Abstract
Two mycobacterial strains with close similarity to the Mycobacterium tuberculosis complex (MTBC) were isolated from cutaneous lesions of patients in the USA and Italy. At the phenotypic level, similarities to the MTBC included slow growth rate, rough morphotype of the unpigmented colonies and nearly identical high-performance liquid chromatography profiles of mycolic acids. In contrast to the MTBC, the strains were niacin- and nitrate-negative, and catalase-positive both at 68°C and in semi-quantitative tests. The clinical isolates were more closely related to M. tuberculosis than to any other known mycobacterium and scored positive with commercial DNA probes (Hologic AccuProbe M. tuberculosis). Both average nucleotide identity and genome-to-genome distance suggested the strains are different from the MTBC. Therefore, given the distinguishing phenotypic and genomic-scale differences, we submit that the strains belong to a new species we have named Mycobacterium decipiens with type strain TBL 1200985T (=ATCC TSD-117T=DSM 105360T).
Keywords: Mycobacterium decipiens, Mycobacterium tuberculosis complex, average nucleotide identity, whole genome sequencing
Previously, a number of mycobacterial species have been reported to demonstrate high-level genotypic relatedness with the Mycobacterium tuberculosis complex (MTBC) and yield false positive results with commercial amplification methods [1] or with DNA probes specific for the MTBC [2, 3]. Here we describe a novel mycobacterium species closely related to the MTBC, but distinct from it.
The first strain, TBL 1200985T, was isolated in 2012 in the USA from a 58-year-old female reporting swelling and pain at the right thumb and wrist for 8 months. Histology revealed granulomatous synovitis and a slowly growing nonpigmented mycobacterium grew by culture of two resected samples. The patient recovered after 18 months of therapy, including a standard anti-tuberculosis regimen which was subsequently adjusted to azithromycin and moxifloxacin [4].
A second strain, FI-16190, was isolated in 2016 from a 5- year-old girl in Italy presenting with abdominal swelling, pain and fever. A biopsy of the intra-abdominal lymph node revealed granulomatous lymphadenitis and yielded mycobacterium in culture.
The adult and child reported that their initial symptom onset was after returning from a vacation in a tropical area, the US Virgin Islands and the Republic of Maldives, respectively. In addition, upon returning from their trips, both the adult and child reported small wounds, at the hand and heel, respectively. An interphalangeal joint culture from the adult patient and an inguinal lymph node tissue culture from the child revealed acid-fast bacilli.
Non-pigmented rough colonies, morphologically compatible with the MTBC, developed in about 20 days at 25 and 37 °C. No pigmentation developed after light exposure. No growth was observed on standard media at 42 °C or on Mac-Conkey agar without crystal violet at 37 °C. Semi-quantitative and thermostable catalase tests were positive; the isolates also exhibited tellurite reduction. Unlike M. tuberculosis, the strains were negative for niacin accumulation, nitrate reduction, Tween 80 hydrolysis, urease activity and β-glucosidase activity [5].
The strains were positive with the MTBC-specific probes (Hologic AccuProbe) [6], while they were identified as non-specified Mycobacterium by GenoType CM (Hain Life- science) [7].
High-performance liquid chromatography profiles of cell-wall mycolic acids [8] of the two strains were obtained using the Sherlock Mycobacteria Identification System (SMIS; midi) and produced a pattern of five major peaks eluting between 7 and 8min (Fig. 1). This profile closely resembles that of M. tuberculosis (Sherlock software similarity index 0.749).
Fig. 1.

Representative mycolic acid patterns of strain TBL 1 200985T and M. tuberculosis. LMMIS, low molecular mass internal standard; HMMIS, high molecular mass internal standard.
The determination of minimum inhibitory concentrations (MICs) of antimicrobials potentially active against slow-growing mycobacteria [9] was performed using commercial plates (Sensititre slomyco, Thermo Fisher Scientific). Both strains showed susceptibility to seven agents: amikacin, clarithromycin, doxycycline (susceptible/intermediate), line-zolid, moxifloxacin, rifabutin and trimethoprim-sulfameth-oxazole, with resistance to rifampicin. Differences in susceptibility to ethambutol and ciprofloxacin were observed between the two strains (Table 1).
Table 1.
Minimum inhibitory concentrations (MICs) of the clinical isolates S, susceptible; I, intermediate; R, resistant.
| Antimicrobial tested | TBL 1200985T
MIC (μg ml −1) (Interpretation) |
FI-16190 MIC (μg ml −1) (Interpretation) |
|---|---|---|
| Amikacin | ≤1 (S) | 4 (S) |
| Ciprofloxacin | 4 (R) | 1 (S) |
| Clarithromycin | 4 (S) | 0.25 (S) |
| Doxycycline | 0.5 (S) | 2 (I) |
| Ethambutol | 4 (I) | 8 (R) |
| Linezolid | ≤1 (S) | ≤1 (S) |
| Moxifloxacin | 0.5 (S) | ≤0.12 (S) |
| Rifabutin | ≤0.25 (S) | ≤0.25 (S) |
| Rifampicin | 2 (R) | 2 (R) |
| Trimethoprim/sulfamethoxazole | 0.25/4.75 (S) | 0.5/9.5 (S) |
The whole genome sequences of both clinical isolates were produced using the Illumina platform and Nextera reagents (Illumina) according to the manufacturer’s protocol. Reads were quality trimmed with TrimGalore and assembled using SPAdes version 3.9.1 software [10]. The genomes of TBL 1200985T and FI-16190 exhibited similar features (Table 2).
Table 2.
Salient genomic features of the Mycobacterium decipiens clinical isolates CDS, coding DNA sequences.
| Strain | Mean coverage | Number of contigs | G+C content (%) | Genome size (bp) | Number of CDS |
|---|---|---|---|---|---|
| TBL 1200985T | 44.9× | 142 | 65.4 | 5 228 000 | 4878 |
| FI-16190 | 44.1× | 176 | 65.5 | 5 432 000 | 4676 |
DNA gene sequences of the entire 16S rRNA gene and of the hypervariable regions of the hsp65 (401 bp) [11] and rpoB (720 bp) genes [12] were extracted from the genomic sequences of the two strains. TBL 1200985T and FI-16190 had identical 16S rRNA and hsp65 sequences while the rpoB sequences differed by only 1 bp. Their 16S sequences were 99.4 % similar to M. tuberculosis, and consequently with all other members of the MTBC, with only seven basepair mismatches. For the hsp65 gene, a large number of species, including MTBC members, were characterized by similarity around 94–95 %, with Mycobacterium intracellulare being the closest (95.3 %). For rpoB, the clinical isolates diverged from every known species, with M. tuberculosis exhibiting the highest resemblance (89.5 and 89.7 % in the two strains).
In addition, the hsp65 gene hypervariable region allowed us to infer the PRA (PCR restriction analysis) pattern of the isolates. The enzyme BstEII produced two restriction fragments of 310 and 116 bp, while HaeIII produced three fragments of 127, 112 and 69 bp. Mycobacterium kumamo-tonense and Mycobacterium gordonae were the species presenting the closest resemblance with this pattern whilst the MTBC patterns were clearly different.
Due to high 16S rRNA gene similarity (99.4 %) between the clinical isolates and the MTBC, the calculation of the average nucleotide identity (ANI) [13] was needed [14]. Both TBL 1200985T and FI-16190 scored lower than the boundary of species (95–96%) in comparison to different members of the MTBC (Table 3 and Fig. S1, available in the online version of this article). As noted in Table 3, one isolate of each of seven species of the MTBC were used for comparison. These species show >99 % similarity to each other, which suggests they represent a single species, as discussed in a recent paper [15]. An analogous result was achieved by calculating the genome-to-genome distance (GGD) [16] (http://ggdc.dsmz.de/), the in silico equivalent of DNA-DNA hybridization (Table 3).
Table 3.
ANI and (GGD) % scores between members of M. tuberculosis complex and Mycobacterium decipiens clinical isolates*
| Accession number |
TBL
1200985T |
M.
tuberculosis |
M.
africanum |
M. bovis
BCG |
M. bovis | ‘M. canettii’ | M. caprae |
M.
microti |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M. tuberculosis H37Rv | NC_000962 | 86.02 (30.20) | ||||||||||||
| M. africanum strain 25 | CP010334 | 86.07 | (30.20) | 99.81 | (96.70) | |||||||||
| M. bovis BCG ATCC 35743 | NZ_CP003494 | 86.04 (30.10) | 99.77 (96.70) | 99.77 (95.70) | ||||||||||
| M. bovis ATCC BAA935 | NZ_CP009449 | 86.10 | (30.20) | 99.79 | (96.60) | 99.78 | (96.40) | 99.75 (96.60) | ||||||
| ‘M. canettii’ CIPT 140010059 | NC_015848 | 86.03 | (30.20) | 99.25 | (91.20) | 99.24 | (91.40) | 99.16 | (89.80) | 99.20 | (90.50) | |||
| M. caprae strain Allgeau | NZ_CP016401 | 86.08 | (30.20) | 99.86 | (97.40) | 99.81 | (97.30) | 99.82 | (96.90) | 99.81 | (97.10) | 99.27 (91.50) | ||
| M. microti strain 12 | CP010333 | 86.05 | (30.10) | 99.85 | (97.10) | 99.82 | (98.00) | 99.75 | (96.10) | 99.78 | (93.50) | 99.27 (91.50) | 99.85 (97.40) | |
| FI-16190 | – | 99.63 | (97.30) | 86.02 | (30.10) | 85.92 | (30.10) | 85.95 | (30.10) | 85.94 | (30.10) | 86.01 (30.10) | 85.96 (30.20) | 85.87 (30.10) |
ANI scores <95 % and GGD scores <70 % are indicative of belonging to different species; ANI scores >96 % and GGD scores >70 % are indicative of belonging to the same species.
To perform phylogenetic analysis, the sequences of closely related mycobacterium species were retrieved from Gen-Bank, aligned with clustal_w [17] and trimmed to start and end at the same nucleotide position. The neighbour-joining method using the Tamura-Nei distance model [18, 19] with 1000 bootstrap replicates was used to reconstruct the trees based on 16S rRNA, hsp65 and rpoB gene sequences individually and concatenated. Mycobacterium abscessus was chosen as an outgroup. Different trees placed TBL 1200985T and FI-16190 within the same clade as M. tuberculosis (Figs 2 and 3, S2 and S3).
Fig. 2.
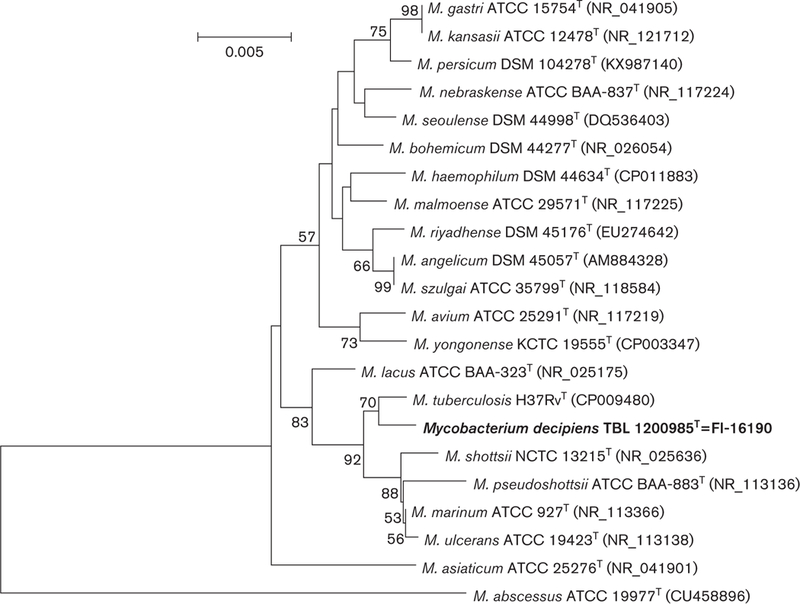
Phylogenetic tree based on 16S rRNA sequences of representative species of the genus Mycobacterium, reconstructed using the neighbour-joining method bootstrapped 1000 times. Bootstrap values >50 are given at nodes. Bar, 0.005 substitutions per nucleo-tide position.
Fig. 3.
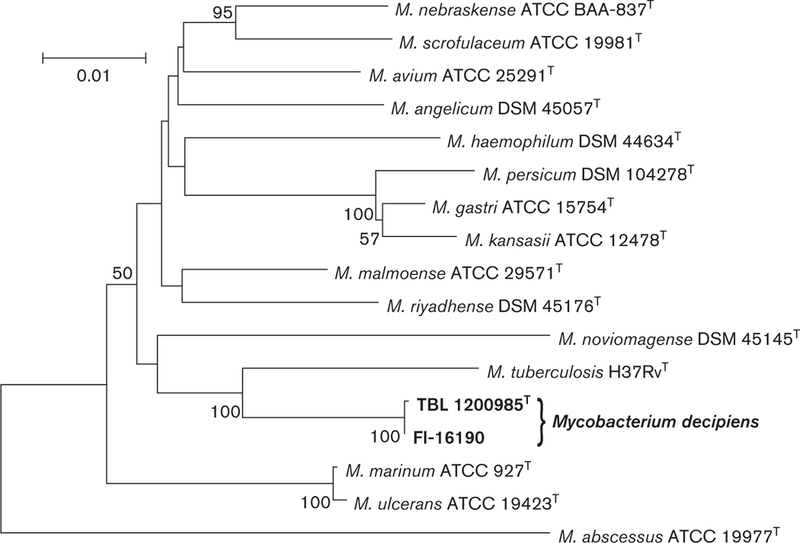
Phylogenetic tree based on concatenated sequences of 16S rRNA, hsp65 and rpoB gene sequences of representative species of the genus Mycobacterium, reconstructed using the neighbour-joining method bootstrapped 1000 times. Bootstrap values >50 are given at nodes. Bar, 0.01 substitutions per nucleotide position.
DESCRIPTION OF MYCOBACTERIUM DECIPIENS SP. NOV.
Mycobacterium decipiens (de.ci’pi.ens L. part. adj. decipiens, deceiver, characterized by deceptive features leading to mis-identification as M. tuberculosis).
Non-motile, non-spore-forming and acid fast. Grows on solid media in approximately 15–20 days at temperatures ranging from 25 to 37 °C, produces rough colonies, and is non-pigmented regardless of exposure to light or dark conditions. The species differs from M. tuberculosis in its inability to accumulate niacin or to reduce nitrates, and in its production of thermostable catalase at high (>45 mm) levels. The commercial AccuProbe misidenti- fied M. decipiens as a member of the MTBC. In the 16S rRNA and rpoB genes, the species is most closely related to the MTBC. The strains were largely susceptible to most antimicrobials, with both being resistant to only rifampicin. The ANI between respective genomes (confirmed by GGD) is supportive of the status of species as independent from the MTBC. The genome sizes and number of coding DNA sequences of the clinical isolates was 5 228 890 bp and 4878 genes (TBL 1200985T), and 5 432 050 bp containing 4676 genes (FI-16190).
The type strain is TBL 1200985T (=ATCC TSD-117T=DSM 105360T).
Supplementary Material
Acknowledgments
Funding information
The authors received no specific grant from any funding agency.
The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA, hsp65 and rpoB gene sequences of strain TBL 1200985 are KF683289, KJ371035 and KJ371034; the rpoB gene sequences of strain FI-16190 is KY657270. The accession number of the shotgun gene sequence of strain TBL 1 200985 is NCXP00000000.
Abbreviations:
- ANI
Average nucleotide identity
- GGD
Genome-to-genome distance
- HMMIS
High molecular mass internal standard
- LMMIS
Low molecular mass internal standard
- MIC
Minimum inhibitory concentration
- MTBC
Mycobacterium tuberculosis complex
Footnotes
Conflicts of interest
The authors declare that there are no conflicts of interest.
References
- 1.Saito H, Iwamoto T, Ohkusu K, Otsuka Y, Akiyama Y et al. Mycobacterium shinjukuense sp. nov., a slowly growing, non-chromo-genic species isolated from human clinical specimens. Int J Syst Evol Microbiol 2011;61:1927–1932. [DOI] [PubMed] [Google Scholar]
- 2.Tortoli E, Rogasi PG, Fantoni E, Beltrami C, de Francisci A et al. Infection due to a novel mycobacterium, mimicking multidrug-resistant Mycobacterium tuberculosis. Clin Microbiol Infect 2010;16: 1130–1134. [DOI] [PubMed] [Google Scholar]
- 3.van Ingen J, Al-Hajoj SA, Boeree M, Al-Rabiah F, Enaimi M et al. Mycobacterium riyadhense sp. nov., a non-tuberculous species identified as Mycobacterium tuberculosis complex by a commercial line-probe assay. Int J Syst Evol Microbiol 2009;59: 1049–1053. [DOI] [PubMed] [Google Scholar]
- 4.Simner PJ, Hyle EP, Buckwalter SP, Branda JA, Brown-Elliott BA et al. Tenosynovitis caused by a novel nontuberculous Mycobacterium species initially misidentified as a member of the Mycobacterium tuberculosis complex. J Clin Microbiol 2014; 52:4414–4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kent PT, Kubica GP. Public Health Mycobacteriology. A Guide for the Level III Laboratory. Atlanta: U.S: Department of Health and Human Services; 1985. [Google Scholar]
- 6.Butler WR, O’Connor SP, Yakrus MA, Gross WM. Cross-reactivity of genetic probe for detection of Mycobacterium tuberculosis with newly described species Mycobacterium celatum. J Clin Microbiol 1994;32:536–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tortoli E, Pecorari M, Fabio G, Messino M, Fabio A. Commercial DNA probes for mycobacteria incorrectly identify a number of less frequently encountered species. J Clin Microbiol 2010; 48:307–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.CDC. Standardized Method for HPLC Identification of Mycobacteria. Atlanta: U.S: Department of Health and Human Services, Public Health Service; 1996. [Google Scholar]
- 9. CLSI. Susceptibility Testing of Mycobacteria, Nocardiae and Other Aerobic Actinomycetes; Approved standard, 2nd ed. M24-A2. Wayne, PA: CLSI; 2011. [PubMed] [Google Scholar]
- 10.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 2012;19:455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Telenti A, Marchesi F, Balz M, Bally F, Böttger EC et al. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J Clin Microbiol 1993;31:175–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adékambi T, Colson P, Drancourt M. rpoB-based identification of nonpigmented and late-pigmenting rapidly growing mycobacteria. J Clin Microbiol 2003;41:5699–5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee I, Ouk Kim Y, Park SC, Chun J. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol 2016;66:1100–1103. [DOI] [PubMed] [Google Scholar]
- 14.Kim M, Oh HS, Park SC, Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol 2014;64:346–351. [DOI] [PubMed] [Google Scholar]
- 15.Riojas MA, McGough KJ, Rider-Riojas CJ, Rastogi N, Hazbón MH. Phylogenomic analysis of the species of the Mycobacterium tuberculosis complex demonstrates that Mycobacterium africanum, Mycobacterium bovis, Mycobacterium caprae, Mycobacterium microti and Mycobacterium pinnipedii are later heterotypic synonyms of Mycobacterium tuberculosis. Int J Syst Evol Microbiol 2018;68:324–332. [DOI] [PubMed] [Google Scholar]
- 16.Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 2013;14:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson JD, Higgins DG, Gibson TJ. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res 1994:11:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 1987;4:406–425. [DOI] [PubMed] [Google Scholar]
- 19.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 2013;30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


