Abstract
Background & objectives:
Azithromycin has been in use as an alternate treatment option for enteric fever even when the guidelines on the susceptibility testing were not available. There is lack of data on susceptibility and mechanisms of resistance of azithromycin in Salmonella Typhi and S. Paratyphi A. The aim of the present study was to determine the azithromycin susceptibility and resistance mechanisms in typhoidal salmonellae isolates archived in a tertiary care centre in north India for a period of 25 years.
Methods:
Azithromycin susceptibility was determined in 602 isolates of S. Typhi (469) and S. Paratyphi A (133) available as archived collection isolated during 1993 to 2016, by disc diffusion and E-test method.PCR was done for ereA, ermA, ermB, ermC, mefA, mphA and msrA genes from plasmid and genomic DNA and sequencing was done to detect mutations in acrR, rplD and rplV genes.
Results:
Azithromycin susceptibility was seen in 437/469 [93.2%; 95% confidence interval (CI), 90.5 to 95.1%] isolates of S. Typhi. Amongst 133 isolates of S. Paratyphi A studied, minimum inhibitory concentration (MIC) of ≤16 mg/l was found in 102 (76.7%; 95% CI, 68.8 to 83.0). MIC value ranged between 1.5 and 32 mg/l with an increasing trend in MIC50 and MIC90 with time. Mutations were found in acrR in one and rplV in two isolates of S. Typhi. No acquired mechanism for macrolide resistance was found.
Interpretation & conclusions:
Azithromycin could be considered as a promising agent against typhoid fever on the basis of MIC distribution in India. However, due to emergence of resistance in some parts, there is a need for continuous surveillance of antimicrobial susceptibility and resistance mechanisms. There is also a need to determine the breakpoints for S. Paratyphi A.
Keywords: Antimicrobial resistance, azithromycin, enteric fever, resistance mechanism, Salmonella, typhoid
Enteric fever is a community-acquired systemic infection commonly caused by Salmonella enterica serovars Typhi and Paratyphi A. Rates of typhoid fever as high as 980 cases per 100, 000 population have been reported from urban slums in Delhi1. Enteric fever can lead to increase in complications and morbidity, if not treated with appropriate antibiotics. Antimicrobial resistance to antityphoidal antibiotics started to emerge in the years following their use in the treatment of typhoid fever2. Due to widespread resistance to traditional first-line drugs such as ampicillin, trimethoprim-sulfamethoxazole and chloramphenicol in 1990s, fluoroquinolone or an extended-spectrum cephalosporin were considered as the main treatment options for enteric fever3. However, soon followed the reports on fluoroquinolone-resistant enterica Typhi (S. Typhi) and S. enterica Paratyphi A (S. Paratyphi A), especially from developing countries4,5. Although cephalosporin susceptibility is retained at nearly 100 per cent, occasional ceftriaxone-resistant strains have been reported from all over the world6.
In the meanwhile, the chloramphenicol susceptible strains have re-emerged, though it is still not a choice of treatment by the clinicians due to earlier reported toxicity of bone marrow depression7. As an alternative to these drugs, azithromycin was in use for uncomplicated cases of typhoid fever since 1990s8. In 2015, Clinical and Laboratory Standards Institute (CLSI) published the recommendation on clinical breakpoints for azithromycin susceptibility in S. Typhi.9 However, there are still no guidelines for the same in S. Paratyphi A9.
Azithromycin is an azalide antimicrobial agent that has been demonstrated to be equivalent to chloramphenicol, fluoroquinolones and extended-spectrum cephalosporins for the treatment of uncomplicated typhoid fever when given for 5-7 days10,11. With the emergence of ciprofloxacin resistance, azithromycin came into use for enteric fever without any supportive evidence in pharmacokinetic and pharmacodynamic studies or laboratory breakpoints in any guidelines10,11. All members of Enterobacteriaceae are intrinsically resistant to macrolides because of their hydrophobic nature, and hence, there is low permeability through the outer membrane, but azithromycin has an enhanced activity for Enterobacteriaceae due to its basic character which favours its high uptake by bacterial cells. In addition, it achieves a higher intracellular concentration of 50-100 times more than the serum level expecting it to act on intracellular pathogens like S. Typhi12. There are some sporadic reports of treatment failure with azithromycin in enteric fever13,14. The main reason could be the lack of guidance for clinical decision making from in vitro susceptibility results.
Although the breakpoints for S. Typhi are defined now, the absence of guidelines for S. Paratyphi A can lead to the suboptimal use of this drug in paratyphoid fever leading to treatment failure and emergence of resistance9,13, azithromycin-resistant strains of S. Paratyphi A have been reported in India leading to the treatment failure14.
The molecular mechanisms conferring resistance to azithromycin in typhoidal salmonellae are yet not fully understood and reported although these mechanisms are well described in non-typhoidal salmonellae15. In this context, there is a need to generate more data on susceptibility and mechanisms of resistance of azithromycin in typhoidal salmonellae. Therefore the present study was carried out to determine the azithromycin susceptibility pattern and resistance mechanisms in a collection of archived isolates of S. Typhi and S. Paratyphi A isolated at the All India Institute of Medical Sciences (AIIMS) hospital, New Delhi, over a period of 25 years from 1993 to 2016 from the patients presented with enteric fever.
Material & Methods
A total of 602 isolates were included in the study, of which 469 (77.9%) were S. Typhi and 133 (22.1%) were S. Paratyphi A. These were the blood culture isolates obtained from enteric fever patients who attended AIIMS hospital during 1993-2016. The blood culture and isolation methods were carried out as per standard protocol. In brief, 5 ml of fresh intravenous blood was inoculated in brain heart infusion broth (Difco, USA) and incubated at 37°C for 18-24 h16. Subcultures were made on MacConkey's agar (HiMedia Laboratories Pvt. Ltd., Mumbai) and 5 per cent sheep blood agar (BioMérieux, France) after 24 h, 48 h and 7 days. All non-lactose-fermenting Gram-negative colonies were identified by standard biochemical tests16. The identification was further confirmed by slide agglutination test using specific antisera (Statens Serum Institute, Copenhagen)16.
Antimicrobial susceptibility testing
Antimicrobial susceptibility was determined by Kirby-Bauer disk diffusion method using azithromycin disks (15 μg) from HiMedia Laboratories Pvt. Ltd., Mumbai17. Minimum inhibitory concentration (MIC) was determined by E-test (Etest®; BioMérieux, France) as per manufacturer's instructions. Interpretation was done as per CLSI 2015 guidelines9. MICs of resistant isolates were further confirmed by broth microdilution18. There is no recommendation for quality control of azithromycin susceptibility in non-fastidious strains of Enterobacteriaceae; so standard recommended strain of Staphylococcus aureus (ATCC 29213) was used as a quality control strain for azithromycin disk diffusion, E-test and broth microdilution. Zone sizes were extrapolated against the MIC values, and a scatterplot diagram was constructed, separately for S. Typhi and S. Paratyphi A.
Determination of macrolide resistance mechanisms: Molecular mechanisms for macrolide resistance were determined in all isolates having MIC ≥16 μg/ml. Some isolates having MIC lower than 4 μg/ml were also included as negative control. Genomic DNA was prepared by QIAmp commercial DNA isolation kit (QIAamp DNA minikit; Qiagen, Hilden, Germany). Plasmids’ extraction was done with QIAGEN Plasmid isolation kit (QIAprep Spin Miniprep, Hilden, Germany). The mutations responsible for intrinsic resistance and the genes conferring acquired resistance mechanisms were explored. PCR and DNA sequencing was used to detect the following macrolide resistance genes using previously described primers15,19,20.
Intrinsic mechanisms for resistance: For intrinsic mutations conferring macrolide resistance, acrR, rlpD and rlpV genes were amplified and sequenced from genomic DNA. Standard S. Typhi strain Ty-2 (ATCC 19430) was used as quality control strain for PCR and sequencing of these genes15,19.
Acquired mechanisms for resistance: For acquired mechanisms of resistance, PCR for ereA, ermA, ermB, ermC, mefA, mphA and msrA genes was done to determine their presence in genomic and plasmid DNA using previously published primers and PCR conditions20. S. aureus ATCC 43300 was used as positive control for ermA. There was no standard ATCC control strain available for ereA, ermB, ermC, mefA, mphA and msrA genes in published literature. Hence, positive controls were identified in house by screening a large collection of clinical isolates for the presence of azithromycin resistance genes. Briefly, a total of 83 azithromycin-resistant strains of S. aureus, Streptococcus pneumoniae, Acinetobacter baumannii, Klebsiella pneumoniae and Enterococcus faecalis available from the clinical bacteriology laboratory of AIIMS were screened by PCR for the presence of resistance determining genes. PCR was performed using published primers and PCR conditions. Of the 88 strains seven were found to be PCR positive for the genes included in the study. Following this, DNA sequencing was performed as described below using the amplified PCR products. The DNA sequences were aligned with the sequences of corresponding genes available at NCBI database (https://www.ncbi.nlm.nih.gov/gene/). The selected specific DNA sequences from designated control strains were submitted to GenBank (NCBI).
PCR and DNA sequencing: PCR reactions were performed in a final reaction volume of 50 μl. The reaction mixture consisted of 39.3 μl of PCR quality water, 5 μl PCR buffer, 1 μl dNTPs (Deoxynucleotide triphosphates), 0.5 μl each of forward and reverse primers and 0.7 μl Taqman DNA polymerse (Thermo Fisher Scientific Inc., USA) in MYCycler thermal cycler (Bio-Rad Laboratories, Inc., USA). Initial denaturation was done at 94°C for eight minutes followed by 35 cycles of denaturation (94°C), annealing (as per annealing temperature of each gene) and extension (72°C) for 60 sec each and a final extension for five minutes at 72°C. To examine the PCR product, 5 μl of the PCR product was mixed with 6X gel loading buffer and loaded along with 100 bp ladder in 1.5 per cent (w/v) agarose gel prepared in 0.5 X TBE (tris-Borate-EDTA) buffer. The gel was examined under ultraviolet light in GEL-DOC system (Bio-Rad Laboratories, Inc., USA).
DNA Sequencing was carried out by Sanger sequencing method, with AmpliTaq Gold DNA polymerase enzyme (Applied Biosystems, USA) using an automated DNA sequencer ABI PRISM® 310 Genetic Analyzer (Applied Biosystems, USA). Forward and reverse DNA sequences for each gene were aligned together and analyzed in Genedoc software v2.6.00221.
Results
Of the 602 isolates tested 469 were S. Typhi and 133 were S. Paratyphi A. All these isolates collected during 1993-2007 were grouped together for analysis due to small numbers being available from each year making it difficult to calculate MIC50 and MIC90 during that time, while from 2008 onwards, analysis was done as per current year.
Antimicrobial susceptibility in S. Typhi: Of the 469 isolates, 437 were azithromycin susceptible [93.2%; 95% confidence interval (CI), 90.5 to 95.1%]. MIC distribution was continuous and the MIC values ranged between 1.5 and 24 μg/ml with MIC50 and MIC9012 μg/ml. MIC distribution peaked at 12 μg/ml for S. Typhi as shown by frequency histogram in Fig. 1. There was a gradual increase in median MIC from 8 to 12 μg/ml, from 2007 to 2016 Table I. Scatterplot analysis of the zones of inhibition vs. MIC to azithromycin showed a linear relation, and the MIC susceptibility breakpoint was found to be corresponding with disk diffusion zone diameter breakpoint (Fig. 2A).
Fig. 1.
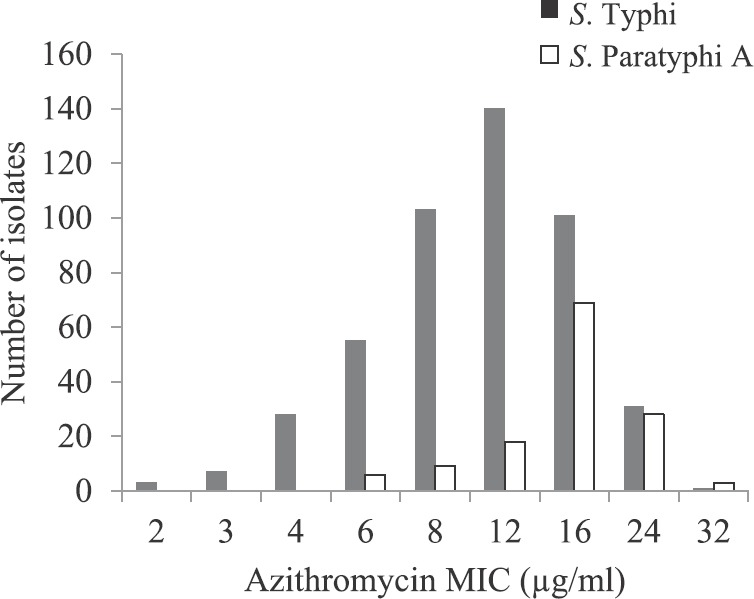
Frequency histogram for azithromycin minimum inhibitory concentration (MIC) in Salmonella Typhi and S. Paratyphi A; distribution curve is skewed towards right for S. Paratyphi A.
Table I.
Temporal comparison of minimum inhibitory concentration 50 (MIC50) and minimum inhibitory concentration 90 (MIC90) for azithromycin in enteric fever isolates
| Year | Salmonella Typhi | Salmonella Paratyphi A | ||||||
|---|---|---|---|---|---|---|---|---|
| Total number of isolates | S % (95% CI) | MIC50 (μg/ml) | MIC90 (μg/ml) | Total number of isolates | S % (95% CI) | MIC50 (μg/ml) | MIC90 (μg/ml) | |
| 1993-2007 | 71 | 98.6 (92.4-99.8) | 8 | 12 | 11 | 100 (74.1-100) | 8 | 12 |
| 2008 | 81 | 92.6 (84.8-96.6) | 6 | 12 | 24 | 75 (55.1-88) | 12 | 16 |
| 2009 | 76 | 84.2 (74.4-90.7) | 8 | 12 | 21 | 76.2 (54.9-89.4) | 8 | 16 |
| 2010 | 98 | 92.9 (85.9-96.5) | 6 | 12 | 23 | 73.9 (53.5-87.4) | 12 | 16 |
| 2011 | 11 | 90.9 (62.3-98.4) | 8 | 12 | 4 | 100 (51.0-100) | 12 | 16 |
| 2012 | 13 | 92.3 (66.7-98.6) | 8 | 12 | 4 | 100 (51.0-100) | 12 | 16 |
| 2013 | 17 | 88.2 (65.7-96.7) | 6 | 12 | 6 | 83.3 (43.6-96.9) | 12 | 16 |
| 2014 | 14 | 92.9 (68.5-98.7) | 8 | 12 | 4 | 100 (51.0-100) | 16 | 16 |
| 2015 | 15 | 100 (79.6-100) | 12 | 16 | 11 | 45.4 (21.3-71.9) | 16 | 24 |
| 2016 | 73 | 98.6 (92.6-99.8) | 12 | 12 | 25 | 72 (52.4-85.7) | 16 | 24 |
| Total | 469 | 93.2 (90.5-95.1) | 12 | 12 | 133 | 76.7 (68.8-83.0) | 16 | 16 |
Antimicrobial susceptibility results are given for disk diffusion method (same results were obtained by all the methods used for susceptibility testing). Isolates from 1993 to 2007 are grouped together for analysis. MIC50, the concentration of drug inhibiting 50% of the isolates; MIC90, the concentration of drug inhibiting 90% of the isolates; CI, confidence interval; S, susceptible
Fig. 2.
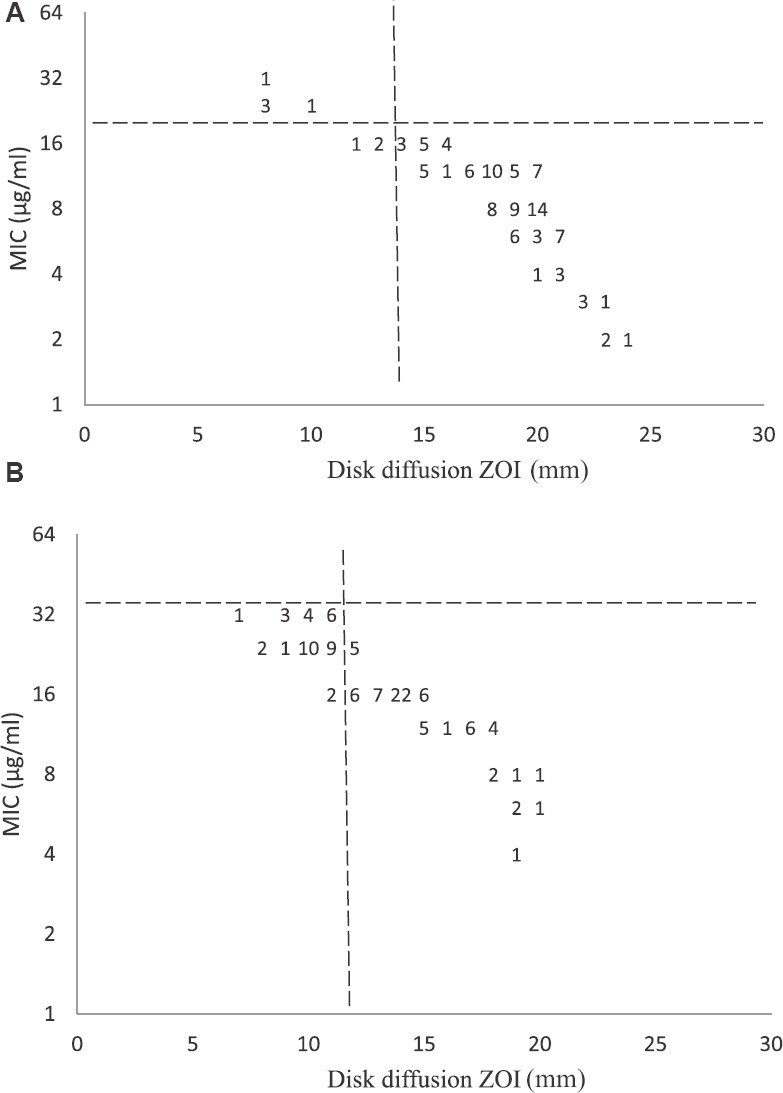
(A). Scatterplot of azithromycin minimum inhibitory concentration (MIC) vs. zone diameter (zone of inhibition, ZOI) for 469 isolates of Salmonella Typhi showing concordance between Clinical and Laboratory Standards Institute (CLSI) breakpoint (ZOI ≥14 mm and MIC ≤16 μl/ml) and determined epidemiological cut-off MIC ≤16 μl/ml), (B). Scatterplot of azithromycin MIC vs. zone diameter for S. paratyphi A, showing lower zone diameter and higher MICdistribution than S. Typhi. Both plots show the relationship between the MIC to azithromycin (y-axis) and the inhibition zone diameters (x-axis). The dotted line denotes the cut-off for susceptible/resistant as per CLSI, 201517.
Antimicrobial susceptibility in S. paratyphi A: In case of S. Paratyphi A, of the 133 isolates tested, MIC ranged between 2 and 32 μg/ml. MIC value of ≤16 μg/ml was found in 102 isolates (76.7%; 95% CI, 68.8 to 83.0). In frequency histogram, the peak for S. Paratyphi A was skewed towards right, showing the higher MIC distribution than S. Typhi (Fig. 1). A gradual increase was seen in MIC50 and MIC90 with time. MIC50 had increased from 8 to 16 μg/ml and MIC90 from 12 to 24 μg/ml, from 2007 to 2016 as shown in Table I. Scatterplot analysis of MIC vs. zone of inhibition showed a linear relationship but distributed towards higher MIC values as shown in Fig. 2B, when compared with S. Typhi.
Molecular mechanisms
A total of 246 isolates were selected for molecular studies for azithromycin resistance, which included those having MIC ≥16 μg/ml. Isolates having MIC lower than 4 μg/ml were also included as negative controls for azithromycin resistance determining mutations. The positive control strains were selected for all the genes by DNA sequencing and the control sequences were submitted to GenBank (NCBI); the accession numbers are given in Table II.
Table II.
Polymerase chain reaction primers and positive control strains along with GenBank accession numbers
| Gene | Band size (bp) | Positive control | GenBank accession number | Reference for primers |
|---|---|---|---|---|
| acrR | 816 | Salmonella enterica Typhi Ty21a | MF150847 | Sharma et al, 201319 |
| rplD | 637 | Salmonella enterica Typhi Ty21a | MF150849 | Gunell et al, 201015 |
| rplV | 394 | Salmonella enterica Typhi Ty21a | MF150850 | Gunell et al, 201015 |
| ereA | 420 | Acinetobacter baumannii-characterized strain | MF095628 | Phuc Nguyen et al, 200920 |
| ermA | 533 | Staphylococcus aureus ATCC 43300 | MF095625 | Phuc Nguyen et al, 200920 |
| ermB | 639 | Escherichia coli-characterized strain | MF095626 | Phuc Nguyen et al, 200920 |
| ermC | 642 | Staphylococcus aureus-characterized strain | MF095627 | Phuc Nguyen et al, 200920 |
| mefA | 345 | Streptococcus pneumoniae-characterized strain | MF095629 | Phuc Nguyen et al, 200920 |
| mphA | 403 | Escherichia coli-characterized strain | MF095624 | Phuc Nguyen et al, 200920 |
| msrA | 384 | Staphylococcus aureus-characterized strain | MF095630 | Phuc Nguyen et al, 200920 |
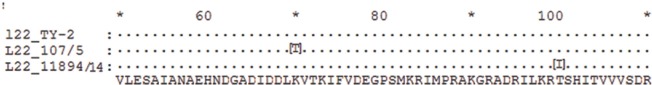
DNA sequencing was done for acrR, rplD and rplV genes to find the mutations responsible for macrolide resistance. Sequence analysis revealed a 6 bp insertion at position 234 acrR in gene, which resulted in insertion of two amino acids: glutamic acid and isoleucine at position 78 in regulator protein of AcrAB efflux pump of S. Typhi (Fig. 3). No mutation was found in rplD gene of any of the isolates. RplV gene had mutations in two isolates of S. Typhi, one had A208 to C transversion resulting in amino acid change from lysine to threonine at position 70 of L22 protein. Fig. 3 depicts the amino acid sequence alignment of mutant rplV sequences with reference strain of S. Typhi ATCC 19430-Ty2. Corresponding amino acid substitutions as a result of mutations are shown in Table III. Another isolate had a mutation C299T leading to change in protein sequence at position 100 from amino acid threonine to isoleucine (Fig. 4). Both these isolates had an MIC of 24 μg/ml. All the mutant sequences were submitted to GenBank; accession numbers are listed in Table III.
Fig. 3.
Protein sequence alignment for acrR of 13876/9 with control sequence from TY2 showing insertion of two amino acids I and E at 78 position relative to first codon of the acrR gene.
Table III.
Molecular mechanism of macrolide resistance found in the isolates
| Isolates ID | Year of isolation | Azithromycin MIC (µg/ml) | Molecular mechanism of resistance found | Amino acid change | GenBank accession number |
|---|---|---|---|---|---|
| 13786/09 | 2009 | 16 | AcrR (insertion 234 ATTGAG) | Insertion at 78- EI | MF150848 |
| 107/5 | 2010 | 24 | RplV (A208C) | Lys70 Thr | MF150851 |
| 11894/14 | 2014 | 24 | RplV (C299T) | Thr100 Ile | MF150852 |
MIC, minimum inhibitory concentration
Fig. 4.
Protein sequence alignment of rplV from mutant strains 107/5 and 11894/14 with control sequence from TY2, showing the amino acid changes at K70 to T and T100 to I.
None of the isolates had acquired macrolide resistance genes ereA, ermA, ermB, ermC, mefA, mphA and msrA in plasmid or genomic DNA.
Discussion
Several randomized clinical trials had established azithromycin as an effective alternative treatment option for uncomplicated typhoid fever, but the treatment was given without any laboratory-supported data in antimicrobial susceptibility due to the absence of any guidelines8. The continued use of azithromycin in patients infected with strains having reduced susceptibility could have been responsible for treatment failures reported13,14 leading to the emergence of azithromycin-resistant strains.
In our study, 93.2 per cent of S. Typhi and 76.7 per cent of S. Paratyphi A were susceptible to azithormycin. Similar results were reported by various studies from India and other parts of the world22,23,24. In contrast to these studies, azithromycin resistance was reported in 34.7 per cent isolates in a study from 2005 to 200824, and clinical non-response was observed in 19 of 36 patients treated with azithromycin when BSAC (British Society for Antimicrobial Susceptibility Testing) guidelines of 201423 were used before the CLSI recommendations25,26,27.
In our study, majority of isolates had azithromycin MIC range from 4 to 12 μg/ml. Comparing susceptibility of S. Typhi and S. Paratyphi A, it was found that MIC values were higher for S. Paratyphi A than S. Typhi. The MIC distribution for S. Paratyphi was skewed to right as has also been observed in other reports from India24 and other parts of the world27. However, the clinical significance of this finding may be difficult to analyze as there are no susceptibility guidelines for S. Paratyphi A in the CLSI or any other international guidelines. Based on our data, a breakpoint of 32 μg/ml was proposed for S. Paratyphi A as has also been suggested by Parry et al23.
In addition to the higher MIC values, MIC50 and MIC90 showed an increase towards the resistance with the time, in case of both S. Typhi and S. Paratyphi A. This could possibly be due to the increased use of this antibiotic imposing increasing selective pressure on the bacteria as observed in our results with 50 per cent of the isolates showing increase in MIC from 8 to 12 μg/ml. This highlights the need for continuous monitoring of antimicrobial susceptibility.
Studies investigating azithromycin resistance mechanisms in Salmonella are scarce. Different mechanisms of macrolide resistance have been identified in Enterobacteriaceae, some of these are located within the bacterial chromosome, namely, target alterations or chromosomal efflux pumps, and others are encoded within transferable elements and able to be transferred among microorganisms28. Mechanisms of azithromycin resistance include the mutations in target genes or efflux pumps and the presence of specific resistance genes (e.g., mphA, mefA, ereA and ermB)28. Although there are multiple mechanisms for macrolide resistance described in Enterobacteriaceae and some in non-typhoidal salmonellae also, there are no studies on azithromycin resistance mechanisms in typhoidal salmonellae15,28.
In this study, mutations associated with azithromycin resistance were found in acrR and rplV genes. Of the total 246 isolates, overexpression of AcrAB efflux pump was observed in a single isolate from 2009, having MIC 16 μg/ml. A 6 base pair insertion in the DNA sequence led to insertion of glutamic acid and isoleucine at position 78, similar to the previous findings by Olliver et al29, in S. Typhimurium. This mutation is responsible for extrusion of macrolides from the bacteria. The role of acrR in resistance was shown in S. Typhi by Nikaido et al30, who observed that inactivation of AcrAB pump resulted in 64-fold decrease in the MIC of erythromycin (from 512 to 8 μg/ml).
Substitutions were also found in rplV gene in two isolates, one isolate of S. Typhi having MIC 24 μg/ml had a mutation at A208C resulting in amino acid change from lysine to glutamine at position 70. The role of this mutation is yet not clear, and there is a need to study the significance at the protein level for expression. In another isolate with azithromycin MIC 24 μg/ml, a C299T mutation was found leading to change in protein sequence at position 100. There is no other study describing the mutation at this position, but the mutation at position 99 (adjacent to Thr100) of L22 protein is known to be associated with macrolide resistance31.
In our study, all isolates were negative for ereA, ereB, ermA, ermB, ermC, mefA, mphA and msrA genes associated with macrolide resistance. None of the S. Paratyphi A isolates included in the study showed any known mechanism of macrolide resistance. Since we looked for only the mechanisms commonly reported in Enterobacteriaceae, we could have missed some resistance determining mechanisms. Furthermore, there were limited number of isolates available for the study from a single hospital. There is a need for monitoring antimicrobial susceptibly and the presence of resistance mechanisms in larger number of S. Typhi and S. Paratyphi A isolates.
In conclusion, the findings of the present study emphasize the need for continuous monitoring of antimicrobial susceptibility of typhoidal salmonellae and investigating the mechanisms underlying azithromycin resistance. More clinical data on the treatment and outcomes may be required before finally recommending azithromycin as empirical antibiotic of choice to treat enteric fever.
Footnotes
Financial support & sponsorship: The study was financially supported by Indian Council of Medical Research. (Code: ECD: 2012-1705/12).
Conflicts of Interest: None.
References
- 1.Ochiai RL, Acosta CJ, Danovaro-Holliday MC, Baiqing D, Bhattacharya SK, Agtini MD, et al. A study of typhoid fever in five Asian countries: Disease burden and implications for controls. Bull World Health Organ. 2008;86:260–8. doi: 10.2471/BLT.06.039818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kapil A, Ayyagari A, Garg RK, Agarwal KC. S. typhi with transferable chloramphenicol resistance isolated in Chandigarh during 1983-87. Indian J Pathol Microbiol. 1994;37:179–83. [PubMed] [Google Scholar]
- 3.Kariuki S, Gordon MA, Feasey N, Parry CM. Antimicrobial resistance and management of invasive Salmonella disease. Vaccine. 2015;33(Suppl 3):C21–9. doi: 10.1016/j.vaccine.2015.03.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dahiya S, Sharma P, Kumari B, Pandey S, Malik R, Manral N, et al. Characterisation of antimicrobial resistance in Salmonellae during 2014-2015 from four centres across India: An ICMR antimicrobial resistance surveillance network report. Indian J Med Microbiol. 2017;35:61–8. doi: 10.4103/ijmm.IJMM_16_382. [DOI] [PubMed] [Google Scholar]
- 5.Harish BN, Menezes GA. Antimicrobial resistance in typhoidal Salmonellae. Indian J Med Microbiol. 2011;29:223–9. doi: 10.4103/0255-0857.83904. [DOI] [PubMed] [Google Scholar]
- 6.Rodrigues C, Kapil A, Sharma A, Devanga Ragupathi NK, Inbanathan FY, Veeraraghavan B, et al. Whole-genome shotgun sequencing of cephalosporin-resistant Salmonella enterica serovar Typhi. Genome Announc. 2017;5 doi: 10.1128/genomeA.01639-16. pii: e01639-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma P, Dahiya S, Balaji V, Kanga A, Panda P, Das R et al. Typhoidal Salmonellae: Use of Multi-locus Sequence Typing to Determine Population Structure. PLoS One. 2016;12:1–13. doi: 10.1371/journal.pone.0162530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tribble D, Girgis N, Habib N, Butler T. Efficacy of azithromycin for typhoid fever. Clin Infect Dis. 1995;21:1045–6. doi: 10.1093/clinids/21.4.1045. [DOI] [PubMed] [Google Scholar]
- 9.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing: 25th international supplement. CLSI document M100-S25. Wayne, PA: CLSI; 2015. [Google Scholar]
- 10.Frenck RW, Jr, Nakhla I, Sultan Y, Bassily SB, Girgis YF, David J. Azithromycin versus ceftriaxone for the treatment of uncomplicated typhoid fever in children. Clin Infect Dis. 2000;31:1134–8. doi: 10.1086/317450. [DOI] [PubMed] [Google Scholar]
- 11.Parry CM, Ho VA, Phuong le T, Bay PV, Lanh MN, Tung le T, et al. Randomized controlled comparison of ofloxacin, azithromycin, and an ofloxacin-azithromycin combination for treatment of multidrug-resistant and nalidixic acid-resistant typhoid fever. Antimicrob Agents Chemother. 2007;51:819–25. doi: 10.1128/AAC.00447-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foulds G, Shepard RM, Johnson RB. The pharmacokinetics of azithromycin in human serum and tissues. J Antimicrob Chemother. 1990;25(Suppl A):73–82. doi: 10.1093/jac/25.suppl_a.73. [DOI] [PubMed] [Google Scholar]
- 13.Molloy A, Nair S, Cooke FJ, Wain J, Farrington M, Lehner PJ, et al. First report of Salmonella enterica serotype Paratyphi A azithromycin resistance leading to treatment failure. J Clin Microbiol. 2010;48:4655–7. doi: 10.1128/JCM.00648-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manesh A, Balaji V, Kumar DR, Rupali P. A case of clinical and microbiological failure of azithromycin therapy in Salmonella enterica serotype Typhi despite low azithromycin MIC. Int J Infect Dis. 2017;54:62–3. doi: 10.1016/j.ijid.2016.11.409. [DOI] [PubMed] [Google Scholar]
- 15.Gunell M, Kotilainen P, Jalava J, Huovinen P, Siitonen A, Hakanen AJ, et al. In vitro activity of azithromycin against nontyphoidal Salmonella enterica. Antimicrob Agents Chemother. 2010;54:3498–501. doi: 10.1128/AAC.01678-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collee JG, Miles RS, Watt B. Tests for the identification of bacteria. In: Collee JG, Fraser AG, Marmion BP, Simmons A, editors. Mackie & McCartney practical medical microbiology. 14th ed. London: Churchill Livingstone; 1996. pp. 131–49. [Google Scholar]
- 17.CLSI document M2-A9. Wayne PA: CLSE; 2006. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing, 16th informational supplements. [Google Scholar]
- 18.10th ed. Wayne, PA: Clinical and Laboratory Standards Institute; 2015. Clinical and Laboratory Standards Institute. Methods for dilution antimicrobial susceptibility tests for bacteria that grow Aerobiacally. Approved standard. CLSI Document M07-A10. [Google Scholar]
- 19.Sharma V, Dahiya S, Jangra P, Das BK, Kumar R, Sood S, et al. Study of the role of efflux pump in ciprofloxacin resistance in Salmonella enterica serotype Typhi. Indian J Med Microbiol. 2013;31:374–8. doi: 10.4103/0255-0857.118898. [DOI] [PubMed] [Google Scholar]
- 20.Phuc Nguyen MC, Woerther PL, Bouvet M, Andremont A, Leclercq R, Canu A, et al. Escherichia coli as reservoir for macrolide resistance genes. Emerg Infect Dis. 2009;15:1648–50. doi: 10.3201/eid1510.090696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicholas KB, Nicholas HB, Jr, DeerfieldDW II. Genedoc: analysis and visualization of genetic variation. Embnew News. 1997;4:14. [Google Scholar]
- 22.Misra R, Prasad KN. Antimicrobial susceptibility to azithromycin among Salmonella enterica Typhi and ParatyphiA isolates from India. J Med Microbiol. 2016;65:1536–9. doi: 10.1099/jmm.0.000390. [DOI] [PubMed] [Google Scholar]
- 23.Parry CM, Thieu NT, Dolecek C, Karkey A, Gupta R, Turner P, et al. Clinically and microbiologically derived azithromycin susceptibility breakpoints for Salmonella enterica serovars Typhi and Paratyphi A. Antimicrob Agents Chemother. 2015;59:2756–64. doi: 10.1128/AAC.04729-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chatterjee S, Eshwara VK, Tellapragada C, Mukhopadhyay C. Azithromycin susceptibility among clinical isolates of Salmonella: Interfacing guidelines with routine practices. Indian J Med Microbiol. 2016;34:397–8. doi: 10.4103/0255-0857.188376. [DOI] [PubMed] [Google Scholar]
- 25.Rai S. Azithromycin zone interpretation for Salmonella: Time to adopt BSAC's zone diameters? Indian J Med Microbiol. 2014;32:465–6. doi: 10.4103/0255-0857.142240. [DOI] [PubMed] [Google Scholar]
- 26.Dutta S, Das S, Mitra U, Jain P, Roy I, Ganguly SS, et al. Antimicrobial resistance, virulence profiles and molecular subtypes of Salmonella enterica serovars Typhi and Paratyphi A blood isolates from Kolkata, India during 2009-2013. PLoS One. 2014;9:e101347. doi: 10.1371/journal.pone.0101347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hassing RJ, Goessens WH, van Pelt W, Mevius DJ, Stricker BH, Molhoek N, et al. Salmonella subtypes with increased MICs for azithromycin in travelers returned to the Netherlands. Emerg Infect Dis. 2014;20:705–8. doi: 10.3201/eid2004.131536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gomes C, Martínez-Puchol S, Palma N, Horna G, Ruiz-Roldán L, Pons MJ, et al. Macrolide resistance mechanisms in Enterobacteriaceae: Focus on azithromycin. Crit Rev Microbiol. 2017;43:1–30. doi: 10.3109/1040841X.2015.1136261. [DOI] [PubMed] [Google Scholar]
- 29.Olliver A, Vallé M, Chaslus-Dancla E, Cloeckaert A. Role of an acrR mutation in multidrug resistance of in vitro-selected fluoroquinolone-resistant mutants of Salmonella enterica serovar Typhimurium. FEMS Microbiol Lett. 2004;238:267–72. doi: 10.1016/j.femsle.2004.07.046. [DOI] [PubMed] [Google Scholar]
- 30.Nikaido H, Basina M, Nguyen V, Rosenberg EY. Multidrug efflux pump AcrAB of Salmonella typhimurium excretes only those beta-lactam antibiotics containing lipophilic side chains. J Bacteriol. 1998;180:4686–92. doi: 10.1128/jb.180.17.4686-4692.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zaman S, Fitzpatrick M, Lindahl L, Zengel J. Novel mutations in ribosomal proteins L4 and L22 that confer erythromycin resistance in Escherichia coli. Mol Microbiol. 2007;66:1039–50. doi: 10.1111/j.1365-2958.2007.05975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]