Abstract
Recent studies have shown that microorganisms may be associated with the onset and development of bladder cancer. The purpose of this study is to identify the common core bacteria associated with bladder cancer. We characterized the urinary microbial profile of the individuals with bladder cancer by 16S rRNA gene sequencing, and the results of 24 bladder cancer samples collected in our laboratory reveal 31 common core bacteria at genera level. In addition, the abundance of four common core bacteria is significantly higher in bladder cancer samples than in samples from nondiseased people analyzed by LEfSe, based on two previous datasets. In particular, the abundance of Acinetobacter is much higher in bladder cancer samples. It has been reported that Acinetobacter is involved not only in biofilm formation but also in the adhesion and invasion of epithelial cells, the spread of bacteria caused by the degradation of phospholipids in the mucosal barrier, and the escape of the host immune response. Thus, Acinetobacter may be related to bladder cancer and is a potential microbial marker of bladder cancer. However, due to the limited number of participants, further studies are needed to better understand the role of microorganisms in bladder cancer to provide novel biomarkers for diagnosis, prognosis, and therapy.
1. Introduction
Bladder cancer is the ninth most common malignant tumor. Every year, more than 430,000 patients are diagnosed with bladder cancer, and over 160,000 people die of bladder cancer globally [1, 2]. In the past few decades, bladder cancer has attracted the attention of scientists for its high incidence and mortality rate. However, the etiology and pathophysiology of bladder cancer remain unclear. It may be caused by genetic mutations and external risk factors, including smoking, exposure to carcinogens, chlorination of drinking water, and cyclophosphamide treatment [3]. In addition to environmental and genetic factors, researchers are increasingly aware that microorganisms in the human body play an important role in maintaining health and developing disease. Microorganisms affect the physiological functions of the human body, such as metabolism, immunity, and hematopoiesis [4]. Transient inflammation is considered part of the body's immune defense against pathogens, but persistent inflammation may promote the development of cancer [5]. Epidemiological studies have shown that chronic inflammation makes people susceptible to various cancers [6]. It is estimated that infections and inflammatory responses are associated with 15% to 20% of all deaths from cancer globally [7]. Studies suggest that microbial dysbiosis may promote the development of some malignant tumors, such as colorectal cancer and breast cancer [8, 9]. Even the links between certain pathogens and cancer have been well established, for example, Helicobacter pylori and gastric cancer [10]. In recent years, urinary microflora in urothelial bladder cancer has been studied. Xu et al. reported that Streptococcus spp. are more abundant in urine from bladder cancer patients (n = 8) compared to healthy individuals (n = 6) [11]. Bucevic et al. found enrichment of some bacteria (e.g., the genera Fusobacterium, Actinobaculum, Facklamia, Campylobacter, and Subdoligranulum and the family Ruminococcaceae) in urine from bladder cancer patients (n = 12) compared to healthy individuals (n = 11) [1]. Wu et al. found enrichment of some bacterial genera (e.g., Acinetobacter, Anaerococcus, and Sphingobacterium) in urine from bladder cancer patients (n = 31) compared to nonneoplastic controls (n = 18) [2]. However, the biomarkers found in the bladder cancer samples of the above three laboratories are different. Therefore, in this study we investigate the common core bacteria in 24 bladder cancer samples collected in our laboratory. We have found that the abundance of some common core bacteria is significantly higher in bladder cancer samples than in samples from nondiseased people (based on two previous datasets [1, 2]), providing insight into the role of the microbiome in bladder cancer.
2. Results and Discussion
2.1. Bladder Cancer Patients and Sequencing Data
Urine samples were collected from 25 patients with bladder cancer. One sample failed to meet the quality standard because of its low sequencing depth. The characteristics of 24 patients with bladder cancer analyzed in this study are presented in Table S1. Eighteen male patients and six female patients were included, ranging from 30 to 86 years. High-throughput sequencing of urine samples from patients with bladder cancer yielded a total of 2,604,140 raw sequences, which were then merged into 1,200,568 paired sequences. After quality filtering and label classification, the number of sequences was reduced to 368,525, with an average read length of 283 base pairs. They were assigned to 4100 operational taxonomic units (OTUs) at 97% similarity by QIIME [12]. Rarefaction curves showed that the 24 samples were sequenced to a sufficient depth, and most samples have captured complete microbial profiles (Figure S1). A total of 26 phyla, 60 classes, 114 orders, 217 families, and 422 bacterial genera were identified, averagely 132.4 genera per sample. A total of 26 phyla, 60 classes, 114 orders, 217 families, and 422 bacterial genera were identified, averagely 132.4 genera per sample. Raw data of bladder cancer and healthy control samples from other laboratories [2, 13] were also analyzed by using QIIME [12]. In the raw data by Wu et al. [2], a total of 34 phyla, 93 classes, 158 orders, 297 families, and 646 bacterial genera were identified, averagely 90 genera per sample. In the raw data by Bucevic et al. [1], a total of 19 phyla, 35 classes, 59 orders, 147 families, and 326 bacterial genera were identified, averagely 73.5 genera per sample.
2.2. Microbial Compositions in Bladder Cancer Samples
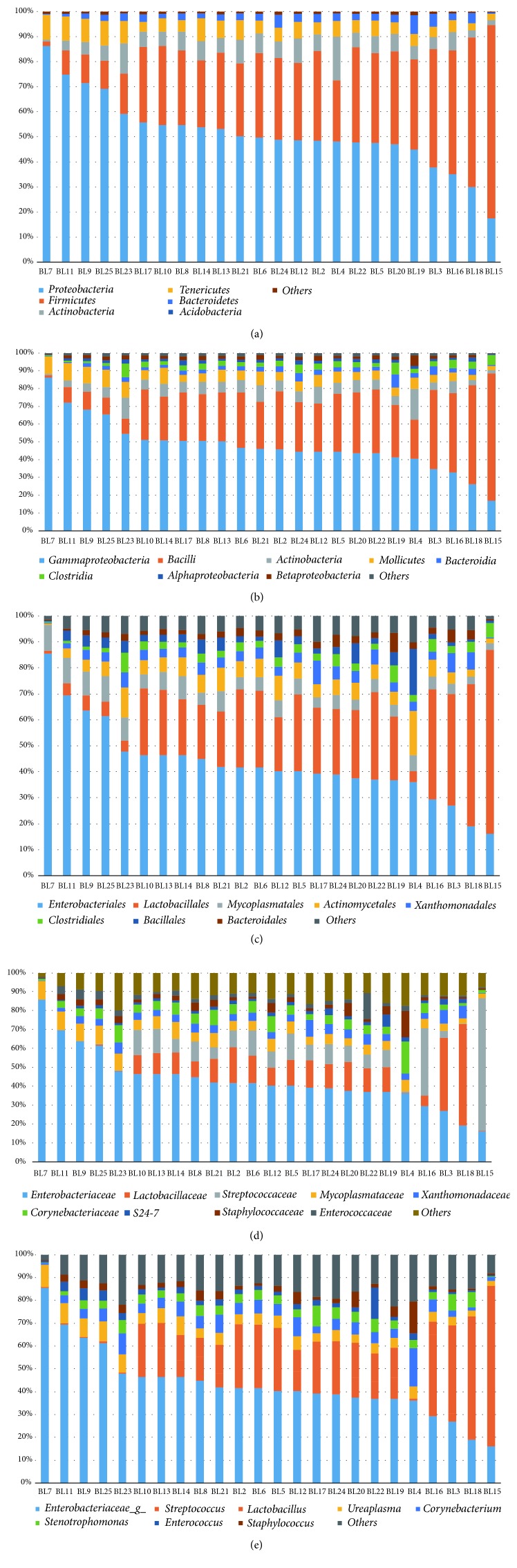
We analyzed the microbial composition in bladder cancer at the levels of phylum, class, order, family, and genus. The five most abundant phyla are Proteobacteria, Firmicutes, Actinobacteria, Tenericutes, and Bacteroidetes (Figure 1(a)). In the study by Wu et al., the top phyla in the bladder cancer samples were Proteobacteria, Firmicutes, Actinobacteria, and Bacteroidetes [2]; and in the study by Bucevic et al., the top phyla in the bladder cancer samples were Firmicutes, Actinobacteria, Bacteroidetes, and Proteobacteria [1]. The phyla Proteobacteria, Firmicutes, Actinobacteria, and Bacteroidetes were abundant in bladder cancer samples from all three laboratories. The most abundant classes in the bladder cancer samples in our laboratory are Gammaproteobacteria, Bacilli, Actinobacteria, Mollicutes, Bacteroidia, Betaproteobacteria, and Clostridia (Figure 1(b)); in the study by Wu et al., the most abundant classes in the bladder cancer samples were Gammaproteobacteria, Bacilli, Actinobacteria, and Betaproteobacteria [2]; and in the study by Bucevic et al., the most abundant classes in the bladder cancer samples were Clostridia, Actinobacteria, Bacteroidia, Bacilli, Epsilonproteobacteria, Gammaproteobacteria, Fusobacteria, and Synergistia [1]. The classes Gammaproteobacteria, Bacilli, and Actinobacteria were abundant in bladder cancer samples from all three studies. The most abundant orders in the bladder cancer samples in our laboratory are Enterobacteriales, Lactobacillales, Mycoplasmatales, Actinomycetales, Xanthomonadales, Clostridiales, Bacillales, and Bacteroidales (Figure 1(c)); in the study by Wu et al., the most abundant orders in the bladder cancer samples were Enterobacteriales, Bacillales, Lactobacillales, Corynebacteriales, and Bacteroidales [2]; in the study by Bucevic et al., the most abundant orders in the bladder cancer samples were Clostridiales, Actinomycetales, Bacteroidales, Lactobacillales, Campylobacterales, Enterobacteriales, Bacillales, and Fusobacteriales [1]. The orders Enterobacteriales, Bacillales, Lactobacillales, and Bacteroidales were abundant in bladder cancer samples from all three laboratories. The most abundant families are Enterobacteriaceae, Lactobacillaceae, Streptococcaceae, Mycoplasmataceae, Xanthomonadaceae, and Corynebacteriaceae (Figure 1(d)). The most abundant genera are Enterobacteriaceae_g_, Streptococcus, Lactobacillus, Ureaplasma, Corynebacterium, Stenotrophomonas, Enterococcus, and Staphylococcus (Figure 1(e)).
Figure 1.
The urinary microbiota of bladder cancer patients. Most abundant taxa are shown at (a) phylum, (b) class, (c) order, (d) family, and (e) genus level.
2.3. The Core Bacteria in Bladder Cancer
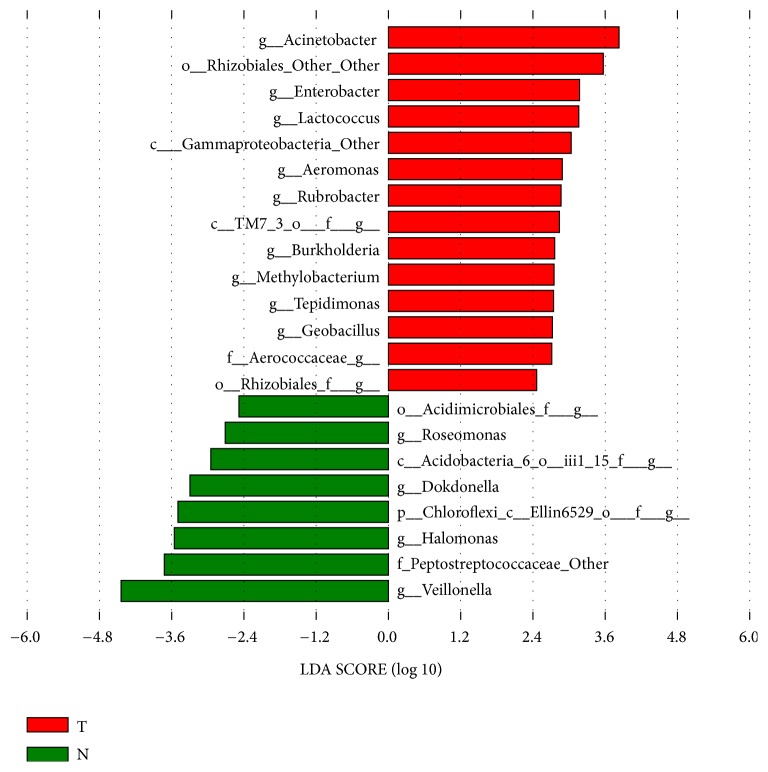
The common bacteria of the 24 bladder cancer samples were analyzed using the online website http://bioinformatics.psb.ugent.be/webtools/Venn/. We found 31 common core bacterial genera in these samples (Figure 2), that is, Clostridiales_f_g, Peptoniphilus, Mycoplasma, Cupriavidus, Lachnospiraceae, Ureaplasma, Delftia, o_Rhizobiales_f_g, Acinetobacter, Enterococcus, Hydrogenophilus, Prevotella, Bacillus, Brevundimonas, f_Enterobacteriaceae_g, Geobacillus, Streptococcus, f_S247_g, Rubrobacter, Bifidobacterium, Finegoldia, Achromobacter, Stenotrophomonas, Actinomyces, Lactobacillus, f_Oxalobacteraceae_g, Sphingomonas, Anaerococcus, Staphylococcus, Corynebacterium, and Sphingobacterium. Some of these common core bacteria may be inherently present in urine, and some may be associated with bladder cancer. In the study by Wu et al., the most abundant genera in the bladder cancer samples were Escherichia, Shigella, Staphylococcus, Streptococcus, Aeromonas, Acinetobacter, Bacteroides, Lactobacillus, Serratia, Proteus, Laceyella, and Fusobacterium [2]. The common core genera Staphylococcus, Streptococcus, Acinetobacter, and Lactobacillus were also abundant in bladder cancer samples from the study by Wu et al. [2]. Moreover, the common core bacteria Acinetobacter, Rubrobacter, Geobacillus, and Rhizobiales (Figure 3) are significantly more abundant in bladder cancer than in control samples (based on two previous datasets [1, 2]). These may include bacteria associated with bladder cancer, but this hypothesis requires further experimental verification.
Figure 2.
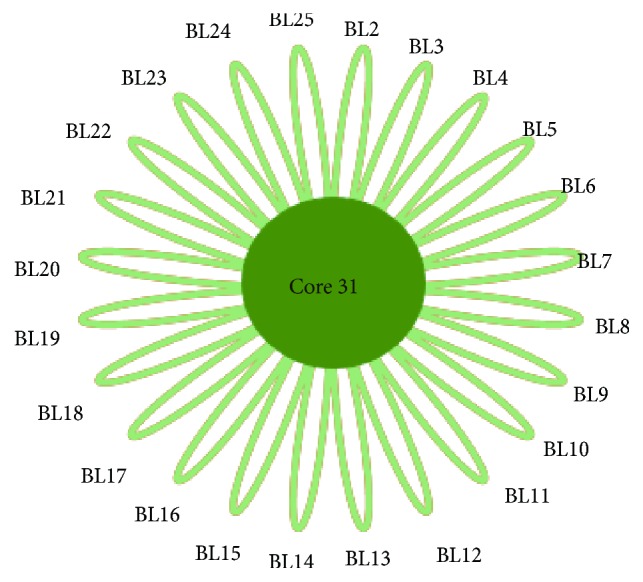
Common core bacteria in 24 bladder cancer samples. There are 31 bacterial genera (Core31) in all of these 24 samples, including Clostridiales_f_g, Peptoniphilus, Mycoplasma, Cupriavidus, Lachnospiraceae, Ureaplasma, Delftia, o_Rhizobiales_f_g, Acinetobacter, Enterococcus, Hydrogenophilus, Prevotella, Bacillus, Brevundimonas, f_Enterobacteriaceae_g, Geobacillus, Streptococcus, f_S247_g, Rubrobacter, Bifidobacterium, Finegoldia, Achromobacter, Stenotrophomonas, Actinomyces, Lactobacillus, f_Oxalobacteraceae_g, Sphingomonas, Anaerococcus, Staphylococcus, Corynebacterium, and Sphingobacterium. BLnum: the number of bladder cancer samples.
Figure 3.
Microbial taxa associated with bladder cancer. The association of microbiota taxa with bladder cancer patients and healthy controls from other laboratories was analyzed by LEfSe. Red indicates taxa enriched in bladder cancer patients, and green indicates taxa enriched in healthy controls.
2.4. Different Abundance of Core Bacteria between Bladder Cancer and Control Samples from Other Laboratories
Linear discriminant analysis effect size (LEfSe) was used to analyze bladder cancer and healthy control samples from other laboratories [2, 13]. Fourteen bacterial genera were found to be significantly higher abundant in bladder cancer samples than in the control group (Figure 3). Among these, four genera including Acinetobacter, Rubrobacter, Geobacillus, and Rhizobiales were also found in the core bacteria of our 24 bladder cancer samples.
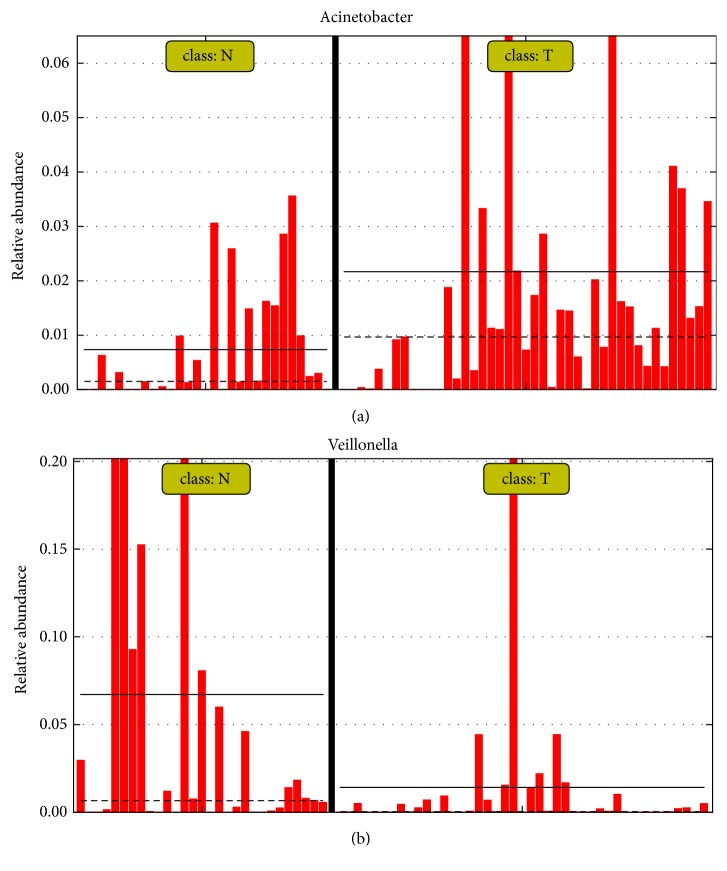
In particular, Acinetobacter abundance was significantly higher in bladder cancer samples than in the control group (based on two previous datasets [1, 2]) (Figures 3 and 4(a)). Interestingly, Acinetobacter was also reported to be significantly more abundant (P = 0.048) in bladder cancer patients in the study by Wu et al. [2]. It was reported that Acinetobacter spp. are among the most abundant Gram-negative bacteria isolated from bovine urine affected by bladder urothelial tumors [14]. In addition, it was reported that the prevalence of Acinetobacter baumannii in gastric cancer patients was significantly higher than in nonulcer dyspepsia and peptic ulcer patients (25% vs. 3.2% and 4.5%) [15]. Acinetobacter is a complex genus, which is associated with nosocomial infections, including urinary tract infections [2]. A. baumannii has been used as model biofilm forming bacteria [16]. Biofilms, in which the microbial communities are embedded in a biopolymer matrix, are especially interesting, because they are the main cause of human bacterial infections and have a strong resistance to antibiotics and host immunity [17–21]. Recent studies have found that bacterial biofilms play a role in the onset and development of various cancers, including stomach cancer [22], colorectal cancer [23], colon cancer [24], prostate cancer [25], and lung cancer [26]. A. baumannii is not only involved in the formation of biofilm but also involved in the adhesion and invasion of epithelial cells, the spread of bacteria caused by the degradation of phospholipids in the mucosal barrier, and the escape of the host immune response [27]. Therefore, Acinetobacter may be related to bladder cancer and is a potential microbial marker of bladder cancer, but a potential causal relationship between Acinetobacter abundance and bladder cancer requires further experimental verification. Interestingly, Veillonella abundance was significantly lower in bladder cancer samples than in the control group (based on two previous datasets [1, 2]); this needs to be further explored in the future (Figures 3 and 4(b)).
Figure 4.
Different relative abundance of (a) Acinetobacter and (b) Veillonella in bladder cancer and healthy control samples.
3. Conclusion
In this study, we characterized the urinary microbial profile of bladder cancer by 16S rRNA gene sequencing. The results reveal 31 common core bacteria from 24 bladder cancer samples collected in our laboratory. In addition, four common core bacteria are significantly more abundant in bladder cancer samples than in nondiseased people (based on two previous datasets [1, 2]). Our study suggests that some common core bacteria, especially Acinetobacter, may be associated with bladder cancer, but the causal relationship is not yet clear. A better understanding of the role of urinary microflora in the onset and development of bladder cancer can provide novel biomarkers for diagnosis and prognosis, as well as more microbial-targeted therapeutic strategies. There are still some limitations of our research. Firstly, a causal relationship between microorganisms and bladder cancer has not been elucidated, and the number of samples is small. Therefore, a larger number of prospective follow-up studies and animal experiments are needed to clarify the role of microorganisms in the onset and development of bladder cancer. Another limitation is that although the 16S rRNA gene sequencing method enables us to detect bacteria that exist in low numbers it cannot detect bacteria at the species level or nonbacterial microorganisms, such as viruses and fungi [5]. The future need to sample at other regions in order to understand the skewed results [28] from different regions by comparing cases and controls. The same problem can happen if samples were processed with different experimental procedures (sample preparation, DNA extraction, PCR, and etc.). The future need to sample at other regions in order to understand the skewed results [28] from different regions by comparing cases and controls.
4. Materials and Methods
4.1. Subject Recruitment and Sample Collection
The research began with the approval of the Ethics Committee of Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences. Urine samples were collected from 25 patients with bladder cancer, 18 males and 7 females, in the First Affiliated Hospital of Zhengzhou University, between January 2017 and March 2017. After the initial suspicion of urinary bladder cancer, the urologist carried out a physical and an ultrasound examination. Then the cancer tissue was removed by the transurethral resection (TUR) approach, and the diagnosis was confirmed by the pathologist after cystoscopy and tumor tissue examination. Urine samples were collected after ultrasound examination and before cystoscopy. The characteristics of bladder cancer patients are given in Table S1. All of the experiments were conducted in accordance with relevant guidelines and regulations, and participants gave written informed consent for urine collection and analysis for research purposes. Clean catch, midstream urine was collected from the bladder cancer patients and stored at −80°C until DNA extraction.
4.2. DNA Isolation from Urine
The genomic DNA of urinary bacteria was extracted using an EasyPure Bacteria Genomic DNA Kit (TRANSGEN). The procedure is as follows: 20 ml urine sample was collected in a 50-ml sterile centrifugal tube and centrifuged at 10,000 rpm for 15 minutes. The supernatant was discarded. The pellet was transferred to a centrifugal tube of 1.5 ml and centrifuged at 13,500 rpm for 10 minutes. The supernatant was discarded. Lysis buffer containing lysozyme was added to the centrifugal tube. The sample was incubated at 37°C for 1 hour and centrifuged at 13,500 rpm for 1 minute, and the supernatant was discarded. The protein fraction was digested by adding protease K at 55°C for 15 minutes. RNaseA was added to digest RNA. Absolute ethanol was added to dehydrate and precipitate the DNA. The solution was added to a centrifugal column and centrifuged at 13,500 rpm for 30 seconds. The effluent was discarded. Washing buffer was used to clean the DNA suspended in the centrifugal column. Finally, DNA was eluted with deionized water. The concentration of DNA was measured with Nanodrop.
4.3. 16S rRNA Gene Amplification and Hi-Seq Sequencing
The V4 region of the 16S rRNA gene was amplified by 515F-806R fusion primers that included a linker and indexing barcodes. The F515 primer (5′TATGGTAATTGTGTGCCAGCMGCCGCGGTAA3′) was used for all of the samples. We added the same linker and different barcode sequences at the 5′ end of the R806 primer (5′AGTCAGTCAGCCGGACTACHVGGGTWTCTAAT3′) (Table S1). PCRs were run in a final volume of 50 μl, containing 2 μl of DNA as template, 2 μl of F515 primer, 2 μl of R806 primer, 4 μl of dNTPs, 4 μl of 25 mmol l-1 MgCl2, 5 μl of 10× Ex Taq buffer, 0.25 μl Taq polymerase (5 U/μl), and 30.75 μl of distilled deionized water. PCR started at 98°C for 1 min, followed by 30 cycles of 98°C for 10 s, 58°C for 30 s, and 72°C for 2 min and one final elongation step at 72°C for 10 min. The PCR products were purified by the SanPrep Column DNA Gel Extraction Kit (Sangon Biotech, Shanghai, China) and preserved in 25 μl sterile water. The concentrations of purified PCR products were determined by NanoDrop (Thermo Fisher Scientific Inc., Waltham, MA, USA); 200 ng of the purified PCR products of each sample was added together in equal amounts and then sent to Illumina Hi-Seq2500 V4 platform of Novogene Genomics Co., Ltd. (Beijing) for sequencing.
4.4. Bioinformatics and Statistical Analyses
Raw data, including bladder cancer and healthy control samples from other laboratories [2, 13], were filtered to eliminate adapter contamination and low quality reads by using QIIME [12]. The filtered sequences were clustered into the OTU with 97% identity, using QIIME [12], and then a representative sequence from each clustered OTU was used to align to the Greengenes Database [29]. QIIME was used to evaluate alpha diversity, which was based on the observed species and the Shannon index. To identify significant differences in bacterial abundance in bladder cancer and healthy control samples from other laboratories [2, 13], taxon summaries at the genus level were reformatted and analyzed by LEfSe [30]. R software was used to find the intersection of the core bacteria of these 24 bladder cancer samples and significantly different bacteria between bladder cancer and healthy control samples.
Acknowledgments
This work was financially supported by the grants of Shenzhen Science and Technology Innovation Committee (JCYJ20160229201759414; JCYJ20170818155011625; JCYJ20160612152651093), respectively; the National Natural Science Foundation of China (31570115).
Data Availability
The datasets generated during the current study are available from the NCBI SRA database with accession numbers SRR8647671.
Disclosure
The funders have no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors' Contributions
Yingfei Ma and Guoqin Mai did experimental design work. Guoqin Mai and Limei Chen conducted experiments. Guoqin Mai, Quan Liu, and Haoran Zhang analyzed the data. Guoqin Mai and Yingfei Ma wrote the manuscript. All authors read and approved the manuscript.
Supplementary Materials
Figure S1: rarefaction curves. It can be seen that the Shannon index of the 24 samples has reached saturation, indicating that the sequencing depth is sufficient, and most samples have captured complete microbial profiles. Table S1: characteristics of 24 patients with bladder cancer.
References
- 1.Popović V. B., Šitum M., Chow C. T., Chan L. S., Roje B., Terzić J. The urinary microbiome associated with bladder cancer. Scientific Reports. 2018;8(1) doi: 10.1038/s41598-018-29054-w.12157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu P., Zhang G., Zhao J., et al. Profiling the urinary microbiota in male patients with bladder cancer in china. Frontiers in Cellular & Infection Microbiology. 2018;8:p. 167. doi: 10.3389/fcimb.2018.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Witjes J. A., Lebret T., Compérat E. M., et al. Updated 2016 EAU guidelines on muscle-invasive and metastatic bladder cancer. European Urology. 2017;72(2):p. e45. doi: 10.1016/j.eururo.2017.02.031. [DOI] [PubMed] [Google Scholar]
- 4.Dzutsev A., Goldszmid R. S., Viaud S., Zitvogel L., Trinchieri G. The role of the microbiota in inflammation, carcinogenesis, and cancer therapy. European Journal of Immunology. 2015;45(1):17–31. doi: 10.1002/eji.201444972. [DOI] [PubMed] [Google Scholar]
- 5.Ainsworth C. MICROBIOME: A bag of surprises. Nature. 2017;551(7679):S40–S41. doi: 10.1038/551S40a. [DOI] [PubMed] [Google Scholar]
- 6.Morgillo F., Dallio M., Corte C. M. D., et al. Carcinogenesis as a result of multiple inflammatory and oxidative hits: a comprehensive review from tumor microenvironment to gut microbiota. Neoplasia (United States) 2018;20(7):721–733. doi: 10.1016/j.neo.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balkwill F., Mantovani A. Inflammation and cancer: back to virchow? The Lancet. 2001;357(9255):539–545. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- 8.Irrazábal T., Belcheva A., Girardin S. E., Martin A., Philpott D. J. The multifaceted role of the intestinal microbiota in colon cancer. Molecular Cell. 2014;54(2):309–320. doi: 10.1016/j.molcel.2014.03.039. [DOI] [PubMed] [Google Scholar]
- 9.Urbaniak C., Gloor G. B., Brackstone M., Scott L., Tangney M., Reida G. The microbiota of breast tissue and its association with breast cancer. Applied and Environmental Microbiology. 2016;82(16):5039–5048. doi: 10.1128/AEM.01235-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunt R. H., Xiao S. D., Megraud F., et al. Helicobacter pylori in developing countries. World gastroenterology organisation global guideline. Journal of Digestive Diseases. 2011;12(5):319–326. doi: 10.1111/j.1751-2980.2011.00529.x. [DOI] [PubMed] [Google Scholar]
- 11.Xu W., Yang L., Lee P., et al. Mini-review: perspective of the microbiome in the pathogenesis of urothelial carcinoma. American Journal of Clinical & Experimental Urology. 2014;2(1):p. 57. [PMC free article] [PubMed] [Google Scholar]
- 12.Caporaso J. G., Kuczynski J., Stombaugh J., et al. QIIME allows analysis of high-throughput community sequencing data. Nature Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Popović V. B., Šitum M., Chow C.-E. T., Chan L. S., Roje B., Terzić J. The urinary microbiome associated with bladder cancer. Scientific Reports. 2018;8(1) doi: 10.1038/s41598-018-29054-w.12157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roperto S., Di Guardo G., Leonardi L., et al. Bacterial isolates from the urine of cattle affected by urothelial tumors of the urinary bladder. Research in Veterinary Science. 2012;93(3):1361–1366. doi: 10.1016/j.rvsc.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 15.Khosravi Y., Dieye Y., Poh B. H., et al. Culturable bacterial microbiota of the stomach of Helicobacter pylori positive and negative gastric disease patients. The Scientific World Journal. 2014;2014:10. doi: 10.1155/2014/610421.610421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gordya N., Yakovlev A., Kruglikova A., et al. Natural antimicrobial peptide complexes in the fighting of antibiotic resistant biofilms: Calliphora vicina medicinal maggots. PLoS ONE. 2017;12(3) doi: 10.1371/journal.pone.0173559.e0173559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leid J. G. Bacterial biofilms resist key host defenses. Microbe. 2009;4(2):66–70. [Google Scholar]
- 18.Høiby N., Bjarnsholt T., Givskov M., Molin S., Ciofu O. Antibiotic resistance of bacterial biofilms. International Journal of Antimicrobial Agents. 2010;35(4):322–332. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 19.Dufour D., Leung V., Lévesque C. M. Bacterial biofilm: structure, function, and antimicrobial resistance. Endodontic Topics. 2012;22(1):2–16. doi: 10.1111/j.1601-1546.2012.00277.x. [DOI] [Google Scholar]
- 20.Römling U., Balsalobre C. Biofilm infections, their resilience to therapy and innovative treatment strategies. Journal of Internal Medicine. 2012;272(6):541–561. doi: 10.1111/joim.12004. [DOI] [PubMed] [Google Scholar]
- 21.Gupta P., Sarkar S., Das B., Bhattacharjee S., Tribedi P. Biofilm, pathogenesis and prevention—a journey to break the wall: a review. Archives of Microbiology. 2016;198(1):1–15. doi: 10.1007/s00203-015-1148-6. [DOI] [PubMed] [Google Scholar]
- 22.Pinto-Santini D., Salama N. R. The biology of Helicobacter pylori infection, a major risk factor for gastric adenocarcinoma. Cancer Epidemiology, Biomarkers & Prevention. 2005;14(8):1853–1858. doi: 10.1158/1055-9965.EPI-04-0784. [DOI] [PubMed] [Google Scholar]
- 23.Collins D., Hogan A. M., Winter D. C. Microbial and viral pathogens in colorectal cancer. The Lancet Oncology. 2011;12(5):504–512. doi: 10.1016/S1470-2045(10)70186-8. [DOI] [PubMed] [Google Scholar]
- 24.Johnson C. H., Dejea C. M., Edler D., et al. Metabolism links bacterial biofilms and colon carcinogenesis. Cell Metabolism. 2015;21(6):891–897. doi: 10.1016/j.cmet.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cohen R. J., Shannon B. A., McNeal J. E., Shannon T., Garrett K. L. Propionibacterium acnes associated with inflammation in radical prostatectomy specimens: a possible link to cancer evolution? The Journal of Urology. 2005;173(6):1969–1974. doi: 10.1097/01.ju.0000158161.15277.78. [DOI] [PubMed] [Google Scholar]
- 26.Zhan P., Suo L.-J., Qian Q., et al. Chlamydia pneumoniae infection and lung cancer risk: a meta-analysis. European Journal of Cancer. 2011;47(5):742–747. doi: 10.1016/j.ejca.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 27.McConnell M. J., Actis L., Pachón J. Acinetobacter baumannii: human infections, factors contributing to pathogenesis and animal models. FEMS Microbiology Reviews. 2013;37(2):130–155. doi: 10.1111/j.1574-6976.2012.00344.x. [DOI] [PubMed] [Google Scholar]
- 28.He Y., Wu W., Zheng H.-M., et al. Regional variation limits applications of healthy gut microbiome reference ranges and disease models. Nature Medicine. 2018;24(10):1532–1535. doi: 10.1038/s41591-018-0164-x. [DOI] [PubMed] [Google Scholar]
- 29.DeSantis T. Z., Hugenholtz P., Larsen N., et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Applied and Environmental Microbiology. 2006;72(7):5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Segata N., Izard J., Waldron L., et al. Metagenomic biomarker discovery and explanation. Genome Biology. 2011;12(6) doi: 10.1186/gb-2011-12-6-r60.R60 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: rarefaction curves. It can be seen that the Shannon index of the 24 samples has reached saturation, indicating that the sequencing depth is sufficient, and most samples have captured complete microbial profiles. Table S1: characteristics of 24 patients with bladder cancer.
Data Availability Statement
The datasets generated during the current study are available from the NCBI SRA database with accession numbers SRR8647671.