Abstract
Purpose
Extremely preterm infants are at increased risk for retinopathy of prematurity (ROP). We previously identified several inflammatory proteins that were expressed early in life and are associated with an increased risk of ROP and several angiogenic and neurotrophic growth factors in the neonatal systemic circulation that are associated with a lower risk of ROP. In this paper, we report the results of a set of analyses designed to test the hypothesis that placental CpG methylation levels of 12 inflammation-, angiogenic-, and neurotrophic-associated genes predict the occurrence of prethreshold ROP in extremely preterm newborns.
Methods
We used placental CpG methylation data from 395 newborns from the Extremely Low Gestational Age Newborns study.
Results
Multivariable regression models revealed that placental DNA methylation of 16 CpG sites representing 8 genes were associated with prethreshold ROP. Specifically, CpG methylation in the serum amyloid A SAA1 and SAA2, brain-derived neurotrophic factor (BDNF), myeloperoxidase (MPO), C-reactive protein (CRP), angiopoietin 1 (ANGPT1), and tumor necrosis factor receptor superfamily member 1B (TNFRSF1B) genes was associated with a lower risk of prethreshold ROP. Conversely, CpG methylation at three probes within tumor necrosis factor receptor superfamily member 1A (TNFRSF1A) and in two alternative probes within the BDNF and ANGPT1 genes was associated with an increased risk of ROP.
Conclusions
CpG methylation may be a useful marker for improving ROP prediction, opening the opportunity for early intervention to lessen disease severity.
Keywords: retinopathy of prematury, methylation, inflammation, angiogenesis, neurotrophic
Retinopathy of prematurity (ROP) predominantly occurs among extremely preterm newborns and is associated with a prominently increased risk for visual disability and blindness later in life.1,2 Treatment is possible but invasive and not without risk.3 Thus, identifying biologic mechanisms that underlie ROP is paramount to the future development of interventions for preventing and/or minimizing the sequalae of ROP.
Until recently, the main pathogenetic paradigm was that ROP is a consequence of the preterm newborn's postnatal exposure to high hemoglobin-oxygen saturations. Specifically, it has been posited that the baby's transition from the low-O2 environment in utero into relatively high ambient oxygen concentrations shortly after birth leads to suppression of retinal vasculogenesis (phase 1). Later on, relatively low retinal oxygen levels lead to vasculogenetic overshoot that can result in retinal hemorrhage and retinal detachment (phase 2).4 Additionally, we have found that prenatal markers of infection and inflammation in the placenta were associated with an increased risk for ROP.5 This has led to the notion of a prenatal prephase of ROP, during which exposure to inflammation while in utero sensitizes the retina to subsequent insults.6 We have also found that elevated inflammation-associated proteins (i.e., C-reactive protein [CRP], serum amyloid A [SAA], myloeperoxidase [MPO], interleukin 6 [IL-6], interleukin 8 [IL-8], tumor necrosis factor receptor 1 [TNF-R1], and tumor necrosis factor receptor 2 [TNF-R2]) measured in the newborn systemic circulation were associated with an increased risk of developing ROP. Conversely, elevated systemic angiogenic and neurotrophic growth factor levels (i.e., neutrophin-4 [NT-4], brain-derived neurotrophic factor [BDNF], and angiopoietin 1 [ANG-1]) were associated with reduced risk.7
Although the placenta is typically discarded after delivery, it is critical for the proper growth and development of the fetus. In addition to controlling the transfer of nutrients and waste between the fetus and mother, the placenta may also serve as a record of in utero processes.8 Placental epigenetic markers are of particular interest, as they have been linked to adverse health outcomes later in life. For example, our group has previously reported that CpG methylation levels of critical neurodevelopment genes in the placenta predict cognitive function in childhood.9,10 We, therefore, set out to test the hypothesis that variation in placental CpG methylation of inflammatory, angiogenic, and neurotrophic growth factor genes is associated with altered risk of ROP.
Methods
The ELGAN Study
The Extremely Low Gestational Age Newborns (ELGAN) study was designed to identify characteristics and exposures that increase the risk of structural and functional neurologic disorders in extremely premature newborns. During the years 2002 to 2004, women delivering before 28 weeks gestation at one of 14 participating institutions in 11 cities across 5 states were asked to enroll in the study. Institutional review boards at each participating institution approved the study procedures and all study procedures followed the tenets of the Declaration of Helsinki.
Mothers were initially approached either upon antenatal admission or shortly after delivery, depending on clinical circumstance and institutional preference. Of the 1509 mothers eligible for inclusion, 1249 (83%) who gave birth to 1506 infants provided informed consent. A total of 1411 placentas were donated to the study, although quantification of DNA methylation was confined to a subset of study participants for whom sufficient placenta tissue was available and who were assessed at 10 years of age (n = 426, 36% of the cohort who were alive at 10 years and 48% of those assessed at 10 years).
Maternal Sociodemographic and Pregnancy Characteristics
After delivery, a trained research nurse interviewed each mother in her native language by using structured data collection procedures. Mothers self-reported their sociodemographic characteristics, including age, race/ethnicity, marital status, and educational attainment, in addition to the sequence of clinical events that led to preterm delivery.
Newborn Characteristics
Gestational ages were estimated according to a hierarchy of the quality of available information. Ideally, we used estimates based on the dates of embryo retrieval or intrauterine insemination for those using assisted reproductive technologies; otherwise, we relied on dating from fetal ultrasounds conducted prior to the 14th week of pregnancy (62%). When these were not available, reliance was placed sequentially on the following: (1) a fetal ultrasound at 14 or more weeks (29%), (2) self-reported last menstrual period without fetal ultrasound (7%), or (3) gestational age as recorded in the log of the neonatal intensive care unit (1%). Anthropometric measures including body weight (in grams) were taken shortly after birth either in the delivery room or upon admission to the neonatal intensive care unit.
Placentas
Delivered placentas were placed in a sterile exam basin and transported to a sampling room. Eighty-two percent of the samples were obtained within 1 hour of delivery. Briefly, the area to be sampled was at the midpoint of the longest distance between the cord insertion and the edge of the placental disk. Once the area was identified, the overlying amnion was lifted with a set of sterile tweezers and cut with sterile scissors. The amnion was gently pulled away from the underlying chorion by using tweezers. The amnion was then snipped open with the scissors and peeled away from the initial site of entry, thus exposing the chorion. With a second set of sterile forceps and scissors, traction was put on the chorion and underlying trophoblast tissue by gently pulling on it. A piece of tissue was removed by cutting at the base of the section with the sterile scissors and placing it into a sterile 2-mL cryovial. This tube was immediately frozen in liquid nitrogen and then stored in a −80°C freezer until shipped. Frozen samples were shipped on a regular basis by using dry ice from the 14 study sites to a central laboratory located in Boston, Massachusetts for storage. In 2015, samples were shipped on dry ice to the University of North Carolina at Chapel Hill where 0.2-g subsections were cut from the frozen tissue, rinsed with 1× PBS to remove any residual red blood cells, and homogenized in Buffer RLT Plus (Qiagen, Valencia, CA, USA).
DNA Extraction and Illumina 850K Methylation Assay
Genomic DNA sequences were isolated with the AllPrep DNA/RNA/miRNA Universal Kit (Qiagen), in accordance with the manufacturer's instructions. DNA was quantified and normalized using the Quant-IT Picogreen assay and subsequently shipped on dry ice to Wayne State University for methylation assays. There, isolated DNA was first bisulfite-converted using the EZ DNA methylation kit (Zymo Research, Irvine, CA, USA). Converted DNA was then hybridized onto the Infinium MethylationEPIC BeadChip (Illumina, San Diego, CA, USA), which interrogates methylation levels at over 850,000 individual probes.
Each probe measured the average methylation level at a single CpG site. Methylation levels were calculated and expressed as β values (β = intensity of the methylated allele [M]/intensity of the unmethylated allele [U] + intensity of the methylated allele [M] + 100), as previously described elsewhere.11 For data filtration, probes with high-detection P values (P > 0.01, n = 810) were considered to be unreliable and removed from analysis, as recommended by the manufacturer. Background correction was performed via the normal-exponential out-of-band (noob) correction method.12 Array data were normalized using functional normalization methodology.13 Batch effects were evaluated using principal component analysis; the first principal component was significantly associated with plate, suggesting plate was a nonnegligible source of variation. We, therefore, removed batch effects using the ComBat procedure, as implemented with the sva package.14 For the purposes of this analysis, we selected 320 normalized β values that corresponded to CpG loci within 12 candidate genes (ANGPT1, BDNF, CRP, IL-6, IL-8, MPO, NTF4, SAA1, SAA2, TNFRSF1A, TNFRSF1B, and TRAF2) that we identified based on the prior literature.7,15
Ophthalmologic Examinations
We defined ROP and its stages according to standards developed by the International Committee for Classification of Retinopathy of Prematurity.16 In keeping with guidelines, the first ophthalmologic examination was within the 31st to 33rd postmenstrual week (i.e., the sum of the newborn's gestational age at birth and chronologic age at the time of the exam).17 Follow-up exams were as clinically indicated until normal vascularization began in zone III, and the most severe ROP stage was recorded. We focused our analyses on prethreshold ROP, defined as any ROP in zone I, ROP stage 2 or 3 with plus disease, or ROP stage 3 without plus disease in zone II.18
Statistical Analysis
Separate mixed-effects Poisson regression models were fit to estimate the relative risk of prethreshold ROP with 95% confidence intervals (CIs). A robust error variance procedure was used to relax variance assumptions.19 The models incorporated three levels with random intercepts to reflect the clustering of multiple births to the same mother nested within the 14 study sites. Our primary predictors of interest were the average methylation level at each of the 320 CpG loci from the candidate genes. Although methylation levels (β values) were initially calculated as a proportion between 0 and 1, we multiplied them by 100 so as to estimate the change in relative risk associated with a 1% increase in methylation. A 1% increase represents a reasonable change given the ranges of our data.
To reduce the possibility that methylation levels were affected by infiltration of inflammatory cells within the placenta, we controlled for acute inflammation status in all models. Samples were considered acutely inflamed if the absolute number of neutrophils in the chorion or decidua exceeded 10,000 mm3. We additionally adjusted for birthweight (continuous, grams) and gestational age (continuous, weeks) as covariates in all models, as both are independent risk factors for ROP.20
Stratified analyses were also performed by the indication for preterm delivery. Prior research by ELGAN investigators has shown preterm deliveries tend to cluster into two distinct groups: those associated with intrauterine inflammation and those associated with aberrant placentation.21 Specifically, intrauterine inflammation-related deliveries are comprised of cases of preterm labor, prelabor premature rupture of membranes, placental abruption, and cervical insufficiency. In contrast, deliveries characterized by placentation aberrations are due to preeclampsia, intrauterine growth restriction, and other fetal indications.
We performed complete case analyses; thus, individuals missing data on placental DNA methylation, prethreshold ROP, or relevant covariates were excluded. This resulted in the exclusion of 7 newborns that were missing data on retinal exams and another 24 that were missing acute chorion/decidua inflammation status, for an analytic sample size of 395 newborns. To correct for multiple comparisons, we calculated the false discovery rate by using the Benjamini-Hochberg procedure and considered q-values < 0.05 to be statistically significant.22 All statistical analyses were conducted using Stata version 15.1 (College Station, TX, USA).
Results
Newborn, maternal, and placental characteristics of the ELGAN subcohort included in the present analysis are provided in Table 1. Fifty-three (13.4%) of the 395 newborns satisfied the criteria for prethreshold ROP upon ophthalmologic examination. Overall, about one-half of the newborns were males (52.7%). Newborns who satisfied the criteria for prethreshold ROP had a median gestational age of 25.1 weeks compared to 26.1 weeks among those who did not. Prethreshold ROP newborns also weighed significantly less at birth (725.1 versus 842.7 grams on average). The majority of births were singleton (70.1%), although our sample did include some twins (26.8%) and even a few triplets (3.0%).
Table 1.
Distribution of Newborn, Maternal, and Placenta Characteristics According to Prethreshold ROP (N = 395)
|
Overall,
N
= 395 |
No ROP,
N
= 342 |
Prethreshold ROP*,
N
= 53 |
|
| Newborn characteristics | |||
| Sex, % | |||
| Female | 47.3 | 47.1 | 49.1 |
| Male | 52.7 | 52.9 | 50.9 |
| Birthweight (grams), Mean (SD) | 827 (187) | 843 (187) | 725 (147) |
| Gestational age (weeks), Median (IQR) | 26 (25–27) | 26 (25–27) | 25 (24–26) |
| Plurality, % | |||
| Singleton | 70.1 | 68.1 | 83.0 |
| Twin | 26.8 | 28.4 | 17.0 |
| Triplet | 3.0 | 3.5 | 0.0 |
| Maternal characteristics | |||
| Race/ethnicity, % | |||
| Non-Hispanic white | 59.2 | 59.9 | 54.7 |
| Non-Hispanic black | 28.4 | 27.8 | 32.1 |
| Non-Hispanic other | 4.3 | 4.4 | 3.8 |
| Hispanic | 8.1 | 7.9 | 9.4 |
| Age (y), Mean (SD) | 29 (7) | 30 (7) | 6.3 |
| Educational attainment, % | |||
| Less than high school/GED | 12.4 | 12.0 | 15.1 |
| High school diploma/GED | 25.1 | 26.9 | 18.9 |
| More than high school/GED | 59.8 | 59.7 | 60.4 |
| Missing | 2.8 | 2.3 | 5.7 |
| Marital status, % | |||
| Single | 23.0 | 22.8 | 24.5 |
| Not married but living with partner | 20.0 | 20.8 | 15.1 |
| Married | 57.0 | 56.4 | 60.4 |
| Placenta characteristics | |||
| Chorion or decidua inflammation, % | |||
| No | 67.6 | 68.4 | 62.3 |
| Yes | 32.4 | 31.6 | 37.7 |
| Delivery characteristics | |||
| Presentation, % | |||
| Intrauterine inflammation | 82.3 | 82.8 | 79.2 |
| Aberrations of placentation | 17.7 | 17.3 | 20.8 |
SD, standard deviation; IQR, interquartile range (25th, 75th percentiles); GED, general education development.
Meets early treatment for ROP (ET-ROP) criteria.
The majority of participants were born to non-Hispanic white mothers, and the average maternal age was 29.4 years. Most mothers had at least some college education and were married. Overall, maternal characteristics were similar between newborns with prethreshold ROP and those without. One in three placentas was found to have acute inflammation in the chorion or decidua upon histologic examinations, but this did not appear to be associated with prethreshold ROP status.
Of the 320 tested CpG probes representing 12 inflammation, angiogenic, and neurotrophic genes, 16 were identified to have CpG methylation significantly associated with prethreshold ROP after adjusting for gestational age, birthweight, and chorion/decidua inflammation (q-values < 0.05; Table 2; Fig. 1). Increasing CpG methylation at 11 probes (cg10266336, cg15484375, cg12907644, cg06979684, cg00473501, cg01565803, cg07694252, cg06740893, cg11151395, cg14619064, and cg22331200) were associated with a lower risk of prethreshold ROP. In contrast, increasing methylation levels in five additional probes (cg14970975, cg05199346, cg07159484, cg15483907, and cg00989220) were associated with an elevated risk of prethreshold ROP. For example, a 1% increase in average methylation at cg00989220 probe within the gene body of TNFRSF1A corresponded to a 39% (95% CI, 14%–70%) greater prethreshold ROP risk.
Table 2.
Differentially Methylated Probes Significantly Related to Prethreshold Retinopathy
|
Gene |
Chromosome |
Probe |
Promoter Region |
Genomic Location |
Relation to CpG Island |
RR for Prethreshold ROP (95% CI) |
q-Value |
| ANGPT1 | 8 | cg07694252 | No | Body | Open sea | 0.97 (0.95, 0.98) | 0.012 |
| cg15483907 | Yes | TSS1500 | Open sea | 1.12 (1.06, 1.19) | 0.005 | ||
| BDNF | 11 | cg06979684 | Varies | 3′ UTR; 1st Exon; Body* | Open sea | 0.94 (0.91, 0.97) | 0.017 |
| cg07159484 | Varies | Body; 5′ UTR; 1st Exon; TSS200; TSS1500† | Island | 1.11 (1.09, 1.13) | <0.001 | ||
| CRP | 1 | cg01565803 | Yes | TSS1500 | Open sea | 0.97 (0.95, 0.98) | 0.026 |
| MPO | 17 | cg00473501 | No | Body | North shore | 0.95 (0.92, 0.97) | 0.002 |
| cg11151395 | No | Body | Island | 0.97 (0.96, 0.98) | 0.004 | ||
| cg14619064 | No | Body | Island | 0.98 (0.97, 0.99) | 0.012 | ||
| cg22331200 | No | Body | Island | 0.98 (0.97, 0.99) | 0.010 | ||
| SAA1 | 11 | cg15484375 | Yes | TSS200 | Open sea | 0.89 (0.83, 0.95) | 0.031 |
| SAA2 | 11 | cg10266336 | Yes | TSS200 | Open sea | 0.83 (0.75, 0.91) | 0.009 |
| cg12907644 | Yes | TSS200 | Open sea | 0.91 (0.85, 0.96) | 0.037 | ||
| TNFRSF1A | 12 | cg00989220 | No | Body | South shore | 1.39 (1.14, 1.70) | 0.025 |
| cg05199346 | No | Body | Open sea | 1.04 (1.02, 1.06) | 0.001 | ||
| cg14970975 | No | Body | Open sea | 1.02 (1.0, 1.03) | 0.009 | ||
| TNFRSF1B | 1 | cg06740893 | No | Body | Island | 0.97 (0.96, 0.98) | 0.017 |
RR, adjusted risk ratios.
Varies by the following isoforms: NM_001143812, NM_001143807, NM_170733, NM_001709, NM_001143814, NM_001143815, NM_001143808, NM_001143805, NM_170735, NM_001143809, NM_001143816, NM_170735, NM_001143811, NM_170734, NM_001143813, NM_001143806, NM_001143810, NR_002832, NM_170732, and NM_170731.
Varies by the following isoforms: NM_170731, NM_170732, NM_001143810, NM_001143806, NM_001143812, NM_001709, NM_001143808, NM_001143811, NM_001143805, NM_001143813, NM_001143815, NM_001143809, NM_170733, NM_001143807, NM_001143810, NM_001143814, NM_170734, and NM_001143811.
Figure 1.
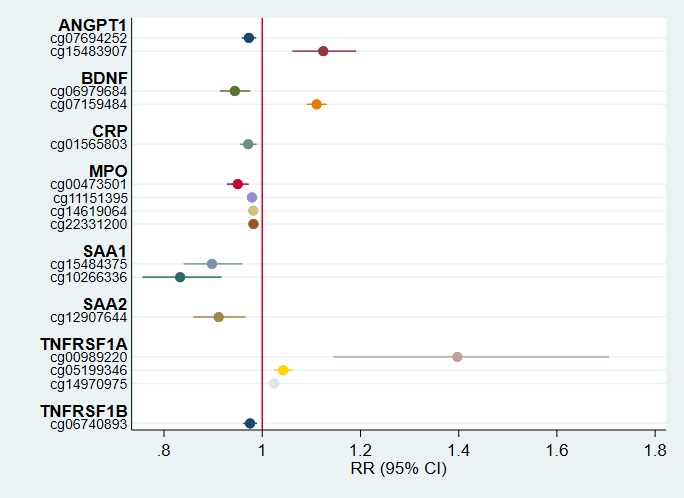
Adjusted risk ratios of prethreshold ROP with 95% CIs. Point estimates represent the relative risk of prethreshold ROP for a 1% increase in CpG methylation at the associated probe holding acute inflammation status, birthweight, and gestational age constant.
Stratified analyses revealed this association was most pronounced among deliveries presenting with intrauterine inflammation (P value for interaction = 0.004). For this subgroup of 325 infants, a 1% increase in methylation of cg00989220 was associated with a 55% (95% CI, 11%–116%) increase in the risk of developing prethreshold ROP (Fig. 2). Among dysfunctional placentation deliveries, the association between cg00989220 methylation and prethreshold ROP incidence was near null (Fig. 3). For all other CpG probes, associations were similar by delivery presentation (P values for interaction > 0.10; Figs. 2 and 3).
Figure 2.
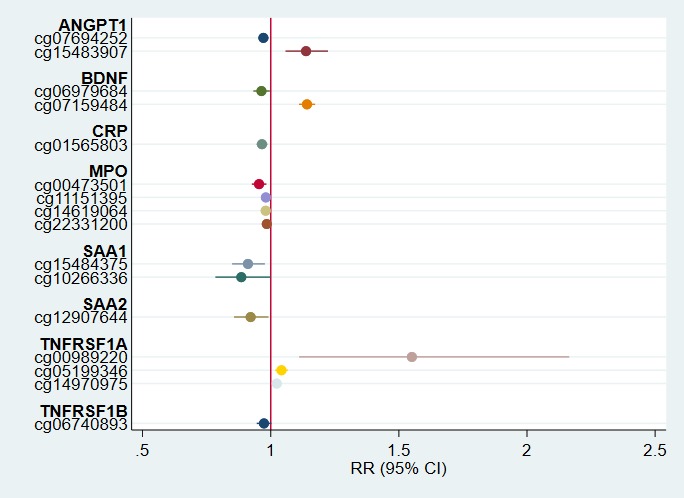
Adjusted risk ratios of prethreshold ROP with 95% CIs among deliveries due to intrauterine inflammation (n = 325). Point estimates represent the relative risk of prethreshold ROP for a 1% increase in CpG methylation at the associated probe among deliveries presenting with intrauterine inflammation (preterm labor, prelabor premature rupture of membranes, placental abruption, or cervical insufficiency). Estimates are adjusted for acute inflammation status, birthweight, and gestational age.
Figure 3.
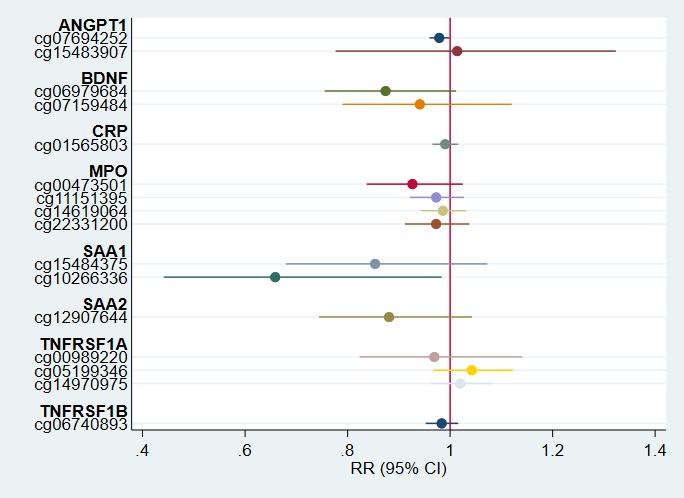
Adjusted risk ratios of prethreshold ROP with 95% CIs among deliveries due to aberrations of placentation (n = 70). Point estimates represent the relative risk of prethreshold ROP for a 1% increase in CpG methylation at the associated probe among deliveries presenting with dysfunctional placentation (preeclampsia, fetal indications, or intrauterine growth restriction). Estimates are adjusted for acute inflammation status, birthweight, and gestational age.
Discussion
In the present study, we set out to identify the association between placental CpG methylation and prethreshold ROP assessed during the first postnatal month among extremely preterm newborns (n = 395). We found differential placental methylation at 16 probes representing 8 unique genes to be associated with prethreshold ROP incidence. This is one of the first studies to demonstrate the potential for early epigenetic marks to improve prediction of ROP development in preterm newborns.
Differential methylation at inflammatory-related genes (namely, ANGPT1, BDNF, CRP, MPO, SAA1, SAA2, TNFRSF1A, and TNFRSF1B) was associated with prethreshold ROP. Promoter CpG methylation is often correlated with reduced gene expression; conversely, methylation of CpG loci within gene bodies tends to correlate with gene activation.23 We found that increasing placental methylation within the TNFRSF1A gene body was strongly related to developing ROP, particularly among infants born following preterm labor, prelabor premature rupture of membranes, placental abruption, or cervical insufficiency, indicating an inflamed in utero environment.21 TNFRSF1A encodes the tumor necrosis factor receptor superfamily 1A protein, one of two receptors for the proinflammatory cytokine tumor necrosis factor-alpha.24 The 1A receptor is recognized for its ability to induce cell death through the recruitment of adaptor proteins; this cascade has even been observed in retinal cells.25,26 If methylation of cg00989220 within the gene body of TNFRSF1A does indeed correlate with increased expression of the receptor protein, this pathway might explain the positive association observed with the development of prethreshold ROP.
The location of the methyl groups (i.e., promoter regions versus gene bodies) and associated differential effects on gene transcription might also explain why both positive and negative associations were observed with ROP risk for ANGPT1 and BDNF methylation. Prior work by ELGAN investigators that have shown higher levels of circulating ANGPT1 and BDNF proteins in the first month of life are associated with an 80% lower likelihood of developing prethreshold ROP.7 ANG-1 promotes vascular maturation and stability and negatively correlates with the amount of vitreous in the eyes of patients with severe ROP.27,28 Our results indicate prenatal methylation of ANGPT1 is likely a driver of the prephase of ROP during which the developing retina becomes sensitized.6 The neurotrophic growth factor BDNF has also been shown to beneficially impact the perinatal vascular system by stabilizing vessels in addition to its widely recognized neuropoietic functions.29 Of the neurotrophic growth factors, BDNF is the most abundant inside the retina, where it is produced by neurons and glial cells.30 Genetic variants in BDNF have been linked to severe ROP in preterm infants.31 In adults with type 2 diabetes, low serum levels of BDNF have been identified as an independent risk factor for retinopathy and for vision-threatening retinopathy in particular.32 BDNF expression, therefore, appears to be an important driver of retinal damage across the lifespan.
We additionally found greater methylation within the gene body of MPO to be associated with a reduced risk of developing the prethreshold ROP. However, a previous study of the ELGAN cohort observed postnatal levels of MPO were positively correlated with an increased risk of the disease.7 Thus, it is unclear precisely how CpG methylation of MPO is related to the development of ROP. Myeloperoxidase is a peroxidase enzyme that catalyzes the formation of reactive oxygen intermediates.33 Although the role of epigenetic regulation of MPO in the development of ROP has yet to be elucidated, increased MPO activity has previously been demonstrated in the eyes of diabetic retinopathy patients.34 Such increased activity is considered indicative of inflammation in the vitreous, suggesting both oxidative stress and inflammatory responses are significant contributors to the pathogenesis of retinopathy. Future mechanistic studies should consider the role and timing of CpG methylation, in addition to other epigenetic mechanisms, such as histone modifications, chromatin structure, or small noncoding RNAs, in the neural and vascular development of the retina.35
This prospective study of prethreshold retinopathy risk has several strengths and adds to the increasing empirical support for the role of angiogenesis, neurogenesis, and inflammation in ROP. This study is among the first study to assess how DNA methylation of the placenta, an ephemeral organ that is most often discarded after birth, is related to ROP. Moreover, this study was comprised of infants born extremely prematurely, 13.4% of whom were affected by this disorder. Thus, we were uniquely positioned to study potential mechanisms of ROP within this high-risk population. Nevertheless, our analysis is also subject to limitations. Notably, methylation patterns are tissue-specific and the placenta likely contains a combination of trophoblasts, mesenchymal, and stromal cells.36 Placentas of babies born prematurely may additionally contain infiltrated neutrophils.37 Without a reference set of DNA methylation data for placental tissue, we were unable to account for differences by cellular composition.38 However, all placenta tissue samples were excised in a standardized fashion and all analyses statistically controlled for acute inflammation status. We, therefore, likely minimized the impact of cellular composition in both the sample collection and analytic phases of our study.
Within the United States, ROP is one of the leading causes of childhood blindness.39 These findings add to the existing knowledge regarding its complex etiology, especially the involvement of inflammation and angiogenic/neurotrophic growth factors. Moreover, the results highlight the potential for early epigenetic marks to predict the development of prethreshold ROP. In the future, DNA methylation in early life could serve as a therapeutic target for reductions in the occurrence or severity of ROP.
Acknowledgments
Supported by the National Institutes of Health: NS040069, HD092374, EY021820, and OD023348.
Disclosure: C.M. Bulka, None; O. Dammann, None; H.P. Santos Jr, None; D.K. VenderVeen, None; L. Smeester, None; R. Fichorova, None; T.M. O'Shea, None; R.C. Fry, None
References
- 1.Hartnett ME, Penn JS. Mechanisms and management of retinopathy of prematurity. N Engl J Med. 2012;367:2515–2526. doi: 10.1056/NEJMra1208129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hellstrom A, Smith LE, Dammann O. Retinopathy of prematurity. Lancet. 2013;382:1445–1457. doi: 10.1016/S0140-6736(13)60178-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan-Ling T, Gole GA, Quinn GE, Adamson SJ, Darlow BA. Pathophysiology, screening and treatment of ROP: a multi-disciplinary perspective. Prog Retin Eye Res. 2018;62:77–119. doi: 10.1016/j.preteyeres.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 4.Chen J, Smith LE. Retinopathy of prematurity. Angiogenesis. 2007;10:133–140. doi: 10.1007/s10456-007-9066-0. [DOI] [PubMed] [Google Scholar]
- 5.Chen ML, Allred EN, Hecht JL, et al. Placenta microbiology and histology and the risk for severe retinopathy of prematurity. Invest Ophthalmol Vis Sci. 2011;52:7052–7058. doi: 10.1167/iovs.11-7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee J, Dammann O. Perinatal infection, inflammation, and retinopathy of prematurity. Semin Fetal Neonatal Med. 2012;17:26–29. doi: 10.1016/j.siny.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holm M, Morken TS, Fichorova RN, et al. Systemic inflammation-associated proteins and retinopathy of prematurity in infants born before the 28th week of gestation. Invest Ophthalmol Vis Sci. 2017;58:6419–6428. doi: 10.1167/iovs.17-21931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maccani MA, Marsit CJ. Epigenetics in the placenta. Am J Reprod Immunol. 2009;62:78–89. doi: 10.1111/j.1600-0897.2009.00716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tilley SK, Martin EM, Smeester L, et al. Placental CpG methylation of infants born extremely preterm predicts cognitive impairment later in life. PLoS One. 2018;13:e0193271. doi: 10.1371/journal.pone.0193271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meakin CJ, Martin EM, Santos HP, Jr,, et al. Placental CpG methylation of HPA-axis genes is associated with cognitive impairment at age 10 among children born extremely preterm. Horm Behav. 2018;101:29–35. doi: 10.1016/j.yhbeh.2018.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bibikova M, Barnes B, Tsan C, et al. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98:288–295. doi: 10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 12.Triche TJ, Jr,, Weisenberger DJ, Van Den Berg D, Laird PW, Siegmund KD. Low-level processing of Illumina Infinium DNA Methylation BeadArrays. Nucleic Acids Res. 2013;41:e90. doi: 10.1093/nar/gkt090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fortin JP, Triche TJ, Jr,, Hansen KD. Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics. 2017;33:558–560. doi: 10.1093/bioinformatics/btw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 15.Leviton A, Ryan S, Allred EN, et al. Antecedents and early correlates of high and low concentrations of angiogenic proteins in extremely preterm newborns. Clin Chim Acta. 2017;471:1–5. doi: 10.1016/j.cca.2017.05.014. [DOI] [PubMed] [Google Scholar]
- 16.International Committee for the Classification of Retinopathy of Prematurity. An international classification of retinopathy of prematurity. Pediatrics. 1984;74:127–133. [PubMed] [Google Scholar]
- 17.American Academy of Pediatrics. Section on Ophthalmology. Screening examination of premature infants for retinopathy of prematurity. Pediatrics. 2001;108:809–811. doi: 10.1542/peds.108.3.809. [DOI] [PubMed] [Google Scholar]
- 18.Early Treatment For Retinopathy Of Prematurity Cooperative Group. Revised indications for the treatment of retinopathy of prematurity: results of the early treatment for retinopathy of prematurity randomized trial. Arch Ophthalmol. 2003;121:1684–1694. doi: 10.1001/archopht.121.12.1684. [DOI] [PubMed] [Google Scholar]
- 19.Zou G. A modified poisson regression approach to prospective studies with binary data. Am J Epidemiol. 2004;159:702–706. doi: 10.1093/aje/kwh090. [DOI] [PubMed] [Google Scholar]
- 20.Alajbegovic-Halimic J, Zvizdic D, Alimanovic-Halilovic E, Dodik I, Duvnjak S. Risk factors for retinopathy of prematurity in premature born children. Med Arch. 2015;69:409–413. doi: 10.5455/medarh.2015.69.409-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McElrath TF, Hecht JL, Dammann O, et al. Pregnancy disorders that lead to delivery before the 28th week of gestation: an epidemiologic approach to classification. Am J Epidemiol. 2008;168:980–989. doi: 10.1093/aje/kwn202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- 23.Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28:1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 24.Costa GN, Vindeirinho J, Cavadas C, Ambrósio AF, Santos PF. Contribution of TNF receptor 1 to retinal neural cell death induced by elevated glucose. Mol Cell Neurosci. 2012;50:113–123. doi: 10.1016/j.mcn.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 25.Tartaglia LA, Ayres TM, Wong GH, Goeddel DV. A novel domain within the 55 kd TNF receptor signals cell death. Cell. 1993;74:845–853. doi: 10.1016/0092-8674(93)90464-2. [DOI] [PubMed] [Google Scholar]
- 26.Genini S, Beltran WA, Aguirre GD. Up-regulation of tumor necrosis factor superfamily genes in early phases of photoreceptor degeneration. PLoS One. 2013;8:e85408. doi: 10.1371/journal.pone.0085408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takagi H. Molecular mechanisms of retinal neovascularization in diabetic retinopathy. Intern Med. 2003;42:299–301. doi: 10.2169/internalmedicine.42.299. [DOI] [PubMed] [Google Scholar]
- 28.Sato T, Shima C, Kusaka S. Vitreous levels of angiopoietin-1 and angiopoietin-2 in eyes with retinopathy of prematurity. Am J Ophthalmol. 2011;151:353–357. doi: 10.1016/j.ajo.2010.08.037. [DOI] [PubMed] [Google Scholar]
- 29.Kermani P, Hempstead B. Brain-derived neurotrophic factor: a newly described mediator of angiogenesis. Trends Cardiovasc Med. 2007;17:140–143. doi: 10.1016/j.tcm.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seki M, Tanaka T, Sakai Y, et al. Müller cells as a source of brain-derived neurotrophic factor in the retina: noradrenaline upregulates brain-derived neurotrophic factor levels in cultured rat Muller cells. Neurochem Res. 2005;30:1163–1170. doi: 10.1007/s11064-005-7936-7. [DOI] [PubMed] [Google Scholar]
- 31.Hartnett ME, Morrison MA, Smith S, et al. Genetic variants associated with severe retinopathy of prematurity in extremely low birth weight infants. Invest Ophthalmol Vis Sci. 2014;55:6194–6203. doi: 10.1167/iovs.14-14841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo M, Liu H, Li SS, Jiang FL, Xu JM, Tang YY. Low serum brain-derived neurotrophic factor but not brain-derived neurotrophic factor gene Val66met polymorphism is associated with diabetic retinopathy in Chinese type 2 diabetic patients. Retina. 2017;37:350–358. doi: 10.1097/IAE.0000000000001132. [DOI] [PubMed] [Google Scholar]
- 33.Aratani Y. Myeloperoxidase: its role for host defense, inflammation, and neutrophil function. Arch Biochem Biophys. 2018;640:47–52. doi: 10.1016/j.abb.2018.01.004. [DOI] [PubMed] [Google Scholar]
- 34.Augustin AJ, Breipohl W, Böker T, Lutz J, Spitznas M. Increased lipid peroxide levels and myeloperoxidase activity in the vitreous of patients suffering from proliferative diabetic retinopathy. Graefes Arch Clin Exp Ophthalmol. 1993;231:647–650. doi: 10.1007/BF00921959. [DOI] [PubMed] [Google Scholar]
- 35.van der Maarel SM. Epigenetic mechanisms in health and disease. Ann Rheum Dis. 2008;67:iii97–iii100. doi: 10.1136/ard.2008.098392. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y, Zhao S. Vascular Biology of the Placenta. San Rafael, CA: Margan & Claypool Life Sciences;; 2010. [PubMed] [Google Scholar]
- 37.Faye-Petersen OM. The placenta in preterm birth. J Clin Pathol. 2008;61:1261–1275. doi: 10.1136/jcp.2008.055244. [DOI] [PubMed] [Google Scholar]
- 38.Houseman EA, Kelsey KT, Wiencke JK, Marsit CJ. Cell-composition effects in the analysis of DNA methylation array data: a mathematical perspective. BMC Bioinformatics. 2015;16:95. doi: 10.1186/s12859-015-0527-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kong L, Fry M, Al-Samarraie M, Gilbert C, Steinkuller PG. An update on progress and the changing epidemiology of causes of childhood blindness worldwide. J AAPOS. 2012;16:501–507. doi: 10.1016/j.jaapos.2012.09.004. [DOI] [PubMed] [Google Scholar]


