Abstract
Background: Tripartite motif-containing protein 11 (TRIM11) is one of the E3 ubiquitin ligases, which is upregulated in several human tumors. Meanwhile, the detailed function of TRIM11 remains unclear in breast cancer cells.
Purpose: The purpose of the present study is to analyze the biological function of TRIM11 and identify its potential signaling pathway in breast cancer cells.
Patients and methods: Thirty five pairs of breast cancer specimens and adjacent-matched noncancerous samples were used to analyse the expression profile of TRIM11. RNA interference was utilized to silence TRIM11 in three breast cancer cell lines (T47D, ZR7530, and BT474) respectively. Meanwhile, overexpression of TRIM11 was induced in one breast cancer cells (MDA-MB-231) by using Lentiviral vector. Moreover, the AKT inhibitor (MK-2206) was used to determine the correlation between TRIM11 and AKT in breast cancer cells.
Results: Our results indicated that TRIM11 was increased in breast cancer tissues. Moreover, TRIM11 was a pro-proliferation regulator in breast cancer cells and participated in the metabolism of glycolysis. Importantly, our results demonstrated that TRIM11 was involved in the AKT/GLUT1 signaling pathway in breast cancer cells.
Conclusion: Present research not only gained a deep understanding of the biological function of TRIM11 but also provided evidences to indicate its possible signaling pathway in breast cancer cells.
Keywords: breast cancer, TRIM11, AKT, GLUT1, HKII
Introduction
Breast cancer is one of the most common malignancies in female all over the world, which is a highly heterogeneous disease.1 Although early detection and treatment benefit to prognosis, the mortality of breast cancer is still very high.2 Currently, the surgical is the major option in the treatment for breast cancer. This treatment not only has negative physical and emotional effects on sufferers but also influences their social activity.3 Hence, enhancing the understanding of the molecule mechanism of breast cancer was urgently needed.
Tripartite motif-containing protein 11 (TRIM11) belongs to the E3 ubiquitin ligase family, which is identified as an oncogene in certain cancers. Previous reports have indicated that TRIM11 promotes the proliferation and invasion of hepatocellular carcinoma cells.4,5 Moreover, TRIM11 has accelerated the progression of colon cancer though increasing the proliferation and suppressing the apoptosis.6 In addition, TRIM11 has also improved the development and metastasis of lung cancer cells.7 However, the precise biological function of TRIM11 remains unclear in breast cancer cells.
It has been confirmed that cancer cells need extra energy to meet the rapid proliferation.8 Therefore, the activity of aerobic glycolysis is often elevated in cancer cells.9 Glucose transporter 1 (GLUT1) has regulated the metabolism of aerobic glycolysis and is upregulated in cancers. Previous reports have demonstrated that GLUT1 promotes the development and metastasis of breast cancer cells.10,11 Therefore, it is vital and valuable to further investigate the molecule network of GLUT1 in breast cancer cells.
The phosphatidylinositol 3-kinase/AKT (PI3K/AKT) signaling pathway plays an important role in the regulation of cell survival and proliferation and closely links to tumorigenesis. Previous reports have demonstrated that the activation of PI3K/AKT signaling pathway promotes the growth and progression of breast cancer.12,13 Moreover, the AKT/GLUT1 signaling pathway plays an essential role in the regulation of proliferation and glycolysis in lung cancer cells.14 However, the biological function of AKT/GLUT1 signaling pathway in breast cancer cells still needed to be further explored.
In this study, we analyzed the function of TRIM11 and identified its potential molecule network in breast cancer cells. TRIM11 was induced silencing and overexpression in breast cancer cells through RNA interference and lentiviral vector, respectively. Our analysis not only gained a deep insight into TRIM11 but also provided evidences in terms to develop a novel approach for the treatment of breast cancer.
Materials and methods
Tissue samples of breast cancer
The number of Thirty five pairs of breast cancer specimens and adjacent-matched noncancerous samples were collected from the Second Affiliated Hospital of Soochow University. Tissues were stored at −80°C after being snap-frozen in liquid nitrogen and used for the following analysis. Our research was approved by the independent ethics committee of the Second Affiliated Hospital of Soochow University and was conducted in accordance with the Declaration of Helsinki. All patients provided written informed consent.
Cell culture
All breast cancer cell lines MDA-MB-231 (ER–; PR–; Her2–), MCF7 (ER+; PR+; Her2–), T47D (ER+; PR+;Her2–), BT474 (ER+; PR+; Her2+) and ZR7530 (ER+; PR-; Her2+) were purchased from cell bank of Shanghai biology institute (Shanghai, China). Cells were planted in DMEM Medium (Trueline, USA) and were cultured in the incubator with the condition of 5% CO2, at 37°C.
RNA extraction and qRT-PCR
TRIzol Reagent kit (Invitrogen, USA) was used to isolate total RNA from all samples. Then, the complementary DNA (cDNA) was synthesized by the cDNA synthesis kit (Fermentas, Canada) following the manufacturer’s introduction. The program of qRT-PCR was set as follows: 95°C for 10 mins followed by 40 cycles of 95°C for 15 s, 60°C for 45 s and normalized to GAPDH. 2−ΔΔCt method was used to calculate the gene relative expression. Three replicates were needed for every data. Primer sequences were presented in the Supplementary material.
Silencing and overexpression of TRIM11
Lentiviral plasmids (pLKO.1) containing three short interfering RNAs (siRNA) targeted to various regions of human TRIM11 (NM_145214.2) and a non-specific siRNA (siNC) were synthesized (Major, Shanghai, China). Meanwhile, pLVX-puro vector contained the full length of human TRIM11 cDNA sequence and the mock plasmid (oeNC) were transiently transfected in breast cancer cells respectively by Lipofectamine 2000 (Invitrogen, USA). Analyses were established at 48 hrs after being transfected. The sequences information of siTRIM11s was listed in Table S1.
Table S1.
Human gene TRIM11 (NM_145214.2) RNAi targeting locus information
| RNAi Targeting Locus | Sequence | |
|---|---|---|
| Name | Locus positon | |
| siTRIM11-1 | 672–690 | GGAGAAGTCACTGGAGCAT |
| siTRIM11-2 | 708–726 | GGATGCGTTGCTGTTCCAA |
| siTRIM11-3 | 746–764 | GCGTCTTGTGGCAGAAGAT |
Western blot
Total protein lysates were isolated from indicated samples by using RIPA lysis buffer (JRDUN, Shanghai, China). BCA protein assay kit (Thermo Fisher, USA) was used to quantify the protein concentration. The protein was fractionated on 10% SDS-PAGE, after that, transferred the gel to a nitrocellulose membrane (Millipore, USA) for 12 hrs. Then, the membrane was blocked with nonfat dry milk (5%) at room temperature for 60 mins. Next, the membranes were probed with the primary antibodies at 4°C overnight. Then, the membranes were probed with the secondary antibody anti-mouse IgG (Beyotime, China) at 37°C for 60 mins. The protein expression value was detected by an enhanced chemiluminescence system (Tanon, China). The detail information of primary antibodies was listed in Table S2.
Table S2.
The primary antibodies information
Cell proliferation
The cell proliferation rate was examined by the cell counting kit-8 (CCK-8) assay kits (SAB, USA). In brief, cells were cultured for 0, 24, 48, and 72 hrs and mixed with the CCK-8 solution (1:10) for each well. Then, cells were incubated for 1 hr. The OD450 nanometer was measured by a Microplate reader (Pulangxin, China). Triplicates independently assays were needed for each time point.
Glucose transport and lactate production assay
Briefly, the glucose analog 2-NBDG (Molecular Probes, OR) was used as a fluorescent probe for determining the activity of Glucose transport. In order to examine the uptake of 2-NBDG, a total of 5×105 cells were seeded in six-well plates. Then, all the cells were pre-incubated in Krebs-Ringer bicarbonate (KRB) buffer (glucose free) for 15 mins after maintaining in a 5% CO2 atmosphere at 37°C for 24 hrs. After that, cells incubated in fresh KRB buffer supplemented with 2-NBDG for 45 mins at 37°C, 5% CO2. Microplate reader (Pulangxin, China) was utilized to examine the OD values at wavelength 485 nm. Moreover, Lactate Assay Kit (njjcbio, China) was utilized to examine the production of lactate in different cells according to the protocol of the manufacturer.
Xenograft model
Experiment was performed in agreement with the independent ethics committee of the Second Affiliated Hospital of Soochow University and was guided by the Institute’s guidelines for animal experiments. Moreover, this research was confirmed by the Institutional Animal Care and Use Committee (IACUC). ZR7530 cells that transfected with siNC or siTRIM11 (n=5×106) were subcutaneously injected into the right flank of 4–6 week old nude mice (n=6 for each group; Shanghai Laboratory Animal Company, China). Tumor length and width were determined every third day for a total of 33 days after injection. Tumor volume was calculated as length × (width2/2).
Statistical analysis
Statistical analyses were performed by using GraphPad Prism software Version 7.0 (CA, USA). All data represented as means ± SEM from three independent experiments. Statistical significance was assessed by the analysis of variance. Statistically significant was accepted as the P -value <0.05.
Results
TRIM11, GLUT1, and HKII were upregulated in breast cancer samples
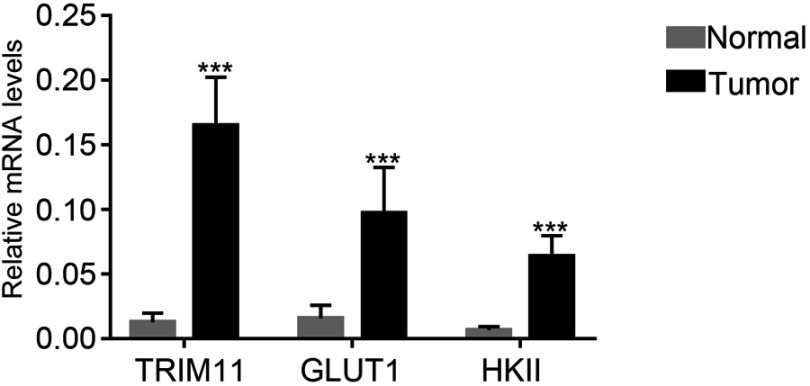
Quantitative real-time PCR (qRT-PCR) was performed to quantify the mRNA expression content of TRIM11, GLUT1, and HKII in 35 pairs of breast cancer tissues and matched para-cancerous samples. As shown in Figure 1, it was clear to identify that the mRNA expression level of TRIM11 was obviously increased in breast cancer tissues compared with that of normal samples. Moreover, both the GLUT1 and HKII were also significantly upregulated in breast cancer tissues.
Figure 1.
TRIM11, GLUT1, and HKII were upregulated in breast cancer tissues.
Note: ***P<0.001 vs normal.
Silencing and overexpression of TRIM11 in breast cancer cells
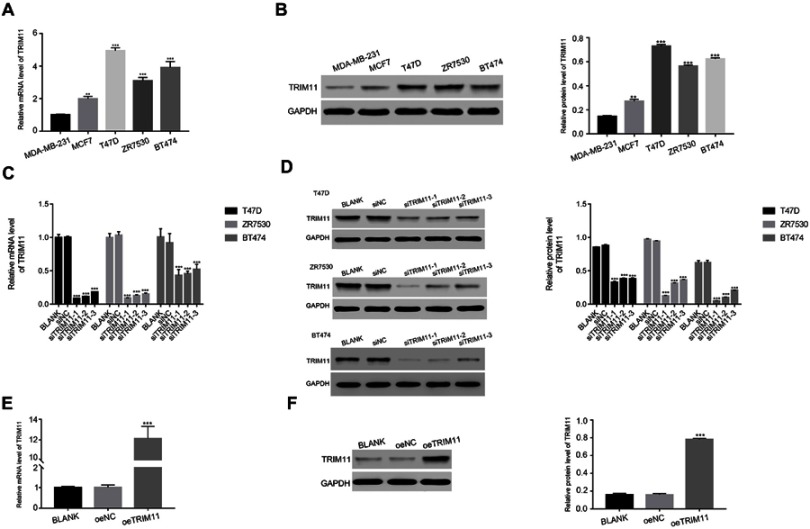
In order to further investigate the function of TRIM11 in breast cancer cells, qRT-RCR, and Western blot were established to quantify the mRNA and protein content of TRIM11 in five breast cancer cell lines, including MDA-MB-231, MCF7, T47D, and ZR7530 and BT474. Clearly, both the relative mRNA and protein content of TRIM11 were remarkably promoted in T47D, ZR7530 and BT474 cells. Meanwhile, the level of TRIM11 was lowest in MDA-MB-231 cells among all cell lines as indicated (Figure 2A and B). Therefore, T47D, ZR7530 and BT474 cells lines were chosen for silencing the expression of TRIM11. Overexpression of TRIM11 was induced in MDA-MB-231 cells.
Figure 2.
Knockdown and overexpression of TRIM11 in breast cancer cells.
Notes: (A) and (B) stand for the mRNA and protein level of TRIM11 in five breast cancer cell lines as indicated. ***P<0.001 vs MDA-MB-231. (C) and (D) stand for the mRNA and protein level of TRIM11 examined in T47D, ZR7530 and BT474 cells that transfected with siNC, siTRIM11-1, siTRIM11-2 or siTRIM11-3, respectively. ***P<0.001 vs siNC. (E) and (F) stand for the mRNA and protein level of TRIM11 examined in MDA-MB-231 that transfected with a mock plasmid (oeNC) and a plasmid overexpressing TRIM11 (oeTRIM11). ***P<0.001 vs oeNC.
To silence the expression of TRIM11 in breast cancer cells, T47D, ZR7530, and BT474 cells were transfected with three siRNAs aiming at human TRIM11 (siTRIM11-1, siTRIM11-2, siTRIM11-3) and a non-specific siRNA (siNC) respectively. The untreated cells were served as the blank control (BLANK). All of the TRIM11 siRNAs were well functioned and deeply inhibited the levels of endogenous TRIM11 (Figure 2C and D). Therefore, we chose siTRIM11-1 and siTRIM11-2 transfected cells in the following analysis.
As for overexpression of TRIM11, a plasmid containing the full length of TRIM11 cDNA (oeTRIM11) was transfected to MDA-MB-231 cells. A mock plasmid (oeNC) was transfected as a negative control. It was easily identified that the content of TRIM11 was significantly upregulated by oeTRIM11 in MDA-MB-231 cells (Figure 2E and F). Therefore, oeTRIM11 transfected cells were used for further analyses.
Silencing of TRIM11 suppressed the proliferation rate of breast cancer cells
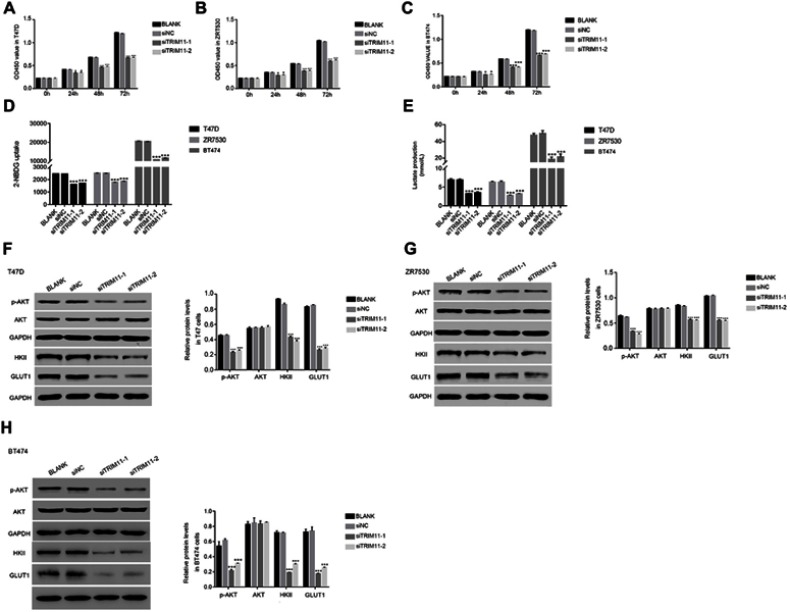
The cell proliferation rate was measured by CCK-8 assay. The proliferation rate of BLANK and siNC transfected cells showed no significant difference in all time point in different cell lines. However, the proliferation rates of T47D, ZR7530, and BT474 cells were suppressed by siTRIM11-1 or siTRIM11-2 at 24 hrs after transfection. Moverover, siTRIM11-1 or siTRIM11-2 showed a stronger effect in inhibiting the proliferation of different cells as indicated at 48 and 72 hrs after transfection. (Figure 3A–C). Therefore, TRIM11 was a pro-proliferation regulator in breast cancer cells.
Figure 3.
Knockdown of TRIM11 suppressed the proliferation and inhibited the glucose transport activity and lactate production in breast cancer cells.
Notes: (A–C) stand for the cell proliferation was detected 0, 24, 48, and 72 hrs after transfection with siNC, siTRIM11-1, and siTRIM11-2 in T47D, ZR7530 and BT474 cells. *P<0.05 vs siNC, ***P<0.001 vs siNC. (D) Glucose transport activity measured using the fluorescent glucose analog 2-NBDG in T47D, ZR7530 and BT474 cells.transfected with siNC, siTRIM11-1, and siTRIM11-2, respectively. ***P<0.001 vs siNC. (E) The production of Lactate in T47D, ZR7530 and BT474 cells transfected with siNC, siTRIM11-1, and siTRIM11-2, respectively. ***P<0.001 vs siNC. (F–H) stand for the protein level of p-AKT,AKT, HKII, and GLUT1 in T47D, ZR7530 and BT474 cells transfected with siNC, siTRIM11-1 and siTRIM11-2, respectively. ***P<0.001 vs siNC.
Glucose transport activity and lactate production were inhibited in siTRIM11-transfected cells
Next, we determined the glucose transport activity and lactate production in TRIM11 siRNAs-transfected cells. Interestingly, the uptake of the glucose analog 2-NBDG was significantly downregulated in siTRIM11-1 or siTRIM11-2 transfected cells (Figure 3D). Meanwhile, the lactate production was also remarkably decreased in siTRIM11-1 or siTRIM11-2 transfected cells (Figure 3E). Therefore, knockdown of TRIM11 deeply suppressed the activity of glycolysis in T47D, ZR7530, and BT474 cells.
Then, the protein content of AKT, p-AKT, GLUT1, and HKII was quantified by Western blot in T47D cells transfected with siTRIM11-1 or siTRIM11-2. The content of p-AKT was significantly reduced in siTRIM11-transfected cell. Therefore, knockdown of TRIM11 significantly inhibited the phosphorylation of AKT in T47D cells. Meanwhile, the level of HKII and GLUT1 was also downregulated in siTRIM11-1 or siTRIM11-2 transfected cells (Figure 3F). These results demonstrated that TRIM11 was positively correlated with HKII and GLUT1 and further indicated that TRIM11 involved in the metabolism of glycolysis in breast cancer cells. Interestingly, the similar results were also obtained in ZR7530 or BT474 cells transfected with siTRIM11-1 or siTRIM11-2 (Figure 3G and H).
The function of TRIM11 was suppressed by the AKT inhibitor
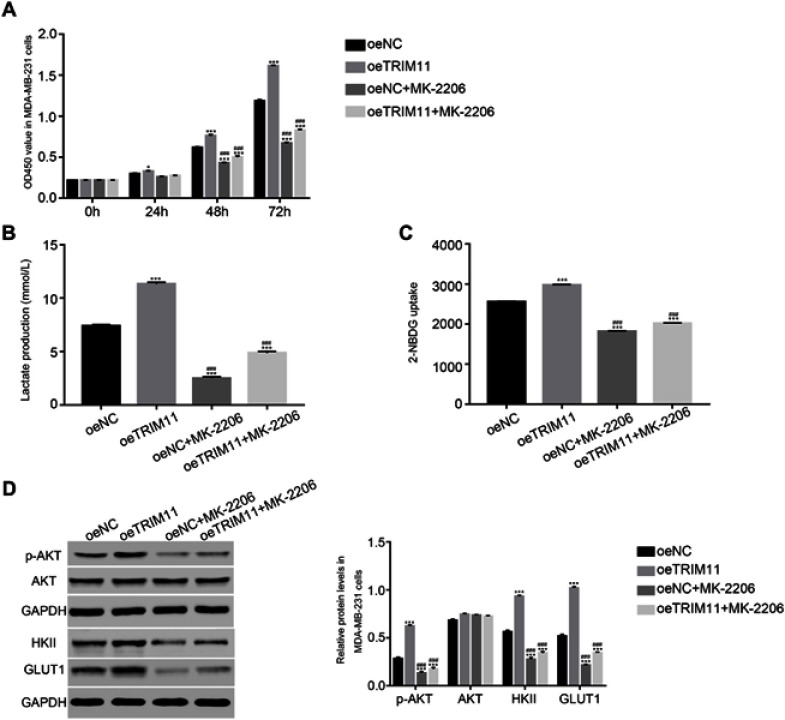
In order to further examine the correlation between TRIM11 and AKT, the oeNC or oeTRIM11 transfected cells were cultured with the AKT inhibitor MK-2206, respectively. At 24 hrs after transfection, the proliferation of oeTRIM11 transfected cells was higher than that of oeNC transfected cells. Moreover, overexpression of TRIM11 remarkably upregulated the proliferation rate of breast cancer cells at 48 and 72 hrs after transfection (Figure 4A). These results demonstrated that overexpression of TRIM11 promoted the proliferation of breast cancer cells. However, this effect was deeply suppressed by the AKT inhibitor MK-2206 in all time points after transfection.
Figure 4.
The function of TRIM11 was suppressed by a specific AKT inhibitor MK-2206.
Notes: (A) Cell proliferation was detected 0, 24, 48, and 72 hrs after transfection with oeNC, oeTRIM11 in MDA-MB-231 cells. *P<0.05 vs oeNC, ***P<0.001 vs oeNC; P<0.001 vs oeTRIM11. (B) Glucose transport activity measured using the fluorescent glucose analog 2-NBDG in MDA-MB-231 cells that transfected with oeNC or oeTRIM11, respectively. ***P<0.001 vs oeNC; P<0.001 vs oeTRIM11. (C) The production of Lactate in MDA-MB-231 cells that transfected with oeNC or oeTRIM11, respectively. ***P<0.001 vs oeNC; P<0.001 vs oeTRIM11. (D) The protein level of p-AKT, AKT, HKII, and GLUT1 in MDA-MB-231 cells that transfected with oeNC or oeTRIM11, respectively. ***P<0.001 vs oeNC; P<0.001 vs oeTRIM11.
In addition, we also investigated the glucose transport activity and lactate production in different cells. Obviously, the glucose transport activity and lactate production were significantly upregulated in oeTRIM11 transfected cells. Interestingly, both of them were remarkably downregulated by the AKT inhibitor MK2206 in oeTRIM11 cultured cells (Figure 4B and C). Meanwhile, we also examined the protein content of p-AKT, AKT, HKII, and GLUT1 in the cells as indicated. As shown in Figure 4D, the phosphorylation of AKT was significantly upregulated in oeTRIM11 transfected cells. Moreover, overexpression of TRIM11 promotes the expression of HKII and GLUT1. Interestingly, the AKT inhibitor deeply suppressed the expression of p-AKT, HKII, and GLUT1 in oeTRIM11 transfected cells. Therefore, TRIM11 might involve in the same pathway with AKT in breast cancer cells.
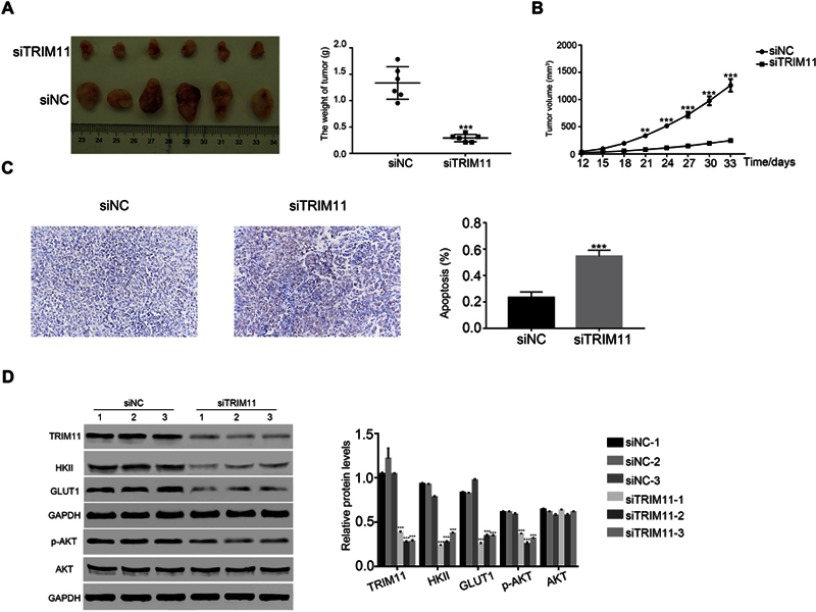
siTRIM11 suppressed the tumorigenicity of breast cancer cells in nude mice
Next, we analyzed the effect of TRIM11 siRNA on tumorigenicity in vivo. The nude mice were hypodermically injected with ZR7530 cells that transfected with siNC or siTRIM11, respectively (n=6 for each group). The formation of tumor was examined every third day for a total of 33 days (beginning at day 12). Both of the injected cells were able to develop tumors. However, the TRIM11 siRNA injected cells deeply suppressed the growth rate of tumor in vivo. The weight and volume of siTRIM11 tumors were significantly downregulated compared with that of siNC tumors (Figure 5A and B). Moreover, the apoptosis ratio of siTRIM11 tumor was much higher than that in siNC tumor (Figure 5C). Further,the protein level of TRIM11 was also deeply decreased in siTRIM11 tumors. Meanwhile, the similar results were also detected in the examination for HKII, GLUT1, and p-AKT (Figure 5C and D). Taken together, all these results indicated that knockdown of TRIM11 inhibited the growth of tumor though regulating the glycolysis metabolism.
Figure 5.
siTRIM11 suppressed the tumorigenicity of breast cancer cells in nude mice.
Notes: (A and B) stand for the tumor weight and volume in nude mice injected with ZR7530 cells transfected with siNC and siTRIM11. **P<0.01 vs siNC, ***P<0.001 vs siNC. (C) TdT-mediated DUTP nick end labeling (TUNEL) assay was performed to examine the apoptosis ratio in siNC or siTRIM11 tumor, ***P<0.001 vs siNC. (D) Protein level of TRIM11, HKII, GLUT1, p-AKT, and AKT was analyzed by Western blot in xenografts from different nude mice as indicated (n=3 for each group), ***P<0.001 vs siNC-1.
Discussion
Breast cancer belongs to the most feared tumors in female all over the world, which displays a high degree of heterogeneity.15 For this reason, the outcomes of traditional therapy (surgery and chemotherapy) methods are not effective enough for breast cancer sufferers. Therefore, deepened the understanding into the molecule network of breast cancer contributes to develop a novel approach for its treatment.
At present research, we systemically analyzed the effect of TRIM11 on breast cancer cells. We induced silencing and overexpression of TRIM11 in breast cancer cells. Results that obtained from these two experimental sections were in agreement. Therefore, our conclusion was more credible.
TRIM11 has been reported as an oncogene in some certain cancers. Previous report has demonstrated that overexpression of TRIM11 induces proliferation and metastasis of lung cancer and gliomas cells.4,7,16 Moreover, silencing of TRIM11 deeply reduced the proliferation and invasion of ovarian cancer cells.17 It has been well elucidated that T47D, ZR7530, and BT474 cells are different types of breast cancer cells.18 In this study, our findings indicated that TRIM11 was a pro-proliferation factor and presented a similar function in different types of breast cancer cells. Therefore, those results further demonstrated that TRIM11 was a crucial proto-oncogene in the pathogenesis of human breast cancer.
Hexokinase II (HK-II) is necessary for the first step in glycolysis process, which is localized in the cytosol and mitochondria.19 Previous report has demonstrated that HKII is overexpressed in various human cancers, which shows the potential value as a therapeutic target in the cancer treatment.20 It has been reported that HKII promotes the activity of glycolytic to meet the energy consumption in the proliferation and progression of breast cancer cells.21 In the present research, the level of HKII was also upregulated in breast cancer tissue and cells. Importantly, these results indicated that HKII was positively correlated with TRIM11 in breast cancer cells. Therefore, TRIM11 might mediate the proliferation of breast cancer cells though regulating HKII.
GLUT1 has been reported as a prognostic biomarker for human breast cancer.22 Previous report has demonstrated that GLUT1 promotes the proliferation and metastasis of breast cancer cells.10 In addition, suppression of GLUT1 has re-sensitized the radio-resistant breast cancer cells to radiation.23 In this study, our results indicated that GLUT1 was positively regulated by TRIM11 in breast cancer cells. Therefore, TRIM11 might promote the metabolism of glycolysis though regulating GLUT1 in breast cancer cells.
Much more importantly, our analysis also suggested the positive correlation between TRIM11 and p-AKT in breast cancer cells. The AKT signaling pathway plays an essential role in the progression of breast cancer cells.24 Moreover, it has been demonstrated that the activity of AKT is positively correlated with the level of GLUT1 in ovarian cancer cells.25 In addition, PI3K/GLUT1 signaling pathway stimulates the glucose uptake and lactate metabolic.26,27 AKT pathway also promotes the expression of HKII and subsequently increased glucose capture.28 The AKT/GLUT1/HKII signaling pathway is reported as the potential target in the prevention of growth and glycolysis activities in lung cancer cells.14 At present research, we found GLUT1 and HKII were also upregulataed in breast cancer tissues and positively correlated with TRIM11. Therefore, our results indicated that TRIM11 was the upstream component in PI3K/GLUT1/HKII signaling pathway in breast cancer cells.
Conclusion
At the present study, we systemically explored the function of TRIM11 in breast cancer cells. This research not only gained a deep understanding of the biological function of TRIM11 but also provided evidences to indicate its possible signaling pathway in breast cancer cells.
Acknowledgments
We acknowledged the support given by The Second Affiliated Hospital of Soochow University for this research.
Author contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this study.
Supplementary materials
Primer sequence information
Homo sapiens tripartite motif containing 11 (TRIM11), mRNA
Primer F 5ʹ CACCTAAGCTGCACAGTTCC 3ʹ
Primer R 5ʹ GGCTGCCTCCTAATTCTTCC 3ʹ
Pos: 2008–2192
Amplified product: Size: 185 bps
Homo sapiens solute carrier family 2 member 1 (GLUT1), mRNA
Primer F 5ʹ TGCAGGAGATGAAGGAAG 3ʹ
Primer R 5ʹ CAATGGTGGCATACACAG 3ʹ
Pos: 939–1151
Amplified product: Size: 213 bps
Homo sapiens hexokinase 2 (HK2), mRNA
Primer F 5ʹ AGGAGGATGAAGGTAGAAATGG 3ʹ
Primer R 5ʹ ACTGGACAATGTGGTCAAAG 3ʹ
Pos: 3491–3667
Amplified product: Size: 277 bps
Homo sapiens glyceraldehyde-3-phosphate dehydrogenase (GAPDH), transcript variant2, mRNA
Primer F 5ʹ AATCCCATCACCATCTTC 3ʹ
Primer R 5ʹ AGGCTGTTGTCATACTTC 3ʹ
Pos: 436-653
Amplified product: Size: 218 bps
References
- 1.Weigelt B, Reis-Filho JS. Histological and molecular types of breast cancer: is there a unifying taxonomy? Nat Rev Clin Oncol. 2009;6(12):718–730. doi: 10.1038/nrclinonc.2009.166 [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer Manag. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- 3.Feiten S, Dünnebacke J, Heymanns J, et al. Breast cancer morbidity. Dtsch Arztebl Online. 2014. doi: 10.3238/arztebl.2014.0537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Z, Xu C, Zhang X, et al. TRIM11 upregulation contributes to proliferation, invasion, and EMT of hepatocellular carcinoma cells. Oncol Res. 2017;25(5):691–699. doi: 10.3727/096504016X14774897404770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu J, Rao J, Lou X, Zhai J, Ni Z, Wang X. Upregulated TRIM11 exerts its oncogenic effects in hepatocellular carcinoma through inhibition of P53. Cell Physiol Biochem. 2017;44(1):255–266. doi: 10.1159/000484678 [DOI] [PubMed] [Google Scholar]
- 6.Yin Y, Liu L, Zhong J, et al. TRIM11, a direct target of miR-24-3p, promotes cell proliferation and inhibits apoptosis in colon cancer. Oncotarget. 2016;7(52):86755–86765. doi: 10.18632/oncotarget.13550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X, Shi W, Shi H, et al. TRIM11 overexpression promotes proliferation, migration and invasion of lung cancer cells. J Exp Clin Cancer Res. 2016;35(1):100. doi: 10.1186/s13046-016-0379-y [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Ra C, Is H, Tw M. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981 [DOI] [PubMed] [Google Scholar]
- 9.Oh S, Kim H, Nam K, Shin I. Silencing of Glut1 induces chemoresistance via modulation of Akt/GSK-3beta/beta-catenin/survivin signaling pathway in breast cancer cells. Arch Biochem Biophys. 2017;636:110–122. doi: 10.1016/j.abb.2017.08.009 [DOI] [PubMed] [Google Scholar]
- 10.Oh S, Kim H, Nam K, Shin I. Glut1 promotes cell proliferation, migration and invasion by regulating epidermal growth factor receptor and integrin signaling in triple-negative breast cancer cells. BMB Rep. 2017;50(3):132–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Venturelli L, Nappini S, Bulfoni M, et al. Glucose is a key driver for GLUT1-mediated nanoparticles internalization in breast cancer cells. Sci Rep. 2016;6:21629. doi: 10.1038/srep21629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9(8):550–562. doi: 10.1038/nrc2664 [DOI] [PubMed] [Google Scholar]
- 13.Saal LH, Johansson P, Holm K, et al. Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proc Natl Acad Sci U S A. 2007;104(18):7564–7569. doi: 10.1073/pnas.0702507104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao X, Lu C, Chu W, et al. MicroRNA-124 suppresses proliferation and glycolysis in non-small cell lung cancer cells by targeting AKT-GLUT1/HKII. Tumour Biol. 2017;39(5):1010428317706215. doi: 10.1177/1010428317706215 [DOI] [PubMed] [Google Scholar]
- 15.Curtis C, Shah SP, Chin SF, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352. doi: 10.1038/nature10983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Di K, Linskey ME, Bota DA. TRIM11 is overexpressed in high-grade gliomas and promotes proliferation, invasion, migration and glial tumor growth. Oncogene. 2013;32(42):5038–5047. doi: 10.1038/onc.2012.531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y, Sun J, Ma J. Proliferation and invasion of ovarian cancer cells are suppressed by knockdown of TRIM11. Oncol Lett. 2017;14(2):2125–2130. doi: 10.3892/ol.2017.6432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dai X, Cheng H, Bai Z, Li J. Breast cancer cell line classification and its relevance with breast tumor subtyping. J Cancer. 2017;8(16):3131–3141. doi: 10.7150/jca.18457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roberts DJ, Miyamoto S. Hexokinase II integrates energy metabolism and cellular protection: akting on mitochondria and TORCing to autophagy. Cell Death Differ. 2015;22(2):248–257. doi: 10.1038/cdd.2014.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thamrongwaranggoon U, Seubwai W, Phoomak C, et al. Targeting hexokinase II as a possible therapy for cholangiocarcinoma. Biochem Biophys Res Commun. 2017;484(2):409–415. doi: 10.1016/j.bbrc.2017.01.139 [DOI] [PubMed] [Google Scholar]
- 21.Sato-Tadano A, Suzuki T, Amari M, et al. Hexokinase II in breast carcinoma: a potent prognostic factor associated with hypoxia-inducible factor-1alpha and Ki-67. Cancer Sci. 2013;104(10):1380–1388. doi: 10.1111/cas.12238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Younes M, Brown RW, Mody DR, Fernandez L, Laucirica R. GLUT1 expression in human breast carcinoma: correlation with known prognostic markers. Anticancer Res. 1995;15(6B):2895–2898. [PubMed] [Google Scholar]
- 23.Zhao F, Ming J, Zhou Y, Fan L. Inhibition of Glut1 by WZB117 sensitizes radioresistant breast cancer cells to irradiation. Cancer Chemother Pharmacol. 2016;77(5):963–972. doi: 10.1007/s00280-016-3007-9 [DOI] [PubMed] [Google Scholar]
- 24.Guerrero-Zotano A, Mayer IA, Arteaga CL. PI3K/AKT/mTOR: role in breast cancer progression, drug resistance, and treatment. Cancer Metastasis Rev. 2016;35(4):515–524. doi: 10.1007/s10555-016-9637-x [DOI] [PubMed] [Google Scholar]
- 25.Phadngam S, Follo C, Castiglioni A, Isidoro C. PTEN dephosphorylates AKT to prevent the expression of GLUT1 on plasmamembrane and to limit glucose consumption in cancer cells. Oncotarget. 2016;7(51):84999–85020. doi: 10.18632/oncotarget.13113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu H, Li X, Luo Z, Liu J, Fan Z. Cetuximab reverses the Warburg effect by inhibiting HIF-1-regulated LDH-A. Mol Cancer Ther. 2013;12(10):2187–2199. doi: 10.1158/1535-7163.MCT-12-1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wieman HL, Wofford JA, Rathmell JC. Cytokine stimulation promotes glucose uptake via phosphatidylinositol-3 kinase/Akt regulation of Glut1 activity and trafficking. Mol Biol Cell. 2007;18(4):1437–1446. doi: 10.1091/mbc.e06-07-0593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hou X, Liu Y, Liu H, et al. PERK silence inhibits glioma cell growth under low glucose stress by blockage of p-AKT and subsequent HK2’s mitochondria translocation. Sci Rep. 2015;5:9065. doi: 10.1038/srep09065 [DOI] [PMC free article] [PubMed] [Google Scholar]