ABSTRACT
Integrating short DNA fragments at the correct leader-repeat junction is key to successful CRISPR-Cas memory formation. The Cas1–2 proteins are responsible to carry out this process. However, the CRISPR adaptation process additionally requires a DNA element adjacent to the CRISPR array, called leader, to facilitate efficient localization of the correct integration site. In this work, we introduced the core CRISPR adaptation genes cas1 and cas2 from the Type I-D CRISPR-Cas system of Synechocystis sp. 6803 into Escherichia coli and assessed spacer integration efficiency. Truncation of the leader resulted in a significant reduction of spacer acquisition levels and revealed the importance of different conserved regions for CRISPR adaptation rates. We found three conserved sequence motifs in the leader of I-D CRISPR arrays that each affected spacer acquisition rates, including an integrase anchoring site. Our findings support the model in which the leader sequence is an integral part of type I-D adaptation in Synechocystis sp. acting as a localization signal for the adaptation complex to drive CRISPR adaptation at the first repeat of the CRISPR array.
Keywords: CRISPR adaptation, CRISPR leader, spacer acquisition, type I-D CRISPR-Cas system
Conserved nucleotide motifs in the CRISPR leader sequence control spacer acquisition levels of CRISPR-Cas systems
INTRODUCTION
Mobile genetic elements (MGEs) such as bacteriophages and conjugative plasmids exert an evolutionary pressure on prokaryotes, demanding bacterial and archaeal cells to frequently update their immunological lines of defense. Prokaryotes evolved an adaptive immune system that relies on the use of clustered regularly interspaced short palindromic repeats (CRISPRs) and their associated proteins (Cas) in order to specifically recognize and destroy predatory elements. Target recognition is mediated by the synthesis of small RNAs (i.e. crRNA), derived from CRISPR arrays, that guide Cas nuclease complexes towards the invading MGE (Barrangou et al. 2007; Brouns et al. 2008; van der Oost et al. 2014; Marraffini, 2015). The adaptive immune response is created in a step termed CRISPR adaptation in which short MGE-derived sequences are inserted between the repeats giving rise to new ‘spacers’ (Amitai and Sorek 2016; Sternberg, Richter and Charpentier 2016; Jackson et al. 2017). Spacer acquisition is carried out by the adaptation proteins Cas1 and Cas2, which are universally encoded in the vast majority of all types and subtypes of the two major classes of CRISPR-Cas systems (Yosef, Goren and Qimron 2012; Koonin, Makarova and Zhang 2017). However, beyond cas1 and cas2, the region adjacent to the CRISPR array (an A-T rich sequence termed leader; McGinn and Marraffini 2019) as well as the repeat sequence itself are required to guide the integration event towards the correct location (Yosef, Goren and Qimron 2012; Goren et al. 2016). The leader sequence contains the promoter necessary to drive transcription of the CRISPR, but importantly also encodes sequences that are recognized by the Cas1–2 complex and other cellular factors. This includes the integration host factor (IHF) which determines the appropriate integration site at the leader-repeat junction in I-E CRISPR-Cas systems (Nuñez et al. 2016). Localizing the correct integration site is a prerequisite for functional interference and helps to increase the immune diversity which limits the emergence of escape phage mutants (van Houte et al. 2016). Leader encoded adaptation signals likely co-evolved with their cognate adaptation proteins in order to support spacer acquisition rates that aid in establishing an efficient immune response while at the same time limiting the potential costs connected to high acquisition rates (e.g. autoimmunity) (Shah and Garrett 2011; Bradde, Mora and Walczak 2019). In the type I-E system, those adaptation signals are found in the sequence 60 bp upstream of the first repeat that ensure efficient spacer integration (Yosef, Goren and Qimron 2012), while the type I-A system requires at least 400 bp of the leader for detectable levels of acquisition (Rollie et al. 2017). The Cas1–2 complex of the type II-A system relies on intrinsic specificity for a short leader-anchoring site adjacent to the first repeat as well as the repeat itself which both are required and sufficient for catalysis of leader proximal spacer integration (Wei et al. 2015; McGinn and Marraffini 2016; Wright and Doudna 2016; Xiao et al. 2017). This large variation in leader length, sequence conservation and host factor requirements is exemplary for the broad diversity of CRISPR-Cas systems and provides insights in how different adaptation modules are optimized towards their respective CRISPR array. Here, we focus on the spacer acquisition rates of a cyanobacterial type I-D CRISPR-Cas system and find that the presence of several conserved sequences in the CRISPR leader enhances the efficiency of spacer integration. By employing sensitive in vivo spacer acquisition assays in a heterologous E. coli host we demonstrate that spacers can be acquired even in the complete absence of the leader. However, efficient spacer uptake requires the conserved 5’ region of the leader. Our results underline the importance of the leader sequence as a non-protein factor that controls the levels of CRISPR adaptation, and suggest interaction of the leader sequence with the Cas1–2 adaptation machinery itself.
MATERIAL AND METHODS
Bacterial strains and growth conditions
E. coli DH5α and BW25113 strains were grown in Lysogeny Broth (LB) at 37ºC and continuous shaking at 180 rpm or grown on LB agar plates (LBA) containing 1.5% (wt/vol) agar. When required, the media were supplemented with 100 µg ml–1 ampicillin and 25 µg ml–1 chloramphenicol (see Table S1 (Supporting Information) for plasmids and corresponding selection markers).
Plasmid construction and transformation
Plasmids used in this study are listed in Table S1 (Supporting Information) . All cloning steps were performed in E. coli DH5α. Primers described in Table S2 (Supporting Information) were used for PCR amplification of the type I-D CRISPR locus (leader-repeat-spacer1) from Synechocystis sp. 6803 cell material using the Q5 high-fidelity Polymerase (New England Biolabs). PCR amplicons were subsequently cloned into the pACYCDuet-1 vector system (Novagen (EMD Millipore) using restriction-ligation cloning. The pCRISPR leader mutants were obtained by PCR-based mutagenesis using primers listed in Table S2 (Supporting Information) . All plasmids were verified by Sanger-sequencing (Macrogen Europe, Amsterdam, The Netherlands). Bacterial transformations were either carried out by electroporation (200 Ω, 25 μF, 2.5 kV) using a ECM 630 electroporator (BTX Harvard Apparatus) or using chemically competent cells prepared according to manufacturer's manual (Mix&Go, Zymo research). Electrocompetent cells were prepared following a protocol adapted from (Gonzales et al. 2013). Transformants were selected on LBA supplemented with appropriate antibiotics.
In vivo spacer acquisition assay
E. coli BW25113 was transformed with pCas1–2 and pCRISPR with varying lengths of the leader sequence (Table S1, Supporting Information). Cultures were inoculated from single colonies and passaged once after 24 hours of growth at 37˚C and continuous shaking at 180 rpm. 200 µL of cells cultured for 48 hours were harvested by centrifugation and resuspended in 50 µL of MilliQ water. Subsequently, 2 µL of cell suspension was subjected to spacer detection PCR using a forward primer annealing in the 3’ end of the CRISPR repeat of pCRISPR but mismatching the first nucleotide of spacer 1 (degenerated primer mix, BN143 + BN144 + BN145) (Heler et al. 2015) and a reverse primer annealing in the vector backbone (BN172) (Table S2, Supporting Information). PCR products were separated on 2% agarose gels and were densitometrically quantified using ImageLab 4.0 (BioRad). Statistical analysis was done using GraphPad Prism 4 to perform one-way ANOVA followed by Dunnett's multiple comparison test. When a higher sensitivity was required, amplicons of expanded pCRISPR arrays were BluePippin (SageScience) size selected and subjected to a second PCR reaction as described previously (Kieper et al. 2018; McKenzie et al.2019).
Sequencing of acquired spacers
BluePippin extracted and re-amplified expanded CRISPR array amplicons were cloned in the pGemT-easy vector (Promega) and Sanger sequenced (Macrogen Europe, Amsterdam, The Netherlands). Using the Geneious 9.0.5 motif search function, the type I-D repeats were annotated in the sequencing reads and the newly acquired spacers extracted. The origin of newly acquired spacers was determined by nucleotide BLAST search against pCas1–2, pCRISPR and the E. coli BW25113 genome.
RESULTS
The leader displays a high degree of conservation
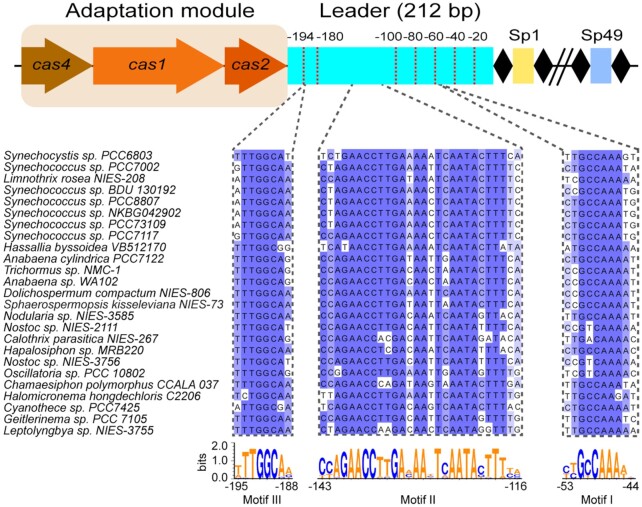
The Cas1–2 adaptation complex is the central element mediating adaptation in almost all CRISPR-Cas systems. It has been proposed that the Cas1 protein co-evolves with its cognate leader as well as the repeat sequence, hence we hypothesized that type I-D Cas1 proteins would recognize conserved motifs within their cognate leader sequences (Shah and Garrett 2011; Alkhnbashi et al. 2016). First, the Cas1 protein of the CRISPR-Cas type I-D system of Synechocystis PCC6803 was used in a BLASTP-search to identify related Cas1 proteins in a variety of different species. Interestingly, most Cas1 proteins that were found were derived from cyanobacterial type I-D systems (Fig. 1). Next, we retrieved the leader sequences (defined as the A-T rich adjacent upstream sequence of the CRISPR array; Jansen et al. 2002) from type I-D systems containing a Cas1 ortholog with at least 60% sequence identity. Below this conservation threshold value we noticed that Cas1 orthologs were more divergent (sequence identity < 40%), and were excluded from the analysis. The 25 selected I-D leader sequences ranged from 202 to 220 bp which represents considerably longer leaders than described for the E. coli type I-E system which are typically shorter than 100 bp (Yosef, Goren and Qimron 2012). We then performed MAFFT alignment of the leaders (Katoh et al. 2002; Katoh and Standley 2016) and identified three regions with more than four consecutive nucleotides that were highly conserved across all the 25 leader sequences (Fig. 1; motifs I-II-III). Interestingly, we found a high degree of conservation at the repeat distal end (II + III) of the leader, while the repeat proximal region displayed more variability with only one conserved motif (I). Altogether, the high conservation of those motifs in the leader sequence suggests that those regions are important for the correct localization of the leader of the CRISPR array, and could serve as recognition signals for the Cas1–2 adaptation complex or host factors to ensure spacer integration at the leader-repeat junction.
Figure 1.
Type I-D arrangement of the adaptation module and the CRISPR array. Downstream of cas2 is the 212 bp leader sequence. Conserved regions obtained from MAFFT alignments of 25 leaders reveal conserved motifs predominantly at the repeat distal end with increasing sequence variability at the repeat proximal end. Sequence conservation is summarized in Weblogo3 depictions (Crooks et al., 2004). Leader truncations from the repeat-distal end for experimental investigation of conserved motifs are indicated with red dashed lines.
Leader motifs stimulate spacer acquisition
To get experimental insight into the previously identified conserved regions, we systematically shortened the leader from the repeat-distal end while leaving the repeat-proximal leader intact. The different CRISPR leader-repeat-spacer1 plasmids were transformed into E. coli K12 cells containing only Cas1 and Cas2. The cas4 gene was omitted because we showed previously that the Cas1–2 adaptation proteins are necessary and sufficient to mediate the acquisition of new spacers (Kieper et al. 2018). After 48 hours of growth, spacer acquisition was assessed by a degenerate primer PCR (McKenzie et al. 2019) and acquisition efficiency was quantified from three independent assays based on the relative difference between the band intensity of the expanded CRISPR amplicon compared to the non-expanded CRISPR array (Xue et al. 2015) (Fig. 2A; Fig. S2, Supporting Information). We observed decreasing adaptation efficiencies depending on the presence or absence of the repeat-distal motifs (Fig. 2A). The highest rate of spacer acquisition was obtained with at least 194 bp of the full-leader sequence (212 and 194 constructs) containing conserved motifs II and III. However, further repeat distal truncations of the leader led to significantly impaired spacer uptake (Fig. 2A). Expansion of the CRISPR array is readily detectable with PCR up to a leader length of 60 bp (preserving only motif I) although with a relative reduction compared to leaders containing motif II and III. Spacer integration with leaders shorter than 60 bp is below the detection limit of the first PCR and can only be detected using a more sensitive second round of PCR (Fig. 2B) as described by McKenzie et al. (2019). With this method, we were able to detect spacer integration even in the absence of the leader. The sequence analysis of spacers that were acquired in the absence of the leader (0 Leader) revealed that the detected integration event gave rise to a single unique spacer (Fig. S1, Supporting Information). This very low spacer diversity indicates that the Cas1–2 adaptation complex is able to integrate spacers at the leader-repeat junction even in absence of the leader sequence, albeit at drastically reduced rates.
Figure 2.
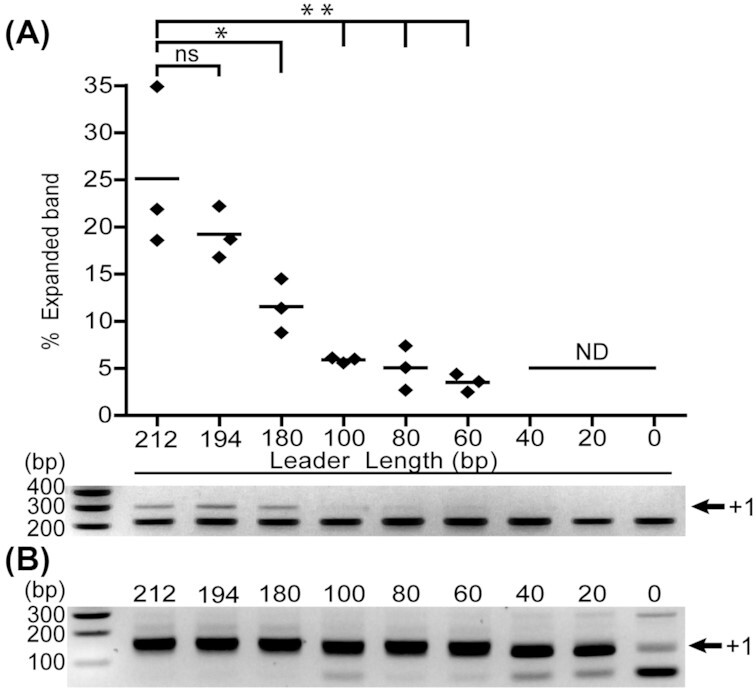
PCR-based detection of spacer acquisition at variable leader length. A—Quantification of expanded CRISPR array band intensity (n = 3). CRISPR adaptation is negatively affected by deletion of motif III that is present in the 212 bp (wild-type) and 194 bp leader (*P < 0.05). Removal of motif II (located in the segment between 180 bp and 100 bp) and motif III significantly reduced (**P < 0.01) acquisition rates close to the detection limit of this PCR. Leaders shorter than 60 bp do not support detectable acquisition (ND). Spacer acquisition rates of the 194 bp leader are not significantly different (ns) from the full 212 bp leader. Statistical significance was calculated using Dunnett's multiple comparisons test. B—Second round of PCR enables the detection of spacer acquisition with leaders shorter than 60 bp or absent leader sequences.
DISCUSSION
During phage infection the integration of novel spacers at the correct site as well as at an appropriate rate is crucial for prokaryotic survival. Recently, it was demonstrated that Cas1–2 can integrate spacers into non-CRISPR genomic regions, however, those non-canonical integration events do not lead to functional spacers that confer CRISPR resistance against sampled invaders (Nivala, Shipman and Church 2018). Therefore, since only acquisitions in the CRISPR array provide the most efficient immune response, Cas1–2 must recognize the correct insertion site. Moreover, spacer integration occurs in a polarized manner at the leader proximal end of the array creating a chronological library of past infections that provides higher levels of protection from the most recently integrated spacer (McGinn and Marraffini 2016). Specificity of the integration reaction towards the cognate CRISPR array might thus be one of the rate limiting factors for rapid and efficient immunization. Here, we demonstrated the importance of conserved leader sequences for naïve acquisition in a minimal I-D CRISPR-Cas system. The alignment of leader sequences from different type I-D systems revealed a conserved region at the repeat distal end as well as a short conserved motif approximately 50 bp upstream of the first repeat, suggesting involvement of those regions in CRISPR array recognition, potentially by the adaptation complex. By systematically truncating the leader from the repeat distal end while leaving downstream sequences intact, we disrupted those leader regions and quantified spacer integration by a semi-quantitative PCR method (Xue et al. 2015). Strikingly, we were able to detect spacer acquisition in vivo even in the complete absence of the leader sequence by using a sensitive detection method. However, the efficiency of spacer integration is drastically reduced in the absence of the leader. Sequencing of the integration event revealed that only a single unique spacer was acquired. In the absence of the leader the type I-D adaptation complex displays baseline adaptation levels, but this low efficiency event only marginally contributes to protection of the population. In contrast, including at least 60 bp upstream of the I-D repeat increased acquisition rates to detectable levels, demonstrating that motif I (5’-GCCAAA-3’) facilitates spacer integration. However, the maximum acquisition rate was only restored when the full leader was provided. Similar results have been obtained in vitro for a Sulfolobus type I-A CRISPR-Cas system that requires at least 400 bp of the leader for detectable acquisition and the full 531 bp leader for maximum adaptation levels (Rollie et al. 2017). Furthermore, in a type I-A system of a related Sulfolobus strain a ∼20 bp deletion within the leader sequence is associated with decreased spacer uptake (Erdmann and Garrett, 2012; Garrett et al. 2015). Our findings are consistent with the observation that deletions of particular leader sequences result in decreased acquisition rates, although future studies are needed to address whether this is caused by the loss of a specific motif, an accumulating effect of deleting several motifs or because a certain spacing between e.g. motif III and the repeat is required. In the type I-E system IHF binds a conserved leader motif called IHF-binding site and induces a 120º bend that brings another conserved motif, the 5’-TTGGT-3’ integrase anchoring site, in proximity to the leader-repeat junction that increases acquisition efficiency by presumably stabilizing the Cas1–2-leader-repeat interaction (Nuñez et al. 2016; Yoganand et al. 2017). Interestingly, motif III (5’-TTGGC-3’) in the type I-D leader strongly resembles the integrase anchoring site described previously. It is plausible that the type I-D Cas1–2 adaptation complex, analogous to the type I-E complex, can recognize this motif to be correctly positioned to integrate novel spacers. However, the E. coli IHF protein is absent from Synechocystis sp. 6803 suggesting that other DNA-binding host factors could be involved in recognizing the conserved region II in the type I-D leader. Overall, our work highlights the importance of the leader sequence for the adaptation stage in the type I-D CRISPR-Cas system. Through evolutionary selection of specific sequences in the leader that likely interact with the adaptation proteins, the integration of new spacers into CRISPR arrays occurs accurately at the first repeat of the CRISPR array improving the chances of prokaryotes to survive predatory invasion.
Supplementary Material
ACKNOWLEDGEMENTS
SJJB likes to thank funding sources FOM [Projectruimte 15PR3188–2]; and European Research Council Stg grant [638707].
Author contributions: SNK, CA and SJJB designed research. SNK and CA performed the research. SNK and CA analyzed data. SNK, CA and SJJB wrote the paper.
Conflicts of interests . None declared.
REFERENCES
- Alkhnbashi OS, Shah SA, Garrett RAet al.. Characterizing leader sequences of CRISPR loci. Bioinformatics. 2016;32:i576–85. [DOI] [PubMed] [Google Scholar]
- Amitai G, Sorek R. CRISPR-Cas adaptation: insights into the mechanism of action. Nat Rev Microbiol. 2016;14:67–76. [DOI] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau Het al.. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–12. [DOI] [PubMed] [Google Scholar]
- Bradde S, Mora T, Walczak AM. Cost and benefits of clustered regularly interspaced short palindromic repeats spacer acquisition. Philos Trans R Soc B Biol Sci. 2019;374:20180095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouns SJJ, Jore MM, Lundgren Met al.. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia J-Met al.. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann S, Garrett RA. Selective and hyperactive uptake of foreign DNA by adaptive immune systems of an archaeon via two distinct mechanisms. Mol Microbiol. 2012;85:1044–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett RA, Shah SA, Erdmann Set al.. CRISPR-cas adaptive immune systems of the sulfolobales: unravelling their complexity and diversity. Life (Basel, Switzerland). 2015;5:783–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales MF, Brooks T, Pukatzki SUet al.. Rapid Protocol for Preparation of Electrocompetent Escherichia coli and Vibrio cholerae. J Vis Exp. 2013;80:50684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goren MG, Doron S, Globus Ret al.. Repeat size determination by two molecular rulers in the type I-E CRISPR array. Cell reports. 2016;16:2811–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heler R, Samai P, Modell JWet al.. Cas9 specifies functional viral targets during CRISPR-Cas adaptation. Nature. 2015;519:199–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SA, McKenzie RE, Fagerlund RDet al.. CRISPR-Cas: adapting to change. Science. 2017;356:eaal5056. [DOI] [PubMed] [Google Scholar]
- Jansen R, Embden JDAV, Gaastra Wet al.. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43:1565–75. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K-iet al.. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. A simple method to control over-alignment in the MAFFT multiple sequence alignment program. Bioinformatics. 2016;32, 1933–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieper SN, Almendros C, Behler Jet al.. Cas4 Facilitates PAM-Compatible Spacer Selection during CRISPR Adaptation. Cell reports. 2018;22:3377–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Makarova KS, Zhang F. Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol. 2017;37:67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA. CRISPR-Cas immunity in prokaryotes. Nature. 2015;526:55. [DOI] [PubMed] [Google Scholar]
- McGinn J, Marraffini LA. CRISPR-Cas systems optimize their immune response by specifying the site of spacer integration. Mol Cell. 2016;64:616–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinn J, Marraffini LA. Molecular mechanisms of CRISPR–Cas spacer acquisition. Nat Rev Microbiol. 2019;17:7–12. [DOI] [PubMed] [Google Scholar]
- McKenzie RE, Almendros C, Vink JNAet al.. Using CAPTURE to detect spacer acquisition in native CRISPR arrays. Nat Protoc. 2019;14:976–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nivala J, Shipman SL, Church GM. Spontaneous CRISPR loci generation in vivo by non-canonical spacer integration. Nature Microbiol. 2018;3:310–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuñez JK, Bai L, Harrington LBet al.. CRISPR immunological memory requires a host factor for specificity. Mol Cell. 2016;62:824–33. [DOI] [PubMed] [Google Scholar]
- Rollie C, Graham S, Rouillon Cet al.. Prespacer processing and specific integration in a Type I-A CRISPR system. Nucleic Acids Res. 2017;46:1007–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SA, Garrett RA. CRISPR/Cas and Cmr modules, mobility and evolution of adaptive immune systems. Res Microbiol. 2011;162:27–38. [DOI] [PubMed] [Google Scholar]
- Sternberg SH, Richter H, Charpentier E. Adaptation in CRISPR-Cas Systems. Mol Cell. 2016;61:797–808. [DOI] [PubMed] [Google Scholar]
- van der Oost J, Westra ER, Jackson RNet al.. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat Rev Microbiol. 2014;12:479–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Houte S, Ekroth AKE, Broniewski JMet al.. The diversity-generating benefits of a prokaryotic adaptive immune system. Nature. 2016;532:385–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Chesne MT, Terns RMet al.. Sequences spanning the leader-repeat junction mediate CRISPR adaptation to phage in Streptococcus thermophilus. Nucleic Acids Res. 2015;43:1749–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AV, Doudna JA. Protecting genome integrity during CRISPR immune adaptation. Nat Struct Mol Biol. 2016;23:876. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Ng S, Nam KH, Ke A. How type II CRISPR–Cas establish immunity through Cas1–Cas2-mediated spacer integration. Nature. 2017;550:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue C, Seetharam AS, Musharova Oet al.. CRISPR interference and priming varies with individual spacer sequences. Nucleic Acids Res. 2015;43:10831–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoganand KNR, Sivathanu R, Nimkar Set al.. Asymmetric positioning of Cas1–2 complex and integration host factor induced DNA bending guide the unidirectional homing of protospacer in CRISPR-Cas type I-E system. Nucleic Acids Res. 2017;45:367–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef I, Goren MG, Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012;40:5569–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.