Abstract
Background
We conducted a diagnostic surveillance study to identify Plasmodium, dengue virus, chikungunya virus, and Orientia tsutsugamushi infections among febrile patients who underwent triage for malaria in the outpatient department at Ispat General Hospital, Rourkela, Odisha, India.
Methods
Febrile patients were enrolled from January 2016–January 2017. Blood smears and small volumes or vacutainers of blood were collected from study participants to carry out diagnostic assays. Malaria was diagnosed using rapid diagnostic tests (RDT), microscopy, and PCR. Dengue, chikungunya, and scrub typhus infections were identified using rapid diagnostic test kits and ELISA.
Results
Nine hundred and fifty-four patients were prospectively enrolled in our study. The majority of patients were male (58.4%) and more than 15 years of age (66.4%). All 954 enrollees underwent additional testing for malaria; a subset of enrollees (293/954) that had larger volumes of plasma available was also tested for dengue, chikungunya and scrub typhus by either RDT or ELISA or both tests. Fifty-four of 954 patients (5.7%) were positive for malaria by RDT, or microscopy, or PCR. Seventy-four of 293 patients (25.3%) tested positive for dengue by either RDT or ELISA, and 17 of 293 patients (5.8%) tested positive for chikungunya-specific IgM by either ELISA or RDT. Ten of 287 patients tested (3.5%) were positive for scrub typhus by ELISA specific for scrub typhus IgM. Seventeen patients among 290 (5.9%) with results for ≥3 infections tested positive for more than one infection. Patients with scrub typhus and chikungunya had high rates of co-infection: of the 10 patients positive for scrub typhus, six were positive for dengue (p = 0.009), and five of 17 patients positive for chikungunya (by RDT or ELISA) were also diagnosed with malaria (p < 0.001).
Conclusions
Dengue, chikungunya and scrub typhus are important etiologies of non-malarial febrile illness in Rourkela, Odisha, and comorbidity should be considered. Routine febrile illness surveillance is required to accurately establish the prevalence of these infections in this region, to offer timely treatment, and to implement appropriate methods of control.
Electronic supplementary material
The online version of this article (10.1186/s12879-019-4161-6) contains supplementary material, which is available to authorized users.
Keywords: Febrile illness, Malaria, Dengue, Chikungunya, Scrub typhus, India, Surveillance
Background
Fever is a condition frequently reported by patients using general medicine outpatient departments in Indian hospitals [1, 2]. When a patient complains of fever, additional clinical symptoms are correlated for diagnosis and treatment. Symptoms are often nonspecific, and delays or inaccuracies in arriving at a clinical diagnosis can lead to inappropriate treatment that may result in fatalities. To establish a specific diagnosis and provide the appropriate treatment regimen, diagnoses of febrile illnesses should be confirmed by laboratory tests [3].
Arthropod-borne infectious diseases such as malaria, dengue, chikungunya, and scrub typhus are significant etiologies of acute febrile illness in India [4, 5]. Malaria is caused by mosquito-borne Plasmodium parasites, with two species, P. vivax and P. falciparum, being responsible for the majority of morbidity and mortality. Plasmodium parasites alternate between human and Anopheles mosquito hosts. Human infection with Plasmodium parasites can cause mild to severe illness, encompassing a wide variety of symptoms including fever, headache, malaise, anemia, sweats, and chills that first appear 7–10 days after a bite from an infected mosquito. Left untreated, symptoms can lead to multi-organ failure, severe anemia, cerebral malaria, coma, and death.
Dengue and chikungunya are viral infections transmitted by the bite of infected Aedes mosquitoes. The dengue virus (DENV) has an incubation of 4–10 days and can cause disease presentation ranging from asymptomatic to severe. Mild ‘dengue fever’ infections are characterized by fever with or without rash, while more severe symptoms include high fever, severe headache with eye pain, myalgia, arthralgia, and rash. Some patients progress to Dengue Hemorrhagic Fever/Dengue Shock Syndrome (DHF/ DSS), which can be fatal. Four distinct serotypes of DENV can cause the disease, and while infection by any one serotype can lead to lifelong immunity against that serotype, it does not lead to long-term protection against the other serotypes; instead, a subsequent infection by a different serotype has a higher risk of developing into severe disease [6, 7].
Chikungunya is caused by a virus (CHIKV) that has an incubation period ranging from 1 to 12 days following the bite of an infected Aedes mosquito. The onset of the disease is characterized by an acute febrile illness, accompanied by rash and severe joint pain that can persist for months [8]. Other symptoms include muscle pain, headache, nausea, and fatigue. Symptoms are often mild in infected individuals and thus the infection may go unrecognized or be misdiagnosed.
Scrub typhus is an acute febrile illness caused by the obligate intracellular bacterium Orientia tsutsugamushi. It is transmitted to humans through the bite of the trombiculid mite, which is both the vector and reservoir for the bacterium. Humans are incidental hosts [9]. Scrub typhus is distinguished by the formation in a subset of patients of a scab-like lesion called an ‘eschar’ at the site of mite feeding. Scrub typhus is often under-diagnosed as its nonspecific clinical features, including high fever, lymphadenopathy, rash, myalgia, and headaches, make it difficult to differentiate from other febrile illnesses.
While there is an existing integrated disease surveillance program in India [10], the true burden of these diseases is likely to be higher than reported [11–13], and may vary by region. In addition, delayed or inaccurate diagnoses are more likely in populations living in inaccessible (e.g., rural or remote) areas of India, where cost and a lack of a well-developed health-care infrastructure are obstacles to the use of sensitive molecular diagnostic tests. Routine surveillance is critical for accurate estimates of disease burden, which in turn drives the implementation of public health policies for prevention, diagnosis, and treatment of common etiologies of infectious disease.
This manuscript describes a pilot project to determine whether chikungunya, dengue, and scrub typhus were common among patients presenting with acute febrile illness who had been triaged for malaria in an outpatient department (OPD) at Ispat General Hospital, Rourkela, in the state of Odisha, India. A second aim of the study was to undertake a comparative assessment of the performance of several standard, commercially available diagnostic tests for these three febrile illnesses. A third aim was to determine if malaria cases were missed during the initial malaria triage by the OPD.
Methods
Study site
Rourkela is located in Sundargarh district close to the northern border of the state of Odisha, India, and has a population of over 0.5 million. It has a tropical climate, with high temperatures and heavy rainfall between June–October and December–January. Subjects were enrolled at the outpatient department (OPD) of Ispat General Hospital (IGH), which is a 685-bed hospital for employees of Rourkela Steel Plant, Steel Authority of India, Inc., and for people living in and around Rourkela. A total of 90,867 patients attended the OPD at IGH Rourkela in 2016, of which 13,534 patients (~ 15%) reported fever.
Study design and inclusion criteria
Between January 2016–January 2017, 954 OPD patients aged 12 months - 70 years (with children categorized as ≤15 years and adults as ≥16 years) who reported having fever up to 48 h prior to enrollment or who were febrile on day of enrollment (body temperature ≥ 37.5 °C) were enrolled in our study. These patients had already been through triage for malaria in the OPD, where probable malaria cases were identified based on presence of high fever, chills, and rigor. Such cases, along with individuals with symptoms of influenza and pregnant women (due to the possible risks associated with enrolling these subjects), were not enrolled in our study. Written consent or assent was obtained from each enrolled patient prior to a physical examination and administration of a detailed clinical questionnaire, and each patient provided a blood sample.
Sample collection and processing
All 954 enrollees were tested for malaria by at least one diagnostic test; a subset of enrollees (293/954) from whom a larger volume of blood was collected was also tested for dengue, chikungunya, and scrub typhus by either RDT, ELISA, or both tests. Patient blood samples were collected either in a microvette (small volume ~ 100 ul) and/or a vacutainer (large volume ~ 3-5 ml) at the time of enrollment and tested by Hemocue to assess haemoglobin levels. Whole blood was used for thin and thick blood smear preparations, as well as malaria RDTs. Microvette and vacutainer samples were centrifuged to separate plasma and red blood cells, and these fractions were stored at − 80 °C before use. DNA was extracted from the red blood cells using QIAamp DNA blood Mini Kits (Qiagen Inc., Valencia, CA).
Diagnostic assays
Malaria: Three tests -- rapid diagnostic test (RDT), blood smear microscopy, and polymerase chain reaction (PCR) -- were used for malaria diagnosis. All 954 participants were tested P. falciparum and P. vivax by RDT (FalciVax, Zephyr Biomedicals, India, a commercially available lateral flow kit that detects P. falciparum histidine-rich protein 2 and P. vivax lactate dehydrogenase) at time of enrollment. All participants also had a thick and thin blood smear made with whole blood from a finger prick, stained with Giemsa and examined by microscopy using a 100x oil immersion objective. Thick smear parasitemia was calculated against 200–500 leukocytes and expressed as parasites per microliter of blood, using the white blood cell (WBC) count if determined, or assuming 8000 WBCs per microliter blood. A semi-nested PCR assay (modified from [14]) targeting the 18S small subunit ribosomal protein gene (SSU rDNA) was used for species-specific molecular detection of four Plasmodium species (P. falciparum, P. vivax, P. malariae, and P. ovale) using DNA extracted from a subset of patients (852/954, 89.3%) from whom a microvette (~100ul) or vacutainer (~ 3-5 ml) of blood had been collected.
Dengue, chikungunya, and scrub typhus: Testing for dengue, chikungunya, and scrub typhus was done on a subset of patients (293/954, 30.7%) enrolled from July 2016 to January 2017 from whom larger volumes of blood were available (since larger volumes of plasma are required for dengue and chikungunya RDTs). A pilot study was first conducted with blood samples from approximately 100 of these 293 patients, to test two commonly used dengue RDTs (Dengue Day 1 test from J. Mitra and Co., India, and SD Bioline Dengue Duo test from Alere Medical, India) and two chikungunya RDTs (Advantage Chikungunya IgM card RDT from J. Mitra and Co., New Delhi, India; and the SD Bioline Chikungunya IgM RDT from Alere Medical, India) [15–17]. The Dengue Day 1 test and the SD Bioline Dengue Duo test recognize the dengue non-structural protein 1 antigen (NS1) and DENV-specific IgM and IgG antibodies in patient sera. The Advantage Chikungunya IgM card RDT and the SD Bioline Chikungunya IgM RDT detect chikungunya-specific IgM in patient sera. The Dengue Day 1 kit and the Advantage Chikungunya IgM card RDTs, both from J. Mitra and Co., worked more efficiently in our hands and were subsequently chosen for all downstream diagnoses.
Since the sensitivity and specificity of ELISA-based diagnostic kits are better than RDTs, we also used ELISA kits to test extracted plasma samples for dengue and chikungunya. The Panbio Dengue Early ELISA kit for detection of NS1 antigen (Alere Medical, India), and the Panbio Dengue IgM Capture ELISA (Alere Medical, India) were used for diagnosis of dengue, and the Chikungunya IgM ELISA kit (SD Bioline) was used for diagnosis of chikungunya.
The Scrub Typhus Detect IgM ELISA kit (InBios International, Inc., Seattle, WA, USA) was used to detect Orientia tsutsugamushi. Optical density values > 0.5 were considered as a positive test result [18, 19].
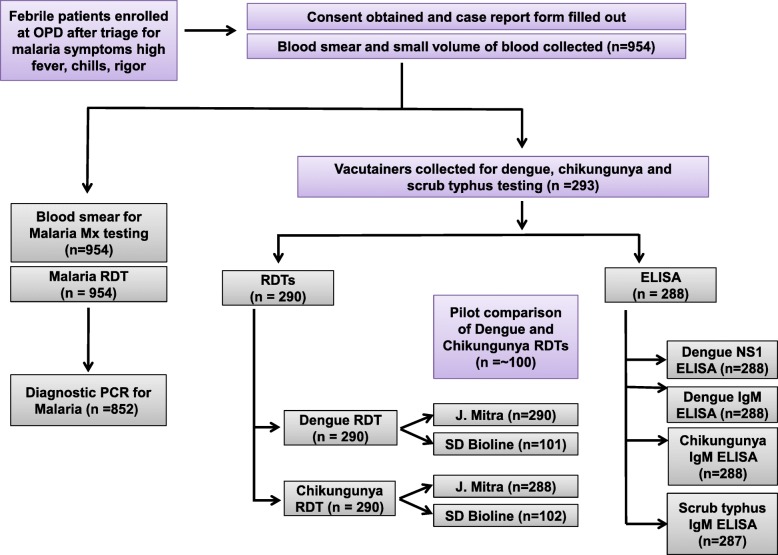
A flowchart describing samples tested for malaria, dengue, chikungunya, and scrub typhus using the above diagnostic tests is shown in Fig. 1.
Fig. 1.
Flowchart of patient testing for febrile illness. A total of 954 patients in the outpatient department at IGH, Rourkela, with reported fever up to 48 h prior to enrollment or on day of enrollment (body temperature ≥ 37.5 °C), were enrolled. All study participants were tested for Plasmodium by RDT and microscopy, and a subset of patients was tested by PCR. Patients from whom a larger volume of blood was available were tested for dengue, chikungunya and scrub typhus by RDT and ELISA
Data management, definitions, and analysis
Patient clinical and demographic questionnaire data were collected on paper case report forms (CRFs) that were entered and uploaded into the CSCMi’s REDCap database [20]. Results for all assays carried out on plasma, blood, and DNA samples were also uploaded into CSCMi REDCap. Data sets were exported into Stata (version 14.2). A patient was considered to have malaria if any malaria test (RDT, microscopy, or PCR) was positive. Based on national guidelines [21] our case definitions for dengue diagnosis were as follows: 1) patients with NS1 or IgG/IgM positive RDT results were considered probable dengue fever cases, and 2) patients with NS1 or IgM positive ELISA results were considered current or past primary or secondary early dengue infection cases. Patients with IgM positive RDT or ELISA results were considered probable chikungunya cases [22]. Patients with febrile illness, with or without eschar, who had plasma positive for IgM (O.D. > 0.5) by ELISA were considered probable scrub typhus cases [18]. The period from June–October was considered the rainy season; age was categorized into ≤15 (child) and ≥ 16 years (adult). Symptoms evaluated for their association with laboratory results included documented fever (body temperature ≥ 37.5 °C at the time of enrollment), headache, aches, chills, vomiting, diarrhea, cough, dizziness, sweating, abdominal pain, chest pain, eye or ear pain, or rash. The association between laboratory test results and symptoms, age, gender, season, and anemia (for the definition see Additional file 1: Table S1) were examined using the chi-square test in univariate and generalized linear regression models for multivariate models with a log link and binomial distribution [23]. A p-value < 0.05 was considered significant. The concordance between diagnostic test results was examined using the McNemar test and visualized using proportional Venn diagrams in Stata (version 14.2).
Results
Patient demographics and clinical presentation
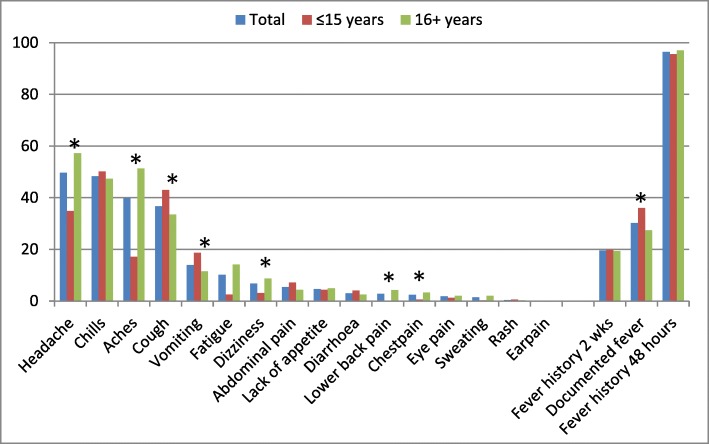
We enrolled 954 participants from patients attending the outpatient clinic at Ispat General Hospital, in Rourkela, Odisha, India from January 2016 to January 2017. A total of 58.4% patients were male and 66.4% > 15 years of age, with a median age of 25 (interquartile range 12–44). Most patients reported having fever in the last 48 h (96.5%) and 276 patients (30.2%) were febrile at the time of enrollment; the use of fever suppressant medication was common (76.6%). The most common clinical symptoms presented by patients included headache (49.7%), chills (48.3%), and aches or pains (39.8%); symptoms differed significantly by age group for eight symptoms, with documented fever, vomiting, and cough more common in children (≤15 years), and chest pain, lower back pain, dizziness, aches, and headache more common among persons aged 16 years and above (Fig. 2). There were no differences in symptoms by gender; documented fever was more common in the rainy season (33.9% vs. 24.9% in dry season, p = 0.004) and chills were more common in the dry season (43.6% in rainy vs. 55.5% in dry, p < 0.001). Sixty-four patients (6.7%) reported being diagnosed with malaria at least once in the past year, and 58 of them (90.6%) reported receiving antimalarial therapy. Anaemia was detected among 38.8% of the study population.
Fig. 2.
Clinical features of study participants. A graphical distribution of clinical features of study participants in the outpatient department at IGH, Rourkela is presented with features on the x-axis and proportion of patients on y-axis. The patients were categorized as children (15 years and below) and adults (16 years and above), with an asterisk to denote statistically significant differences between children and adults (*p = < 0.05 comparing children vs. adults)
Malaria
Fifty-four out of 954 patients (5.7%) were positive for P. falciparum, P. vivax, or P. malariae by at least one diagnostic method (RDT, microscopy, or PCR): 46 by PCR (46/852, 5.4%), 30 by RDT (30/954, 3.1%) and 25 by microscopy (25/954, 2.6%; Table 1). Among the 852 patients tested by all three tests, three patients were positive by RDT but not by another malaria test: two of these (both with P. falciparum) had a history of antimalarial treatment within the last month, but this was absent in the third patient who had both P. falciparum and P. vivax (Table 2). No patient was positive by microscopy and negative by both other tests (Table 2). Three patients were positive by RDT and microscopy but not by PCR, with species matching (2 P. vivax and 1 P. falciparum). Twenty-three patients were positive by PCR but not by RDT or microscopy (sub-patent infections, all P. falciparum); when only considering microscopy, 25 patients had submicroscopic infections (positive by PCR, but negative by microscopy, 1 P. vivax, and 24 P. falciparum, one of each was also detected by RDT). A proportional Venn diagram (Fig. 3) shows the concordance between the malaria diagnostic test results.
Table 1.
Plasmodium infections among OPD patients as determined by RDT, microscopy and PCR, Rourkela 2016–2017
| Test | Total tested | P. falciparum positive (%) | P. vivax positive (%) | P. malariae positive (%) | Any species positive (%) |
|---|---|---|---|---|---|
| RDT | 954 | 20 (2.1) | 11 (1.2) | 0 (0) | 30 (3.1) |
| Microscopy | 954 | 16 (1.7) | 9 (0.9) | 0 (0) | 25 (2.6) |
| PCR | 852 | 39 (4.6) | 6 (0.7) | 1 (0.1) | 46 (5.4) |
Table 2.
Agreement of malaria tests among 852 OPD patients with all three test results available, Rourkela 2016–2017
| RDT | Microscopy | PCR | Number (%) | Species | Report of recent malaria treatment |
|---|---|---|---|---|---|
| Positive | Negative | Negative | 3 (0.4) | 2 Pf & 1 Pf + Pv | 2 Pf in previous month |
| Positive | Positive | Negative | 3 (0.4) | 1 Pf & 2 Pv | 1 Pv 4 months before |
| Positive | Negative | Positive | 2 (0.2) | 1 Pf & 1 Pv | 1 Pf in previous month |
| Positive | Positive | Positive | 20 (2.3) | 14 Pf, 5 Pv, 1 Pm* | 2 Pf and 1 Pv in previous month |
| Negative | Positive | Negative | 0 | ||
| Negative | Positive | Positive | 1 (0.1) | 1 Pf | None |
| Negative | Negative | Positive | 23 (2.7)† | All Pf | None |
| Negative | Negative | Negative | 800 (93.9) | 30 (3.8%) in previous month |
Abbreviations: Pf: P. falciparum, Pv: P. vivax, Pm: P. malariae, RDT: rapid diagnostic test, PCR: polymerase chain reaction
*P. malaria by PCR, P. vivax by RDT and microscopy
† Sub-patent malaria
Fig. 3.
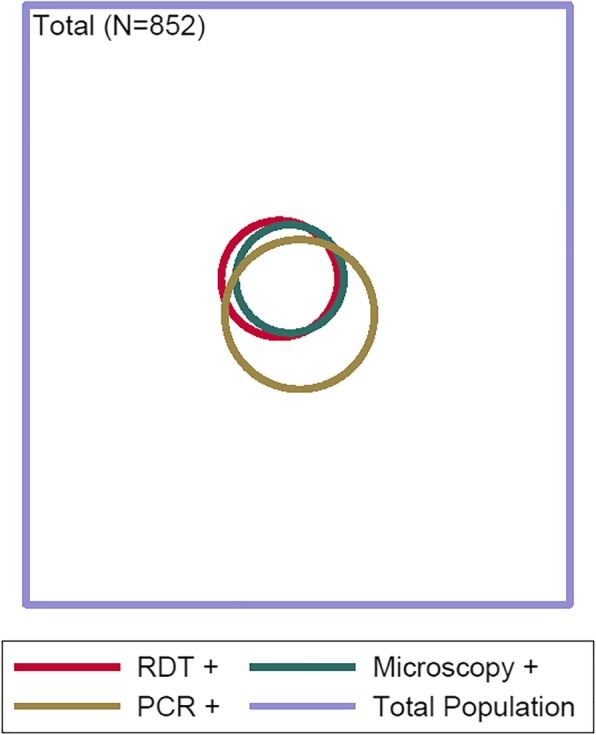
Venn diagram of malaria test results among OPD patients, Rourkela, 2016–2017. A proportional Venn diagram representing Plasmodium positivity among 852 study participants each tested by three different assays: RDT, microscopy and PCR. Each circle represents positives by the different assays, with the numbers also shown in Table 2
Twenty patients were positive by all three tests (14 P. falciparum) with only one species discrepancy whereby RDT and microscopy detected P. vivax but PCR detected P. malariae. Mixed Plasmodium species (defined as more than one species detected by the same test) were identified in only one patient (P. falciparum and P. vivax). When using PCR as the gold standard, RDT had a sensitivity of 47.8% and a specificity of 99.3%; for microscopy this was 45.7 and 99.6%, respectively. By microscopy, men were more likely to be positive for malaria (3.9% vs. 1.4% among women, p = 0.029), whereas by PCR adults were more likely to be positive (6.5% vs. 3.0% among children, p = 0.036); sub-patent infections or Plasmodium species were not associated with age, gender, or season. Vomiting and lack of body aches were associated with malaria for all tests, e.g., for RDT, vomiting was 3.80 times more likely (95% CI 1.77–8.16, p = 0.001) among patients with a positive RDT compared to patients without, whereas the prevalence ratio of report of body ache was 0.35 (95% CI 0.15–0.82, p = 0.016).
Dengue, chikungunya, and scrub typhus
Seventy-four patients of 293 patients tested were positive (25.3%) for dengue by either RDT or ELISA (Table 3). Thirty-seven patients out of 285 patients tested were positive (13%) for NS1 antigen by both RDT and ELISA, and 8 patients out of 285 patients tested were positive (2.8%) for dengue virus (DENV)-specific IgM by both RDT and ELISA (Table 3). Ten patients of 288 tested were positive (3.5%) for both NS1 antigen and DENV-specific IgM by ELISA. Men were more likely to be positive than women for dengue IgM by the J. Mitra RDT (13/169 or 7.7% vs. 3/121 or 2.5%, p = 0.06). Rainy season was associated with a positive result for the IgM ELISA (p = 0.004) and NS1- specific ELISA (p < 0.001) tests and the J. Mitra rapid test for NS1 (p < 0.001). Documented fever was more common among patients who tested positive for NS1 antigen, but not for the other tests used (documented fever was 56.4% among 39 patients positive for NS1 antigen by RDT, and 24.4% among 246 negative patients, p < 0.001). A proportional Venn diagram (Fig. 4) shows the concordance between the dengue diagnostic test results.
Table 3.
Detection of dengue and chikungunya by RDT and ELISA in the 293 subjects for whom sufficient plasma was available
| Antigen | Type of test | Manufacturer (no. of patients tested) | Positive cases | % Positivity | Concordance between tests |
|---|---|---|---|---|---|
| Dengue NS1 (n = 293) | RDT | J. Mitra (n = 290) | 39 | 13.4 |
NS1 RDT: J. Mitra vs. SD Bioline: 100% same results, n = 101 |
| SD Bioline (n = 101) | 13 | 12.9 | |||
| ELISA | Panbio (n = 288) | 41 | 14.2 |
J. Mitra RDT vs. Panbio ELISA (NS1): McNemar test, n = 285, exact p-value 0.688 |
|
| Dengue IgM (n = 293) | RDT | J. Mitra (n = 290) | 16 | 5.5 |
IgM RDT: J. Mitra vs. SD Bioline: McNemar test, N = 101, exact p-value 0.625 |
| SD Bioline (n = 101) | 1 | 1 | |||
| ELISA | Panbio (n = 288) | 35 | 12.2 |
J. Mitra RDT vs. Panbio ELISA (NS1): McNemar test, n = 285, exact p-value 0.688 |
|
| Dengue IgG (n = 290) | RDT | J. Mitra (n = 290) | 8 | 2.8 |
IgG RDT: J. Mitra vs. SD Bioline: 100% same results, n = 101 |
| SD Bioline (n = 101) | 2 | 2 | |||
| Dengue NS1 or Dengue IgG or Dengue IgM | RDT or ELISA | J. Mitra or SD Bioline or Panbio (n = 293) | 74 | 25.3 | Not applicable |
| chikungunya IgM (n = 293) | RDT | J. Mitra (n = 288) | 8 | 2.8 |
IgM RDT: J. Mitra vs. SD Bioline: McNemar test, n = 100, exact p-value 0.250 |
| SD Bioline (n = 102) | 0 | 0 | |||
| ELISA | Panbio (n = 288) | 15 | 5.2 |
J. Mitra RDT vs. Panbio IgM ELISA: McNemar test, n = 283, exact p-value 0.022 |
|
| RDT or ELISA | J. Mitra or SD Bioline or Panbio (n = 293) | 17 | 5.8 | Not applicable |
Fig. 4.

Venn diagram of select dengue test results among OPD patients, Rourkela, 2016–2017. A proportional Venn diagram representing dengue positivity among 288 study participants tested by three different assays: J. Mitra NS1 RDT, NS1 ELISA, and DENV-specific IgM ELISA. The numbers in each compartment of the Venn diagram are depicted on the graph
Eight patients out of 290 tested were positive (2.8%) for chikungunya virus (CHIKV)-specific IgM by either the J. Mitra or SD Bioline RDT, and 15 of 288 patients tested were positive (5.2%) by ELISA for CHIKV-specific IgM. Seventeen of 293 patients tested positive (5.8%) by either ELISA or RDT for CHIKV-specific IgM, while 6 of 285 patients tested were positive (2.1%) by both CHIKV-specific IgM RDT and ELISA (Table 3). No significant associations were detected between being positive for chikungunya and gender, age, season, or symptoms, but persons positive for chikungunya by any test were more likely to be anemic (see Additional file 1: Table S1).
Of 287 samples tested, 10 (3.5%) were positive for Orientia tsutsugamushi-specific IgM antibodies by ELISA. Of these, 3 samples had an O.D. value > 1 and were likely to have active or recent infection; O.D. values between 0.5 and 1, observed in 7 patients in our study, may be indicative of early or past infections (Table 4). Scrub typhus was only detected among persons > 15 years during the rainy season; patients positive for scrub typhus were more likely to be anemic (Additional file 1: Table S1).
Table 4.
Characteristics and O.D. values of study participants positive for scrub typhus by ELISA
| Subject | O.D. value | Age | Sex | Month of sample collection |
|---|---|---|---|---|
| IS0585 | 2.531 | 48 | Male | August |
| IS0697 | 1.113 | 37 | Female | September |
| IS0527 | 1.049 | 35 | Female | August |
| IS0642 | 0.716 | 26 | Female | August |
| IS0595 | 0.702 | 62 | Male | August |
| IS0743 | 0.685 | 16 | Male | September |
| IS0477 | 0.678 | 23 | Male | July |
| IS0593 | 0.669 | 45 | Female | August |
| IS0525 | 0.599 | 32 | Male | August |
| IS0783 | 0.598 | 40 | Male | September |
*All samples tested for scrub typhus were collected between July 2016 and January 2017
Co-infections
Of the 290 patients tested for at least three infections, 105 (36.2%) were positive for at least one pathogen, 17 (6.1%) for two, and one patient (0.4%) for three pathogens (Table 5) by any test for malaria, dengue, chikungunya, or scrub typhus. This last patient was positive for dengue, chikungunya, and P. falciparum. Eight out of ten patients positive for scrub typhus were also positive for dengue, chikungunya or malaria; six of these were dengue positive by any test (p = 0.009 for association between scrub typhus and dengue). Almost half of chikungunya positive patients (by any test) were positive for dengue, malaria or scrub typhus (8/17 or 47%); five of these were malaria positive (p < 0.001 for the association between malaria detected by any test and chikungunya, N = 280, 2 sub-patent malaria infections). No association was seen between dengue and chikungunya positives (4 co-infections with dengue positives among 17 chikungunya positive patients or 23.5%, versus 69 dengue positives among persons negative for chikungunya or 25.8%, p = 0.83). Patients tested for at least three infections were more likely to be adults and enrolled in the rainy season compared to patients tested for malaria only (p < 0.001 for both). All patients positive for multiple infections visited in the rainy season versus 93.2% of the persons with one detected infection (82/88) and 69.2% (128/185) of the patients with no infection detected (p < 0.001). No significant differences were detected by age, gender or symptoms compared to patients with a single or no infection; however, anemia was higher among the 17 patients positive for ≥2 infections (58.3%), compared to 26.8% in patients positive for one infection, and 22.1% in patients where none of these four infections were detected (N = 266, p = 0.018, Additional file 1: Table S1).
Table 5.
Characteristics of 17 outpatients with multiple infections among 290 patients with at least three tests, Rourkela, 2016–2017
| Tests with positive results | ||||||||
|---|---|---|---|---|---|---|---|---|
| Patient | Positive for | CHIKV | DENV | Pf | Scrub typhus | Age | Gender | Month |
| IS0596 | CHIKV, DENV, Pf | J. Mitra IgM RDT | J. Mitra IgM RDT | RDT, BS & PCR | NA | 35 | Male | August |
| IS0525 | Scrub typhus, Pf | NA | NA | RDT, BS & PCR | IgM ELISA | 32 | Male | August |
| IS0642 | Scrub typhus, DENV | NA | IgM ELISA & NS1 ELISA | NA | IgM ELISA | 26 | Female | August |
| IS0585 | Scrub typhus, DENV | NA | IgM ELISA | NA | IgM ELISA | 48 | Male | August |
| IS0783 | Scrub typhus, DENV | NA | IgM ELISA | NA | IgM ELISA | 40 | Male | September |
| IS0697 | Scrub typhus, DENV | NA | NS1 ELISA | NA | IgM ELISA | 37 | Female | September |
| IS0595 | Scrub typhus, DENV | NA | J. Mitra IgM RDT | NA | IgM ELISA | 62 | Male | August |
| IS0743 | Scrub typhus, DENV | NA | J. Mitra IgM RDT | NA | IgM ELISA | 16 | Male | September |
| IS0477 | Scrub typhus, CHIKV | IgM ELISA | NA | NA | IgM ELISA | 23 | Male | July |
| IS0742 | DENV, Pf | NA | NS1 ELISA | PCR | NA | 40 | Female | September |
| IS0731 | CHIKV, Pf |
IgM ELISA & J. Mitra IgM RDT |
NA | RDT, BS & PCR | NA | 42 | Male | September |
| IS0849 | CHIKV, Pf |
IgM ELISA & J. Mitra IgM RDT |
NA | RDT, BS & PCR | NA | 16 | Male | October |
| IS0695 | CHIKV, Pf |
IgM ELISA & J. Mitra IgM RDT |
NA | PCR | NA | 69 | Male | September |
| IS0674 | CHIKV, Pf |
IgM ELISA & J. Mitra IgM RDT |
NA | RDT | NA | 14 | Male | September |
| IS0848 | CHIKV, DENV | IgM ELISA | IgM ELISA | NA | NA | 14 | Female | October |
| IS0815 | CHIKV, DENV | IgM ELISA |
NS1 ELISA, J. Mitra IgM RDT, J. Mitra IgG RDT |
NA | NA | 24 | Male | October |
| IS0586 | CHIKV, DENV | J. Mitra IgM RDT |
NS1 ELISA, IgM ELISA, J. Mitra NS1 RDT |
NA | NA | 37 | Male | August |
Pf: P. falciparum, CHIKV: chikungunya virus, DENV: Dengue virus, NA: Not applicable, RDT: rapid diagnostic test, BS: blood smear (microscopy), PCR: polymerase chain reaction
Discussion
A summary of the few hospital-based febrile illness surveillance studies that have occurred in India is shown in Table 6. Of note is a 5-year acute febrile illness (AFI) study conducted by Manipal Centre for Virology Research (MCVR) in collaboration with the U.S. Centers for Disease Control [4]. Researchers at MCVR recruited patients at 27 hospital-based sentinel sites across India and tested them for bacterial and viral pathogens starting in June 2014. Of the 1483 in-patient and out-patient cases recruited in Odisha until July 2017, 595 (40%) were diagnosed positive for one of the tests, with malaria as the major etiology (12.4%) followed by scrub typhus (3.2%), leptospirosis (2.4%), influenza (1.4%), rotavirus (1.2%), and dengue (0.7%). The higher malaria prevalence in the MCVR study may have been observed because the RT-PCR used for parasite detection is more sensitive than regular PCR.
Table 6.
Summary of results from six febrile illness studies from India, 2007–2017
| Study | Location and year | Population and sample size | Select tests and results | Results (n, %) |
|---|---|---|---|---|
| Chrispal 2010 | Christian medical college, Vellore; 2007 |
≥16 years, febrile for 5–21 days, N = 398 in-patients |
1) Thin blood smear for malaria 2) Dengue IgM-IgG ELISA 3) Scrub typhus IgM ELISA 4) Blood culture or Typhidot for Salmonella 5) Leptospirosis IgM ELISA 6) Spotted fever IgM ELISA 7) Hantavirus IgM and IgG |
Malaria (68, 17.1%) Dengue (28, 7.0%) Scrub typhus (189, 47.5%) Enteric fever (32, 8.0%) Leptospirosis (12, 3.0%) Spotted fever (7, 1.8%) Hantavirus (1, 0.3%) |
| Mittal 2015 |
Himalayan Institute of Medical Sciences, Dehradun, Dec 2012- Nov 2013 |
> 18 years, febrile for 5–14 days, N = 2547 in-patients |
1) Malaria microscopy, RDT 2) Scrub typhus IgM ELISA 3) Dengue NS1/ IgM RDT 4) Leptospira IgM RDT 5) Widal Ag kit for Salmonella 6) Anti HEV IgM EIA 7) Anti HAV IgM EIA |
Malaria (175, 6.8%); Scrub typhus (367, 14.4%); Dengue (956, 37.5%); Leptospirosis (0.14%); Enteric fever (424, 16.5%); Hepatitis A (1.9%); Hepatitis E (1.4%); Undetermined 11%. Mixed infections (48, 1.9%) |
| Abhilash 2016 | Christian medical college, Vellore; Oct 2012- Sep 2013 |
≥16 years, febrile for 3–14 days, N = 1258 Both in-patients and out-patients |
1) Thin blood smear for malaria 2) Dengue IgM-IgG ELISA 3) Scrub typhus IgM ELISA 4) Blood culture for Salmonella, Widal 5) Leptospirosis IgM ELISA |
Malaria (131, 10.4%) Dengue (386, 30.6%) Scrub typhus (452, 35.9%) Enteric fever (47, 3.7%) Leptospirosis (8, 0.6%) Undetermined (220, 17.4%) |
| Morch 2017 |
7 hospitals in India, April 2011- Nov 2012 |
≥5 years, N = 1564, in-patients |
1) Malaria PCR, RDT, microscopy 2) Dengue RDT, IgM ELISA 3) chikungunya IgM ELISA 4) Leptospirosis IgM ELISA 5) Scrub typhus IgM ELISA 6) Blood culture for bacterial infections |
Malaria (268, 17%) Dengue (244, 16%) chikungunya (98, 6%) Leptospirosis (116, 7%) Scrub typhus (159, 10%) Bacteremia (124, 8%) |
| Robinson 2018 | BJ Medical College, Pune; 2013–2015 |
> 6 months, fever or complaint of fever, N = 1725 in-patients |
1) Malaria RDT, microscopy 2) Dengue NS1 ELISA, IgM 3) chikungunya IgM RDT 4) Influenza RDT 5) Leptospirosis IgM |
Malaria (102, 6%) Dengue (252, 15%) chikungunya (35, 2%) Influenza (13, 0.8%) Leptospirosis (18, 1%) Mixed mosquito borne infections (23, 1%) Undetermined (965, 56%) |
| MCVR report 2017 | 27 hospital-based sentinel sites, June 2014–July 2017 |
N = 27,586 in-patients |
1) PCR for Influenza 2) Dengue IgM, IgG ELISA, PCR 3) Scrub typhus IgM ELISA and PCR 4) Leptospirosis IgM ELISA, PCR, MAT 5) Malaria RDT, PCR 6) chikungunya IgM ELISA, PCR |
Influenza (4118, 15%) Dengue (1898, 7%) Scrub typhus (1177, 4%) Leptospirosis (1107, 4%) Malaria (953, 3%) chikungunya (371, 1%) |
In our study, we enrolled 954 patients over 12 months from January 2016 to January 2017 from an outpatient clinic in Ispat General Hospital in Rourkela, Odisha, India, who had first undergone triage for malaria by OPD staff, and we carried out diagnostic tests for febrile illnesses known to be prevalent in the area based on hospital data. We detected dengue in approximately one in eight patients by any ELISA-based diagnostic test, and positivity rates for chikungunya and scrub typhus were low (< 6%). Co-infections were commonly detected for scrub typhus and chikungunya; for example, dengue was detected in six out of the ten patients with scrub typhus and malaria detected in five of the 17 patients with chikungunya. Below we discuss each of the four diseases in depth.
Malaria
There were 1,090,724 cases of malaria and 331 deaths reported in India in 2016, of which 449,467 cases (41.2%) and 77 deaths (23.3%) were reported from Odisha (NVBDCP). These case numbers are likely underestimates since only ~ 8% of cases may be detected by the surveillance system, as noted by the World Health Organization (W.H.O.) [24]. Even going by these low estimates, India accounts for ~ 6% of all malaria cases and 51% of P. vivax cases globally. Malaria transmission occurs in both urban and rural India and is especially high among tribal populations in the mountainous, inaccessible regions of eastern and northeastern India [25].
Of the three commonly used methods for malaria diagnosis (RDTs, microscopy, and PCR), microscopy remains the standard for malaria diagnosis in India. In Asian countries, males have a higher risk of malaria (as diagnosed by microscopy) compared to females (as diagnosed by microscopy) [26]; the reason is not completely clear but may relate to differences in exposure by gender. PCR-based methods are more sensitive, but require expensive equipment that is unavailable in most regions. RDTs, while less sensitive and specific than microscopy, are available and used for first-line malaria diagnosis in remote regions. Our results confirm findings of previous studies across India of higher sensitivity of PCR over microscopy when used for detection of Plasmodium [27, 28]. Submicroscopic malaria has been reported to be more common among older people and possibly related to the development of immunity, and also with the recent use/suboptimal use of antimalarial drugs [29]. Three patients were positive by RDT and negative by microscopy and PCR: the antigens produced by recently-cleared malaria parasites can persist in the blood after treatment for a period of time, explaining the positive RDT in the presence of a negative PCR test [30]. In our previous treatment studies, we have noted that for P. falciparum (although not for P. vivax), some RDTs can be positive up to 28 days post treatment, whereas the PCR test result for the same patient was negative in 1 week. No patient identified as Plasmodium positive by microscopy was negative by both RDT and PCR. However, there were 23 sub-patent infections among the 46 patients positive for Plasmodium by PCR. Sub-patent infections are of concern, because malaria infections not detected by microscopy or RDT may not be treated appropriately; patients may continue to carry the parasite and act as a pool for further transmission. Overall, our finding that 54 out of 954 patients (5.7%) were positive for P. falciparum, P. vivax, or P. malariae by at least one diagnostic method despite all 954 patients having undergone malaria triage in the OPD leads us to recommend the use of malaria diagnostic test(s) rather than relying upon patient symptoms.
Dengue
The incidence of dengue has grown dramatically around the world in recent decades. The WHO predicts an incidence of 50–100 dengue million cases and ~ 20,000 dengue-related deaths per year, and nearly 40% of the world’s population lives in areas with a risk of DENV transmission [31]. India is estimated to contribute 34% of the total global burden of dengue [32], and suffered several outbreaks of severe dengue over the last few years [33–36]. There were 129,166 reported cases with 245 deaths in 2016 (NVBDCP). Dengue is endemic in Odisha since 2010, and there has been a four-fold increase in the number of reported cases, from 2450 in 2015 to 8380 in 2016, accounting for 6.5% of the total cases in India [35].
Dengue diagnosis in our study was carried out by detection of either the NS1 antigen or antibodies specific to DENV envelope proteins. We used two techniques for detection of DENV: RDTs to detect NS1, DENV-specific IgG and IgM, and ELISA targeting either NS1 or DENV-specific IgM. NS1 antigen is present in the bloodstream immediately after onset of fever for up to 18 days, and detection of NS1 can thus be used as a proxy for early infection. DENV-specific IgM antibodies may be present in the serum as early as 3–5 days following onset of fever, and persist for several months, in the case of a primary infection, but appear earlier in a secondary infection. IgG antibodies appear 14 days after a primary dengue infection, and can persist for life, but appear within the first couple of days following onset of symptoms in a secondary infection.
We observed a 100% concordance of Dengue NS1 positivity between 101 samples tested by RDTs from two different manufacturers (J. Mitra and SD Bioline) in India, with a positivity rate of ~ 13%. A slightly larger but comparable number of samples (~ 14.2%) were positive when tested by ELISA for NS1 antigen. Four samples were NS1 antigen positive by ELISA but not RDT, and two samples were positive by RDT but not ELISA. There was a significant difference in the number of samples positive by ELISA for Dengue NS1 versus DENV-specific IgM, as expected since these methods test chronologically different stages of the infection, with NS1 antigen presence in the serum preceding DENV-specific IgM by 3–5 days. However, there were significantly fewer samples identified as positive for DENV-specific IgM by RDT than ELISA (20%), which may be due to the increased sensitivity of the capture ELISA IgM over the RDT. Only 2 samples were positive for DENV-specific IgG by the SD Bioline RDT but they were also positive by the J. Mitra RDT. There was also a large variation in positivity in our dengue test results, ranging from 1 to 14.2% by any single test, and 25.3% positive across all dengue tests combined, so it is likely that some positives will be missed if just one diagnostic test is used. Dengue prevalence rates varied from 7 to 38% across other hospital-based febrile illness studies in India and depending upon the assay used; the highest prevalence was observed in a study that used RDTs for detection of DENV (Table 6).
Chikungunya
Chikungunya transmission is concentrated in Southeast Asia, the Americas, Pacific Islands, and Africa. The virus has a high infection rate, with up to 75% of a population infected in areas of virus circulation [37]. Early documented outbreaks of chikungunya in India in the 1960s and 1970s were followed by a major outbreak in Kenya in 2004, which spread to the Indian subcontinent and southeast Asia by 2010 [8, 38]. Since then, there has been continuous transmission of chikungunya in India [8, 39], with an increase in number of cases in India from 27,553 in 2015 to 64,057 in 2016, although in Odisha the number of cases decreased from 81 to 51 during the same period (NVBDCP).
A definitive diagnosis can be accomplished using three laboratory tests: virus isolation, serology, and PCR. We used both an immunochromatography-based RDT as well as an ELISA assay to measure chikungunya-specific IgM levels in patient plasma. It is important to note that chikungunya-specific IgM is usually detected in the plasma from day 4–7 following the onset of symptoms, so we may have missed early infections. In our study, 2.8% patients tested were positive for chikungunya-specific IgM by the RDT (J. Mitra and Co.), but twice as many (5.2%) were found to be positive by the more sensitive ELISA based assay. We did not have the resources to confirm the test results with virus culture and RT-PCR, so it is unclear which test result is more reliable, and thus, further examination of these results is warranted. Similar proportions of chikungunya IgM-positive individuals (2–6%) were observed in other hospital-based studies from India (Table 6) [4, 5, 11].
Scrub typhus
Scrub typhus is a re-emerging infection in India in that its prevalence has been increasing in recent years across various parts of the country [11]. It is underreported [11, 40], and prevalence data are not easily available as routine field and hospital-based surveillance has only recently begun [41]. It rose to prominence as a dominant cause of fever among soldiers along the Indo-Burmese border during World War II, and the Indo-Pakistan border in the 1990s [42]. India belongs to the ‘tsutsugamushi triangle’, the postulated zone of highest prevalence of scrub typhus cases [42], and the illness has been found to be prevalent ubiquitously across the country [40, 43–47].
Scrub typhus diagnosis may be carried out by one or more of the following methods: IgM and IgG ELISA to test for the presence of anti-O. tsutsugamushi antibodies in patient serum, and PCR targeting O. tsutsugamushi antigenic genes. We used the Scrub Typhus Detect ELISA kit from InBios international to identify IgM antibodies targeting the O. tsutsugamushi-derived recombinant 56-kDa antigen, as an indicator of exposure to the bacterium. We observed a positive result in 10/287 (3.5%) patient sera tested. Other hospital-based surveillance studies in India have identified scrub typhus as a frequent etiology of acute febrile illness, with frequencies varying from 9 to 48% [4, 11, 44, 48, 49] (Table 6). We were unable to test for the presence of Orientia bacteria by PCR and did not collect convalescent sera from patients to test an increase in antibody titers by IFA. We may also have missed some scrub typhus cases if the plasma sample was collected too early in the infection. Eschar was not observed in any patients.
Co-infections
DENV and CHIKV are both transmitted by the Aedes mosquito, leading to significant overlap in areas of circulation of these viruses. Several studies report co-infections of dengue and chikungunya in India [15, 50–52]. In our study, we found a statistically significant association between chikungunya and malaria, and between scrub typhus and dengue, but not between chikungunya and dengue. The number of co-infections with malaria as one of the infections is likely an underestimate due to our patient pool having undergone triage for malaria on the basis of presumptive malaria symptoms of high fever, chills and rigor.
Anemia
Anemia was associated with chikungunya, scrub typhus, and multiple infections (Additional file 1: Table S1). Although anemia is a frequent reported effect of malaria, only a borderline association with malaria as diagnosed by RDT was detected. It is possible that anemia was not associated with malaria because our study sample set was derived from an outpatient population with a high prevalence of anemia (39%), and the etiology of anemia is diverse.
Future studies
Our pilot study testing for four common infectious diseases could be expanded to include additional components. For example, conducting viral culture or RT-PCR-based assays would increase the robustness of our studies. Apart from malaria, dengue, chikungunya, and scrub typhus, there are other etiologies for febrile illness prevalent in Odisha that we could test for in the future, including the bacterial illnesses leptospirosis, melioidosis, and typhoid fever, and the viral illnesses influenza and Japanese encephalitis.
Conclusions
Of the three infectious diseases (dengue, chikungunya, and scrub typhus) we tested patients for after initial triage for malaria cases by the OPD, dengue was the most common infection detected (25.3% of all patients tested by either RDT or ELISA). Our results suggest that using more than one dengue diagnostic test is important since available diagnostic tests have different sensitivities and capture different time points in an infection. Our results also recommend using diagnostic tests to identify the malaria parasite rather than relying upon patient symptoms. Although other infections were less common, physicians should keep the possibility of co-morbidity in mind when treating patients. Routine febrile illness surveillance is required to accurately establish the prevalence of these infections in this region, to offer timely treatment, implement appropriate methods of control, and ultimately inform policy.
Additional file
Table S1. A. Anemia of participants for each diagnostic test. B. Anemia of participants according to number of infections*. The association of anemia with patients tested for malaria, dengue, chikungunya and scrub typhus by every diagnostic test used is presented in supplemental Table 1, and the association between anemia and number of infections detected in a patient is presented in supplemental Table 2. (DOCX 29 kb)
Acknowledgements
We thank Steven Sullivan for editing and proofing the manuscript, and the people of Rourkela, Sundargarh District, Odisha.
Authors’ contributions
Study conception and design: PNR, SM, JC, SS. Acquisition of data: PNR, SC, SZA, AD, PB, RRO, GP, SS. Data analysis and interpretation: PNR, SC, SZA, AD, AVE, SS. Data Entry: PB, PN, SA. Statistical analysis: PNR and AVE. Manuscript writing: PNR, AVE, JC. All authors contributed to revision of the manuscript and approved the final version of the manuscript.
Funding
Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (NIH) under Award Number U19AI089676 as part of the International Centers for Excellence in Malaria Research. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The funding agency had no role in the design of the study or collection, analysis, and interpretation of data, or in writing the manuscript.
Availability of data and materials
Data generated and analyzed during this study are available through this published article and its supplementary information files. In addition, data have been submitted to the EuPathDB clinical database ClinEpiDB (https://clinepidb.org/), part of the EuPathDB project [53].
Ethics approval and consent to participate
Ethical approval to conduct this study was obtained from the Ethics Committees of Ispat General Hospital, India and the Institutional Review Board of New York University (Study #i10–00173). All project staff completed Protection of Human Research Subjects training prior to beginning the study, and clinical samples were collected after written or verbal informed consent was obtained from participants. For adult participants, written consent was obtained from literate patients, and oral consent documented by a thumb print was obtained from illiterate patients (approved by the ethical committees). For child participants, assent was obtained from the participant, in addition to written or oral consent from their parent or legal guardian.
Consent for publication
Not Applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Pavitra N. Rao, Email: pavitra.rao@gmail.com
Anna Maria van Eijk, Email: amvaneijk@gmail.com.
Sandhya Choubey, Email: drsandhyarajivmaharshi@gmail.com.
Syed Zeeshan Ali, Email: syedzeeshanali9@gmail.com.
Aditee Dash, Email: draditeedash@gmail.com.
Punam Barla, Email: punamrkl26@gmail.com.
Rajshri Rani Oraon, Email: rajshri86@gmail.com.
Gautam Patel, Email: surajit.goutam@rediffmail.com.
P. Nandini, Email: pnandini18@gmail.com
Subrata Acharya, Email: sulekh.liki@gmail.com.
Sanjib Mohanty, Email: sanjib.mohanty54@gmail.com.
Jane M. Carlton, Email: jane.carlton@nyu.edu
Sanghamitra Satpathi, Email: sanghamitra.satpathi@gmail.com.
References
- 1.John TJ, Dandona L, Sharma VP, Kakkar M. Continuing challenge of infectious diseases in India. Lancet. 2011;377(9761):252–269. doi: 10.1016/S0140-6736(10)61265-2. [DOI] [PubMed] [Google Scholar]
- 2.Salvi S, Apte K, Madas S, Barne M, Chhowala S, Sethi T, Aggarwal K, Agrawal A, Gogtay J. Symptoms and medical conditions in 204 912 patients visiting primary health-care practitioners in India: a 1-day point prevalence study (the POSEIDON study) Lancet Glob Health. 2015;3(12):e776–e784. doi: 10.1016/S2214-109X(15)00152-7. [DOI] [PubMed] [Google Scholar]
- 3.Robinson ML, Manabe YC. Reducing uncertainty for acute febrile illness in resource-limited settings: the current diagnostic landscape. Am J Trop Med Hyg. 2017;96(6):1285–1295. doi: 10.4269/ajtmh.16-0667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Annual report of hospital based surveillance of acute febrile illness in India in.: Manipal Centre for Virus Research; 2017.
- 5.Robinson ML, Kadam D, Khadse S, Balasubramanian U, Raichur P, Valvi C, Marbaniang I, Kanade S, Sachs J, Basavaraj A, et al. Vector-borne disease is a common cause of hospitalized febrile illness in India. Am J Trop Med Hyg. 2018;98(5):1526–1533. doi: 10.4269/ajtmh.17-0571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simmons CP, Farrar JJ, Nguyen v V, Wills B. Dengue. N Engl J Med. 2012;366(15):1423–1432. doi: 10.1056/NEJMra1110265. [DOI] [PubMed] [Google Scholar]
- 7.Dengue and severe dengue. http://www.who.int/news-room/fact-sheets/detail/dengue-and-severe-dengue. Accessed 23 June 2018.
- 8.Zeller H, Van Bortel W, Sudre B. Chikungunya: its history in Africa and Asia and its spread to new regions in 2013-2014. J Infect Dis. 2016;214(suppl 5):S436–S440. doi: 10.1093/infdis/jiw391. [DOI] [PubMed] [Google Scholar]
- 9.Walker DH. Scrub typhus - scientific neglect, ever-widening impact. N Engl J Med. 2016;375(10):913–915. doi: 10.1056/NEJMp1608499. [DOI] [PubMed] [Google Scholar]
- 10.Integrated Disease Surveillance Programme. http://idsp.nic.in/. Accessed 23 June 2018.
- 11.Morch K, Manoharan A, Chandy S, Chacko N, Alvarez-Uria G, Patil S, Henry A, Nesaraj J, Kuriakose C, Singh A, et al. Acute undifferentiated fever in India: a multicentre study of aetiology and diagnostic accuracy. BMC Infect Dis. 2017;17(1):665. doi: 10.1186/s12879-017-2764-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dhingra N, Jha P, Sharma VP, Cohen AA, Jotkar RM, Rodriguez PS, Bassani DG, Suraweera W, Laxminarayan R, Peto R, et al. Adult and child malaria mortality in India: a nationally representative mortality survey. Lancet. 2010;376(9754):1768–1774. doi: 10.1016/S0140-6736(10)60831-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kakkar M. Dengue fever is massively under-reported in India, hampering our response. BMJ. 2012;345:e8574. doi: 10.1136/bmj.e8574. [DOI] [PubMed] [Google Scholar]
- 14.Rubio JM, Benito A, Roche J, Berzosa PJ, Garcia ML, Mico M, Edu M, Alvar J. Semi-nested, multiplex polymerase chain reaction for detection of human malaria parasites and evidence of Plasmodium vivax infection in Equatorial Guinea. Am J Trop Med Hyg. 1999;60(2):183–187. doi: 10.4269/ajtmh.1999.60.183. [DOI] [PubMed] [Google Scholar]
- 15.Saswat T, Kumar A, Kumar S, Mamidi P, Muduli S, Debata NK, Pal NS, Pratheek BM, Chattopadhyay S, Chattopadhyay S. High rates of co-infection of dengue and chikungunya virus in Odisha and Maharashtra, India during 2013. Infect Genet Evol. 2015;35:134–141. doi: 10.1016/j.meegid.2015.08.006. [DOI] [PubMed] [Google Scholar]
- 16.Mitra S, Choudhari R, Nori H, Abhilash KP, Jayaseelan V, Abraham AM, Cherian AO, Prakash JA, Muliyil J. Comparative evaluation of validity and cost-benefit analysis of rapid diagnostic test (RDT) kits in diagnosis of dengue infection using composite reference criteria: a cross-sectional study from South India. J Vector Borne Dis. 2016;53(1):30–36. [PubMed] [Google Scholar]
- 17.Rao MR, Padhy RN, Das MK. Prevalence of dengue viral and malaria parasitic co-infections in an epidemic district, Angul of Odisha, India: an eco-epidemiological and cross-sectional study for the prospective aspects of public health. J Infect Public Health. 2016;9(4):421–428. doi: 10.1016/j.jiph.2015.10.019. [DOI] [PubMed] [Google Scholar]
- 18.Rahi M, Gupte MD, Bhargava A, Varghese GM, Arora R. DHR-ICMR guidelines for diagnosis & management of Rickettsial diseases in India. Indian J Med Res. 2015;141(4):417–422. doi: 10.4103/0971-5916.159279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Varghese GM, Janardhanan J, Mahajan SK, Tariang D, Trowbridge P, Prakash JA, David T, Sathendra S, Abraham OC. Molecular epidemiology and genetic diversity of Orientia tsutsugamushi from patients with scrub typhus in 3 regions of India. Emerg Infect Dis. 2015;21(1):64–69. doi: 10.3201/eid2101.140580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42(2):377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.National Guidelines for Clinical Management of Dengue Fever. http://www.searo.who.int/india/publications/national_guidelines_clinical_management_dengue1.pdf?ua=1. Accessed 23 June 2018.
- 22.National Guidelines for Clinical Management of Chikungunya. https://nvbdcp.gov.in/WriteReadData/l892s/77728737401531912419.pdf. Accessed 28 Oct 2016.
- 23.Cummings P. Methods for estimating adjusted risk ratios. Stata J. 2009;9(2):175–196. doi: 10.1177/1536867X0900900201. [DOI] [Google Scholar]
- 24.World malaria report 2017. http://www.who.int/malaria/publications/world-malaria-report-2017/report/en/.
- 25.World malaria report 2017. http://www.who.int/malaria/publications/world-malaria-report-2017/report/en/. Accessed 27 June 2018.
- 26.Pathak S, Rege M, Gogtay NJ, Aigal U, Sharma SK, Valecha N, Bhanot G, Kshirsagar NA, Sharma S. Age-dependent sex bias in clinical malarial disease in hypoendemic regions. PLoS One. 2012;7(4):e35592. doi: 10.1371/journal.pone.0035592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dhangadamajhi G, Kar SK, Ranjit MR. High prevalence and gender bias in distribution of Plasmodium malariae infection in central East-Coast India. Trop Biomed. 2009;26(3):326–333. [PubMed] [Google Scholar]
- 28.Haanshuus CG, Chandy S, Manoharan A, Vivek R, Mathai D, Xena D, Singh A, Langeland N, Blomberg B, Vasanthan G, et al. A high malaria prevalence identified by PCR among patients with acute undifferentiated fever in India. PLoS One. 2016;11(7):e0158816. doi: 10.1371/journal.pone.0158816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okell LC, Bousema T, Griffin JT, Ouedraogo AL, Ghani AC, Drakeley CJ. Factors determining the occurrence of submicroscopic malaria infections and their relevance for control. Nat Commun. 2012;3:1237. doi: 10.1038/ncomms2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dalrymple U, Arambepola R, Gething PW, Cameron E. How long do rapid diagnostic tests remain positive after anti-malarial treatment? Malar J. 2018;17(1):228. doi: 10.1186/s12936-018-2371-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.CDC: Dengue. 2018.
- 32.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, et al. The global distribution and burden of dengue. Nature. 2013;496(7446):504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siddiqui O, Chakravarti A, Abhishek KS. Dengue: lessons of an outbreak. J Clin Diagn Res. 2016;10(6):DC01–DC04. doi: 10.7860/JCDR/2016/19238.7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dar L, Broor S, Sengupta S, Xess I, Seth P. The first major outbreak of dengue hemorrhagic fever in Delhi, India. Emerg Infect Dis. 1999;5(4):589–590. doi: 10.3201/eid0504.990427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mutheneni SR, Morse AP, Caminade C, Upadhyayula SM. Dengue burden in India: recent trends and importance of climatic parameters. Emerg Microbes Infect. 2017;6(8):e70. doi: 10.1038/emi.2017.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chakravarti A, Arora R, Luxemburger C. Fifty years of dengue in India. Trans R Soc Trop Med Hyg. 2012;106(5):273–282. doi: 10.1016/j.trstmh.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 37.CDC Yellow Book 2018. https://wwwnc.cdc.gov/travel/yellowbook/2018/infectious-diseases-related-to-travel/chikungunya. Accessed 21 Jan 2017.
- 38.Pialoux G, Gauzere BA, Jaureguiberry S, Strobel M. Chikungunya, an epidemic arbovirosis. Lancet Infect Dis. 2007;7(5):319–327. doi: 10.1016/S1473-3099(07)70107-X. [DOI] [PubMed] [Google Scholar]
- 39.Jain J, Nayak K, Tanwar N, Gaind R, Gupta B, Shastri JS, Bhatnagar RK, Kaja MK, Chandele A, Sunil S. Clinical, serological, and Virological analysis of 572 chikungunya patients from 2010 to 2013 in India. Clin Infect Dis. 2017;65(1):133–140. doi: 10.1093/cid/cix283. [DOI] [PubMed] [Google Scholar]
- 40.Khan SA, Bora T, Chattopadhyay S, Jiang J, Richards AL, Dutta P. Seroepidemiology of rickettsial infections in Northeast India. Trans R Soc Trop Med Hyg. 2016;110(8):487–494. doi: 10.1093/trstmh/trw052. [DOI] [PubMed] [Google Scholar]
- 41.Bonell A, Lubell Y, Newton PN, Crump JA, Paris DH. Estimating the burden of scrub typhus: a systematic review. PLoS Negl Trop Dis. 2017;11(9):e0005838. doi: 10.1371/journal.pntd.0005838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu G, Walker DH, Jupiter D, Melby PC, Arcari CM. A review of the global epidemiology of scrub typhus. PLoS Negl Trop Dis. 2017;11(11):e0006062. doi: 10.1371/journal.pntd.0006062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kalal BS, Puranik P, Nagaraj S, Rego S, Shet A. Scrub typhus and spotted fever among hospitalised children in South India: clinical profile and serological epidemiology. Indian J Med Microbiol. 2016;34(3):293–298. doi: 10.4103/0255-0857.188315. [DOI] [PubMed] [Google Scholar]
- 44.Mittal G, Ahmad S, Agarwal RK, Dhar M, Mittal M, Sharma S. Aetiologies of acute undifferentiated febrile illness in adult patients - an experience from a tertiary care hospital in northern India. J Clin Diagn Res. 2015;9(12):DC22–DC24. doi: 10.7860/JCDR/2015/11168.6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sinha P, Gupta S, Dawra R, Rijhawan P. Recent outbreak of scrub typhus in North Western part of India. Indian J Med Microbiol. 2014;32(3):247–250. doi: 10.4103/0255-0857.136552. [DOI] [PubMed] [Google Scholar]
- 46.Sivarajan S, Shivalli S, Bhuyan D, Mawlong M, Barman R. Clinical and paraclinical profile, and predictors of outcome in 90 cases of scrub typhus, Meghalaya, India. Infect Dis Poverty. 2016;5(1):91. doi: 10.1186/s40249-016-0186-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stephen S, Sangeetha B, Ambroise S, Sarangapani K, Gunasekaran D, Hanifah M, Somasundaram S. Outbreak of scrub typhus in Puducherry & Tamil Nadu during cooler months. Indian J Med Res. 2015;142(5):591–597. doi: 10.4103/0971-5916.171289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abhilash KP, Jeevan JA, Mitra S, Paul N, Murugan TP, Rangaraj A, David S, Hansdak SG, Prakash JA, Abraham AM, et al. Acute undifferentiated febrile illness in patients presenting to a tertiary Care Hospital in South India: clinical Spectrum and outcome. J Glob Infect Dis. 2016;8(4):147–154. doi: 10.4103/0974-777X.192966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chrispal A, Boorugu H, Gopinath KG, Chandy S, Prakash JA, Thomas EM, Abraham AM, Abraham OC, Thomas K. Acute undifferentiated febrile illness in adult hospitalized patients: the disease spectrum and diagnostic predictors - an experience from a tertiary care hospital in South India. Trop Dr. 2010;40(4):230–234. doi: 10.1258/td.2010.100132. [DOI] [PubMed] [Google Scholar]
- 50.Afreen N, Deeba F, Khan WH, Haider SH, Kazim SN, Ishrat R, Naqvi IH, Shareef MY, Broor S, Ahmed A, et al. Molecular characterization of dengue and chikungunya virus strains circulating in New Delhi, India. Microbiol Immunol. 2014;58(12):688–696. doi: 10.1111/1348-0421.12209. [DOI] [PubMed] [Google Scholar]
- 51.Chahar HS, Bharaj P, Dar L, Guleria R, Kabra SK, Broor S. Co-infections with chikungunya virus and dengue virus in Delhi, India. Emerg Infect Dis. 2009;15(7):1077–1080. doi: 10.3201/eid1507.080638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taraphdar D, Sarkar A, Mukhopadhyay BB, Chatterjee S. A comparative study of clinical features between monotypic and dual infection cases with chikungunya virus and dengue virus in West Bengal, India. Am J Trop Med Hyg. 2012;86(4):720–723. doi: 10.4269/ajtmh.2012.11-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aurrecoechea C, Barreto A, Basenko EY, Brestelli J, Brunk BP, Cade S, Crouch K, Doherty R, Falke D, Fischer S, et al. EuPathDB: the eukaryotic pathogen genomics database resource. Nucleic Acids Res. 2017;45(D1):D581–D591. doi: 10.1093/nar/gkw1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. A. Anemia of participants for each diagnostic test. B. Anemia of participants according to number of infections*. The association of anemia with patients tested for malaria, dengue, chikungunya and scrub typhus by every diagnostic test used is presented in supplemental Table 1, and the association between anemia and number of infections detected in a patient is presented in supplemental Table 2. (DOCX 29 kb)
Data Availability Statement
Data generated and analyzed during this study are available through this published article and its supplementary information files. In addition, data have been submitted to the EuPathDB clinical database ClinEpiDB (https://clinepidb.org/), part of the EuPathDB project [53].