for survival (Table II). Univariate Cox analysis also indicated an association between metastasis and survival for patients with GC (Table II; P<0.001). Considering that TNM contains metastasis as a component in this staging system, it was excluded in the Cox multivariate regression analysis. Patients with low miR-125a-5p expression exhibited a 2-fold increased risk of fatal outcome compared with those with high miR-125a-5p expression. Collectively, the data suggested that miR-125a-5p may be a potential prognostic biomarker for patients with GC.
Table II.
Univariate and multivariate Cox regression analysis of risk factors for patients with gastric carcinoma.
| Univariate Cox analysis | Multivariate Cox analysis | |||
|---|---|---|---|---|
| Variable | HR (95% CI) | P-value | HR (95% CI) | P-value |
| Sex (female vs. male) | 0.920 (0.553–1.530) | 0.748 | ||
| Age (≥60 vs. <60 years) | 1.184 (0.760–1.847) | 0.455 | ||
| Differentiation (poor vs. well) | 1.102 (0.607–2.000) | 0.749 | ||
| Ki67 (high vs. low) | 1.502 (0.190–11.892) | 0.610 | ||
| Metastasis (yes vs. no) | 3.200 (2.040–5.021) | <0.001 | ||
| Tumor size (≥5 vs. <5 cm) | 1.704 (1.090–2.660) | 0.019 | 1.286 (0.808–1.237) | 0.288 |
| TNM stage (III and IV vs. I and II) | 2.064 (1.655–4.098) | <0.001 | 2.550 (1.595–4.077) | <0.001 |
| microRNA-125a-5p (high vs. low) | 0.528 (0.338–0.825) | 0.005 | 0.497 (0.314–0.789) | 0.003 |
CI, confidence interval; HR, hazard ratio.
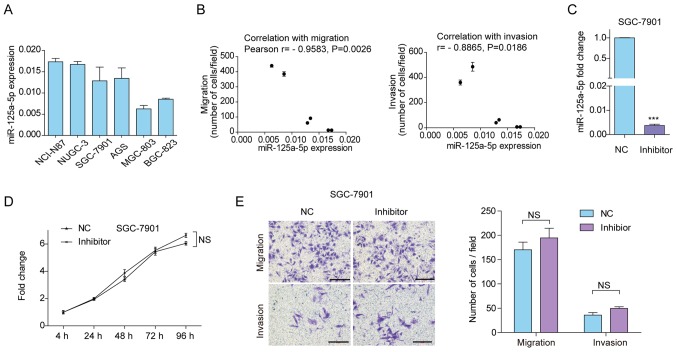
miR-125a-5p expression levels in GC cell lines
In order to determine suitable cell lines to perform functional assays to assess the effects of miR-125a-5p overexpression and inhibition, its expression was evaluated in six GC cell lines. Four GC cell lines, including NCI-N87, NUGC-3, SGC-7901 and AGS, had relatively high expression, while MGC-803 and BGC-823 had relatively low expression (Fig. 3A). Therefore, SGC-7901 cells were selected to assess the effects of miR-125a-5p silencing, and MGC-803 cells were selected to assess the effects of miR-125a-5p overexpression. Previously, we evaluated the migration and invasion ability of these gastric cancer cells by Transwell assays (20). The correlation between miR-125a-5p expression levels and migration or invasion ability was subsequently analyzed in GC cells. As indicated in Fig. 3B, the expression level of miR-125a-5p was negatively correlated with cell migration (r=−0.9583, P=0.0026) and invasion (r=−0.8865; P=0.0186) ability.
Figure 3.
Downregulation of miR-125a-5p expression in SGC-7901 cells. (A) Analysis of miR-125a-5p expression in six gastric carcinoma cell lines. (B) Scatter graph indicating the miR-125a-5p expression, migrated and invasive cell number of each cell line. (C) miR-125a-5p expression in SGC-7901 cells transfected with inhibitors or NC. ***P<0.001 vs. NC. (D) Analysis of cell proliferation using the Cell Counting Kit-8 assay. (E) Analysis of cell migration and invasion using the Transwell assay. Scale bar, 500 mm. Data are presented as the mean ± standard deviation. Three individual experiments with at least three replicates were performed. miR, microRNA; NC, negative control; NS, not significant.
Inhibition of miR-125a-5p fails to enhance the viability, migration and invasion of GC cells in vitro
As it was hypothesized that miR-125a-5p may act as a tumor suppressor in GC, the phenotypic changes were evaluated following the downregulation of miR-125a-5p by miRNA inhibitors in SGC-7901 cells with a relatively higher endogenous expression (Fig. 3A). RT-qPCR analysis demonstrated 99.6% transfection efficiency in response to treatment using miRNA inhibitors (Fig. 3C). However, there was no significant effect on cell viability in response to treatment using miR-125a-5p inhibitors as assessed using CCK8 assay (Fig. 3D). The results of the Transwell assay demonstrated no significant changes in the migratory or invasive ability of SGC-7901 cells transfected with miR-125a-5p inhibitors and NC (Fig. 3E). These results suggest that miR-125a-5p silencing has no effect on SGC-7901 cell proliferation, migration and invasion in vitro.
Ectopic overexpression of miR-125a-5p fails to inhibit cell viability, migration and invasion in vitro
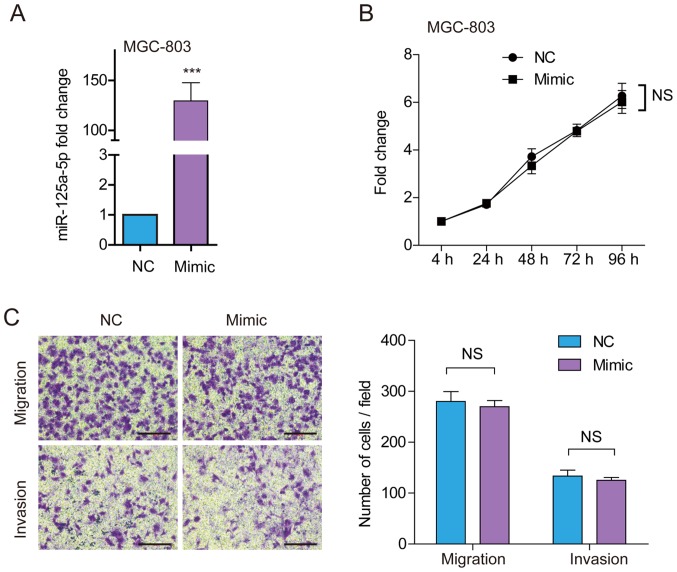
To further confirm the aforementioned results on viability and the migratory and invasive ability of GC cells, MGC-803 cells, which showed relatively lower endogenous miR-125a-5p expression among the six GC cell lines, were treated with miR-125a-5p mimics. RT-qPCR analysis demonstrated that miR-125a-5p expression was increased 129.4-fold in cells transfected with miR-125a-5p mimics (Fig. 4A). Consistently with the aforementioned results, there was no significant difference in cell viability (Fig. 4B), cell migration or invasion (Fig. 4C) in MGC-803 cells transfected with miR-125a mimics and NC. Therefore, miR-125a-5p overexpression has no effect on MGC-803 cell proliferation, migration and invasion in vitro.
Figure 4.
Treatment of MGC-803 cells using miR-125a-5p mimics. (A) Analysis of miR-125a-5p expression in MGC-803 cells transfected with miR-125a-5p mimics or NC. (B) Analysis of cell proliferation using the Cell Counting Kit-8 assay. (C) Analysis of cell migration and invasion using the Transwell assay. Scale bar, 500 mm. Data are presented as the mean ± standard deviation. Three individual experiments with at least three replicates were performed. ***P<0.001 vs. NC. miR, microRNA; NC, negative control; NS, not significant.
Discussion
The hsa-miR-125a/let-7e/miR-99b cluster is located within the 19q13.2–19q13.4 region (21). Further understanding of hsa-miR-125a/let-7e/miR-99b cluster biogenesis and function is required. In physiological conditions, high levels of all cluster members or miR-125a-5p are essential in maintaining the phenotype of mouse hematopoietic stem/progenitor cells (22). Furthermore, a single cluster member, miR-125a is sufficient to induce the amplification of hematopoietic stem cells, suggesting that miR-125a/let-7e/miR-99b cluster members differ in their expression patterns and functions, although they are derived from a common RNA precursor (23). Increased expression of these miRNAs is essential for osteoclast differentiation from primary human monocytes (24). These studies suggest the important role of miR-125a/let-7e/miR-99b cluster members in normal cell differentiation.
The miR-125a/let-7e/miR-99b cluster or its members may be involved in human pathological processes. miR-99b/let-7e/miR-125a cluster may promote metastasis in esophageal squamous cell carcinoma (ESCC) (25). Overexpression of this miRNA cluster was demonstrated to induce ESCC cell migration and invasion in vitro, and experimental metastasis in vivo (25). The miR-125a/let-7e/miR-99b cluster members were also shown to be upregulated in synovial sarcomas, however only two cluster members, let-7e and miR-99b, promoted cancer cell proliferation (26). Similarly, two cluster members, miR-99b and miR-125a, were previously implicated in the pathogenesis of cystic fibrosis (21). Decreased expression level of miR-125a, but not of miR-99b or let-7e, was identified in hepatocellular carcinoma biopsies compared with normal liver tissues, and predicted poor prognosis of patients with hepatocellular carcinoma (27). miR-125a-5p has been reported to be a potential tumor suppressor in lung carcinoma cells (28), retinoblastoma (29) and cervical carcinoma (30). All of these studies support that these miRNAs may have diverse functions in disease. In accordance with the aforementioned studies, the results of the present study demonstrated that the expression of miR-125a-5p was downregulated in GC tissues compared with adjacent normal tissues.
Consistent with previous research (31–34), the results of the present study identified decreased expression of miR-125a-5p in GC tissues compared with adjacent normal tissues, and decreased miR-125a-5p expression predicted poor OS in patients with GC. From the univariate analysis results of Cox proportional hazard model analysis, higher tumor size, advanced TNM stage and low miR-125a-5p expression were associated with reduced survival of patients with GC. Furthermore, advanced TNM stage and low miR-125a-5p expression were found to be independent prognostic factors associated with survival, as assessed using multivariate Cox's analysis, which is in accordance with a previous study (30).
Decreased expression levels of miR-125a-5p were reported to be associated with enhanced malignant potential, including tumor size, tumor invasion, liver metastasis and poor prognosis (32). In the present study, the association between low expression of miR-125a-5p and tumor size was established, however miR-125a-5p was not associated with distant metastasis. Differences in sample size or distribution may contribute to the outcomes of the study. Consistent with a previous study (27), the results of the present study demonstrated that low expression level of miR-125a-5p was associated with high Ki67 expression, a cellular marker for proliferation in GC; however, evaluation of Ki67 expression in cells transfected with miR125a-5p mimics or inhibitors remains to be investigated.
However, in vitro experiments demonstrated that miR-125a-5p mimics and inhibitors had no effect on GC cell proliferation, migration and invasion. These results differ from a previous report (34), where proliferation, migration and invasion of AGS and MGC-803 cells treated with pre-miR-125a were evaluated. Since miR-125a-3p, a known tumor suppressor in GC (35), derives from the 3′-arm of the same precursor (pre-miR-125a), these inhibitory effects on GC cell proliferation, migration and invasion may be mediated by miR-125a-3p, miR-125a-5p or even both. In the present study, MGC-803 cells were transfected with miR-125a-5p mimics (specifically altering miR-125a-5p expression) and SGC-7901 cells were transfected with miR-125a-5p inhibitors. However, there were no significant changes in cell proliferation, migration and invasion. Previously, it had been reported that low expression of miR-125a-5p was able to promote the paracrine vascular endothelial growth factor A signaling pathway in GC, while the latter increased Akt phosphorylation in endothelial cells and, thereby, promoted tumor angiogenesis (36). Therefore, even though there were no changes in GC cell proliferation, migratory and invasive ability in vitro, it is possible that miR-125a-5p functions as a tumor suppressor in vivo by regulating the tumor microenvironment, including effects on tumor angiogenesis. It is still possible that miR-125a-5p may regulate stemness (37), a critical feature of tumor formation, just like the tumor size. Therefore, in vivo experiments assessing the effect of miR-125a-5p on tumor growth and metastasis are required. TargetScan indicates that there are 3,991 candidate target genes for miR-125a-3p; however, which ones are real targets of miR-125a-5p requires further investigation using expression correlation and dual-luciferase reporter assays.
The probe sequences of miR-125a were checked in TGCA, which were matched to the chromosome 19 plus strand from 56888319-56888404. This indicates that the miR-125a expression data obtained from TCGA includes both miR-125a-3p and miR-125a-5p expression. Only the miR-125a-5p expression was analyzed in the cohort in the present study. The expression of miR-125a-3p expression in the GC tissues should be investigated in future studies.
Taken together, the results of the present study support that reduced miR-125a-5p expression is associated with larger tumor size, high Ki67 expression and poor prognosis in patients with GC. Therefore, miR-125a-5p may have prognostic value as a tumor biomarker for patients with GC.
Acknowledgements
Not applicable.
Funding
This work was supported by the grants from ‘San Ming’ Project of Shenzhen city, China (no. SZSM 201612051).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
GL performed most experiments, including the RNA extraction, quantitative PCR analysis and cell transfection. SA and JH participated in the in vitro study and acquired cell viability data. GQL designed the experiments, coordinated the project and wrote the manuscript. All authors have read and approved the final version of this manuscript.
Ethics approval and consent to participate
Written informed consents were obtained from all patients participated in the present study and this work was approved by the Peking University Cancer Hospital and Institute Ethical Committee (approval no. 2017KT79).
Patient consent for publication
Written informed consent for publication was obtained from all patients who participated in the present study.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Zong L, Abe M, Seto Y, Ji J. The challenge of screening for early gastric cancer in China. Lancet. 2016;388:2606. doi: 10.1016/S0140-6736(16)32226-7. [DOI] [PubMed] [Google Scholar]
- 4.Svoronos AA, Engelman DM, Slack FJ. OncomiR or Tumor suppressor? The duplicity of microRNAs in cancer. Cancer Res. 2016;76:3666–3670. doi: 10.1158/0008-5472.CAN-16-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsai MM, Wang CS, Tsai CY, Huang HW, Chi HC, Lin YH, Lu PH, Lin KH. Potential diagnostic, prognostic and therapeutic targets of MicroRNAs in human gastric cancer. Int J Mol Sci. 2016;17(pii):E945. doi: 10.3390/ijms17060945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.da Silva Oliveira KC, Thomaz Araujo TM, Albuquerque CI, Barata GA, Gigek CO, Leal MF, Wisnieski F, Rodrigues Mello Junior FA, Khayat AS, de Assumpção PP, et al. Role of miRNAs and their potential to be useful as diagnostic and prognostic biomarkers in gastric cancer. World J Gastroenterol. 2016;22:7951–7962. doi: 10.3748/wjg.v22.i35.7951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishiguro H, Kimura M, Takeyama H. Role of microRNAs in gastric cancer. World J Gastroenterol. 2014;20:5694–5699. doi: 10.3748/wjg.v20.i19.5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riquelme I, Letelier P, Riffo-Campos AL, Brebi P, Roa JC. Emerging role of miRNAs in the drug resistance of gastric cancer. Int J Mol Sci. 2016;17:424. doi: 10.3390/ijms17030424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu HH, Lin WC, Tsai KW. Advances in molecular biomarkers for gastric cancer: miRNAs as emerging novel cancer markers. Expert Rev Mol Med. 2014;16:e1. doi: 10.1017/erm.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shin VY, Ng EK, Chan VW, Kwong A, Chu KM. A three-miRNA signature as promising non-invasive diagnostic marker for gastric cancer. Mol Cancer. 2015;14:202. doi: 10.1186/s12943-015-0473-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu T, Wang LN, Li W, Zuo QF, Li MM, Zou QM, Xiao B. Downregulation of miR-491-5p promotes gastric cancer metastasis by regulating SNAIL and FGFR4. Cancer Sci. 2018;109:1393–1403. doi: 10.1111/cas.13583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang C, Zhang CD, Ma MH, Dai DQ. Three-microRNA signature identified by bioinformatics analysis predicts prognosis of gastric cancer patients. World J Gastroenterol. 2018;24:1206–1215. doi: 10.3748/wjg.v24.i11.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z, Ji F. Downregulation of microRNA-17-5p inhibits drug resistance of gastric cancer cells partially through targeting p21. Oncol Lett. 2018;15:4585–4591. doi: 10.3892/ol.2018.7822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi R, Chiang VL. Facile means for quantifying microRNA expression by real-time PCR. Biotechniques. 2005;39:519–525. doi: 10.2144/000112010. [DOI] [PubMed] [Google Scholar]
- 15.Wang S, Han H, Hu Y, Yang W, Lv Y, Wang L, Zhang L, Ji J. MicroRNA-130a-3p suppresses cell migration and invasion by inhibition of TBL1XR1-mediated EMT in human gastric carcinoma. Mol Carcinog. 2018;57:383–392. doi: 10.1002/mc.22762. [DOI] [PubMed] [Google Scholar]
- 16.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Yang D, Li R, Wang H, Wang J, Li Y, Wang H, Wang W, Liu Z. Clinical significance of tumor necrosis factor receptor 2 in middle and lower thoracic esophageal squamous cell carcinoma. Oncol Lett. 2018;16:2971–2978. doi: 10.3892/ol.2018.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoon J, Chung YE, Lim JS, Kim MJ. Quantitative assessment of mesorectal fat: New prognostic biomarker in patients with mid-to-lower rectal cancer. Eur Radiol. 2019;29:1240–1247. doi: 10.1007/s00330-018-5723-5. [DOI] [PubMed] [Google Scholar]
- 20.Wang S, Han H, Hu Y, Yang W, Lv Y, Wang L, Zhang L, Ji J. SLC3A2, antigen of mAb 3G9, promotes migration and invasion by upregulating of mucins in gastric cancer. Oncotarget. 2017;8:88586–88598. doi: 10.18632/oncotarget.19529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Endale Ahanda ML, Bienvenu T, Sermet-Gaudelus I, Mazzolini L, Edelman A, Zoorob R, Davezac N. The hsa-miR-125a/hsa-let-7e/hsa-miR-99b cluster is potentially implicated in Cystic Fibrosis pathogenesis. J Cyst Fibros. 2015;14:571–579. doi: 10.1016/j.jcf.2015.02.011. [DOI] [PubMed] [Google Scholar]
- 22.Gerrits A, Walasek MA, Olthof S, Weersing E, Ritsema M, Zwart E, van Os R, Bystrykh LV, de Haan G. Genetic screen identifies microRNA cluster 99b/let-7e/125a as a regulator of primitive hematopoietic cells. Blood. 2018;119:377–387. doi: 10.1182/blood-2011-01-331686. [DOI] [PubMed] [Google Scholar]
- 23.Guo S, Lu J, Schlanger R, Zhang H, Wang JY, Fox MC, Purton LE, Fleming HH, Cobb B, Merkenschlager M, et al. MicroRNA miR-125a controls hematopoietic stem cell number. Proc Natl Acad Sci USA. 2010;107:14229–14234. doi: 10.1073/pnas.0913574107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de la Rica L, Garcia-Gomez A, Comet NR, Rodríguez-Ubreva J, Ciudad L, Vento-Tormo R, Company C, Álvarez-Errico D, García M, Gómez-Vaquero C, Ballestar E. NF-κB-direct activation of microRNAs with repressive effects on monocyte-specific genes is critical for osteoclast differentiation. Genome Biol. 2015;16:2. doi: 10.1186/s13059-014-0561-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma J, Zhan Y, Xu Z, Li Y, Luo A, Ding F, Cao X, Chen H, Liu Z. ZEB1 induced miR-99b/let-7e/miR-125a cluster promotes invasion and metastasis in esophageal squamous cell carcinoma. Cancer Lett. 2017;398:37–45. doi: 10.1016/j.canlet.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 26.Hisaoka M, Matsuyama A, Nagao Y, Luan L, Kuroda T, Akiyama H, Kondo S, Hashimoto H. Identification of altered MicroRNA expression patterns in synovial sarcoma. Genes Chromosomes Cancer. 2011;50:137–145. doi: 10.1002/gcc.20837. [DOI] [PubMed] [Google Scholar]
- 27.Lu G, Ma Y, Jia C, Yang H, Xie R, Luo P, Chai L, Cai H, Cai M, Lv Z, et al. Reduced miR-125a levels associated with poor survival of patients with hepatocellular cancer. Oncol Lett. 2017;14:5952–5958. doi: 10.3892/ol.2017.6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhong L, Sun S, Shi J, Cao F, Han X, Chen Z. MicroRNA-125a-5p plays a role as a tumor suppressor in lung carcinoma cells by directly targeting STAT3. Tumour Biol. 2017;39:1010428317697579. doi: 10.1177/1010428317697579. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Xue C, Zhu X, Zhu X, Xian H, Huang Z. Suppression of microRNA-125a-5p upregulates the TAZ-EGFR signaling pathway and promotes retinoblastoma proliferation. Cell Signal. 2016;28:850–860. doi: 10.1016/j.cellsig.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 30.Qin X, Wan Y, Wang S, Xue M. MicroRNA-125a-5p modulates human cervical carcinoma proliferation and migration by targeting ABL2. Drug Des Devel Ther. 2015;10:71–79. doi: 10.2147/DDDT.S93104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao Y, Tan S, Tu Y, Zhang G, Liu Y, Li D, Xu S, Le Z, Xiong J, Zou W, et al. MicroRNA-125a-5p inhibits invasion and metastasis of gastric cancer cells by targeting BRMS1 expression. Oncol Lett. 2018;15:5119–5130. doi: 10.3892/ol.2018.7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishida N, Mimori K, Fabbri M, Yokobori T, Sudo T, Tanaka F, Shibata K, Ishii H, Doki Y, Mori M. MicroRNA-125a-5p is an independent prognostic factor in gastric cancer and inhibits the proliferation of human gastric cancer cells in combination with trastuzumab. Clin Cancer Res. 2011;17:2725–2733. doi: 10.1158/1078-0432.CCR-10-2132. [DOI] [PubMed] [Google Scholar]
- 33.Fassan M, Pizzi M, Realdon S, Balistreri M, Guzzardo V, Zagonel V, Castoro C, Mastracci L, Farinati F, Nitti D, et al. The HER2-miR125a5p/miR125b loop in gastric and esophageal carcinogenesis. Hum Pathol. 2013;44:1804–1810. doi: 10.1016/j.humpath.2013.01.023. [DOI] [PubMed] [Google Scholar]
- 34.Xu Y, Huang Z, Liu Y. Reduced miR-125a-5p expression is associated with gastric carcinogenesis through the targeting of E2F3. Mol Med Rep. 2014;10:2601–2608. doi: 10.3892/mmr.2014.2567. [DOI] [PubMed] [Google Scholar]
- 35.Hashiguchi Y, Nishida N, Mimori K, Sudo T, Tanaka F, Shibata K, Ishii H, Mochizuki H, Hase K, Doki Y, Mori M. Down-regulation of miR-125a-3p in human gastric cancer and its clinicopathological significance. Int J Oncol. 2012;40:1477–1482. doi: 10.3892/ijo.2012.1363. [DOI] [PubMed] [Google Scholar]
- 36.Dai J, Wang J, Yang L, Xiao Y, Ruan Q. miR-125a regulates angiogenesis of gastric cancer by targeting vascular endothelial growth factor A. Int J Oncol. 2015;47:1801–1810. doi: 10.3892/ijo.2015.3171. [DOI] [PubMed] [Google Scholar]
- 37.Chen J, Ouyang H, An X, Liu S. miR-125a is upregulated in cancer stem-like cells derived from TW01 and is responsible for maintaining stemness by inhibiting p53. Oncol Lett. 2019;17:87–94. doi: 10.3892/ol.2018.9587. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.