Abstract
The development of an effective HIV vaccine continues to be a major health challenge since, so far, only the RV144 trial has demonstrated a modest clinical efficacy. Recently, the targeting of the 12 highly conserved protease cleavage sites (PCS1–12) has been presented as a strategy seeking to hamper the maturation and infectivity of HIV. To pursue this line of research, and because peptide antigens have low immunogenicity, we have included these peptides in engineered nanoparticles, aiming at overcoming this limitation. More specifically, we investigated whether the covalent attachment of a PCS peptide (PCS5) to polysaccharide-based nanoparticles, and their coadministration with polyinosinic:polycytidylic acid (poly(I:C)), improved the generated immune response. To this end, PCS5 was first conjugated to two different polysaccharides (chitosan and hyaluronic acid) through either a stable or a cleavable bond and then associated with an oppositely charged polymer (dextran sulfate and chitosan) and poly(I:C) to form the nanoparticles. Nanoparticles associating PCS5 by ionic interactions were used in this study as the control formulation. In vivo, all nanosystems elicited high anti-PCS5 antibodies. Nanoparticles containing PCS5 conjugated and poly(I:C) seemed to induce the strongest activation of antigen-presenting cells. Interestingly, T cell activation presented different kinetics depending on the prototype. These findings show that both the nanoparticle composition and the conjugation of the HIV peptide antigen may play an important role in the generation of humoral and cellular responses.
Keywords: HIV vaccine, peptide antigen, polysaccharide, nanovaccine, poly(I:C), nanoparticle, antigen encapsulation
With almost 2 million newly infected individuals per year, HIV continues to be one of the most important global health challenges.1 Despite the efforts dedicated to the discovery of an effective vaccine against this virus, the most positive results achieved so far are those reported in the RV144 vaccine (Phase III clinical trial), which conferred a modest 30% protection against the infection.2 This protection was associated with the ability of the vaccine to generate weakly neutralizing and non-neutralizing antibody responses.3 It is also worth noting that the efficacy of this trial declined from 60 to 30% from 42 to 60 months after administration, which further underscores the need for HIV vaccine designs that would efficiently induce protective and long-lasting immune responses. This efficiency depends, to a great extent, on the adequate combination of antigen and adjuvant.4,5
Considering how the natural immunity to HIV-1 works, the peptide sequences around the protease cleavage sites (PCS) have been proposed as targets for a candidate vaccine against HIV. The protease is an essential enzyme for the HIV virus because its function is to cleave specific proteins, such as Gag, Gag-Pol, and Nef, that are essential for the maturation of HIV virions and, thus, crucial to its infectivity.6−8 The proteolysis of these proteins must take place on the 12 cleavage sites in a controlled and sequential way, and the interruption of this process, even in one single site, may interrupt virus maturation and, therefore, stop virus infection.6,7,9 In addition, these sites are highly conserved among HIV-1 viruses, which could help overcome the problems associated with the HIV virus high-sequence diversity and rapid mutation rates.
A wide variety of nanosystems are currently under evaluation for their ability to deliver vaccine immunogens to protect the antigens from degradation and facilitate their internalization by the target immune cells.10 In addition, it has been reported that the response to antigen-loaded nanocarriers may be influenced by their composition and physicochemical properties.10−12 Among these nanocarriers, chitosan (CS)-based nanosystems have been widely employed as antigen carriers through both parenteral and mucosal routes.13 In fact, CS is a biodegradable and biocompatible polymer, with a FDA GRAS status, which has already been approved for dietary use and as a wound-dressing material.14,15 Our group has originally reported the development of CS nanoparticles (NPs)16 and nanocapsules (NCs)17 and assessed their potential to generate humoral responses against different antigens, such as the recombinant hepatitis B surface antigen (rHBsAg), the tetanus toxoid, and IutA antigen from E. coli.18−21 Other studies have also shown promising results when using CS-based nanosystems in order to enhance T cell responses.22,23
A critical aspect that has not been completely elucidated is the influence of the antigen–nanocarrier interaction on the nature of the immune response generated. In some studies, the association of antigens to nanocarriers has relied on simple physical entrapment techniques or the establishment of ionic or hydrophobic forces,24−26 whereas in others the association involved the chemical conjugation of the antigens to the nanosystems.27−37 In the later studies, chemical conjugation was shown to increase the humoral response when compared to other types of antigen association.31,32,36,37 More specifically, a study performed for a malaria antigen showed that these improved humoral responses were caused by an increased interaction of the antigen with the B-cell receptor when the antigen was covalently attached to the surface of lipid NCs.32 Similarly, the covalent attachment of an HIV protein antigen to virus-like particles resulted in an efficient generation of the specific humoral and CD8+ T-cell responses.30
On the other hand, over the past decade it has become evident that the coencapsulation of antigens and adjuvants, such as the Toll-like receptor (TLR) agonists, further improves the performance of antigen-loaded nanosystems in vaccination.10 Furthermore, it has been reported that these TLR molecules are able to direct the immune response toward either humoral or cellular profiles.20,26,38
On the basis of this background information, the primary objective of this work was to evaluate if the chemical linkage of an SIV PCS peptide antigen (PCS5) to polysaccharide NPs and the inclusion of poly(I:C), could improve its immunogenicity over the association by ionic forces to NPs. We hypothesized that the conjugation of the antigen to the polysaccharides might prevent its release until the particles are engulfed and processed by antigen presenting cells (APCs), thereby dually improving and prolonging the induced immune responses. CS and hyaluronic acid (HA) were the polysaccharides of choice to conjugate the peptide antigen PCS5. In addition, the TLR3 agonist poly(I:C) was included in these two nanosystems to stimulate a cellular response against the antigen. As the control formulation, PCS5 was entrapped by ionic forces in NPs of CS and dextran sulfate (DS), which were previously reported as good antigen carriers for PCS peptide antigens.39 Polymer–PCS5 conjugates and the resulting NPs were characterized in terms of their chemical composition and structural organization. Finally, their capacity to activate the immune system, regarding antibody production, T cell and APCs activation, and generation of central and effector memory T cells in mice, was studied.
Results and Discussion
Developing an effective HIV vaccine has proved to be particularly challenging due to the special characteristics of this virus. On the one hand, the virus infects CD4+ T cells, which have an important role in the development of the adaptive immune response. On the other hand, it mutates rapidly, leading to a deleterious activation of the immune system, which results in the generation of ineffective humoral and cellular responses. In addition, the different virus strains are highly variable, a fact that complicates the generation of broad and protective immune responses.5 Our approach for the design of an HIV vaccine has been to use the peptide sequences that overlap the 12 HIV PCS as antigens. For the generation of infective virions, these HIV PCS sites have to be cleaved sequentially; thus, if a single proteolytic reaction is disrupted, the whole process could fail. An additional advantage of the PCS peptide antigens relies on the fact that these PCS are highly conserved regions among HIV-1 viruses, and therefore, the mutation of the virus will not diminish the activity of the vaccine.6−9 In a prior approach, these peptide antigens were entrapped into CS/DS NPs and administered to nonhuman primates, which had already been primed with the PCS1–12 encoding plasmids cloned into viral vectors. The results showed a significant increase in the IgG responses against PCS after nasal boosting with the nanoparticles.39
Based on these preliminary findings, we sought to further improve the immunogenicity of our PCS antigens by exploring different NPs-based formulation strategies to deliver the peptide antigen. For this purpose, the PCS5 antigen was selected for its association to the nanosystems, based on the premise that one cleavage error could interrupt virus maturation. PCS5 was first conjugated to two different polysaccharides, CS and HA, and the resulting conjugate was used to form NPs by ionic interaction with an oppositely charged polymer (CS–PCS5/DS and CS/HA–PCS5 NPs). The TLR3 agonist, poly(I:C), was also associated with these NPs in order to evaluate its effect on the cellular immune response generated by the NPs. The already reported CS/DS NPs with PCS5 physically entrapped were used as the control formulation. These strategies were based on the hypothesis that conjugation of PCS5 peptide antigen to the matrix-forming polymer would facilitate the intracellular delivery of the antigen with the subsequent improvement of the humoral and cellular immune responses. In addition, poly(I:C) was expected to boost the activation of APCs, effector and memory T cells.
Development of Peptide-Loaded Polysaccharide NPs
Polysaccharides, particularly CS, have attracted significant attention as biomaterials for the design of antigen delivery carriers.13,40,41 Our group has originally reported the development of NPs and NCs made of CS16,17 for the delivery of a variety of antigens (e.g., tetanus toxoid, rHBsAg, influenza, E. coli antigen, and also peptide antigens) through different routes of administration.18,19,24,42,43 Other authors have also disclosed the utility of nanosystems made of DS13,44−46 or HA,47−52 both negatively charged polysaccharides, to induce immune responses. In most of these reports, the association of the antigen to the polymeric nanocarrier was based on a simple entrapment process, normally driven by ionic interactions between the polymers and the antigen. However, in this work our objective was to covalently conjugate the PCS5 peptide antigen to the NPs in order to achieve a more sustained and improved presentation of the antigen to the immune cells.
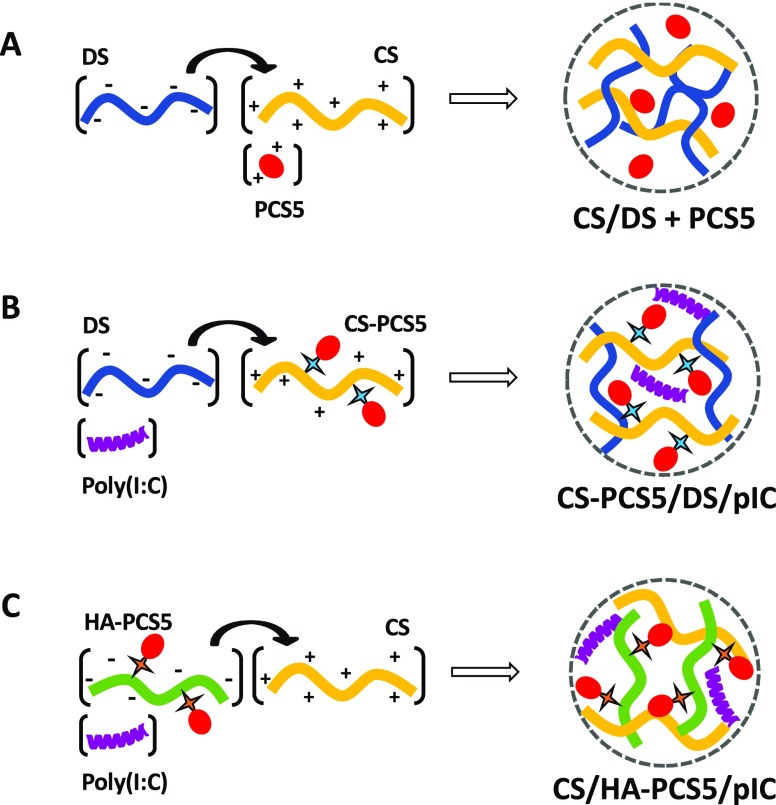
Apart from selecting the biomaterials to form the antigen nanocarriers, we also chose an adjuvant, the TLR3 agonist poly(I:C). This molecule mimics viruses dsRNA and activates TLR3 inducing robust cytokine and chemokine responses.43,53−55 With these components, we designed three different nanosized systems, where the antigenic peptide PCS5 was either (i) entrapped into the CS/DS NPs, as the control formulation, (ii) covalently linked to CS, or (iii) covalently linked to HA; prior to the formation of the NPs by adjusting the ionic interaction of CS with DS or HA (Figure 1).
Figure 1.
Composition of the different prototypes developed in this work. Schematic representation of the preparation process for the three developed nanosystems. (A) CS/DS + PCS5 NPs, (B) CS–PCS5/DS/pIC NPs, and (C) CS/HA–PCS5/pIC NPs. Key: CS, chitosan; DS, dextran sulfate; PCS5, protease cleavage site 5; NPs, nanoparticles; pIC, poly(I:C) (polyinosinic:polycytidylic acid); HA, hyaluronic acid.
Preparation of CS/DS + PCS5 NPs
Using the preparation method for CS/DS NPs previously described by our laboratory,39 PCS5 was easily entrapped within CS/DS nanoparticles by simply adjusting the ionic interaction of the cationic peptide (PCS5) and the cationic polysaccharide (CS) with the negatively charged polysaccharide (DS) (Figure 1A). The mass ratio 1:3 CS:DS resulted in adequate physicochemical properties, association efficiency (80 ± 8%), and antigen loading (8%). This apparent excess of DS was required to complex efficiently the cationic peptide. The resulting NPs had a particle size of approximately 120 nm, a low polydispersity, and a negative ζ-potential (Table 1).
Table 1. Physicochemical Characterization of the Three PCS5-Loaded Nanosystemsa.
| nanosystems | particle size (nm) | PDI | ζ-potential (mV) | antigen loading (%) | pIC loading (%) |
|---|---|---|---|---|---|
| CS/DS + PCS5 | 119 ± 7 | 0.15 | –50 ± 3 | 7.9 | N/A |
| CS–PCS5/DS/pIC | 141 ± 6 | 0.17 | +29 ± 3 | 4.9 | 0.3 |
| CS/HA–PCS5/pIC | 211 ± 15 | 0.07 | +30 ± 1 | 4.6 | 0.05 |
Mean ± S.D., n ≥ 3. Key: CS, chitosan; DS, dextran sulfate; PCS5, protease cleavage site 5; NPs, nanoparticles; pIC, poly(I:C); HA, hyaluronic acid; PDI, polydispersity index.
Preparation of CS–PCS5/DS/pIC NPs
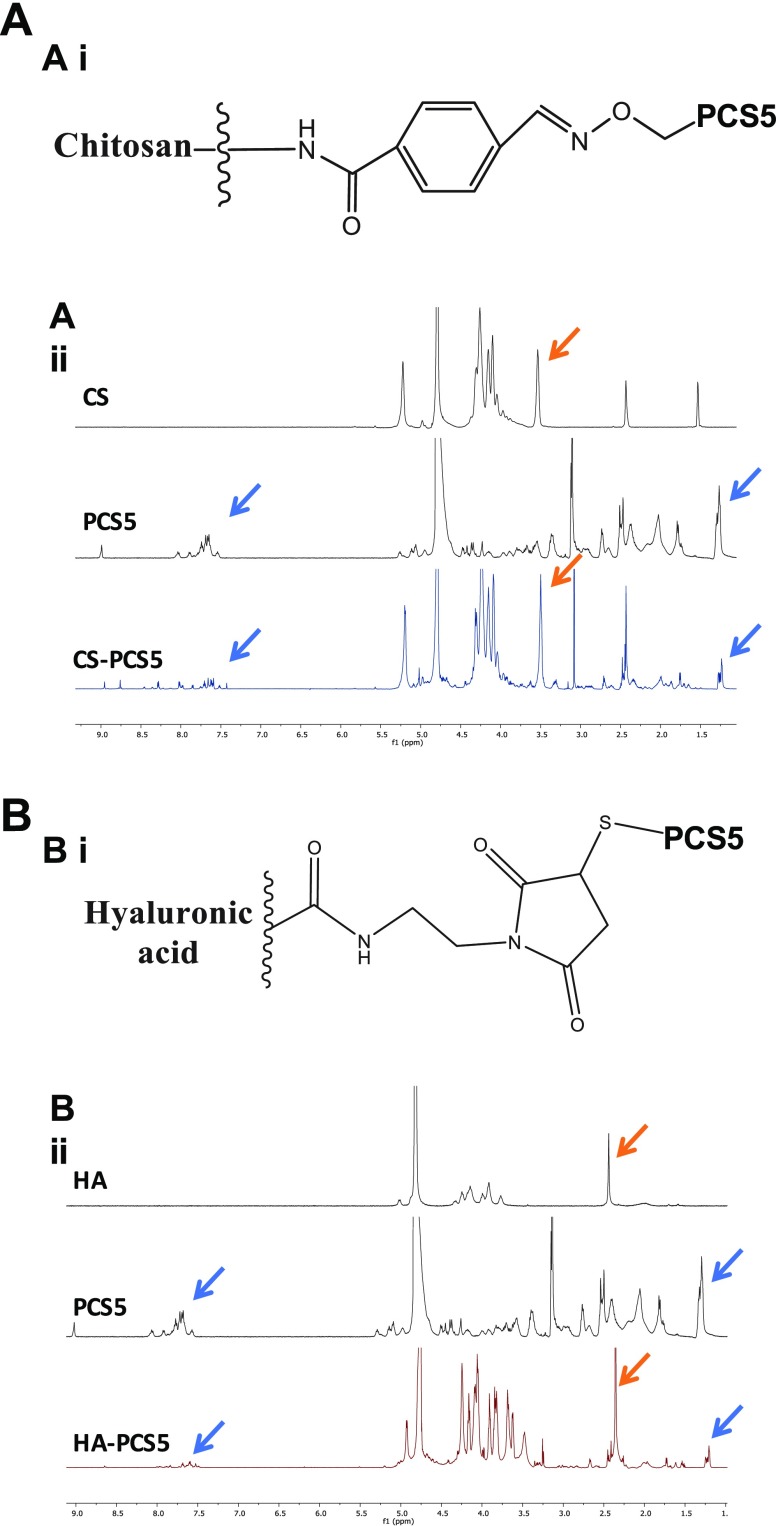
PCS5 peptide was conjugated to CS by an oxime bond, as schematically represented in Figure 2Ai. 1H NMR analysis led to the identification of the signal peaks of the aromatic rings and isopropyl groups of PCS5 (in blue), together with specific signal peaks of CS’s glucosamine units (orange arrows), which confirms the chemical conjugation of peptide and polymer. The substitution degree, assessed using a colorimetric assay, was of 2.6 mol of PCS5 per mol of CS (14% in weight).
Figure 2.
Chemical characterization of polymer–PCS5 conjugates. (A) CS–PCS5 final conjugate, (i) schematically represented, (ii) 1H NMR of CS, PCS5, and CS–PCS5 conjugate. Blue arrows indicate the signals from the aromatic rings and isopropyl groups of PCS5 and orange arrows the glucosamine units of CS, both observed in the NMR of the conjugate. (B) HA–PCS5 conjugate (i) schematically represented, (ii) 1H NMR of HA, PCS5, and HA–PCS5 conjugate. These results corroborated the presence of both the PCS5 characteristic signals (blue arrows) and the acetyl groups of HA (orange arrows) in the conjugate. Key: CS, chitosan; PCS5, protease cleavage site 5; HA, hyaluronic acid.
The CS–PCS5 conjugate was then used to form NPs through ionic interaction with DS. In addition, poly(I:C) was also incorporated into the NPs to increase its immunostimulatory properties. NPs with adequate final antigen loading (5%), and poly(I:C) loading (0.3%) could be obtained when the CS:DS (w/w) ratio was 2:1 (Figure 1B). The resulting NPs had a particle size of 140 nm, with low polydispersity and positive surface charge (Table 1). This positive charge was attributed to a certain excess of CS conjugated with the peptide, since increasing amounts of DS caused a decrease in the surface charge (Supporting Information, Figure S1A).
Preparation of CS/HA–PCS5/pIC NPs
The conjugation of PCS5 to HA was achieved through the formation of a thioether bond, which has been reported to be cleaved in glutathione-rich environments, such as the cytosol.56 In this case, thiol–maleimide chemistry was employed to link PCS5 to HA (Supporting Information, Figure S2). The results of the 1H NMR and DOSY analyses allowed us to evaluate the peptide conjugation reaction and its yield. As shown in Figure 2Bii, the NMR spectrum of the conjugate exhibits the characteristic peaks of PCS5 (blue arrows), and those corresponding to the acetyl groups of HA (orange arrows). The substitution degree calculated by NMR was of 3.8 mol of PCS5 per mol of HA (15.9% in weight). On the other hand, DOSY analysis showed a diffusion coefficient for the conjugate of 9.5 × 10–8 cm2/s, similar to the one for HA (8.8 × 10–8 cm2/s) and much lower than the one observed for the free PCS5 (8.4 × 10–7 cm2/s) (Supporting Information, Figure S3), confirming the conjugation of the peptide to the polymer.
Prior to the formation of the NPs using the HA–PCS5 conjugate, a screening of blank CS/HA NPs was conducted to select the most adequate polymer ratio combination (Supporting Information, Figure S1B). A surface charge switch from positive to negative values was observed when the mass ratio CS:HA decreased. For this reason, a mass ratio CS/HA 1:1 was selected to form these NPs, since their positive surface charge was expected to allow the association of the negatively charged poly(I:C) to the system (Figure 1C). The final NPs had a particle size of approximately 200 nm and a positive surface charge of +30 mV (Table 1). Adequate loading values for PCS5 (5%) and poly(I:C) (0.05%) were also achieved (Table 1).
Morphological Analysis of the Nanosystems
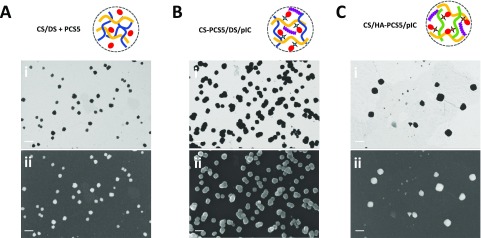
Field emission scanning electron microscopy (FESEM) images from the NPs were taken for each nanosystem using the STEM and InLens detectors (Figure 3). In all cases, a spherical particle shape could be observed, as well as diameters similar to the ones reported by DLS analysis (Table 1).
Figure 3.
Morphological characterization of the developed nanosystems. Field emission scanning electron microscopy (FESEM) images of (A) CS/DS + PCS5 NPs, (B) CS–PCS5/DS/pIC NPs, and (C) CS/HA–PCS5/pIC NPs with the (i) STEM and (ii) InLens detectors. Scale bar: 200 nm. Key: CS, chitosan; DS, dextran sulfate; PCS5, protease cleavage site 5; pIC, poly(I:C) (polyinosinic:polycytidylic acid); HA, hyaluronic acid; STEM, scanning transmission electron microscopy; InLens, immersion lens.
Structural Distribution of PCS5 in CS/HA–PCS5/pIC NPs
XPS analysis was also used to characterize the surface composition of this nanocarrier. The amount of sulfur present on CS/HA–PCS5/pIC NPs (1.15%) was higher than the one observed in blank CS/HA/pIC NPs (0.28%), which indicates that PCS5 (2.05% of sulfur) must be present on the external layers of the NPs (Supporting Information, Table S1). Similarly, the ratios C/O and C/N determined for CS/HA–PCS5/pIC NPs (3.19 and 9.31, respectively) were closer to those of PCS5 (3.50 and 3.66) than to those observed in the blank CS/HA/pIC NPs (1.87 and 12.86), further confirming the prevalent presence of PCS5 on the surface of the NPs.
On the other hand, the binding energy of an element is very sensitive to its chemical environment, and it can be used as the fingerprint of a compound. Thus, we compared the required energy to extract one electron from the carbon 1s orbital of PCS5, CS/HA/pIC, and CS/HA–PCS5/pIC NPs (Supporting Information, Figure S4). The obtained profiles indicated that the binding energies of PCS5 alone and CS/HA–PCS5/pIC were very similar, as additional proof that PCS5 was present on the surface of the NPs. This structural organization could facilitate the presentation of the antigen to B cells, and increase the humoral response against it, as already reported for similar antigen presentations.32,34
PCS5 Release When Covalently Attached to NPs
In the present work, we also compared the influence of different covalent attachments in the generation of the immune response. In this regard, a noncleavable bond (oxime bond, in the CS–PCS5 conjugate) and a cleavable one (thioether bond, in HA–PCS5 conjugate) were evaluated. In the case of the CS–PCS5 conjugate (Figure 2Ai), the oxime bonds have been reported as highly stable linkages at physiological pHs.57−59 Therefore, this linkage would be the model for a long-lasting attachment of the peptide antigen, which would only be released after being processed by APCs.
On the other hand, thioether bonds are known to undergo retro-Michael reactions in the presence of free thiols,33,56,60 which are presented in great amounts in the cytosol of cells as part of glutathione (GSH) molecules. To demonstrate this hypothesis, NPs were incubated in phosphate buffered saline (PBS), alone or with a 10 mM GSH concentration for up to 8 h at 37 °C. As shown in Figure 4, almost no PCS5 was detected upon incubation in PBS, however, upon incubation in a GSH-rich medium, PCS5 was immediately released from the NPs. These results indicate that the release of the peptide is triggered by GSH and led us to speculate that the peptide will not be released in the extracellular medium, but only in the intracellular compartments where high concentrations of GSH are present.61,62
Figure 4.
PCS5 release from CS/HA–PCS5/pIC NPs in a thiol-rich PBS solution (GSH 10 mM) in comparison to only PBS. Key: PBS, phosphate buffer saline; PCS5, protease cleavage site 5; GSH, glutathione.
Freeze-Drying Preserves NP Properties and Allows Long-Term Storage
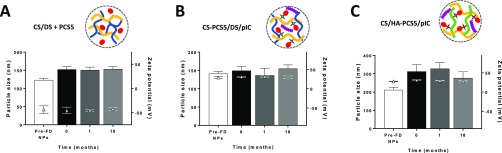
A key feature of vaccine formulations is their stability during storage. Thus, in order to improve the long-term stability of the antigen-loaded NPs, the different formulations were freeze-dried using different cryoprotectants. The results of the screening of the freeze-drying conditions led to the selection of trehalose 7% for both CS/DS + PCS5 and CS/HA–PCS5/pIC NPs and 4% for the CS–PCS5/DS/pIC NP formulation. These conditions guaranteed the preservation of the original properties of the formulations (Figure 5). When stored at 4 °C, these freeze-dried formulations maintained PDI and surface charge stable for at least 18 months, while particle size slightly increased.
Figure 5.
Freeze-dried stability of the developed nanosystems. Evolution of the particle size and the surface charge of the freeze-dried (A) CS/DS + PCS5 NPs, (B) CS–PCS5/DS/pIC NPs and (C) CS/HA–PCS5/pIC NPs during storage. Key: CS, chitosan; DS, dextran sulfate; PCS5, protease cleavage site 5; NPs, nanoparticles; pIC, poly(I:C) (polyinosinic:polycytidylic acid); HA, hyaluronic acid; FD, freeze-dried.
In Vivo Antibody Responses
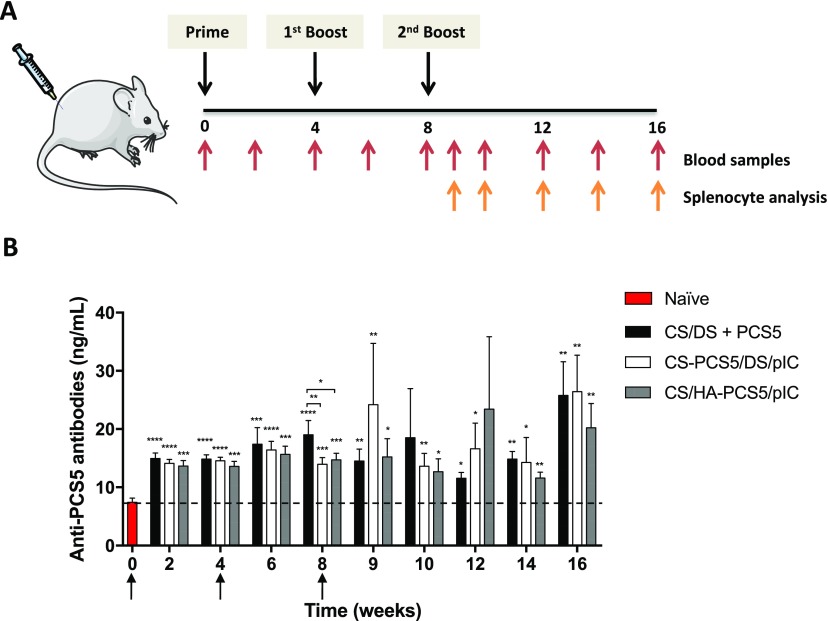
To evaluate the in vivo activity of the different PCS5-loaded prototypes, freeze-dried formulations were resuspended in water, and 50 μL of this suspension (containing a PCS5 dose of 5 μg) was intramuscularly injected to BALB/c mice. A group of nonvaccinated animals was used as control. Animals were vaccinated three times, as shown in Figure 6A.
Figure 6.
In vivo studies in mice. (A) Schematic representation of the study design and timeline of the analysis. (B) Anti-PCS5 antibody levels after the intramuscular administration of the three nanoformulations CS/DS–PCS5 (black bars), CS–PCS5/DS/pIC (white bars), and CS/HA–PCS5/pIC (gray bars) to 50 mice per group. Values represent mean ± SEM (n ≥ 5). Statistical comparison between groups was done using a Mann–Whitney test. Significant statistical differences are represented as * (p < 0.05), ** (p < 0.01), *** (p < 0.001), and **** (p < 0.0001) for comparison between groups and to naïve mice. Mouse and syringe images in (A) were reproduced from Servier Medical Art under a Creative Commons Attribution 3.0 Unported License, https://creativecommons.org/licenses/by/3.0. Key: NPs, nanoparticles; CS, chitosan; DS, dextran sulfate; PCS5, protease cleavage site 5; pIC, poly(I:C) (polyinosinic:polycytidylic acid); HA, hyaluronic acid.
Figure 6B shows the evolution of the anti-PCS5 IgG levels over time. Overall, the IgG responses elicited by the three different NP formulations increased significantly over time, reaching their maximum values at the latest time point in the experiment (week 16). At this time point, the amount of anti-PCS5 antibodies detected in all NPs was three times higher than the levels detected in unvaccinated mice. This increasing and prolonged immunogenic response is of particular interest in the design of an HIV vaccine because persistent levels of antibodies are essential for an effective vaccination.63 This response is also in agreement with previous data reported by our group that showed the capacity of the antigen-loaded NPs to induce significant IgG levels up to 28 or 37 weeks after immunization.20,25 Considering these previous results, it is possible that the antigen-loaded NPs could still produce important levels of antibodies beyond the 16 weeks considered in our study.
In addition to eliciting a strong and maintained immune response, the results of this study also indicate that the covalent linkage between the peptide and the NPs does not have an effect on the humoral responses. This is in contrast with previous findings where antigen conjugation improved humoral response when compared to simple ionic interactions.31,32
In Vivo Cellular Activation
To evaluate the changes in T cell subsets and the cellular activation of different immune cells upon vaccination, splenocytes from naïve or NP-vaccinated mice were stained and analyzed for different T cell and monocyte/macrophage phenotyping markers. Multicolor flow cytometry gating strategy used for phenotyping is summarized in the Supporting Information, Figure S5.
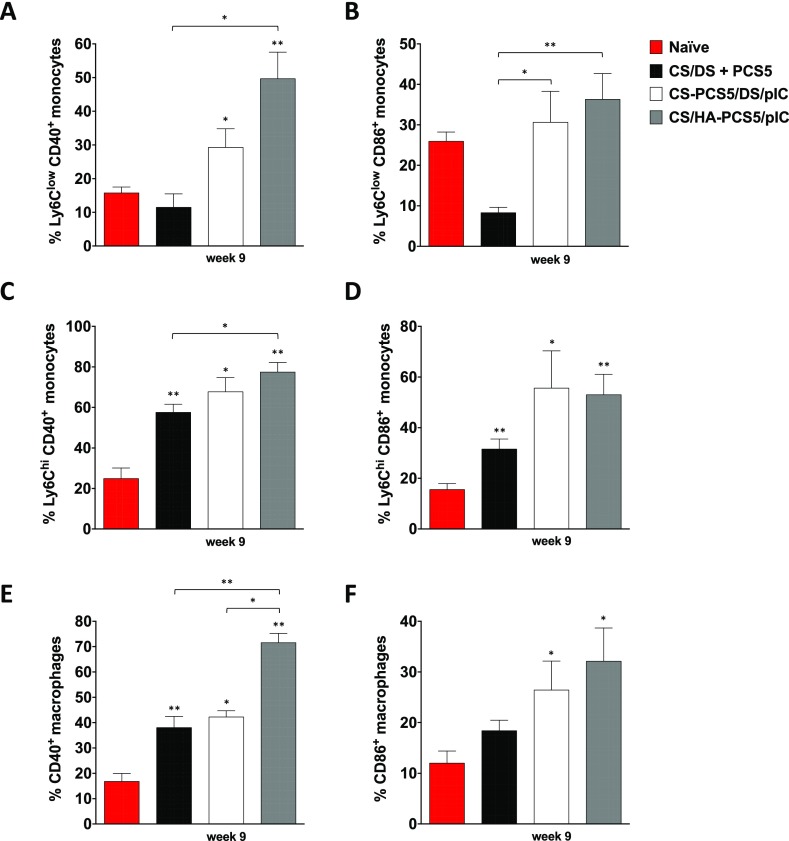
Monocytes and macrophages are APCs that process and present pathogen-derived antigens to T cells. Depending on their function, murine monocytes can be further divided as Ly6chi and Ly6clow, with the former being the classical pro-inflammatory and phagocytic monocytes and the latter the nonclassical or patrolling monocytes.64−66 Thus, the greater the activation of these APCs, the higher the capacity to recall other cellular populations and to stimulate the immune system. Splenocytes were stained with markers for phenotyping monocytes subsets Ly6chi and Ly6clow and markers for characterizing macrophages (CD11b+ CD11c– F4/80+) to identify the different subsets (Supporting Information, Figure S5A). To investigate if the nanosystems were able to activate these subpopulations, the expression of the costimulatory signals CD40+ and CD86+, both involved in the T-cell activation process,67−69 was measured. Regarding CD40+ expression, the highest activation of Ly6chi, Ly6clow, and macrophages by CS/HA–PCS5/pIC NPs took place 9 weeks post prime (Figure 7). However, this expression decreased slightly during the subsequent weeks (Supporting Information, Figure S6). For CS/DS + PCS5 and CS–PCS5/DS/pIC NPs, the number of monocytes and macrophages expressing CD40+ was lower than in the prototype containing HA, but the response remained for a longer period of time. In the case of CD86+ monocytes and macrophages, the values mirrored the ones for CD40+, with the difference that at later time points the values were similar to those found in naïve mice (Supporting Information, Figure S6). Overall, the prototypes of CS/HA–PCS5/pIC NPs and CS–PCS5/DS/pIC NPs elicited the highest stimulation of the monocyte/macrophage lineage (Figure 7, and Supporting Information, Figure S6), which might be caused by the covalent conjugation of the peptide antigen, in accordance with our hypothesis. In addition, we cannot discard the potential role of poly(I:C) in the stimulation process.70
Figure 7.
Monocyte and macrophage expression of costimulatory factors. CD40+ and CD86+ expression in (A, B) Ly6Clow monocytes; (C, D) Ly6Chigh monocytes, and (E, F) macrophages at 9 weeks post prime was quantified by multicolor flow cytometry of splenocytes obtained from nontreated naïve (red bars) and NP-vaccinated mice: CS/DS + PCS5 (black bars), CS–PCS5/DS/pIC (white bars), or CS/HA–PCS5/pIC (gray bars). Values represent mean ± SEM (n ≥ 3). Statistical comparison between groups was done using a Mann–Whitney test. Significant statistical differences are represented as * (p < 0.05) and ** (p < 0.01) for comparison between groups and to naïve mice. Key: NPs, nanoparticles; CS, chitosan; DS, dextran sulfate; PCS5, protease cleavage site 5; pIC, poly(I:C) (polyinosinic:polycytidylic acid); HA, hyaluronic acid.
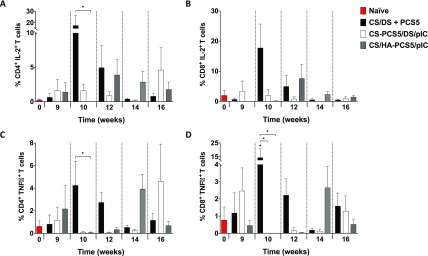
In a second set of experiments and in order to further assess the activation of T cells, the secretion of two cytokines that influence anti-HIV responses, i.e., interleukin 2 (IL-2) and tumor necrosis factor α (TNFα),71 was measured. IL-2 is involved in T-cell proliferation and expansion,72 while TNFα is a pro-inflammatory cytokine that participates in innate and adaptive immune responses.73−75 We also aimed at evaluating the kinetics of T cell activation over time, bearing in mind that an immediate high T cell response would not necessary translate into the best protection. The results showed that the three prototypes were able to activate both CD4+ and CD8+ T cells with different cytokine secretion kinetics. The highest overall secretion was observed for CS/DS + PCS5 NPs at 10 weeks. In the case of CS–PCS5/DS/pIC NPs, the highest secretion of both IL-2 and TNFα was observed at 9 weeks and 16 weeks post prime (Figure 8), whereas in the case of CS/HA–PCS5/pIC NPs, this activation was more sustained at 12–16 weeks (Figure 8). This pattern could be caused by the different release profiles of the antigen from the NPs, as previously discussed. It is also worth mentioning that delayed T cell activation observed here has also been recently described for an mRNA-based vaccine,76 a result that raises questions regarding the ideal T cell activation profile. The way this T cell activation profile correlates with efficacy will be a matter of further investigation, preferably in larger animal models.
Figure 8.
CD4+ and CD8+ T-cell activation. IL-2 and TNFα secretion in (A, C) CD4+ and (B, D) CD8+ T cells was quantified by multicolor flow cytometry of T-cells derived from splenocytes from nontreated naïve (red bars) and NP-vaccinated mice: CS/DS + PCS5 (black bars), CS–PCS5/DS/pIC (white bars) or CS/HA–PCS5/pIC (gray bars). Values represent mean ± SEM (n ≥ 3). Statistical comparison between groups was done using a Mann–Whitney test. Significant statistical differences are represented as * (p < 0.05) for comparison between groups and naïve mice. Key: NPs, nanoparticles; CS, chitosan; DS, dextran sulfate; PCS5, protease cleavage site 5; pIC, poly(I:C) (polyinosinic:polycytidylic acid); HA, hyaluronic acid.
Besides effector T cells, the generation of memory T cells by vaccines is also important to guarantee long-term responses against infections.77 Depending on the expression of the homing marker L-selectin (CD62L), CD4+ and CD8+ memory T cells can be further divided as central memory T cells (TCM; CD44+ CD62L+) and effector memory T cells (TEM; CD44+ CD62L–). Regarding their role in the immune response, TCM are important for the stimulation of dendritic cells and B cells and also because they help expand the effector T cell subsets once they have encountered the antigen. On the other hand, in the same situation, TEM rapidly convert to effector cells to fight against the infection.78,79 Hence, our objective was to assess if the three nanoformulations under study were able to increase both types of memory T cells, thereby ensuring a good immune response upon infection. The results in Figure 9 indicate that the three prototypes generated a modest number of memory T cells for up to several weeks after the last boost. In general, the highest proportions of memory T cells corresponded to CS/HA–PCS5/pIC NPs and CS/DS + PCS5 NPs, both at shorter and later time points.
Figure 9.
Change in CD4+ and CD8+ central memory and effector memory T cells. The amount of CD4+ and CD8+ T cell subsets expressing (A, C) CD44+ CD62L– (T effector memory – TEM) and (B, D) CD44+ CD62L+ (T central memory – TCM)) was quantified by multicolor flow cytometry of splenocytes from naïve (red bars) and NP-vaccinated mice: CS/DS + PCS5 (black bars), CS–PCS5/DS/pIC (white bars) or CS/HA–PCS5/pIC (gray bars). Values represent mean ± SEM (n ≥ 3). Statistical comparison between groups was done using a Mann–Whitney test. Significant statistical differences are represented as * (p < 0.05) and ** (p < 0.01) for comparison between groups. Key: NPs, nanoparticles; CS, chitosan; DS, dextran sulfate; PCS5, protease cleavage site 5; pIC, poly(I:C) (polyinosinic:polycytidylic acid); HA, hyaluronic acid.
Overall, the three prototypes of NPs showed the ability to generate important antibody responses (Figure 6B), whereas the ones containing PCS5 conjugated (CS/HA–PCS5/pIC NPs and CS–PCS5/DS/pIC NPs) activated APCs in a higher extent (Figure 7). In terms of cellular activation, all prototypes were able to increase the secretion of important cytokines such as IL-2 and TNFα, although the production kinetics varied depending on the prototype (Figure 8). The implications of these secretion patterns in the protection against infection is a subject that has to be further analyzed.
Conclusions
In the present study, we engineered different polysaccharide-based NPs loaded with an HIV peptide antigen candidate, PCS5. The results showed that different factors, such as the attachment of the antigen (ionic interactions and cleavable and noncleavable conjugations), the presence of immunomodulatory molecules such as poly(I:C), or the nature of the polysaccharides (CS, DS, or HA) could importantly influence the type of the elicited immune response. All of the nanosystems showed the ability to induced humoral responses against the antigen, while for the kinetics of the effector T cell responses varied depending on the prototype. In summary, composition, antigen attachment, and adjuvants are important design aspects that need to be considered when developing nanovaccines. Further in vivo studies would be needed to evaluate whether these humoral and cellular responses would translate into efficient protection in larger animal models.
Materials and Methods
Materials
Good manufacturing practice grade chitosan (CS) (chlorhydrate salt, molecular weight (MW) 42.7 kDa and an 88% deacetylation degree) was purchased from HMC+ (Germany). Pharmaceutical grade dextran sulfate (DS), (sodium salt, MW 8 kDa) was obtained from Sigma-Aldrich SAFC (USA). Pharmaceutical grade sodium hyaluronate (HA) (MW 57 kDa) was purchased from Lifecore Biomedical (USA). Poly(I:C) HMW was acquired from InvivoGen (USA), and high purity α,α-trehalose dihydrate was purchased from Pfanstiehl (USA).
HIV PCS5 peptide (GPWGKKPRNFPMAQVHQGLM, MW 2280 Da and >95% purity), with or without a terminal cysteine residue, was purchased from GenScript (USA).
MES hydrate, N-(2-aminoethyl)maleimide trifluoroacetate salt (ASEM), N-hydroxysuccinimide (NHS), and N-(3-(dimethylamino)propyl)-N-ethylcarbodiimide hydrochloride (EDC) were obtained from Sigma-Aldrich (USA).
Bio-Plex Pro Magnetic COOH Beads and Bio-Plex Amine Coupling Kit were purchased from Bio-Rad (USA). Phycoerythrin-labeled mouse antimonkey IgG was obtained from Southern Biotech (USA), and PCS5 monoclonal antibody was donated by National Microbiology Laboratory (Canada).
Screening of Blank Nanoparticles
The preparation method for all of the NPs in this study was ionic complexation, based on the interactions between the oppositely charged polymers: CS and DS or CS and HA.
For CS/DS NPs, mass ratios from 3:1 to 1:3 were screened. Equal volumes (0.825 mL) of an aqueous solution of CS (concentration from 1.875 to 0.625 mg/mL) and aqueous solution of DS (concentration from 0.625 to 1.875 mg/mL) were mixed under mild magnetic stirring. The solutions were kept under magnetic stirring for 10 min.
In the case of CS/HA NPs, mass ratios from 1.5:1 to 1:4 were screened. Equal volumes (0.550 mL) of an aqueous solution of CS (concentrations from 2.40 to 0.80 mg/mL) and aqueous solution of HA (concentration from 1.60 to 3.20 mg/mL) were mixed under mild magnetic stirring. The solution was stirred for an additional 10 min.
Synthesis of CS/DS + PCS5 NPs
CS/DS NPs at a 1:3 mass ratio were prepared as previously described39 but with materials of a higher quality grade. Briefly, 0.055 mL of PCS5 in aqueous solution (4 mg/mL) was added to 0.770 mL of a CS aqueous solution (0.67 mg/mL) under mild magnetic stirring. After being stirred for 5 min, 0.825 mL of a DS aqueous solution (1.875 mg/mL) was also added. The solution was stirred for an additional 5 min, and the formulation was kept for 10 min without stirring prior to characterization.
Determination of PCS5 Association Efficiency
CS/DS + PCS5 NPs were isolated by size-exclusion chromatography (SEC) using CentriPure P10 gel filtration columns (emp Biotech GmbH, Germany). The columns were first washed and stabilized with 15 mL of ultrapure water, and then, 1 mL of NPs was transferred to the column and allowed to enter the gel bed completely. After the void volume was discarded, 2 mL of ultrapure water were added to the column, and the isolated NPs were collected. The association efficiency (AE) was calculated by comparing the amount of PCS5 associated with the NPs with the initial amount added to prepare the NPs (eq 1). For this purpose, isolated NPs from SEC were treated to a final KCl concentration of 2 M to break the ionic interactions between the components of the NPs. Samples were analyzed by ultra performance liquid chromatography (UPLC) on an Acquity H-UPLC Class system with a Tunable UV (TUV) detector (Waters Corporation, USA) equipped with an Aeris 3.6 μm Widepore XB-C18 LC 100 × 2.1 mm Column (Phenomenex, USA). Mobile phase A consisted of 0.1% trifluoroacetic acid (v/v) in ultrapure water and phase B of 0.1% trifluoroacetic acid (v/v) in acetonitrile HPLC gradient. The column temperature was set at 30 °C, and the run from 10% to 100% of phase B in 5 min. The amount of PCS5 in each sample was calculated using a standard curve generated with known concentrations of the peptide (6.5–100 μg/mL, r2 = 0.99).
| 1 |
Preparation of CS–PCS5/DS/pIC NPs
The conjugate CS–PCS5 was purchased from Pepscan. PAoa-Ahx-PCS5-OH was synthesized using standard solid-phase peptide chemistry through Fmoc/tBu chemistry and subsequently purified to specifications using reversed-phase preparative HPLC.
On the other hand, CS was dissolved in buffer (10 mM phosphate buffer at pH 6.5), while N-succinimidyl 4-formylbenzoate (S-4FB) was dissolved in dry dimethyl sulfoxide (DMSO) to generate a stock of 30 mg/mL. A 50 μL portion of the S-4FB stock was added dropwise to the CS solution (10 mg/mL in phosphate buffer), and the reaction mixture was stirred overnight to allow for the coupling of the S-4FB linker to the free amine residues in the CS. This derivatized CS was then desalted using dialysis against phosphate buffer. Next, a peptide aqueous solution at pH 6 was added to the activated CS solution and allowed to conjugate overnight with agitation. The resulting conjugate was then subjected to dialysis against phosphate buffer to remove the unbound peptide.
The ratio of PCS5/CS was determined by a colorimetric assay. First, 2-hydrazinopyridine was added to react with the S-4FB incorporated to CS. Following incubation at 37 °C for 30 min, the chromophoric bis-arylhydrazone obtained was quantified at 350 nm. The difference between the amount of free linker before and after PCS5 conjugation was considered the value of the substitution degree. CS, PCS5, and CS–PCS5 samples were diluted in D2O water and analyzed in a Varian Inova 750 spectrometer for 1H NMR, processed using MestreNova (MESTRELAB).
For the preparation of these NPs, a mass ratio 2:1 CS/DS was selected. A 0.0385 mL portion of an aqueous solution of the modified CS (8.8 mg/mL) was added to a CS aqueous solution of 0.909 mg/mL to obtain a solution containing 2 mg/mL of CS and 0.2 mg/mL of PCS5. Simultaneously, 2.5 μL of poly(I:C) (1 mg/mL) was then added to 0.250 mL of an aqueous solution of DS (1 mg/mL), and this solution was poured over 0.250 mL of the previous CS solution (2 mg/mL CS and 0.2 mg/mL of PCS5) under magnetic stirring for 10 min.
Synthesis of CS/HA–PCS5/pIC NPs
The PCS5 peptide was linked to HA by a thiol–maleimide conjugation reaction, adapting a recently described protocol.80 First, HA was diluted in MES buffer (0.1 N, pH 6) to a concentration of 2 mg/mL, and ASEM, NHS, and EDC in the same buffer solution were added at a concentration 6-, 14-, and 143-fold times higher than HA, respectively. The solution was kept under magnetic stirring for 4 h at room temperature and then dialyzed for 24 h to remove free reactives (SnakeSkin, cellulose membrane MW 7 kDa, Thermo, Spain). In a second step, 0.228 mL of PCS5 with a terminal cysteine residue in aqueous solution (5 mg/mL) were poured over 5 mL of the solution of HA–ASEM (1 mg/mL) in buffer (MES 0.1 N, NaCl 100 mM) under magnetic stirring. The solution was kept under stirring for 4 h and then dialyzed for 24 h. Finally, the modified HA solution was frozen at −20 °C and freeze-dried.
NMR analyses of HA, PCS5, and HA–PCS5 were conducted to characterize the link and determine the substitution degree and the yield. For this purpose, freeze-dried samples were resuspended in D2O and analyzed using a Varian Inova 750 spectrometer for 1H NMR and diffusion-ordered spectroscopy (DOSY). Then the NMR spectra were processed using MestreNova (MESTRELAB). The degree of substitution (DS) was calculated as the amount of PCS5 conjugated per amount of HA, while the yield was determined by comparing the amount of PCS5 conjugated to the total amount added to the reaction (eq 2).
| 2 |
NPs were formed at a 1:1 CS/HA mass ratio. Aqueous solutions of either HA or HA–PCS5 at 2 mg/mL were prepared, and the ratio between the two solutions adjusted to a final PCS5 peptide concentration of 0.20 mg/mL. Poly(I:C) was also added as a 1 mg/mL aqueous solution to a final concentration of 2 μg/mL. Later, 0.55 mL of this HA and poly(I:C) solution (2 mg/mL, 2 μg/mL) were poured over 0.55 mL of a CS aqueous solution (2 mg/mL) under magnetic stirring and kept stirring for 10 min.
Nanoparticle Characterization
The mean size and polydispersity index (PDI) of the NPs were characterized by dynamic light scattering (DLS). The zeta potential values of the NPs were determined by Laser Doppler Anemometry (LDA), measuring the mean electrophoretic mobility after a 10-times dilution of the NPs in ultrapure water. Both properties were measured using a Zetasizer NanoZS (Malvern Instruments, United Kingdom). The measurements were performed at 25 °C with a detection angle of 173°.
The PCS5 peptide loading capacity was determined by calculating the real amount of PCS5 associated with the NPs (theoretical mass × AE%) relative to the real mass of the formulation, represented in eq 3. Poly(I:C) loading values were calculated according to eq 4.
| 3 |
| 4 |
Morphological Analysis
Morphological analysis of the suspension of NPs was conducted by field emission scanning electron microscopy (FESEM) (Zeiss Gemini Ultra Plus, Germany). NPs were diluted 1:100 in water and then with the same volume of phosphotungstic acid (2% in water). A volume of 1 μL of sample was placed on a copper grid with carbon films and allow to dry. Then the grids were washed with water and analyzed under the microscope once dried. STEM and immersion lens (InLens) detectors were used to observe the samples.
Nanoparticle Surface Analysis by X-ray Photoelectron Spectroscopy (XPS)
The surface of CS/HA/pIC and CS/HA–PCS5/pIC NPs was analyzed to confirm the presence of PCS5. A droplet of each NP suspension was placed on a silicon wafer, used as sample holder, and then allowed to dry in a desiccator overnight. For the reference materials (CS, HA, and PCS5), a small quantity of powder was pressed onto a conductive double side adhesive tape on the standard sample holder.
Samples were analyzed by angle-resolved XPS using the ESCALAB 250 iXL instrument (Thermo Scientific K-Alpha ESCA, Thermo Scientific, UK), and photoelectrons were collected from a takeoff angle of 45◦ relative to the sample surface. Monochromatic X-ray source Al Kα (1486.6 eV) was used for experiments and spectra were acquired at 10–10 mbar. Surface elemental composition was determined using the standard Scofield photoemission cross section. The binding energies positions on unsputtered surfaces were calibrated by setting the C1s photopeak corresponding to aliphatic carbon at 285.0 eV. The atomic concentrations were determined from the XPS peak areas using the Shirley background subtraction technique and the Scofield sensitivity factors.
PCS5 Release in Glutathione-Rich Media
CS/HA–PCS5/pIC NPs were incubated at 37 °C up to 8 h in PBS with a 10 mM concentration of reduced glutathione (GSH). In parallel, as a control, NPs were incubated in PBS under the same conditions. At the different time-points, NPs were centrifuged for 10 min at 16000 g to break the NPs and collect the HA–PCS5 from the supernatant. Later, 1 N HCl was added to the supernatant to lower the pH to 2, in order to avoid interactions between the free peptide (pI 11) and the conjugate HA–PCS5 (pKa 3), and to prevent that any remaining GSH kept reacting. Samples were analyzed by UPLC, as described for PCS5, but with a run from 10% to 50% of phase B in 20 min. The amount of free PCS5 peptide and conjugated PCS5 in each sample was calculated using a standard curve generated with known concentrations of the peptide and conjugate (6–50 μg/mL, r2 = 0.99).
Freeze-Drying
Nanoparticles were frozen in the presence of the cryoprotectant agent trehalose at −80 °C for at least 2 h and then freeze-dried (Genesis 25 EL, S.P Industries, USA). Lyophilization was done at a temperature ranging from −40 to +20 °C, applying a progressive vacuum from 200 to 20 mTorr. After this process, NPs were resuspended in water and their physicochemical properties determined by DLS and LDA as previously described.
Animal Care
Female BALB/c mice 6–8 weeks old were obtained from Charles River Laboratories (Wilmington, MA. USA) and housed at the Veterinary Technical Service unit of National Microbiology Laboratory (Winnipeg, Manitoba). A maximum of five animals were housed per cage in a level 2 facility. All animals used in the project were treated in a humane manner in accordance with the Principles of the Canadian Council on Animal Care contained in the “Guide to the Care and Use of Experimental Animals”. All animal procedures were performed by highly trained personnel using approved techniques. All mice were anesthetized by inhalation of 3–5% isofluorane, and all possible efforts were made to minimize the pain and discomfort of the animals. Euthanization of animals was conducted using an excess amount of the anesthetic.
Murine Experiments
Eighty female BALB/c mice were divided into four categories. Five unvaccinated mice (week 0) were used as a control. The rest of the mice were divided into three groups of 25 animals each. Mice were intramuscularly injected with 50 μL of the resuspended freeze-dried NPs (CS/DS + PCS5, CS/HA–PCS5/pIC and CS–PCS5/DS/pIC) at 0, 4, and 8 weeks. Each group of vaccinated animals was kept in a separate cage and monitored every 24 h for feeding, water drinking, and weight loss.
Plasma Antibody Quantification
Plasma was separated from whole blood collected from mice using saphenous bleeding, by centrifugation at 2000 g for 10 min. Mouse plasma samples were collected at 2, 4, 8, 9, 10, 12, 14, and 16 weeks following vaccination, and the concentrations of PCS5-specific plasma IgG antibodies were quantified using a previously published protocol.39 Briefly, 20 μg of PCS5 was coupled to 1.25 × 106 Bio-Plex Pro Magnetic COOH Beads using a Bio-Plex Amine Coupling Kit. 50 μL of plasma (1:80 diluted) were incubated with 2,500 beads. SIV-specific IgG was detected with phycoerythrin-labeled mouse antimonkey IgG at 5 μg/mL. Bead fluorescence intensities were acquired on the Bio-Plex 200 system and converted to concentrations as previously described.39
Splenocytes Response Quantification
Five mice per group were sacrificed at 0 (naïve mice) 9, 10, 12, 14, and 16 weeks (vaccinated mice), and their spleens were collected. Splenocytes were purified by straining of spleens through a 40 μm nylon mesh. The resulting mixtures were suspended in R10 media containing RPMI-1640 complemented with 10% fetal bovine serum (FBS) and 2% antibiotic–antimycotic solutions. The splenocytes were then frozen in freezing media consisting of 95% FBS and 5% DMSO. Half a million splenocytes were transferred into 12 × 75 mm polypropylene BD Falcon tubes for flow cytometry antibody labeling. Alternatively, 5.0 × 105 splenocytes were stimulated separately with a 5 μg/mL suspension of a pool of PCS5 peptides: PCS5-1 (GPWGKKPRNFPMAQVHQGLM), PCS5-2 (YGQMPRQTGGFFRPWSMGKE), or PCS5-3 (KPRNFPMAQVHQGLM) peptides. Peptide stimulated and unstimulated splenocytes were washed using PBS with 2% FBS at 1500 rpm for 5 min. The washed splenocytes were surface stained with either a T-cell or monocyte/macrophage cocktails of antibodies for 30 min in the dark and at room temperature. The stained cells were washed using 5% PBS followed by addition of 150 μL of BD Cytofix/Cytoperm and a 10 min incubation in the dark. Following the incubation, permeabilized cells were washed using 1× BD Permwash solution followed by intracellular cytokine staining (ICS) for 30 min in the dark. After ICS, cells were washed using 1× BD PermWash and then run on LSRII flow cytometer. Data acquisition was done using BD FACSDiva software and analyzed using FlowJo (Treestar, Inc., USA).
Statistics
Data analysis was performed using GraphPad Prism version 7.0 (GraphPad Inc.). Comparison of group data was performed with a Mann–Whitney test. Data was expressed as the mean ± standard error of the mean (SEM). p values of 0.05 or less were considered statistically significant.
Acknowledgments
This work was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (Award No. R01AI111805, Subaward No. 41795-02) and by the European Union’s Horizon 2020 research program (NanoPilot project, Grant Agreement No. 646142). T.G.D. acknowledges a predoctoral FPU grant from the Spanish Ministry of Education, Culture and Sports (Grant No. FPU14/05866). We also thank Pepscan for kindly providing all of the chemical information regarding the synthesis and characterization of the CS–PCS5 conjugate.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsnano.8b07662.
Additional material includes the screening of ratios of blank NPs, synthesis of HA–PCS5 conjugate, and DOSY characterization, XPS results of the CS/HA–PCS5/pIC NPs, and additional in vivo results regarding cell gating and monocyte and macrophage activation at weeks 10–16 post prime (PDF)
Author Contributions
⊥ T.G.D. and R.W.O. contributed equally to this work.
The authors declare no competing financial interest.
Supplementary Material
References
- UNAIDS Data 2018 http://www.unaids.org/sites/default/files/media_asset/unaids-data-2018_en.pdf (accessed Sep 19, 2018).
- Rerks-Ngarm S.; Pitisuttithum P.; Nitayaphan S.; Kaewkungwal J.; Chiu J.; Paris R.; Premsri N.; Namwat C.; de Souza M.; Adams E.; Benenson M.; Guruathan S.; Tartaglia J.; McNeil J. G.; Francis D. P.; Stablein D.; Birx D. L.; Chunsuttiwat S.; Khamboonruang C.; Thongcharoen P.; et al. Vaccination with ALVAC and AIDSVAX to Prevent HIV-1 Infection in Thailand. N. Engl. J. Med. 2009, 361, 2209–2220. 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- Kim J. H.; Excler J.-L.; Michael N. L. Lessons from the RV144 Thai Phase III HIV-1 Vaccine Trial and the Search for Correlates of Protection. Annu. Rev. Med. 2015, 66, 423–437. 10.1146/annurev-med-052912-123749. [DOI] [PubMed] [Google Scholar]
- Haynes B. F. New Approaches to HIV Vaccine Development. Curr. Opin. Immunol. 2015, 35, 39–47. 10.1016/j.coi.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Chen C. Role of Nanotechnology in HIV/AIDS Vaccine Development. Adv. Drug Delivery Rev. 2016, 103, 76–89. 10.1016/j.addr.2016.02.010. [DOI] [PubMed] [Google Scholar]
- Luo M.; Capina R.; Daniuk C.; Tuff J.; Peters H.; Kimani M.; Wachihi C.; Kimani J.; Ball T. B.; Plummer F. A. Immunogenicity of Sequences around HIV-1 Protease Cleavage Sites: Potential Targets and Population Coverage Analysis for a HIV Vaccine Targeting Protease Cleavage Sites. Vaccine 2013, 31, 3000–3008. 10.1016/j.vaccine.2013.04.057. [DOI] [PubMed] [Google Scholar]
- Li H.; Omange R. W.; Plummer F. A.; Luo M. A Novel HIV Vaccine Targeting the Protease Cleavage Sites. AIDS Res. Ther. 2017, 14, 51. 10.1186/s12981-017-0174-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.; Omange R. W.; Czarnecki C.; Correia-Pinto J. F.; Crecente-Campo J.; Richmond M.; Li L.; Schultz-Darken N.; Alonso M. J.; Whitney J. B.; Plummer F. A.; Luo M. Mauritian Cynomolgus Macaques with M3M4MHC Genotype Control SIVmac251 Infection. J. Med. Primatol. 2017, 46, 137–143. 10.1111/jmp.12300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan A. H.; Zack J. A.; Knigge M.; Paul D. A.; Kempf D. J.; Norbeck D. W.; Swanstrom R. Partial Inhibition of the Human Immunodeficiency Virus Type 1 Protease Results in Aberrant Virus Assembly and the Formation of Noninfectious Particles. J. Virol. 1993, 67, 4050–4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dacoba T. G.; Olivera A.; Torres D.; Crecente-Campo J.; Alonso M. J. Modulating the Immune System through Nanotechnology. Semin. Immunol. 2017, 34, 78–102. 10.1016/j.smim.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gause K. T.; Wheatley A. K.; Cui J.; Yan Y.; Kent S. J.; Caruso F. Immunological Principles Guiding the Rational Design of Particles for Vaccine Delivery. ACS Nano 2017, 11, 54–68. 10.1021/acsnano.6b07343. [DOI] [PubMed] [Google Scholar]
- Irvine D. J.; Hanson M. C.; Rakhra K.; Tokatlian T. Synthetic Nanoparticles for Vaccines and Immunotherapy. Chem. Rev. 2015, 115, 11109–11146. 10.1021/acs.chemrev.5b00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordeiro A. S.; Alonso M. J.; de la Fuente M. Nanoengineering of Vaccines Using Natural Polysaccharides. Biotechnol. Adv. 2015, 33, 1279–1293. 10.1016/j.biotechadv.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Fuentes M.; Alonso M. J. Chitosan-Based Drug Nanocarriers: Where Do We Stand?. J. Controlled Release 2012, 161, 496–504. 10.1016/j.jconrel.2012.03.017. [DOI] [PubMed] [Google Scholar]
- Shariatinia Z. Pharmaceutical Applications of Chitosan. Adv. Colloid Interface Sci. 2019, 263, 131–194. 10.1016/j.cis.2018.11.008. [DOI] [PubMed] [Google Scholar]
- Calvo P.; Remuñán-López C.; Vila-Jato J. L.; Alonso M. J. Novel Hydrophilic Chitosan-Polyethylene Oxide Nanoparticles as Protein Carriers. J. Appl. Polym. Sci. 1997, 63, 125–132. . [DOI] [Google Scholar]
- Prego C.; Torres D.; Alonso M. J. Chitosan Nanocapsules: A New Carrier for Nasal Peptide Delivery. J. Drug Delivery Sci. Technol. 2006, 16, 331–337. 10.1016/S1773-2247(06)50061-9. [DOI] [Google Scholar]
- Prego C.; Paolicelli P.; Díaz B.; Vicente S.; Sánchez A.; González-Fernández Á.; Alonso M. J. Chitosan-Based Nanoparticles for Improving Immunization against Hepatitis B Infection. Vaccine 2010, 28, 2607–2614. 10.1016/j.vaccine.2010.01.011. [DOI] [PubMed] [Google Scholar]
- Vila A.; Sánchez A.; Janes K.; Behrens I.; Kissel T.; Vila-Jato J. L.; Alonso M. J. Low Molecular Weight Chitosan Nanoparticles as New Carriers for Nasal Vaccine Delivery in Mice. Eur. J. Pharm. Biopharm. 2004, 57, 123–131. 10.1016/j.ejpb.2003.09.006. [DOI] [PubMed] [Google Scholar]
- Vicente S.; Peleteiro M.; Díaz-Freitas B.; Sanchez A.; González-Fernández Á.; Alonso M. J. Co-Delivery of Viral Proteins and a TLR7 Agonist from Polysaccharide Nanocapsules: A Needle-Free Vaccination Strategy. J. Controlled Release 2013, 172, 773–781. 10.1016/j.jconrel.2013.09.012. [DOI] [PubMed] [Google Scholar]
- Crecente-Campo J.; Lorenzo-Abalde S.; Mora A.; Marzoa J.; Csaba N.; Blanco J.; González-Fernández Á.; Alonso M. J. Bilayer Polymeric Nanocapsules: A Formulation Approach for a Thermostable and Adjuvanted E. Coli Antigen Vaccine. J. Controlled Release 2018, 286, 20–32. 10.1016/j.jconrel.2018.07.018. [DOI] [PubMed] [Google Scholar]
- Zupančič E.; Curato C.; Paisana M.; Rodrigues C.; Porat Z.; Viana A. S.; Afonso C. A. M.; Pinto J.; Gaspar R.; Moreira J. N.; Satchi-Fainaro R.; Jung S.; Florindo H. F. Rational Design of Nanoparticles towards Targeting Antigen-Presenting Cells and Improved T Cell Priming. J. Controlled Release 2017, 258, 182–195. 10.1016/j.jconrel.2017.05.014. [DOI] [PubMed] [Google Scholar]
- Shi G.-N.; Zhang C.-N.; Xu R.; Niu J.-F.; Song H.-J.; Zhang X.-Y.; Wang W.-W.; Wang Y.-M.; Li C.; Wei X.-Q.; Kong D.-L. Enhanced Antitumor Immunity by Targeting Dendritic Cells with Tumor Cell Lysate-Loaded Chitosan Nanoparticles Vaccine. Biomaterials 2017, 113, 191–202. 10.1016/j.biomaterials.2016.10.047. [DOI] [PubMed] [Google Scholar]
- Vicente S.; Diaz-Freitas B.; Peleteiro M.; Sanchez A.; Pascual D. W.; Gonzalez-Fernandez A.; Alonso M. J. A Polymer/Oil Based Nanovaccine as a Single-Dose Immunization Approach. PLoS One 2013, 8, 2–9. 10.1371/journal.pone.0062500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Aramundiz J. V.; Presas E.; Dalmau-Mena I.; Martínez-Pulgarín S.; Alonso C.; Escribano J. M.; Alonso M. J.; Csaba N. S. Rational Design of Protamine Nanocapsules as Antigen Delivery Carriers. J. Controlled Release 2017, 245, 62–69. 10.1016/j.jconrel.2016.11.012. [DOI] [PubMed] [Google Scholar]
- Li A. V.; Moon J. J.; Abraham W.; Suh H.; Elkhader J.; Seidman M. A.; Yen M.; Im E.-J.; Foley M. H.; Barouch D. H.; Irvine D. J. Generation of Effector Memory T Cell-Based Mucosal and Systemic Immunity with Pulmonary Nanoparticle Vaccination. Sci. Transl. Med. 2013, 5, 1–11. 10.1126/scitranslmed.3006516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J. T.; Keller S.; Manganiello M. J.; Cheng C.; Lee C.-C.; Opara C.; Convertine A.; Stayton P. S. PH-Responsive Nanoparticle Vaccines for Dual-Delivery of Antigens and Immunostimulatory Oligonucleotides. ACS Nano 2013, 7, 3912–3925. 10.1021/nn305466z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloat B. R.; Sandoval M. A.; Hau A. M.; He Y.; Cui Z. Strong Antibody Responses Induced by Protein Antigens Conjugated onto the Surface of Lecithin-Based Nanoparticles. J. Controlled Release 2010, 141, 93–100. 10.1016/j.jconrel.2009.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A.; Yan W.; Miller K. R.; O’Carra R.; Woodward J. G.; Mumper R. J. Tresyl-Based Conjugation of Protein Antigen to Lipid Nanoparticles Increases Antigen Immunogenicity. Int. J. Pharm. 2010, 401, 87–92. 10.1016/j.ijpharm.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caivano A.; Doria-Rose N. A.; Buelow B.; Sartorius R.; Trovato M.; D’Apice L.; Domingo G. J.; Sutton W. F.; Haigwood N. L.; De Berardinis P. HIV-1 Gag P17 Presented as Virus-Like Particles on the E2 Scaffold from Geobacillus Stearothermophilus Induces Sustained Humoral and Cellular Immune Responses in the Absence of IFNγ Production by CD4+ T Cells. Virology 2010, 407, 296–305. 10.1016/j.virol.2010.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson D. S.; Platt V. M.; Cao L.; Venditto V. J.; Szoka F. C. Antibody Response to Polyhistidine-Tagged Peptide and Protein Antigens Attached to Liposomes via Lipid-Linked Nitrilotriacetic Acid in Mice. Clin. Vaccine Immunol. 2011, 18, 289–297. 10.1128/CVI.00425-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J. J.; Suh H.; Li V.; Ockenhouse C. F.; Yadava A.; Irvine D. J. Enhancing Humoral Responses to a Malaria Antigen with Nanoparticle Vaccines That Expand Tfh Cells and Promote Germinal Center Induction. Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 1080–1085. 10.1073/pnas.1112648109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molino N. M.; Anderson A. K. L.; Nelson E. L.; Wang S. W. Biomimetic Protein Nanoparticles Facilitate Enhanced Dendritic Cell Activation and Cross-Presentation. ACS Nano 2013, 7, 9743–9752. 10.1021/nn403085w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temchura V. V.; Kozlova D.; Sokolova V.; Überla K.; Epple M. Targeting and Activation of Antigen-Specific B-Cells by Calcium Phosphate Nanoparticles Loaded with Protein Antigen. Biomaterials 2014, 35, 6098–6105. 10.1016/j.biomaterials.2014.04.010. [DOI] [PubMed] [Google Scholar]
- Liu Q.; Zhang C.; Zheng X.; Shao X.; Zhang X.; Zhang Q.; Jiang X. Preparation and Evaluation of Antigen/N-Trimethylaminoethylmethacrylate Chitosan Conjugates for Nasal Immunization. Vaccine 2014, 32, 2582–2590. 10.1016/j.vaccine.2014.03.041. [DOI] [PubMed] [Google Scholar]
- Liu Q.; Zheng X.; Zhang C.; Shao X.; Zhang Q.; Jiang X. Conjugating Influenza A (H1N1) Antigen to N-Trimethylaminoethylmethacrylate Chitosan Nanoparticles Improves the Immunogenicity of the Antigen after Nasal Administration. J. Med. Virol. 2015, 87, 1807–1815. 10.1002/jmv.24253. [DOI] [PubMed] [Google Scholar]
- Bale S.; Goebrecht G.; Stano A.; Wilson R.; Ota T.; Tran K.; Ingale J.; Zwick M. B.; Wyatt R. T. Covalent Linkage of HIV-1 Trimers to Synthetic Liposomes Elicits Improved B Cell and Antibody Responses. J. Virol. 2017, 91, e00443–17. 10.1128/JVI.00443-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasturi S. P.; Kozlowski P. A.; Nakaya H. I.; Burger M. C.; Russo P.; Pham M.; Kovalenkov Y.; Silveira E. L. V.; Havenar-Daughton C.; Burton S. L.; Kilgore K. M.; Johnson M. J.; Nabi R.; Legere T.; Sher Z. J.; Chen X.; Amara R. R.; Hunter E.; Bosinger S. E.; Spearman P.; et al. Adjuvanting a Simian Immunodeficiency Virus Vaccine with Toll-Like Receptor Ligands Encapsulated in Nanoparticles Induces Persistent Antibody Responses and Enhanced Protection in TRIM5α Restrictive Macaques. J. Virol. 2017, 91, e01844–16. 10.1128/JVI.01844-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.; Nykoluk M.; Li L.; Liu L. R.; Omange R. W.; Soule G.; Schroeder L. T.; Toledo N.; Kashem M. A.; Correia-Pinto J. F.; Liang B.; Schultz-Darken N.; Alonso M. J.; Whitney J. B.; Plummer F. A.; Luo M. Natural and Cross-Inducible Anti-SIV Antibodies in Mauritian Cynomolgus Macaques. PLoS One 2017, 12, 1–20. 10.1371/journal.pone.0186079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J.; Kantoff P. W.; Wooster R.; Farokhzad O. C. Cancer Nanomedicine: Progress, Challenges and Opportunities. Nat. Rev. Cancer 2017, 17, 20–37. 10.1038/nrc.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose F.; Wern J. E.; Gavins F.; Andersen P.; Follmann F.; Foged C. A Strong Adjuvant Based on Glycol-Chitosan-Coated Lipid-Polymer Hybrid Nanoparticles Potentiates Mucosal Immune Responses against the Recombinant Chlamydia Trachomatis Fusion Antigen CTH522. J. Controlled Release 2018, 271, 88–97. 10.1016/j.jconrel.2017.12.003. [DOI] [PubMed] [Google Scholar]
- Vila A.; Sánchez A.; Tobío M.; Calvo P.; Alonso M. J. Design of Biodegradable Particles for Protein Delivery. J. Controlled Release 2002, 78, 15–24. 10.1016/S0168-3659(01)00486-2. [DOI] [PubMed] [Google Scholar]
- Correia-Pinto J. F.; Csaba N.; Schiller J.; Alonso M. J. Chitosan-Poly (I:C)-PADRE Based Nanoparticles as Delivery Vehicles for Synthetic Peptide Vaccines. Vaccines 2015, 3, 730–750. 10.3390/vaccines3030730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L.; Cohen J. A.; Broaders K. E.; Beaudette T. T.; Fréchet J. M. J. Mannosylated Dextran Nanoparticles: A PH-Sensitive System Engineered for Immunomodulation through Mannose Targeting. Bioconjugate Chem. 2011, 22, 949–957. 10.1021/bc100596w. [DOI] [PubMed] [Google Scholar]
- Sharma S.; Mukkur T. K.; Benson H. A.; Chen Y. Enhanced Immune Response Against Pertussis Toxoid by IgA-Loaded Chitosan–Dextran Sulfate Nanoparticles. J. Pharm. Sci. 2012, 101, 233–244. 10.1002/jps.22763. [DOI] [PubMed] [Google Scholar]
- Perica K.; Tu A.; Richter A.; Bieler J. G.; Edidin M.; Schneck J. P. Magnetic Field-Induced T Cell Receptor Clustering by Nanoparticles Enhances T Cell Activation and Stimulates Antitumor Activity. ACS Nano 2014, 8, 2252–2260. 10.1021/nn405520d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baaten B. J. G.; Tinoco R.; Chen A. T.; Bradley L. M. Regulation of Antigen-Experienced T Cells: Lessons from the Quintessential Memory Marker CD44. Front. Immunol. 2012, 3, 1–12. 10.3389/fimmu.2012.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Termeer C. C.; Hennies J.; Voith U.; Ahrens T.; M. Weiss J.; Prehm P.; Simon J. C. Oligosaccharides of Hyaluronan Are Potent Activators of Dendritic Cells. J. Immunol. 2000, 165, 1863–1870. 10.4049/jimmunol.165.4.1863. [DOI] [PubMed] [Google Scholar]
- Ke C.; Wang D.; Sun Y.; Qiao D.; Ye H.; Zeng X. Immunostimulatory and Antiangiogenic Activities of Low Molecular Weight Hyaluronic Acid. Food Chem. Toxicol. 2013, 58, 401–407. 10.1016/j.fct.2013.05.032. [DOI] [PubMed] [Google Scholar]
- Fan Y.; Sahdev P.; Ochyl L. J.; Akerberg J. J.; Moon J. J. Cationic Liposome–Hyaluronic Acid Hybrid Nanoparticles for Intranasal Vaccination with Subunit Antigens. J. Controlled Release 2015, 208, 121–129. 10.1016/j.jconrel.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L.; Cao F.; Liu X.; Wang H.; Zhang C.; Sun H.; Wang C.; Leng X.; Song C.; Kong D.; Ma G. Hyaluronic Acid-Modified Cationic Lipid–PLGA Hybrid Nanoparticles as a Nanovaccine Induce Robust Humoral and Cellular Immune Responses. ACS Appl. Mater. Interfaces 2016, 8, 11969–11979. 10.1021/acsami.6b01135. [DOI] [PubMed] [Google Scholar]
- González-Aramundiz J. V.; Peleteiro Olmedo M.; González-Fernández Á.; Alonso M. J.; Csaba N. S. Protamine-Based Nanoparticles as New Antigen Delivery Systems. Eur. J. Pharm. Biopharm. 2015, 97, 51–59. 10.1016/j.ejpb.2015.09.019. [DOI] [PubMed] [Google Scholar]
- Correia-Pinto J. F.; Peleteiro M.; Csaba N.; González-Fernández Á.; Alonso M. J. Multi-Enveloping of Particulated Antigens with Biopolymers and Immunostimulant Polynucleotides. J. Drug Delivery Sci. Technol. 2015, 30, 424–434. 10.1016/j.jddst.2015.08.010. [DOI] [Google Scholar]
- Hafner A. M.; Corthésy B.; Merkle H. P. Particulate Formulations for the Delivery of Poly(I:C) as Vaccine Adjuvant. Adv. Drug Delivery Rev. 2013, 65, 1386–1399. 10.1016/j.addr.2013.05.013. [DOI] [PubMed] [Google Scholar]
- Park H.; Adamson L.; Ha T.; Mullen K.; Hagen S. I.; Nogueron A.; Sylwester A. W.; Axthelm M. K.; Legasse A.; Piatak M.; Lifson J. D.; McElrath J. M.; Picker L. J.; Seder R. A. Polyinosinic-Polycytidylic Acid Is the Most Effective TLR Adjuvant for SIV Gag Protein-Induced T Cell Responses in Nonhuman Primates. J. Immunol. 2013, 190, 4103–4115. 10.4049/jimmunol.1202958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin A. D.; Kiick K. L. Tunable Degradation of Maleimide-Thiol Adducts in Reducing Environments. Bioconjugate Chem. 2011, 22, 1946–1953. 10.1021/bc200148v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao J.; Tam J. P. Unprotected Peptides as Building Blocks for the Synthesis of Peptide Dendrimers with Oxime, Hydrazone, and Thiazolidine Linkages. J. Am. Chem. Soc. 1995, 117, 3893–3899. 10.1021/ja00119a001. [DOI] [Google Scholar]
- Cho H.; Daniel T.; Buechler Y. J.; Litzinger D. C.; Maio Z.; Putnam A.-M. H.; Kraynov V. S.; Sim B.-C.; Bussell S.; Javahishvili T.; Kaphle S.; Viramontes G.; Ong M.; Chu S.; GC B.; Lieu R.; Knudsen N.; Castiglioni P.; Norman T. C.; Axelrod D. W.; et al. Optimized Clinical Performance of Growth Hormone with an Expanded Genetic Code. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 9060–9065. 10.1073/pnas.1100387108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pifferi C.; Berthet N.; Renaudet O. Cyclopeptide Scaffolds in Carbohydrate-Based Synthetic Vaccines. Biomater. Sci. 2017, 5, 953–965. 10.1039/C7BM00072C. [DOI] [PubMed] [Google Scholar]
- Yang J.; Li J.; Li X.; Wang X.; Yang Y.; Kawazoe N.; Chen G. Nanoencapsulation of Individual Mammalian Cells with Cytoprotective Polymer Shell. Biomaterials 2017, 133, 253–262. 10.1016/j.biomaterials.2017.04.020. [DOI] [PubMed] [Google Scholar]
- Meng F.; Hennink W. E.; Zhong Z. Reduction-Sensitive Polymers and Bioconjugates for Biomedical Applications. Biomaterials 2009, 30, 2180–2198. 10.1016/j.biomaterials.2009.01.026. [DOI] [PubMed] [Google Scholar]
- Cheng R.; Feng F.; Meng F.; Deng C.; Feijen J.; Zhong Z. Glutathione-Responsive Nano-Vehicles as a Promising Platform for Targeted Intracellular Drug and Gene Delivery. J. Controlled Release 2011, 152, 2–12. 10.1016/j.jconrel.2011.01.030. [DOI] [PubMed] [Google Scholar]
- Lewis G. K.; DeVico A. L.; Gallo R. C. Antibody Persistence and T-Cell Balance: Two Key Factors Confronting HIV Vaccine Development. Proc. Natl. Acad. Sci. U. S. A. 2014, 111, 15614–15621. 10.1073/pnas.1413550111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auffray C.; Sieweke M. H.; Geissmann F. Blood Monocytes: Development, Heterogeneity, and Relationship with Dendritic Cells. Annu. Rev. Immunol. 2009, 27, 669–692. 10.1146/annurev.immunol.021908.132557. [DOI] [PubMed] [Google Scholar]
- Auffray C.; Fogg D.; Garfa M.; Elain G.; Join-Lambert O.; Kayal S.; Sarnacki S.; Cumano A.; Lauvau G.; Geissmann F. Monitoring of Blood Vessels and Tissues by a Population of Monocytes with Patrolling Behavior. Science 2007, 317, 666–670. 10.1126/science.1142883. [DOI] [PubMed] [Google Scholar]
- Yang J.; Zhang L.; Yu C.; Yang X.-F.; Wang H. Monocyte and Macrophage Differentiation: Circulation Inflammatory Monocyte as Biomarker for Inflammatory Diseases. Biomark. Res. 2014, 2, 1. 10.1186/2050-7771-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout R. D.; Suttles J. The Many Roles of CD40 in Cell-Mediated Inflammatory Responses. Immunol. Today 1996, 17, 487–492. 10.1016/0167-5699(96)10060-I. [DOI] [PubMed] [Google Scholar]
- Grewal I. S.; Flavell R. A. CD40 and CD154 in Cell-Mediated Immunity. Annu. Rev. Immunol. 1998, 16, 111–135. 10.1146/annurev.immunol.16.1.111. [DOI] [PubMed] [Google Scholar]
- Thompson C. B. Distinct Roles for the Costimulatory Ligands B7–1 and B7–2 in T Helper Cell Differentiation?. Cell 1995, 81, 979–982. 10.1016/S0092-8674(05)80001-7. [DOI] [PubMed] [Google Scholar]
- Song H.; Huang P.; Niu J.; Shi G.; Zhang C.; Kong D.; Wang W. Injectable Polypeptide Hydrogel for Dual-Delivery of Antigen and TLR3 Agonist to Modulate Dendritic Cells In Vivo and Enhance Potent Cytotoxic T-Lymphocyte Response against Melanoma. Biomaterials 2018, 159, 119–129. 10.1016/j.biomaterials.2018.01.004. [DOI] [PubMed] [Google Scholar]
- Lucey D. R.; Clerici M. Type 1 and Type 2 Cytokine Dysregulation in Human Infectious, Neoplastic, and Inflammatory Diseases. Clin. Microbiol. Rev. 1996, 9, 532–562. 10.1128/CMR.9.4.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams M. A.; Tyznik A. J.; Bevan M. J. Interleukin-2 Signals During Priming Are Required For Secondary Expansion of CD8+ Memory T Cells. Nature 2006, 441, 890–893. 10.1038/nature04790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G.; Goeddel D. V. TNF-R1 Signaling: A Beautiful Pathway. Science 2002, 296, 1634–1635. 10.1126/science.1071924. [DOI] [PubMed] [Google Scholar]
- Vallabhapurapu S.; Karin M. Regulation and Function of NF-ΚB Transcription Factors in the Immune System. Annu. Rev. Immunol. 2009, 27, 693–733. 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- Arango Duque G.; Descoteaux A. Macrophage Cytokines: Involvement in Immunity and Infectious Diseases. Front. Immunol. 2014, 5, 1–12. 10.3389/fimmu.2014.00491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyo N.; Vogel A. B.; Buus S.; Erbar S.; Wee E. G.; Sahin U.; Hanke T. Efficient Induction of T Cells against Conserved HIV-1 Regions by Mosaic Vaccines Delivered as Self-Amplifying MRNA. Mol. Ther.--Methods Clin. Dev. 2019, 12, 32–46. 10.1016/j.omtm.2018.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams M. A.; Bevan M. J. Effector and Memory CTL Differentiation. Annu. Rev. Immunol. 2007, 25, 171–192. 10.1146/annurev.immunol.25.022106.141548. [DOI] [PubMed] [Google Scholar]
- Esser M. T.; Marchese R. D.; Kierstead L. S.; Tussey L. G.; Wang F.; Chirmule N.; Washabaugh M. W. Memory T Cells and Vaccines. Vaccine 2003, 21, 419–430. 10.1016/S0264-410X(02)00407-3. [DOI] [PubMed] [Google Scholar]
- Sallusto F.; Geginat J.; Lanzavecchia A. Central Memory and Effector Memory T Cell Subsets: Function, Generation, and Maintenance. Annu. Rev. Immunol. 2004, 22, 745–763. 10.1146/annurev.immunol.22.012703.104702. [DOI] [PubMed] [Google Scholar]
- Teijeiro-Valiño C.; Novoa-Carballal R.; Borrajo E.; Vidal A.; Alonso-Nocelo M.; de la Fuente Freire M.; Lopez-Casas P. P.; Hidalgo M.; Csaba N.; Alonso M. J. A Multifunctional Drug Nanocarrier for Efficient Anticancer Therapy. J. Controlled Release 2019, 294, 154–164. 10.1016/j.jconrel.2018.12.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.