Abstract
Background
DNA aneuploidy has attracted growing interest in clinical practice. Nevertheless, its prognostic value in gastric cancer patients remains controversial. This meta-analysis aims to explore the impact of DNA ploidy status on the survival of gastric cancer patients.
Methods
We used PubMed and Web of Science databases to retrieve relevant articles. The correlation between DNA aneuploidy and the clinicopathological features of gastric cancer, such as stage, depth of invasion (T), lymph node metastasis (N), distant metastasis (M), differentiation (G), tumor types (Lauren classification) and overall survival (OS) were evaluated. Hazard ratios (HRs) with corresponding 95% confidence intervals (CIs) were collected carefully from each article OS was presented with HRs. The relationships between DNA aneuploidy and each characteristic were analyzed using risk ratios (RR) and a 95% confidence interval (CI). Significance was established using P < 0.05. Funnel plot was conducted to detect the publication bias.
Results
After careful selection, 25 studies involving 3449 cases were eligible for further analyses. Patients with DNA aneuploidy were considered at risk of more advanced stages (stage III-IV vs. stages I-II, RR = 1.23; 95% CI, 1.07 to 1.42; P = 0.003), lymph node metastasis (N+ vs. N-: RR = 1.43; 95% CI, 1.12 to 1.82, P = 0.004), and intestinal tumor type (intestinal vs. diffuse: RR = 1.45; 95% CI, 1.02 to 2.06; P = 0.04). And an adverse relation was observed between DNA aneuploidy and tumor differentiation. While no association was found between DNA aneuploidy and distant metastasis (P = 0.42) nor depth of tumor invasion (P = 0.86). Regarding overall survival, aneuploid tumors were associated with worse survival in all patients (P < 0.00001).
Conclusions
We found that DNA aneuploidy was an important predictor for gastric cancer patients, and should be used as a potential biomarker for further classification in gastric cancer.
Electronic supplementary material
The online version of this article (10.1186/s12885-019-5869-9) contains supplementary material, which is available to authorized users.
Keywords: Aneuploidy, Gastric cancer, Survival
Background
Gastric cancer was the third leading cause of cancer-related death, and was responsible for 723,000 deaths all over the world [1]. At present, proliferation markers such as the histopathological TNM (tumor-node-metastasis) classification and Lauren classification have been extensively adopted for predicting survival in gastric cancer [2]. Nonetheless, there is a large variation in survival of gastric cancer patients with similar TNM stage or Lauren classification. Therefore, it is required to explore more precise markers at a molecular level to increase the precision of prognosis prediction for individual patients [3]. For decades there has been a growing interest in chromosomal instability, which was defined by DNA aneuploidy, to classify gastric carcinoma into molecular subtype groups [1].
DNA aneuploidy was found in the majority (70–90%) of cancer cells, and was defined as the set of an abnormal number of chromosomes in cells [4, 5]. DNA aneuploidy reflected a high genotypic instability, leading to malignant behavior of cells. Numerous articles which investigated DNA aneuploidy in various cancers have been published. Its role in non-small cell lung cancer, colorectal cancer and breast cancer seems clear. It was observed that patients with DNA aneuploid tumor had worse clinical outcomes in non-small cell lung cancer, colorectal cancer and breast cancer [6–9].
Previous studies [10–34] which focused on the correlation between DNA aneuploidy and survival in gastric cancer reported conflicting results. Consequently, we conducted this meta-analysis to explore the prognostic value of DNA aneuploidy status in gastric cancer patients.
Methods
Search strategy and study selection
Our meta-analysis was conducted following the Preferred Reported Items for Systematic Reviews and Meta- Analysis (PRISMA) statement [35]. We searched relevant articles from two databases (PubMed, and Web of Science March 20th 2018). Articles were identified using the following keywords: ‘DNA ploidy/aneuploidy’, ‘gastric cancer/carcinoma’ and ‘survival’. There was no restriction of English language during our search.
Articles were picked out on the basis of inclusion criteria as follows: 1. sufficient information of baseline clinicopathological characteristics of gastric cancer patients; 2. the research focused on the association between overall survival (OS) and DNA aneuploidy; 3. proven histological diagnosis of gastric cancer prior to anticancer therapies; 4. DNA aneuploidy status were presented specificly; 5. value of Hazard ratio (HR) with a 95% confidence interval (CI). Alternatively, the Kaplan-Meier (K-M) curve or a Cox regression model was provided..
Literature retrieval and selection was cautiously performed by two authors (JX and RZ). To avoid overlap of patient cohort, we included the latest studies with largest samples from the same organization. All authors reached consensus and made final decision through thorough discussion.
Data extraction
Two authors (JX, RZ) independently extracted the useful data from all eligible articles. The collected information was listed as follows: the last name of the first author, publication date, sample size, mean or median age of population, DNA ploidy methods, specimens, disease stage, follow-up period, treatments and OS. The risk ratios (RRs) of DNA aneuploidy to tumor stage, depth of invasion (T), lymph node metastasis (N), distant metastasis (M), differentiation (G), tumor types (intestinal and diffuse, based on Lauren classification), and HRs of OS with 95% CIs were checked. If the value of HRs were not provided straightforwardly, survival data were estimated using K-M curves by Engauge Digitizer 4.1.
Quality assessment of primary studies
The Newcastle-Ottawa Scale (NOS) scale was occupied to assess the quality of included papers.
Statistical analysis
We conducted statistical analyses on the basis of the Cochrane Collaboration Guidelines. The pooled RRs were estimated by the Mantel-Haenszel’s method, whereas the pooled HRs were calculated by the inverse variance method. The homogeneity assumption was investigated by Cochran’s Q statistic. The Higgins’ I2 index was calculated to evaluate inconsistency among the included studies. If the heterogeneity was absent, the pooled RR and HRs were calculated by the fixed-effect model (χ2P < 0.100 and I2 < 50%). Otherwise the meta-analysis was performed by employing the DerSimonian random-effects model [36, 37]. Significance was identified if the value of P was lower than 0.05. Egger’s tests were generated to detect potential publication bias (P > 0.05 was considered representative of no statistically significant publication bias) [38]. All the statistical calculations of meta-analyses were performed by RevMan 5.3. And Egger’s tests were carried out through Stata11.0. All the P values were two-sided.
Results
Literature search
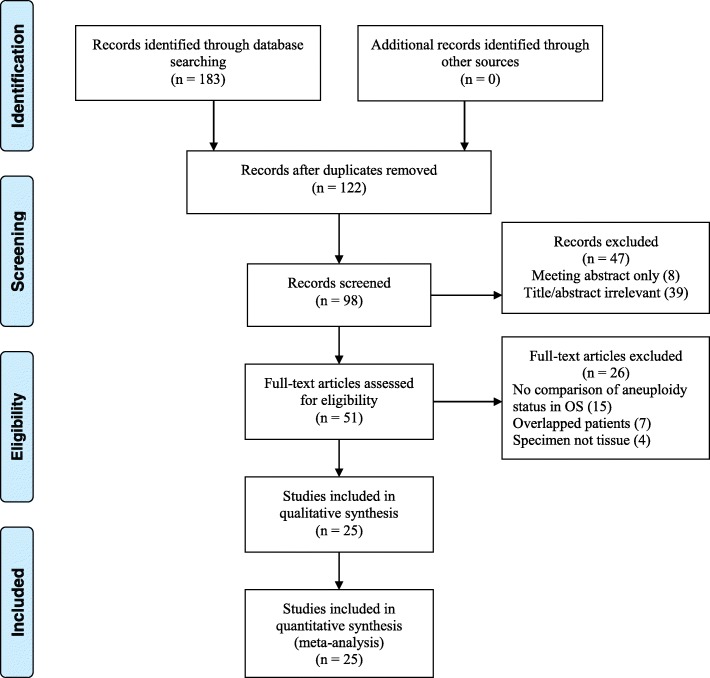
According to the search strategy, 98 studies were identified for full text review, in which 25 studies [10–34] possessed sufficient data for following meta-analyses (Fig. 1). The main features of each study were shown in Table 1.
Fig. 1.
PRISMA flow diagram. OS, overall survival
Table 1.
Characteristics of included studies
| Publications | No. of patients | Age (year) | Method | Specimen | Stage | Follow-up (month) |
Treatment | neclei | Multiple sample form one case | NOS |
|---|---|---|---|---|---|---|---|---|---|---|
| Ballantyne 1987 [10] | 77 | 67 (43–88)a | FCM | PES | NA | 36 | Surgery | > 20,000 | NA | 7 |
| Sasaki 1989 [11] | 70 | 58.1 (33–80)b | FCM | FTS | NA | 48 | Surgery | 10,000 | NA | 7 |
| Wyatt 1989 [12] | 76 | 66 (40–88)a | FCM | PES | NA | 60 | Surgery | NA | NA | 8 |
| Baretton 1991 [13] | 125 | 66.8 ± 11.3b | FCM | PES | NA | 108 | Surgery | NA | NA | 8 |
| Filipe 1991 [14] | 116 | 64 (31–87)a | FCM | PES | NA | 108 | Surgery | > 10,000 | NA | 8 |
| Kimura 1991 [15] | 270 | NA | FCM | PES | I-IV | 60 | Surgery | > 10,000 | NA | 8 |
| Kakeji 1993 [16] | 93 | NA | NA | NA | I-IV | 60 | Surgery | 100 | No | 8 |
| Suh 1993 [17] | 103 | 55.9 (29–70)b | FCM | PES | I-IV | 24 | Surgery | > 10,000 | NA | 7 |
| Flyger 1995 [18] | 97 | 67 (23–85)a | FCM | PES | NA | 60 | Surgery | 10,000 | NA | 8 |
| Tsuchiya 1995 [19] | 127 | NA | FCM | PES | NA | 96 | Surgery+CT chemotherapy | NA | No | 8 |
| Victorzon 1996 [21] | 242 | NA | NA | NA | I-IV | 120 | Surgery | NA | NA | 8 |
| Sakusabe 1996 [20] | 216 | 56.7 (24–86)b | FCM | PES | III | 60 | Surgery | 10,000 | No | 8 |
| Imada 1997 [22] | 88 | 62.4b | FCM | FTS | NA | 36 | Surgery+CT chemotherapy | > 10,000 | Yes | 7 |
| Omejc 1997 [23] | 76 | NA | FCM | PES | I-IIIB | 47 (14–67)a | Surgery | NA | Yes | 8 |
| Abad 1998 [24] | 76 | 70b | FCM | PES | I-IV | 43a | NA | 10,000 | Yes | 7 |
| Hirose 1998 [25] | 142 | 59b | Cytofluorometry | PES | I-IV | 132 | Surgery | NA | NA | 8 |
| Danesi 2000 [26] | 137 | 66.3b | FCM | PES | I-III | 80.8 (42.9–111.5)a | Surgery | 20,000 | NA | 8 |
| Ikeguchi 2000 [27] | 97 | 58 ± 13b | FCM | PES | II | 60 | Surgery | 10,000 | NA | 8 |
| Russo 2001 [28] | 69 | NA | FCM | PES | I-IV | 95b | Surgery | NA | Yes | 8 |
| Wu 2005 [29] | 60 | 59.6a | FCM | NA | NA | 60 | Surgery | NA | No | 8 |
| Nesi 2007 [30] | 115 | 65.6 ± 11.3b | FCM | PES | I-IV | 84 | Surgery | > 20,000 | Yes | 8 |
| Wiksten 2008 [31] | 337 | NA | FCM | PES | NA | 150 (52.8–249.6)a | Surgery | > 10,000 | No | 8 |
| Belien 2009 [32] | 221 | 71 (34–96)a | FCM | PES | I-IV | 21a | Surgery | NA | No | 7 |
| Syrios 2012 [33] | 212 | 61a | ICM | PES | IV | 65 | Surgery+CT | 200–300 | NA | 8 |
| Nishimura 2017 [34] | 207 |
A: 64.8 ± 10.9b D: 62.1 ± 13.5b |
LSC | PES | I-IV | 60 | RA | NA | NA | 8 |
A aneuploidy, D diploidy, PES paraffin embedded specimen, FTS fresh tissue specimen, FCM flow cytometry, ICM image cytometry, SCM static cytometry, LSC laser scanning cytometry, NA not available; a: median; b: mean; CT: chemotherapy
3449 gastric patients diagnosed between 1968 and 2006 were evaluated for our meta-analyses. The population of patients in each study ranged from 60 to 337, with the follow-up period varying from 21 months to 20 years. Additional file 1: Table S1 displayed the specific characteristics of each meta-analysis. The definitions of DNA aneuploidy in these studies shared some common points as follows: DNA index (DI) reflected the ratio of DNA content in G0/1 cells to the reference G0/1 diploid peak. If a cell population displayed only one G0/G1 peaks (DI = 1), tumor was considered diploidy. Otherwise tumor with additional G0/G1 peaks was aneuploidy.
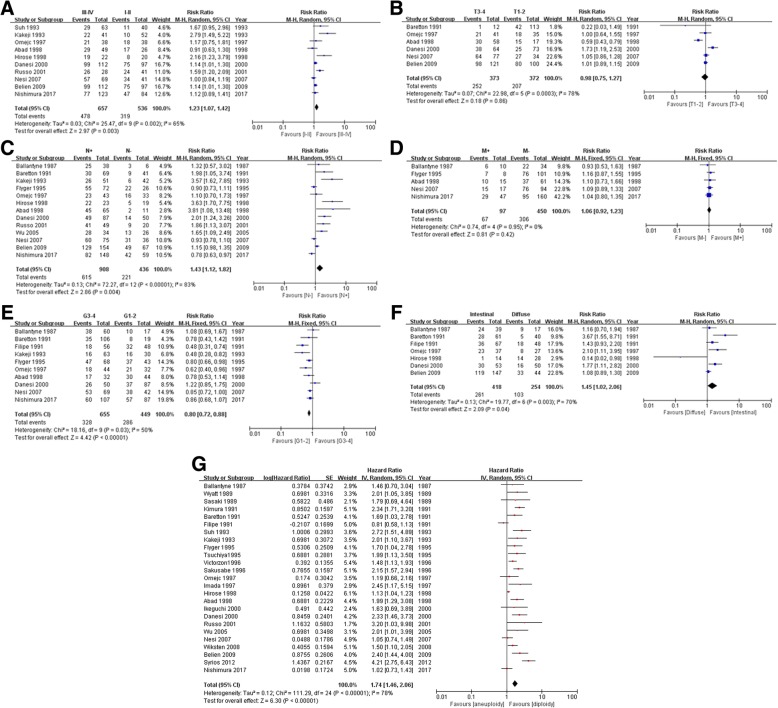
DNA aneuploidy and tumor stage
There were ten studies examining the association between DNA aneuploidy rate and tumor pTNM stages. It could be seen from Fig. 2a that DNA aneuploidy was more frequent in patients with stage III-IV gastric cancer than those with stages I-II tumors (RR = 1.23; 95% CI, 1.07 to 1.42; P = 0.003).
Fig. 2.
a Forest Plot of DNA aneuploidy rates in stage III-IV tumor vs. I-II tumor. b Forest Plot of DNA aneuploidy rates in T3-4 tumor vs. T1-2 tumor. c Forest Plot of DNA aneuploidy rates in tumor with lymph node metastasis vs. tumor without lymph node metastasis. d Forest Plot of DNA aneuploidy rates in tumor with distant metastasis vs. tumor without distant metastasis. e Forest Plot of DNA aneuploidy rates in G3-4 tumor vs. G1-2 tumor. f Forest Plot of DNA aneuploidy rates in intestinal tumor vs. diffuse tumor. g Forest Plot of DNA aneuploidy for overall survival
DNA aneuploidy and depth of tumor invasion
Six studies reported the relationship between DNA aneuploidy and the depth of invasion. However, no association was found between DNA aneuploidy and depth of gastric cancer invasion (T3–4 vs. T1–2: RR = 0.98; 95% CI, 0.75 to 1.27; P = 0.86; Fig. 2b).
DNA aneuploidy and lymph node metastasis
The correlation between DNA aneuploidy and lymph node metastasis was mentioned in thirteen studies. The pooled RR indicated that aneuploidy was more frequently observed in lymph node positive tumor than in lymph node negative ones (RR = 1.43; 95% CI, 1.12 to 1.82, P = 0.004; Fig. 2c).
DNA aneuploidy and distant metastasis
Nevertheless, there was no evident relation between aneuploidy and distant metastasis (P = 0.42; Fig. 2d) from five relevant studies. The RR of patients with distant metastatic tumor compared to those without distant metastatic cancer was 1.06 (Fig. 2d).
DNA aneuploidy and tumor differentiation
A total of ten studies with 1104 patients investigated data concerning the association between DNA aneuploidy status and tumor differentiation. The meta-analysis implied that DNA aneuploidy was remarkably more frequent in G1–2 tumors than in G3–4 tumors (G3–4 vs. G1–2: RR = 0.8; 95% CI, 0.72 to 0.88; P < 0.00001; Fig. 2e).
DNA aneuploidy and tumor type
There were seven articles which studied the DNA aneuploidy frequency in intestinal compared with diffuse gastric cancer. The pooled analysis from these articles suggested a higher presence rate of DNA aneuploidy in intestinal tumors than in diffuse tumors (RR = 1.45; 95% CI, 1.02 to 2.06; P = 0.04; Fig. 2f).
Meta-analysis for overall survival
For the entire gastric cancer population, the OS of patients with DNA aneuploid tumors was worse than that of patient with DNA diploid ones (25 studies; HR = 1.74; 95% CI, 1.46 to 2.06; P < 0.00001; Fig. 2g).
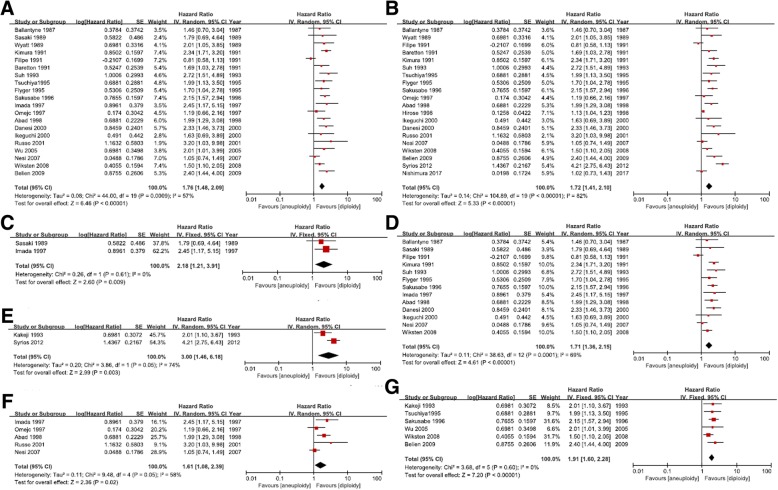
Meta-analysis for different measurements of DNA aneuploidy
Twenty studies utilized flow cytometry for DNA aneuploidy detection. The pooled analysis based on them exhibited a pronounced relation between DNA aneuploidy and gastric cancer survival (HR = 1.76; 95%CI, 1.48 to 2.09; P < 0.00001; Fig. 3a).
Fig. 3.
a Forest Plot of DNA aneuploidy for OS using flow cytometry. b Forest Plot of DNA aneuploidy for OS using paraffin embedded tissue. c Forest Plot of DNA aneuploidy for OS using fresh tissue specimen. d Forest Plot of DNA aneuploidy for OS: cell number>10000. e Forest Plot of DNA aneuploidy for OS: cell number<1000. f Forest Plot of DNA aneuploidy for OS: multiple samples per tumor. g Forest Plot of DNA aneuploidy for OS: single sample per tumor
A total of 20 studies used paraffin embedded tissues (PFS), while only 2 studies used fresh tissues (FTS). DNA aneuploidy status in both specimens showed significant prognostic value for gastric cancer patients (PFS: P < 0.00001, Fig. 3b; FTS: P = 0.009, Fig. 3c).
We analyzed survival data not only for studies with large scale of cell numbers (nuclei more than 10,000, Fig. 3d), but also for studies with small scale of cell numbers (nuclei less than 1000, Fig. 3e). The P values for both groups were less than 0.01.
Tumors contain massive heterogeneity including of aneuploidy. Multiple samples per tumor were supposed to be tested in order to give an overall picture of the genomic stability. Detailed information was listed in Table 1. Five studies used multiple samples (HR = 1.61; 95%CI, 1.08 to 2.39; P = 0.02; Fig. 3f), six studies used single sample (HR = 1.91; 95%CI, 1.60 to 2.28; P < 0.00001; Fig. 3g), while others gave no specific information. No matter what kind of sample was used, DNA aneuploidy was still significantly associated with worse survival.
Publication bias
Funnel plots were used to detect the publication bias which did not reveal remarkable asymmetry. In addition, there was also no considerable publication bias detected by Egger’s tests. The P-values for Egger’s test of each category were listed in Additional file 1: Table S1.
Discussion
The analysis of DNA ploidy is based on the evaluation of cell viability, which exhibits the percent of DNA in S phase cells. Nevertheless, various factors, e.g., the overlap between diploid cells and aneuploid cells, background fragments, and insufficient number of cells, might disturb the evaluation of S phase. Therefore, the assessment of DNA aneuploidy is usually applied for the analysis of DNA content. Although DNA aneuploidy was supposed to play a crucial role in varies cancers, its prognostic value in gastric cancer was still under debate. To the best of our knowledge, our study is the first meta-analysis exploring the prognostic value of aneuploidy in gastric cancer patients.
Recently, antitumor strategy mainly relies on the TNM system and histologic classification which are common prognostic factors for the disease-free survival and overall survival in cancer patients. However, patients sharing the same clinicopathologic characteristics may present various clinical outcomes. It has been suggested that the measurement of DNA nuclear content via flow cytometry may help distinguish gastric cancer patients who have different disease relapse risk and cancer-related death risk [39–41]. Nevertheless, the results of previous studies were often influenced by the heterogeneity in the composition and size of the samples. From our study, the prognostic value of DNA aneuploidy in GC is clearer.
DNA aneuploid tumors were usually regarded as high metastatic risk tumors with aggressive behaviors [42, 43]. We did not, however, find any associations between depth of tumor invasion and distant metastases. Likewise, there is a negative association between aneuploidy and tumor differentiation. It may suggest that the malignant potential of a cell clone does not directly rely on the DNA amount. Moreover, only one sample was extracted from each tumor patient, which may lead to a relatively low incidence of DNA aneuploidy.
Our study revealed that DNA aneuploidy was remarkably correlated with advanced TNM staging and lymph-node metastases. This further indicated that DNA aneuploidy played a critical role in tumor progression and aggressiveness. This also suggested the combination of these three factors were crucial to predict the clinical course of gastric cancer patients.
Diffuse tumor was supposed to have a poorer prognosis compared with intestinal tumor [44]. Interestingly our results show a remarkable correlation between DNA aneuploidy and intestinal histotype rather than the diffuse type. Such heterogeneity may suggest that in these two different tumor types, DNA aneuploidy have various biological and clinical prognostic implications. According to Wyatt et al.’s study, intestinal tumor patients with DNA aneuploidy suffered from shorter survival [12]. Further studies of this patient population are required to validate this association.
There were some advantages of our meta-analyses. For example, our data processing was based on sufficient literature retrieval and eligible statistical methods. Moreover, we adopted NOS scale to evaluate all the included articles, and found that the quality of their data was relatively high. Nevertheless, some limitations in our study should be mentioned. Firstly, our analyses were based on published studies instead of individual data. Secondly, the included studies were not randomized work. Some of them were prospective studies, while the rest were retrospective studies. Thirdly, the enrolled patients were diagnosed from 1968 to 2006. In this period, the treatment for gastric cancer improved greatly. Fourthly, the methodological change in aneuploidy measurements across these years is a confounding factor. Fifthly, the methodological differences in each study might affect the final conclusion. Subgroup analyses on DNA aneuploidy measurements, cell numbers and specimen preparations were performed. We found that these factors did not influence the association between DNA aneuploidy and clinical outcomes of gastric cancer. Sixthly, stomach contains acid and enzymes which might affect the sample preparations for DNA aneuploidy detection. This factor could affect the obtained data in each study. To avoid this, some of the studies specified the time limit to fix the tissues. And others used deep site or multiple sites of tumor for preparation. However, a standard is required in the future experiments. Our meta-analysis provide an indication of the prognostic value of DNA aneuploidy, we hope our study could encourage future studies to draw a standard guideline in this aspect.
Conclusion
In summary, here we reported that DNA aneuploidy might serve as an important biomarker for gastric cancer. DNA aneuploidy could provide detailed prognostic information for different tumor type and stage. Although further experiments will be required to provide more evidence, our findings will be helpful for personal treatment strategy in gastric cancer patients.
Additional file
Table S1. Outcomes of each category. (DOCX 17 kb)
Acknowledgements
We thank Dr. Laiming Wei for helpful discussion.
Abbreviations
- CI
Confidence interval
- G
Differentiation
- HR
Hazard ratio
- K-M
Kaplan-Meier
- M
Distant metastasis
- N
Lymph node metastasis
- NOS
Newcastle-Ottawa Scale
- OS
Overall survival
- PRISMA
Preferred Reported Items for Systematic Reviews and Meta- Analysis
- RR
Risk ratios
- T
Depth of invasion
- TNM
Tumor-node-metastasis
Authors’ contributions
JX drafted the protocol and wrote the paper. GS revised the protocol. JX and RZ coordinated the identification of studies. JX and RZ conducted the study selection and the data extraction. LF and SG independently assessed the risk of bias. JX, RZ, LF and SG performed statistical analyses. WW, XL, LD, ZJ, ZZ, JN, JD, WP and KG revised the manuscript. All authors commented and approved the final manuscript.
Funding
The present study was supported by the National Natural Science Foundation of China (Grant No.81572430 and No.81872047). This funding source paid the open access fee, and had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
Not applicable. Our study is a systematic meta-analysis all of the data were related to the studies listed in in the Table 1.
Ethics approval and consent to participate
Not applicable. Because our study is a systematic meta-analysis, the ethics approval and consent to participate is not relevant to our article type.
Consent for publication
Not applicable. There are no details on individuals reported within the manuscript, so we don’t have the consent for publication.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jing Xu and Ruolin Zhu contributed equally to this work.
Contributor Information
Jing Xu, Email: xujing@ahmu.edu.cn.
Ruolin Zhu, Email: 323218032@qq.com.
Lulu Fan, Email: 87062716@qq.com.
Shangqing Ge, Email: 464586242@qq.com.
Wei Wei, Email: wwei1213@126.com.
Xiaoqiu Li, Email: xiaoqiuli@ahmu.edu.cn.
Liangshan Da, Email: roger093@163.com.
Zhenya Jia, Email: 790606302@qq.com.
Zhiyan Zhao, Email: zhaozy1101@126.com.
Jie Ning, Email: 5383966@qq.com.
Jie Da, Email: dajie213@126.com.
Wanren Peng, Email: wanrenpeng820@163.com.
Kangsheng Gu, Email: 13805692145@163.com.
Guoping Sun, Email: sungp@ahmu.edu.cn.
References
- 1.Cancer Genome Atlas Research Network Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513(7517):202. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palli D, Bianchi S, Cipriani F, et al. Reproducibility of histologic classification of gastric cancer. Br J Cancer. 1991;63(5):765. doi: 10.1038/bjc.1991.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matboli M, El-Nakeep S, Hossam N, et al. Exploring the role of molecular biomarkers as a potential weapon against gastric cancer: a review of the literature. World J Gastroenterol. 2016;22(26):5896. doi: 10.3748/wjg.v22.i26.5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bignold LP, Coghlan B, Jersmann H. David Paul Hansemann: chromosomes and the origin of the cancerous features of tumor cells. Anal Cell Pathol. 2009;31(1):61. doi: 10.3233/CLO-2009-0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weaver BAA, Cleveland DW. Does aneuploidy cause cancer? Curr Opin Cell Biol. 2006;18(6):658–667. doi: 10.1016/j.ceb.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 6.Giam M, Rancati G. Aneuploidy and chromosomal instability in cancer: a jackpot to chaos. Cell Div. 2015;10:3. doi: 10.1186/s13008-015-0009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choma D, Daures JP, Quantin X, et al. Aneuploidy and prognosis of non-small-cell lung cancer: a meta-analysis of published data. Br J Cancer. 2001;85(1):14. doi: 10.1054/bjoc.2001.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laubert T, Freitag-Wolf S, Linnebacher M, et al. Stage-specific frequency and prognostic significance of aneuploidy in patients with sporadic colorectal cancer—a meta-analysis and current overview. Int J Color Dis. 2015;30(8):1015–1028. doi: 10.1007/s00384-015-2259-x. [DOI] [PubMed] [Google Scholar]
- 9.Xu J, Huang L, Li J. DNA aneuploidy and breast cancer: a meta-analysis of 141,163 cases. Oncotarget. 2016;7(37):60218. doi: 10.18632/oncotarget.11130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ballantyne KC, James PD, Robins RA, et al. Flow cytometric analysis of the DNA content of gastric cancer. Br J Cancer. 1987;56(1):52. doi: 10.1038/bjc.1987.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sasaki K, Takahashi M, Hashimoto T, et al. Flow cytometric DNA measurement of gastric cancers: clinico-pathological implication of DNA ploidy. Pathol Res Pract. 1989;184(6):561–566. doi: 10.1016/S0344-0338(89)80160-8. [DOI] [PubMed] [Google Scholar]
- 12.Wyatt JI, Quirke P, Ward DC, et al. Comparison of histopathological and flow cytometric parameters in prediction of prognosis in gastric cancer. J Pathol. 1989;158(3):195–201. doi: 10.1002/path.1711580305. [DOI] [PubMed] [Google Scholar]
- 13.Baretton G, Carstensen O, Schardey M, et al. DNA-ploidy and survival in gastric carcinomas: a flow-cytometric study. Virchows Archiv A. 1991;418(4):301–309. doi: 10.1007/BF01600158. [DOI] [PubMed] [Google Scholar]
- 14.Filipe MI, Rosa J, Sandey A, et al. Is DNA ploidy and proliferative activity of prognostic value in advanced gastric carcinoma? Hum Pathol. 1991;22(4):373–378. doi: 10.1016/0046-8177(91)90085-4. [DOI] [PubMed] [Google Scholar]
- 15.Kimura H, Yonemura Y. Flow cytometric analysis of nuclear DNA content in advanced gastric cancer and its relationship with prognosis. Cancer. 1991;67(10):2588–2593. doi: 10.1002/1097-0142(19910515)67:10<2588::aid-cncr2820671031>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 16.Kakeji Y, Maehara Y, Korenaga D, et al. Prognostic significance of tumor-host interaction in clinical gastric cancer: relationship between DNA ploidy and dendritic cell infiltration. J Surg Oncol. 1993;52(4):207–212. doi: 10.1002/jso.2930520402. [DOI] [PubMed] [Google Scholar]
- 17.Suh KS, Min SK. Flow cytometric DNA analysis of gastric Cancer. J Korean Med Sci. 1993;8(5):348–354. doi: 10.3346/jkms.1993.8.5.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flyger HL, Christensen IJ, Thorup J, et al. DNA aneuploidy in gastric carcinoma flow cytometric data related to survival, location, and histopathologic findings. Scand J Gastroenterol. 1995;30(3):258–264. doi: 10.3109/00365529509093274. [DOI] [PubMed] [Google Scholar]
- 19.Tsuchiya A, Kikuchi Y, Ando Y, et al. Correlation between expression of sialosyl-T antigen and survival in patients with gastric cancer. BJS. 1995;82(7):960–962. doi: 10.1002/bjs.1800820734. [DOI] [PubMed] [Google Scholar]
- 20.Sakusabe M, Kodama M, Sato Y, et al. Clinical significance of DNA ploidy pattern in stage III gastric cancer. World J Surg. 1996;20(1):27–31. doi: 10.1007/s002689900005. [DOI] [PubMed] [Google Scholar]
- 21.Victorzon M, Lundin J, Haglund C, et al. A risk score for predicting outcome in patients with gastric cancer, based on stage, sialyl-Tn immunoreactivity and ploidy—a multivariate analysis. Int J Cancer. 1996;67(2):190–193. doi: 10.1002/(SICI)1097-0215(19960717)67:2<190::AID-IJC7>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 22.Imada T, Yamamoto Y, Fukuzawa K, et al. Flow cytometric analysis of nuclear DNA heterogeneity in gastric cancer. Jpn J Clin Oncol. 1997;27(4):221–226. doi: 10.1093/jjco/27.4.221. [DOI] [PubMed] [Google Scholar]
- 23.Omejc M, Repše S, Bračko M. DNA flow cytometry in gastric carcinoma: implication in patients with potentially curative resection. J Surg Oncol. 1997;65(4):237–241. doi: 10.1002/(sici)1096-9098(199708)65:4<237::aid-jso3>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 24.Abad M, Rincon MR, Silva I, et al. DNA aneuploidy by flow cytometry is an independent prognostic factor in gastric cancer. Anal Cell Pathol. 1998;16(4):223–231. doi: 10.1155/1998/158243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirose K, Iida A, Yamaguchi A, et al. Prognostic value of DNA ploidy and proliferating cell nuclear antigen in gastric cancer. Oncology. 1998;55(4):300–306. doi: 10.1159/000011867. [DOI] [PubMed] [Google Scholar]
- 26.Danesi DT, Spanò M, Fabiano A, et al. Flow cytometric DNA ploidy, p53, PCNA, and c-erbB-2 protein expressions as predictors of survival in surgically resected gastric cancer patients. Cytometry Part A. 2000;42(1):27–34. doi: 10.1002/(sici)1097-0320(20000215)42:1<27::aid-cyto5>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 27.Ikeguchi M, Oka S, Saito H, et al. Nuclear accumulation of p53 protein in gastric cancer strongly correlates with enlargement of nuclear area of cancer cells. Oncol Rep. 2000;7(3):579–663. doi: 10.3892/or.7.3.579. [DOI] [PubMed] [Google Scholar]
- 28.Russo A, Bazan V, Migliavacca M, et al. DNA aneuploidy and high proliferative activity but not K-ras-2 mutations as independent predictors of clinical outcome in operable gastric carcinoma. Cancer. 2001;92(2):294–302. doi: 10.1002/1097-0142(20010715)92:2<294::aid-cncr1322>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 29.Wu YF, Xu HM, Chen JQ. Evaluation of the correlations of cell proliferation activity with lymphatic and vascular invasion and prognosis in gastric carcinoma. Zhonghua Zhong Liu Za Zhi. 2005;27(8):492–495. [PubMed] [Google Scholar]
- 30.Nesi G, Bruno L, Saieva C, et al. DNA ploidy and S-phase fraction as prognostic factors in surgically resected gastric carcinoma: a 7-year prospective study. Anticancer Res. 2007;27(6C):4435–4441. [PubMed] [Google Scholar]
- 31.Wiksten JP, Lundin J, Nordling S, et al. Comparison of the prognostic value of a panel of tissue tumor markers and established clinicopathological factors in patients with gastric cancer. Anticancer Res. 2008;28(4C):2279–2287. [PubMed] [Google Scholar]
- 32.Belien JAM, Buffart TE, Gill AJ, et al. Gross genomic damage measured by DNA image cytometry independently predicts gastric cancer patient survival. Br J Cancer. 2009;101(6):1011. doi: 10.1038/sj.bjc.6605266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Syrios J, Sougioultzis S, Xynos ID, et al. Survival in patients with stage IV noncardia gastric cancer-the influence of DNA ploidy and helicobacter pylori infection. BMC Cancer. 2012;12(1):264. doi: 10.1186/1471-2407-12-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishimura S, Oki E, Ando K, et al. High ubiquitin-specific protease 44 expression induces DNA aneuploidy and provides independent prognostic information in gastric cancer. Cancer Med. 2017;6(6):1453–1464. doi: 10.1002/cam4.1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6(7):e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DerSimonian R, Laird N. Meta-analysis in clinical trials revisited. Contemp Clin Trials. 2015;45:139–145. doi: 10.1016/j.cct.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 38.Egger M, Smith GD, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. Bmj. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Danova M, Riccardi A, Mazzini G, et al. Flow cytometric analysis of paraffin-embedded material in human gastric cancer. Anal Quant Cytol Histol. 1988;10(3):200–206. [PubMed] [Google Scholar]
- 40.Johnson H, Belluco C, Masood S, et al. The value of flow cytometric analysis in patients with gastric cancer. Arch Surg. 1993;128:314. doi: 10.1001/archsurg.1993.01420150070013. [DOI] [PubMed] [Google Scholar]
- 41.Esteban F, Garcia R, Rodriguez R, et al. DNA content by flow cytometry in gastric carcinoma: pathology, ploidy and prognosis. Hepatogastroenterology. 1999;46(27):2039–2043. [PubMed] [Google Scholar]
- 42.Korenaga D, Saito A, Baba H, et al. Cytophotometrically determined DNA content, mitotic activity, and lymph node metastasis in clinical gastric cancer. Surgery. 1990;107(3):262–267. [PubMed] [Google Scholar]
- 43.Macartney JC, Camplejohn RS, Powell G. DNA flow cytometry of histological material from human gastric cancer. J Pathol. 1986;148(4):273–277. doi: 10.1002/path.1711480403. [DOI] [PubMed] [Google Scholar]
- 44.Lauren P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. Apmis. 1965;64(1):31–49. doi: 10.1111/apm.1965.64.1.31. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Outcomes of each category. (DOCX 17 kb)
Data Availability Statement
Not applicable. Our study is a systematic meta-analysis all of the data were related to the studies listed in in the Table 1.