Short abstract
IL‐4 induced IL‐10‐producing CD8+ T cells possess suppressive function both in vitro and in vivo.
Keywords: differentiation, transcription factor, suppression
Abstract
CD8+ T cells play an important role in immune regulation and effective immune responses against tumor cells, viral infection, and intracellular pathogens. In this report, using tiger or 10BiT mice, we defined a population of IL‐10‐producing CD8+ T cells that were induced by IL‐4. These IL‐10+CD8+ T cells possessed a strong inhibitory effect on the CD4+ T cell proliferation in an IL‐10‐dependent and cell contact‐dependent fashion. In comparison with IL‐10−CD8+ T cells, IL‐10+CD8+ T cells expressed an array of Th2‐like cytokines (IL‐4, IL‐5), perforin, and granzymes, as well as the cell cycle regulatory protein Cdkn2a. Interestingly, knockdown of cdkn2a using siRNA reduced IL‐4‐induced IL‐10 production significantly. Furthermore, CD8+ T cells from Cdkn2a−/− mice produced a significantly lower amount of IL‐10, and the effect was limited to CD8+ T cells but not observed in CD4+ T cells and APCs. Finally, IL‐10+CD8+ T cells played a protective role in the TNBS‐induced murine colitis model, indicating a critical role of this population of CD8+ T cells in regulatory immune responses. Taken together, we have defined a population of IL‐10‐producing CD8+ Tregs induced by IL‐4 and mediated by Cdkn2a.
Abbreviations
- 10BiT
Il10 bacterial artificial chromosome in‐transgene mice
- Blimp1
B lymphocyte‐induced maturation protein‐1
- Cdkn2a
cyclin‐dependent kinase inhibitor 2a
- Cdkn2a−/− mice
B6.129‐cyclin‐dependent kinase inhibitor 2a tm1Rdp mice
- Foxp3
forkhead box P3
- GITR
glucocorticoid‐induced TNFR
- IBD
inflammatory bowel disease
- iTreg
inducible regulatory T cell
- N.C.
control small interfering RNA
- PD‐1
programmed death‐1
- rm
recombinant mouse
- siRNA
small interfering RNA
- Sp1/3
specificity protein 1/3
- Tc
cytotoxic T lymphocyte
- tiger
IL‐10 IRES GFP‐enhanced reporter mice
- TNBS
2,4,6‐trinitrobenzenesulfonic acid
- Treg
regulatory T cell
Introduction
Lymphocyte heterogeneity is required for optimal immune responses against pathogens and self‐homeostasis. Several functionally distinct subsets of CD8+ T cells have been defined. Traditional Tc1 and Tc2 CD8+ T cells play overlapping and different roles in tumor immunity [1], viral infection [2], and some allergic diseases [3]. Tc17 cells can be generated in vitro and in vivo and exert critical functions in tumor rejection and viral clearance [4, 5]. CD8+ Tregs play an important role in maintaining immune self‐homeostasis and resistance to autoimmune diseases. CD8+CD122+ Tregs prevent and cure naïve CD4+ T cell‐induced IBD [6]. Moreover, transferring CD8+CD28− T cells into CD8‐deficient mice can suppress development of experimental autoimmune encephalomyelitis [7]. CD8+ IL‐10‐producing T cells were found in several murine disease models, including coronavirus‐induced encephalitis [8], acute influenza virus infection [9], and Salmonella and Leishmania infection [10]. Also, they were found in human HIV‐1 infection [11] and chronic hepatitis C virus infection [12].
IL‐10 is a multifunctional cytokine produced by a variety of cell types, including Th2 cells, DCs, activated macrophages, B cells, and mast cells [13]. Recent reports have demonstrated that IL‐10 is indispensable for Treg function [14] and suppresses proinflammatory T cell immunity [15]. Several groups, including us, have established that IL‐10 is critical for maintaining the suppressive function of Tregs in arthritis [16] and colitis [17]. IL‐10 suppresses TNF‐α production by macrophages [18], Th1 cell cytokine production, and T cell proliferation [19]. IL‐10 restrains Th17 cell‐mediated pathology [20] and CD45RBlo cell‐mediated colitis [21]. Although Tregs [13] and type 1 Treg [22], macrophages [13], and various immune cells were found to be important sources of IL‐10, the function of IL‐10+CD8+ T cells in inflammation remains to be investigated.
A number of transcription factors involved in regulating IL‐10 expression in CD4+ T cells and CD8+ T cells have been defined. GATA‐3 [23] and Blimp1 [24] mainly regulate IL‐10 expression through a specific signal pathway in CD4+ or CD8+ T cells. Several transcription factors have been found to regulate IL‐10 expression through different mechanisms in macrophages or monocytes. c‐Maf [25], stat1 [26], and stat3 [27] have been reported to be involved in IL‐10 transcription regulation in the LPS signal transduction pathway, whereas Sp1 [28] or Sp3 [28] directly binds to some specific motifs of the IL‐10 promoter to alter IL‐10 mRNA levels. However, the comprehensive transcription networks that are responsible for IL‐10 production and their regulation by the cytokine environments are still unknown.
The cdkn2a locus in mouse encodes two distinct tumor‐suppressor proteins: p19ARF and p16INK4a [29]. p19ARF mainly regulates p53 in response to aberrant growth or oncogenic stresses, such as c‐Myc activation [29], whereas p16INK4a, which has been mutated or deleted in several tumor tissues [30, 31], plays an important role in regulating the cell cycle. Cdkn2a−/− mice [32] are susceptible to tumor generation and growth. However, whether p19ARF and p16INK4a have any roles in T cell differentiation is unclear.
The TNBS‐induced murine colitis is an experimental model to be used to study the pathogenesis and therapy of human IBD. In this model, a combination of TNBS and colonic proteins induces excessive production of IFN‐γ by Th1 cells, which then activates monocytes/macrophages to produce a series of cytokines and chemokines, such as TNF‐α and IL‐6 [33]. Down‐regulation of IL‐10 is also observed in this model, indicating an immunomodulatary role of cytokines in the pathogenesis of such an animal model [34].
In this report, we demonstrated that CD8+ T cells could be induced to produce IL‐10 in the presence of IL‐4 upon activation. These CD8+ Tregs suppressed CD4+ T cell proliferation in vitro through IL‐10‐ and cell contact‐dependent mechanisms. Furthermore, we determined that the cell cycle regulatory protein Cdkn2a controlled IL‐4‐induced IL‐10 production in CD8+ T cells. Finally, these CD8+ T cells played a protective role in TNBS‐induced murine colitis in vivo.
MATERIALS AND METHODS
Mice
10BiT mice, in which Thy1.1 expression is used to indicate IL‐10 gene expression [35], were generated by Dr. Casey Weaver from University of Alabama and kindly provided by Dr. Susan Kaech from Yale School of Medicine. Tiger mice, in which GFP‐positive cells are used to indicate cells expressing IL‐10 [10] and CD4dnIL‐10Rα mice [21], were kindly provided by Dr. Richard Flavell from Yale School of Medicine. OT‐1 mice (OVA257–264/H‐2Kb‐restricted) were crossed with 10BiT mice to obtain 10BiT.OT‐1 dual‐transgenic mice. Cdkn2a−/− mice [32] were purchased from the NCI Mouse Repository (National Cancer Institute, Frederick, MD, USA) and were crossed with 10BiT mice to obtain Cdkn2a−/−.10BiT mice. C57BL/6J (B6 WT) mice were purchased from the Academy of Military Medical Science (Beijing, China). All animals were maintained under specific pathogen‐free conditions at Nankai University. All experiments were performed in accordance with guidelines for animal care, created by Nankai University Experimental Animal Ethics Committee.
Reagents
rmIL‐2 and rmIL‐4 and OVA257–264 peptide (SIINFEKL) were purchased from R&D Systems (Minneapolis, MN, USA). rmGM‐CSF and rmM‐CSF were purchased from PeproTech (London, UK). Purified IFN‐γ mAb (clone XMG1.2) and purified CD210 mAb (clone 1B1.3a) were from eBioscience (San Diego, CA, USA). GolgiStop, anti‐mouse CD3 mAb (clone 145‐2C11) and anti‐mouse CD28 mAb (clone PV1) were purchased from BD Biosciences (San Jose, CA, USA). FITC‐conjugated anti‐mouse CD11c (clone N418), PE‐conjugated anti‐mouse Thy1.1 (clone OX‐7), PE‐conjugated anti‐mouse CD4 (clone RM4‐5), PE‐conjugated anti‐mouse CD69 (clone H1.2F3), PE‐conjugated anti‐mouse CD122 (clone 5H4), PE‐conjugated anti‐mouse CD152 (CTLA‐4; clone UC10‐4B9), PE‐conjugated anti‐mouse CD357 (GITR; clone DTA‐1), PE‐conjugated anti‐mouse/rat/human Foxp3 (clone 150D), PE‐conjugated anti‐mouse ICOS (clone 7E.17G9), PE‐conjugated anti‐mouse I‐A/I‐E (clone M5/114.15.2), PE‐conjugated anti‐mouse F4/80 (clone BM8), PE‐conjugated rat IgG2b, κ isotype control antibody (clone RTK4530), PE‐conjugated Rat Ig2a, κ isotype control antibody (clone RTK2758), PE‐conjugated mouse IgG1, κ isotype control antibody (clone 150D), allophycocyanin‐conjugated anti‐mouse CD8α (clone 53‐6.7), and allophycocyanin‐conjugated anti‐mouse Thy1.1 (OX‐7) were from BioLegend (San Diego, CA, USA). FITC‐conjugated anti‐mouse CD11b (clone M1/70) was from Sungene Biotech (Tianjin, China). PMA, ionomycin, CFSE, TNBS, Con A (C5275), and LPS were purchased from Sigma Chemical (St. Louis, MO, USA). Lipofectamine 2000 was purchased from Invitrogen (Carlsbad, CA, USA). Microarray analysis was performed by CapitalBio (Beijing, China).
CD8+ T cell differentiation
Naïve CD8+ T cells (CD8+CD62LhiCD44lo) were sorted from tiger or 10BiT mice and cultured with soluble anti‐CD3 (2 μg/ml) or Con A (2.5 μg/ml) together with APCs in the presence of IL‐4 (10 ng/ml) and IL‐2 (2 ng/ml) for 48 h. After expansion for 2 days, cells were restimulated with PMA (50 ng/ml) and ionomycin (1 μg/ml) for 6 h, and GFP+ or Thy1.1+ T cells were detected by flow cytometry. APCs were from bone marrow‐derived DCs [10] or fixed splenocytes treated with mitomycin C, as described previously [36].
CD4+ T cell differentiation
In Th2 conditions, naïve CD4+ T cells (2.5×106/ml), sorted from reporter mice, were cultured with immobilized anti‐CD3 (10 μg/ml), soluble anti‐CD28 (1 μg/ml), IL‐2 (2 ng/ml), IL‐4 (10 ng/ml), and anti‐IFN‐γ (10 μg/ml) for 48 h. After expansion for another 48 h, cells were restimulated for 6 h by PMA and ionomycin.
In Treg conditions, naïve CD4+ T cells (2.5×106/ml) were cultured with immobilized anti‐CD3 (10 μg/ml), soluble anti‐CD28 (1 μg/ml), IL‐2 (2 ng/ml), and TGF‐β (5 ng/ml) for 72 h [37].
Cell coculture assay
Naïve CD8+ T cells from 10BiT mice were activated with Con A together with APCs in the presence of IL‐2 and IL‐4 for 48 h. After expansion for 2 days, cells were restimulated with PMA and ionomycin as described above. Cells were sorted into CD8+Thy1.1+/Thy1.1− T cells (2.5×105/well), which were then cocultured separately with CFSE‐labeled, naïve CD4+ T cells (2.5×105/well) from B6 WT or CD4dnIL‐10Rα mice (the ratio was 1:1) in 96‐well flat‐bottom plates and mixed with APCs, IL‐2, and soluble anti‐CD3 (2 μg/ml) in the culture media. After 72 h, cells were harvested; the CFSE profiles of the CD4+ T cells were analyzed by flow cytometry.
A 96‐well Transwell plate with 0.4 μm pore size was also used (Corning, Corning, NY, USA) for the coculture as described above. CFSE‐labeled B6 WT naïve CD4+ T cells (2.5×105/well), mixed with APCs, IL‐2, and soluble anti‐CD3 (2 μg/ml) in the culture media, were placed in the lower chamber alone or together with sorted CD8+Thy1.1+T cells or with CD8+Thy1.1+T cells (2.5×105/well) in the upper chamber. Cells were cultured for 72 h, and CD4+ T cells in the lower chambers were collected and stained with PerCP‐Cy5.5‐conjugated anti‐mouse CD4, and CFSE dilution was analyzed by flow cytometry.
ELISA
Mouse IL‐10 ELISA kits were purchased from BioLegend, and ELISA was performed according to the manufacturerˈs protocol.
Real‐time PCR
Total RNA was extracted from cells using TRIzol Reagent (Invitrogen) and reverse‐transcribed by QuantScript RT Kit (Tiangen, Beijing, China). mRNA expression was quantified by real‐time PCR. SYBR PreMix HotMaster Taq (Tiangen) was used in real‐time PCR. The primer sequences used were listed in Supplemental Table 1.
Microarray analysis
For global gene expression analysis, sorted CD8+GFP+/GFP− T cells were lysed by TRIzol Reagent (Invitrogen). Microarray analysis was performed as described in the CapitalBio manual. Microarray data are available in the Gene Expression Omnibus database (Accession Number GSE37739; www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE37739).
siRNA transfection and detection
FAM‐conjugated siRNAs were synthesized by GenePharma (Shanghai, China). The specific siRNA sequences used were listed in Supplemental Table 2, and Lipofectamine 2000 reagent (Invitrogen) was used for transfection. siRNA or N.C. was transfected into mouse splenocytes at a final concentration of 20 nM. Transfection was performed together with cells culturing in the presence of Con A, IL‐2, and IL‐4 for 48 h, and then cells transfected with FAM‐conjugated siRNA (FAM+ cells) were sorted. Gene‐silencing efficiency was detected by real‐time PCR.
TNBS‐induced colitis
Colitis was induced by administration of TNBS in B6 WT mice on Day 0, as described previously [38]. Mice were i.v.‐injected with GFP+CD8+ (1×106 cells/mouse) or GFP−CD8+ (1×106 cells/mouse) T cells or PBS vehicle, 24 h before and 24 h after TNBS administration. Mice treated with 50% ethanol were used as a negative control. Mice were weighed daily and killed on Day 5. Colon tissues were harvested, fixed for H&E staining, and RNA‐extracted for IFN‐γ mRNA level analysis. Histopathological scoring rules of colon tissue sections were as described [39].
Flow cytometry
A BD FACSCalibur (BD Biosciences) instrument was used for flow cytometry analysis; the software was CellQuest Pro (BD Biosciences) or FlowJo (Tree Star, Ashland, OR, USA). BD FACSAria (BD Biosciences) was used for cell sorting; the software was FACSDiva (BD Biosciences).
Statistics
Statistical significance was evaluated by a two‐tailed unpaired Studentˈs t‐test using InStat version 3.06 software for Windows (GraphPad, San Diego, CA, USA).
RESULTS
IL‐4 promotes CD8+ T cell differentiation into IL‐10‐producing cells
To define the role of IL‐4 in CD8+ T cell differentiation, naïve (CD44loCD62Lhi) CD8+ T cells from tiger mice, in which GFP‐positive cells are used to indicate cells expressing IL‐10, were cultured with Con A or soluble anti‐CD3 in the presence of IL‐4. We demonstrated that IL‐4 induced a significantly higher number of GFP+CD8+ T cells under Con A and anti‐CD3 conditions (Fig. 1A). In our preliminary studies, this effect was IL‐4 dose‐dependent (data not shown). The induction of IL‐10 by IL‐4 was independent of IFN‐γ (Fig. 1B). Interestingly, using CD8+ T cells from 10BiT.OT‐1 transgenic mice, in which Thy1.1 expression is used to indicate IL‐10 gene expression, we determined that IL‐4 also induced antigen‐responding IL‐10 production by CD8+ T cells (Fig. 1C). To confirm the correlation between GFP and IL‐10 expression level, GFP+ and GFP−CD8+ T cells were sorted from the culture, and the expression level of IL‐10 was analyzed by real‐time PCR. Indeed, GFP+CD8+ T cells expressed a significantly higher level of IL‐10 (Fig. 1D), which was confirmed further by detection of IL‐10 protein levels using ELISA (Fig. 1E). Interestingly, under the same culture conditions, CD4+ T cells expressed a significantly lower level of IL‐10 (Fig. 1F), and these two reporter mice (tiger and 10BiT) showed similar results. Therefore, both of these reporter mice were used in our following studies. Our results present new evidence that CD8+ T cells could be induced to produce IL‐10 by IL‐4 upon activation.
Figure 1.
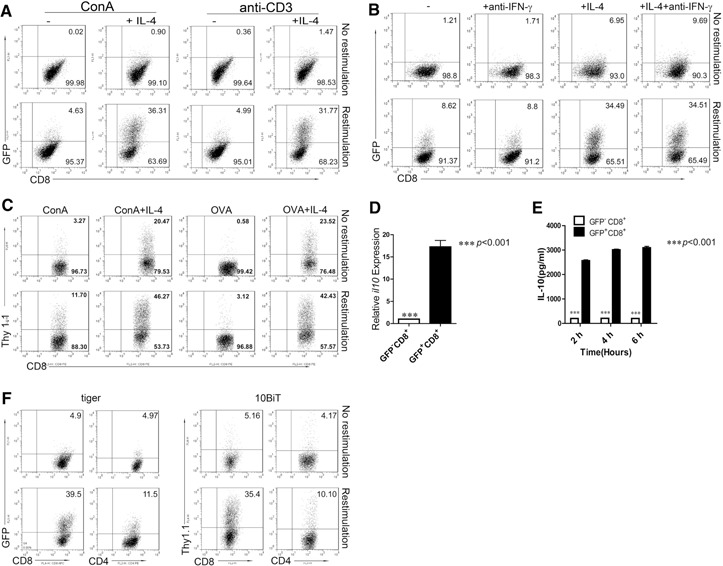
IL‐4 promotes CD8+ T cell differentiation into IL‐10‐producing cells. (A) Naïve CD8+ T cells from tiger mice were sorted, mixed with DCs (5:1 ratio), and stimulated with Con A (2.5 μg/ml) + IL‐2 (2 ng/ml) or activated with soluble anti‐CD3 (2 μg/ml) + IL‐2 in the absence (“−”) or presence of IL‐4 (+IL‐4; 10 ng/ml) for 48 h and then expanded for 2 days. Cells were then restimulated for 6 h with PMA and ionomycin and stained for CD8. One of three independent experiments is shown. (B) Naïve CD8+ T cells from tiger mice were stimulated with Con A + IL‐2 (“−”) in the presence or absence of IL‐4 or anti‐IFN‐γ (10 μg/ml) for 48 h and then expanded for 2 days. Cells were then restimulated for 6 h with PMA and ionomycin and stained for CD8. One of three independent experiments is shown. (C) Splenocytes from 10BiT.OT‐1 mice were stimulated with Con A or OVA257–264 (1 μg/ml) and IL‐2 in the presence or absence of IL‐4 for 48 h and then expanded for 2 days. Cells were then restimulated as described above and stained for CD8 and Thy1.1. One of three reproducible experiments is shown. (D) GFP+CD8+ T cells and GFP−CD8+ T cells from A were sorted for IL‐10 mRNA quantification by real‐time PCR. Data are shown as the mean + sem of three replicate measurements from three independent experiments. ***P < 0.001. (E) GFP+CD8+ T cells and GFP−CD8+ T cells were sorted and cultured without IL‐2. Supernatants were collected at different time‐points for IL‐10 ELISA. Data are shown as mean + sem of triplicate of three experiments performed. ***P < 0.001. (F) Naïve CD4+ and CD8+ T cells were sorted from tiger and 10BiT reporter mice, cultured as described above, and analyzed by flow cytometry. One representative experiment of three is shown. FL1/2‐H, Fluorescence 1/2‐height; APC, allophycocyanin.
Characterization of IL‐4‐induced, IL‐10‐producing CD8+ T cells
To characterize these IL‐4‐induced, IL‐10‐producing CD8+ T cells, a series of surface markers was analyzed by taking advantage of GFP+/GFP− CD8+ T cells (Fig. 2A). GFP+CD8+ T cells showed an activated phenotype, with higher expression of CD44, CD25, and CD69. These GFP+CD8+ T cells also expressed higher levels of ICOS and PD‐1. Moreover, these cells expressed higher levels of IL‐7R and IL‐15R, indicating a stronger ability for survival of these CD8+ T cells. Whereas these cells did not express Foxp3, no differences in the expression of GITR, CTLA4, and TGF‐β were observed between GFP+CD8+ T and GFP−CD8+ T cells.
Figure 2.
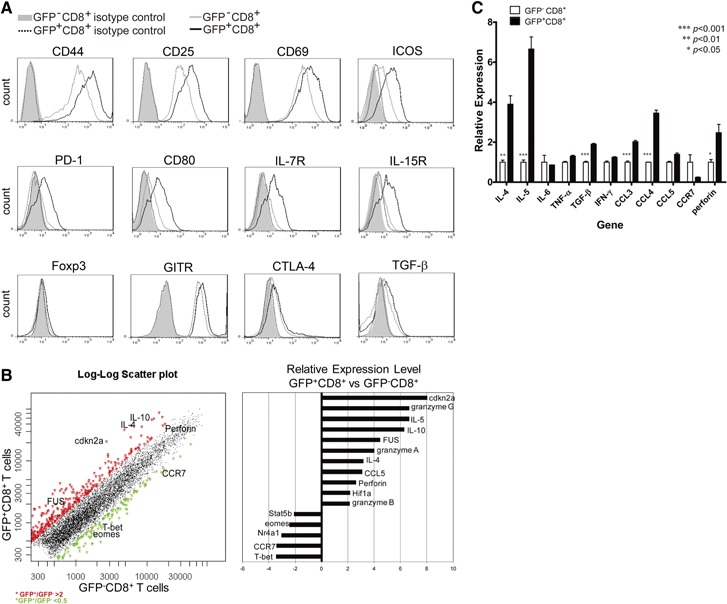
Characteristics of IL‐4‐induced, IL‐10‐producing CD8+ T cells. (A) Naïve CD8+ T cells from tiger mice were cultured as in Fig. 1, and GFP−CD8+ T cells (gray, solid‐line histograms) or GFP+CD8+ T cells (black, solid‐line histograms) were gated, and surface molecules, as indicated, were analyzed. Isotype‐matched control antibody staining for GFP−CD8+ T cells (gray‐filled histograms) or GFP+CD8+ T cells (black, dotted‐line histograms) were also shown. One typical staining of three is shown. (B) Gene expression analysis comparing GFP+CD8+ with GFP−CD8+ T cells from the above cultures. (C) GFP+CD8+ and GFP−CD8+ T cells were sorted from the above cultures, and cDNA was prepared for analyzing the expression levels of indicated genes. Data represent mean + sem of three combined experiments: *P < 0.05; **P < 0.01; ***P < 0.001. eomes, Eomesodermin; FUS, fused in sarcoma; T‐bet, T‐box expressed in T cells; Hif‐1a, hypoxia‐inducible factor 1α; Nr4a1, nuclear receptor subfamily 4 group A member 1.
To compare the broad gene profiles between these two types of CD8+ T cells under the identical conditions, GFP+ and GFP−CD8+ T cells were sorted, and cDNA microarray was performed. A series of functional genes and transcription factors was found to be differentially expressed between these two populations of CD8+ T cells (Fig. 2B). To confirm the expression profiles, cDNA samples prepared from these two cell populations were used for real‐time PCR analysis. IL‐10‐producing CD8+ T cells were confirmed to express higher levels of IL‐4, IL‐5, CCL3, CCL4, and perforin (Fig. 2C).
IL‐10‐producing CD8+ T cells suppress CD4+ T cell proliferation through IL‐10‐ and cell contact‐dependent mechanisms
Based on the fact that IL‐10 is a suppressive cytokine, we sought to determine the suppressive function of IL‐10‐producing CD8+ T cells. Naïve CD8+ T cells from 10BiT mice were activated with Con A together with APCs in the presence of IL‐2 and IL‐4 to induce Thy1.1+ expression. Cultured CD8+ T cells were sorted into Thy1.1+ and Thy1.1− cells, followed by the coculture experiment. The proliferation of naïve CD4+ T cells in the presence of Thy1.1+CD8+T cells were overlaid with those cultured with Thy1.1−CD8+ T cells. Indeed, Thy1.1+ CD8+ T cells dramatically suppressed proliferation of CD4+ T cells (Fig. 3A). Interestingly, addition of an anti‐IL‐10R antibody blocked the suppression (Fig. 3B). The suppression effect was also impaired when using CD4+ T cells isolated from CD4dnIL‐10Rα mice [21], in which CD4+ T cells express a high level of IL‐10R but without the cytoplasmic domain for signaling through these receptors as the responders (Fig. 3C). Our results thus demonstrate a critical role of IL‐10 in mediating the suppressive effect of these IL‐10‐producing CD8+ T cells.
Figure 3.
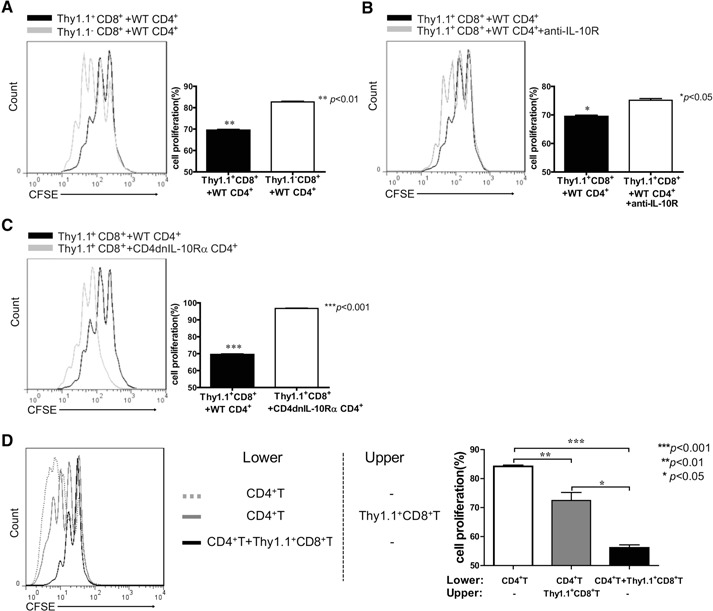
IL‐10‐producing CD8+ T cells suppress CD4+ T cell proliferation in IL‐10‐ and cell contact‐dependent mechanisms. (A) Naïve CD4+ T cells from B6 WT mice were sorted, labeled with CFSE, mixed with APCs (5:1 ratio), and stimulated with soluble anti‐CD3 (2 μg/ml) + IL‐2 (2 ng/ml) in the presence of sorted Thy1.1+CD8+ or Thy1.1−CD8+ T cells, produced as described in Materials and Methods. After 72 h, cells were harvested and stained for CD4. CFSE profiles of the CD4+ T cells were analyzed. The proliferation of naïve CD4+ T cells in the presence of Thy1.1+CD8+T cells (black‐line histogram) were overlaid with those cultured with Thy1.1−CD8+ T cells (gray‐line histogram). One representative experiment of three is shown. Cell proliferation was analyzed according to CFSE dilution. Data are representative of three independent experiments (mean+sem; **P<0.01). (B) In the parallel culture as described above (A), sorted Thy1.1+CD8+ T cells and responders were cultured in the presence (gray‐line histogram) or absence (black‐line histogram) of anti‐IL‐10R mAb. The black‐line histograms in A and B show the identical data that represent proliferation of naïve CD4+ T cells from B6 WT mice cocultured with Thy1.1+CD8+ T cells. One representative experiment of three is shown. Data represent three independent experiments (mean+sem; *P<0.05). (C) Responder cells from B6 WT (black‐line histogram) or CD4dnIL‐10Rα (gray‐line histogram) mice were cocultured with sorted Thy1.1+CD8+ T cells. One representative experiment of three is shown. Data represent three independent experiments (mean+sem; ***P<0.001). (D) Naïve CD4+ T cells (2.5×105/well) mixed with APCs (5:1 ratio), IL‐2, and soluble anti‐CD3 (2 μg/ml) were cultured in the lower chambers of transwell plates, and sorted Thy1.1+CD8+ T cells (2.5×105/well) were cultured in the lower (black, solid‐line histogram) or upper chambers (gray, solid‐line histogram). The gray dotted‐line histogram is the control of naïve CD4+ T cells without effector cells. One typical experiment of three is shown. Data represent three independent experiments (mean+sem; *P<0.05; **P<0.01; ***P<0.001).
To further determine whether the suppressive effect of IL‐10‐producing CD8+ T cells was cell contact‐dependent, transwell plates were used to separate the effector cells and responders. The separation indeed reduced the suppression effect, indicating a contact‐dependent mechanism (Fig. 3D). Overall, IL‐10‐producing CD8+ T cells suppressed CD4+ T cell proliferation through IL‐10‐ and cell contact‐dependent mechanisms.
Cdkn2a is a critical factor in mediating the IL‐4‐induced IL‐10 expression
To define the key transcription factors that might mediate the induction of IL‐10 by IL‐4, several siRNA sequences were screened to knock down those transcription factors that were up‐regulated in GFP+CD8+ T cells based on the cDNA microarray results (Fig. 2B). The efficiency of these siRNAs reached >70%, as confirmed by real‐time PCR (Fig. 4A). Strikingly, only knockdown of cdkn2a and blimp1 significantly reduced IL‐4‐induced IL‐10 expression (Fig. 4B). To examine whether Cdkn2a and blimp1 had any interaction, the expression of blimp1 or Cdkn2a in the opposite siRNA‐treated cells was analyzed. Interestingly, no direct effect on the expression level of IL‐10 induced by one factor was caused by the opposite factor (Fig. 4C).
Figure 4.
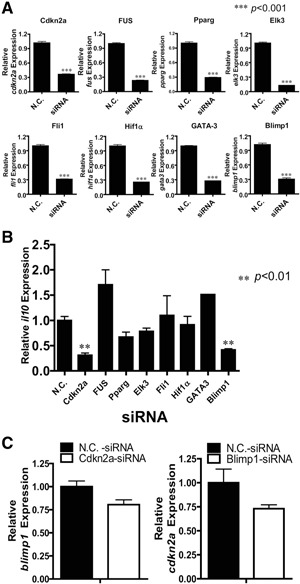
Cdkn2a is critical for IL‐4‐induced IL‐10 production. (A) Splenocytes from B6 WT mice were transfected with FAM‐conjugated siRNAs and cultured under the Con A + IL‐2 + IL‐4 condition for 48 h; cells transfected with siRNA (FAM+ cells) were sorted and lysed for real‐time PCR to detect the expression level of the indicated gene. The cDNA was also used to analyze the mRNA levels of IL‐10 by real‐time PCR (B). Data are shown as mean + sem of three independent experiments performed; **P < 0.01; ***P < 0.001. (C) Splenocytes from B6 WT mice were transfected with Cdkn2a or blimp1 siRNA and cultured with Con A + IL‐2 + IL‐4 for 48 h; FAM+ cells were sorted for real‐time PCR analysis of blimp1 and Cdkn2a, respectively. Data are representative of at least three independent experiments (mean+sem). No statistical difference was observed. Pparg, Peroxisome proliferator‐activated receptor γ; Elk3, a member of ETS oncogene family; Fli1, Friend leukemia integration 1 transcription factor.
Cdkn2a significantly regulates the IL‐4‐induced IL‐10 production in CD8+ T cells
To further study the role of Cdkn2a in IL‐4‐induced IL‐10 expression, naïve CD8+ T cells were sorted from B6 WT and Cdkn2a−/− mice, and we demonstrated by real‐time PCR that CD8+ T cells expressed significantly less IL‐10 in the absence of Cdkn2a (Fig. 5A). This conclusion was confirmed further by detection of IL‐10 protein levels using ELISA (Fig. 5B). Interestingly, IL‐10 expression by iTregs and Th2‐differentiated CD4+ T cells was not affected (Fig. 5C and D). Moreover, IL‐10 expression from splenic macrophages and CD11c+ DCs triggered by LPS was also not affected (Fig. 5E), and the same result was also obtained from the bone marrow‐derived DCs and macrophages (data not shown). Taken together, our results thus define a unique role for Cdkn2a in CD8+ T cell differentiation toward IL‐10 production induced by IL‐4.
Figure 5.
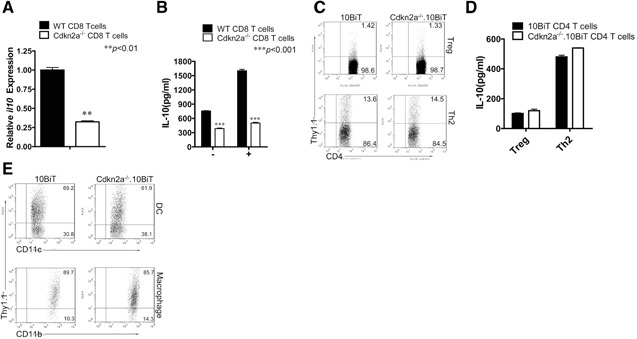
Cdkn2a is required for IL‐10 expression only in CD8+ but not CD4+ T cells, macrophages, and DCs. (A) Naïve CD8+ T cells from B6 WT (black bar) and Cdkn2a−/− (white bar) mice were sorted and stimulated with immobilized anti‐CD3 (10 μg/ml) and soluble anti‐CD28 (1 μg/ml) in the presence of IL‐2 and IL‐4 for 48 h, expanded 2 days. Cells were then restimulated by PMA and ionomycin for 6 h and then analyzed IL‐10 expression by real‐time PCR. Data are representative of three independent experiments (mean+sem; **P<0.01). (B) Supernatants from the parallel, naïve CD8+ T cell, cultured in A at 96 h (“−”, no restimulation) and 6 h after restimulation (“+”), were collected for IL‐10 detection by ELISA. Results from three independent experiments were combined and presented (mean+sem; ***P<0.001). (C) Naïve CD4+ T cells were sorted from 10BiT and Cdkn2a−/−.10BiT mice, cultured under iTreg conditions or Th2 conditions, as described in Materials and Methods. Cells were then analyzed by flow cytometry. Data were representative of three independent experiments. (D) Supernatants from iTreg and Th2 cultures of C were collected and used for IL‐10 detection by ELISA. Data (mean+sem) from three combined experiments were shown. No statistical difference was observed. (E) Splenocytes from 10BiT mice or Cdkn2a−/−.10BiT mice cultured with LPS (1 μg/ml) for 24 h. Cells were then analyzed by flow cytometry. Data are representative of three independent experiments.
IL‐10‐producing CD8+ T cells play a protective role in a TNBS‐induced murine colitis model
To define the regulatory role of IL‐10‐producing CD8+ T cells in vivo, a TNBS‐induced colitis model was adapted with different populations of CD8+ T cells transferred at different time‐points of disease induction, as described in Materials and Methods. Mice injected with GFP+CD8+ T cells recovered their body weight more rapidly than mice treated with GFP−CD8+ T cells or PBS at the later stage of colitis (Days 4 and 5; Fig. 6A). The reduced inflammation in the GFP+CD8+ T cell‐treated group was also confirmed by colon histopathology (Fig. 6B). With the use of a standard scoring system, the GFP+CD8+ T cell‐treated group reduced the severity of the disease significantly (Fig. 6C). Finally, colon lengths of GFP+CD8+ T cell‐treated mice were also obviously longer than those in the GFP−CD8+ T cells or PBS‐injected groups and had no significant differences from those in the control group (Fig. 6D). These results indicate that IL‐10‐producing CD8+ T cells play a protective role in the TNBS‐induced murine colitis model.
Figure 6.
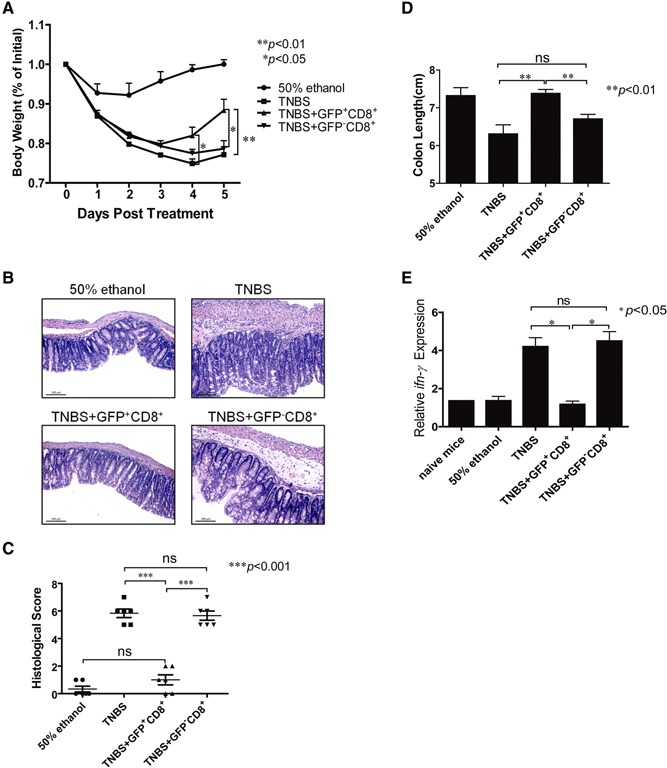
IL‐10‐producing CD8+ T cells play a protective role in TNBS‐induced murine colitis model. Sex‐ and age‐matched B6 WT mice were intrarectally administrated with TNBS (130 mg/kg body weight; n=11 for each group). Mice were transferred with GFP+CD8+ (1×106 cells/mouse), GFP−CD8+ (1×106 cells/mouse), or PBS vehicle through i.v. injection at 24 h prior to and 24 h post‐TNBS administration. Mice treated with 50% ethanol were used as negative control. (A) Mice were weighed daily after treatment for 5 days. One representative of three experiments was shown (mean+sem; *P<0.05;**P<0.01). (B and C) Mice were killed at Day 5 post‐TNBS induction. Colon tissues from four groups of mice were fixed for H&E staining to determine disease severity. Representative sections were shown in B, and histopathological scores were shown in C (mean±sem; ***P<0.001). Tissue sections were observed by Leica DM3000, and photos were obtained by the Leica Application Suite V3 software. Data were representative of three independent experiments (original magnification, ×200; original scale bars=100 μm). (D) Colon lengths were compared among different groups at Day 5 after treatment. One representative experiment of three was shown (mean+sem; **P<0.01). (E) RNA was extracted from colon tissues, and the cDNA was then prepared for analyzing the expression levels of ifn‐γ among different groups. One representative experiment of three was shown (mean+sem; *P<0.05).
To investigate further the mechanisms underlying the protective role of IL‐10‐producing CD8+ T cells in TNBS‐induced colitis model, we analyzed IFN‐γ mRNA expression levels in colon tissues among different groups. The results showed that IFN‐γ mRNA levels in the GFP+CD8+ T cell‐treated group were significantly lower than that of GFP−CD8+ T cells or PBS groups, which were similar to the control group and naïve mice (Fig. 6E). Taken together, IL‐10‐producing CD8+ T cells possibly protect mice from TNBS‐induced colitis through inhibition of IFN‐γ production.
DISCUSSION
CD8+ T cells play an important role in protective immune responses against intracellular viral, bacterial infection, and tumor cells [1, 2, 40, 41, 42]. Similar to CD4+ T cells, CD8+ T cells have divergent, functional subsets, according to their cytokine profiles, including Tc1, Tc2, Tc17, and Tregs [43, 44]. However, less attention has been paid to the study of differentiation and transcriptional‐controlling mechanisms of CD8+ Tregs. In this study, we observed that CD8+ T cells were induced to produce IL‐10 by IL‐4 and controlled by the cell cycle regulatory protein Cdkn2a.
IL‐4 is a critical cytokine in the induction of CD4+ T cell Th2 differentiation, and these CD4+ T cells produce IL‐4, IL‐5, IL‐13, and IL‐10 cytokines [45, 46]. It has also been reported that together with IL‐12, IL‐4 also induces IL‐10 production by CD8+ T cells [47]. Interestingly, we demonstrated that together with IL‐2, IL‐4 was sufficient to induce naïve CD8+ T cells to differentiate into IL‐10‐producing cells (Fig. 1). Either IL‐2 alone or IL‐4 alone failed to induce significant levels of IL‐10 (data not shown). In our preliminary experiments, cells cultured in the absence of IL‐2 failed to survive and grow. We therefore speculated that IL‐2 served as the growth and surviving factor in our system, and IL‐4 played a critical role in inducing IL‐10 production by CD8+ T cells. Also, this process was dependent on TCR signaling‐mediated activation, as even cytokine alone in the absence of T cell activation could lead to cell death (data not shown). Furthermore, this process was not affected by the presence of endogenous IFN‐γ, as neutralizing IFN‐γ did not change the expression level of IL‐10 (Fig. 1). Our results thus provide an additional pathway for inducing IL‐10 production by CD8+ T cells.
One of the key findings in our study was the identification of the specific role of Cdkn2a in CD8+ T cell differentiation, especially in IL‐4‐induced IL‐10 secretion. Although quite a few signaling factors were up‐regulated upon IL‐4 treatment, only knockdown of cdkn2a and blimp1 suppressed IL‐10 expression significantly (Fig. 4), and it was evident that these two pathways had no direct interaction (Fig. 4). The critical role of Cdkn2a was confirmed further using Cdkn2a−/− mice, and the effect was restricted to CD8+ T cells but was not identified in CD4+ T cells or APCs (Fig. 5). Interestingly, knockdown of gata3 failed to affect IL‐4‐induced IL‐10 expression (Fig. 4), implying that Cdkn2a is not downstream of GATA3. It is unclear whether Cdkn2a is downstream of phosphorylated Stat6 or directly downstream of TCR signaling. To the best of our knowledge, this is the first report to define the role of this cell cycle regulatory protein in CD8+ T cell differentiation, and further studies are required to explore the function of Cdkn2a in other cytokine‐induced differentiation of CD8+ T cells and their detailed signaling pathways.
Defining the function of IL‐10‐producing T cells is a challenge. Recently, several reporter mice have been made to use GFP or Thy1.1 to indicate IL‐10‐expressing cells, such as tiger mice [10] and 10BiT mice [35]. By taking advantage of IL‐10 GFP or Thy1.1 reporter mice, we demonstrated that IL‐10‐producing CD8+ T cells possessed a strong, suppressive function in vitro via IL‐10‐ and cell contact‐dependent mechanisms (Fig. 3) and in vivo through suppressing IFN‐γ production (Fig. 6). Interestingly, these IL‐4‐induced, IL‐10‐producing CD8+ T cells expressed suppressing molecules (PD‐1, CTLA‐4), as well as a high level of Tc2 cytokines (Fig. 2). Several subsets of CD8+ Tregs have been identified, such as CD8+CD122+ Tregs [6], CD8+CD28− T cells [7], and Qa‐1‐restricted CD8+ T cells [48]. It is interesting to define whether this population of IL‐10‐producing CD8+ T cells is a new subset of CD8+ Tregs. Further validation would be needed to study the gene expression profiles and functions of these IL‐4‐induced, IL‐10‐producing CD8+ T cells.
It has been shown that IL‐10 suppresses Th1‐mediated inflammation in several animal models [49, 50, 51, 52]. TNBS‐induced colitis is a typical Th1‐mediated inflammation [53]. We demonstrated a protective role of these IL‐10‐producing CD8+ T cells in this colitis model (Fig. 6). Interestingly, colon tissue in mice injected with IL‐10+CD8+ T cells showed a significantly decreased level of IFN‐γ (Fig. 6), implying a possible mechanism of these IL‐10+CD8+ T cells in TNBS‐induced colitis through suppressing the Th1 response. Further studies are in progress to test the detailed molecular mechanisms of this population of CD8+ Tregs in the TNBS‐induced colitis model and its function in other inflammatory models.
In summary, we have defined a population of CD8+ Tregs that was induced by IL‐4 and controlled by the cell cycle regulatory protein Cdkn2a, which is the first time it was reported to be involved in T cell differentiation. These IL‐10‐producing CD8+ T cells played a protective role in TNBS‐induced colitis. Further studies to understand the pathophysiological function of this subset of CD8+ T cells will enable us to facilitate the effective CD8+ T cells in protective immune responses against pathogens and tumors.
AUTHORSHIP
Y.Z. and H.Z. designed the project, performed experiments, collected and analyzed the data, and wrote the manuscript. Y.S. performed mouse model and real‐time PCR. J.H. helped to design the project and write the manuscript. X.Q. performed cell culture and arranged the figures. X.Z. performed flow cytometry analysis. Z.W., P.W., and Z. Yao supervised and coordinated this work. S.M.K., C.T.W., and R.A.F. kindly provided transgenic mice. Z. Yin and L.Z. designed the overall research and helped to write the manuscript. All authors have read, commented on, and approved the final manuscript.
DISCLOSURES
The authors have no financial or commercial conflicts of interest.
Supporting information
Supplementary data
ACKNOWLEDGMENTS
This work was supported by grants from the National Science Foundation of China (31170858 and 30007101), International S&T Cooperation Program of China (2010DFB34000) and was supported by 111 project (B08011).
We thank Dr. Mark Bartlam for manuscript revision.
Footnotes
SEE CORRESPONDING EDITORIAL ON PAGE 1097
Contributor Information
Liqing Zhao, Email: yinzn@nankai.edu.cn.
Zhinan Yin, Email: yinzn@nankai.edu.cn.
REFERENCES
- 1. Dobrzanski, M. J. , Reome, J. B. , Hollenbaugh, J. A. , Dutton, R. W. (2004) Tc1 and Tc2 effector cell therapy elicit long‐term tumor immunity by contrasting mechanisms that result in complementary endogenous type 1 antitumor responses. J. Immunol. 172, 1380–1390. [DOI] [PubMed] [Google Scholar]
- 2. Cerwenka, A. , Morgan, T. M. , Harmsen, A. G. , Dutton, R. W. (1999) Migration kinetics and final destination of type 1 and type 2 CD8 effector cells predict protection against pulmonary virus infection. J. Exp. Med. 189, 423–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gluck, J. , Rogala, B. , Rogala, E. , Oles, E. (2007) Allergen immunotherapy in intermittent allergic rhinitis reduces the intracellular expression of IL‐4 by CD8+ T cells. Vaccine 26, 77–81. [DOI] [PubMed] [Google Scholar]
- 4. Garcia‐Hernandez Mde, L. , Hamada, H. , Reome, J. B. , Misra, S. K. , Tighe, M. P. , Dutton, R. W. (2010) Adoptive transfer of tumor‐specific Tc17 effector T cells controls the growth of B16 melanoma in mice. J. Immunol. 184, 4215–4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Intlekofer, A. M. , Banerjee, A. , Takemoto, N. , Gordon, S. M. , Dejong, C. S. , Shin, H. , Hunter, C. A. , Wherry, E. J. , Lindsten, T. , Reiner, S. L. (2008) Anomalous type 17 response to viral infection by CD8+ T cells lacking T‐bet and eomesodermin. Science 321, 408–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Endharti, A. T. , Okuno, Y. , Shi, Z. , Misawa, N. , Toyokuni, S. , Ito, M. , Isobe, K. , Suzuki, H. (2011) CD8 + CD122+ regulatory T cells (Tregs) and CD4+ Tregs cooperatively prevent and cure CD4+ cell‐induced colitis. J. Immunol. 186, 41–52. [DOI] [PubMed] [Google Scholar]
- 7. Najafian, N. , Chitnis, T. , Salama, A. D. , Zhu, B. , Benou, C. , Yuan, X. , Clarkson, M. R. , Sayegh, M. H. , Khoury, S. J. (2003) Regulatory functions of CD8 + CD28‐ T cells in an autoimmune disease model. J. Clin. Invest. 112, 1037–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Trandem, K. , Zhao, J. , Fleming, E. , Perlman, S. (2011) Highly activated cytotoxic CD8 T cells express protective IL‐10 at the peak of coronavirus‐induced encephalitis. J. Immunol. 186, 3642–3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sun, J. , Madan, R. , Karp, C. L. , Braciale, T. J. (2009) Effector T cells control lung inflammation during acute influenza virus infection by producing IL‐10. Nat. Med. 15, 277–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kamanaka, M. , Kim, S. T. , Wan, Y. Y. , Sutterwala, F. S. , Lara‐Tejero, M. , Galan, J. E. , Harhaj, E. , Flavell, R. A. (2006) Expression of interleukin‐10 in intestinal lymphocytes detected by an interleukin‐10 reporter knockin tiger mouse. Immunity 25, 941–952. [DOI] [PubMed] [Google Scholar]
- 11. Elrefaei, M. , Barugahare, B. , Ssali, F. , Mugyenyi, P. , Cao, H. (2006) HIV‐specific IL‐10‐positive CD8+ T cells are increased in advanced disease and are associated with decreased HIV‐specific cytolysis. J. Immunol. 176, 1274–1280. [DOI] [PubMed] [Google Scholar]
- 12. Abel, M. , Sene, D. , Pol, S. , Bourliere, M. , Poynard, T. , Charlotte, F. , Cacoub, P. , Caillat‐Zucman, S. (2006) Intrahepatic virus‐specific IL‐10‐producing CD8 T cells prevent liver damage during chronic hepatitis C virus infection. Hepatology 44, 1607–1616. [DOI] [PubMed] [Google Scholar]
- 13. Couper, K. N. , Blount, D. G. , Riley, E. M. (2008) IL‐10: the master regulator of immunity to infection. J. Immunol. 180, 5771–5777. [DOI] [PubMed] [Google Scholar]
- 14. Liu, H. , Hu, B. , Xu, D. , Liew, F. Y. (2003) CD4 + CD25+ regulatory T cells cure murine colitis: the role of IL‐10, TGF‐β, and CTLA4. J. Immunol. 171, 5012–5017. [DOI] [PubMed] [Google Scholar]
- 15. Netea, M. G. , Sutmuller, R. , Hermann, C. , van der Graaf, C. A. , van der Meer, J. W. , van Krieken, J. H. , Hartung, T. , Adema, G. , Kullberg, B. J. (2004) Tolllike receptor 2 suppresses immunity against Candida albicans through induction of IL‐10 and regulatory T cells. J. Immunol. 172, 3712–3718. [DOI] [PubMed] [Google Scholar]
- 16. Min, S. Y. , Hwang, S. Y. , Park, K. S. , Lee, J. S. , Lee, K. E. , Kim, K. W. , Jung, Y. O. , Koh, H. J. , Do, J. H. , Kim, H. , Kim, H. Y. (2004) Induction of IL‐10‐producing CD4 + CD25+ T cells in animal model of collagen‐induced arthritis by oral administration of type II collagen. Arthritis Res. Ther. 6, R213–R219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Murai, M. , Turovskaya, O. , Kim, G. , Madan, R. , Karp, C. L. , Cheroutre, H. , Kronenberg, M. (2009) Interleukin 10 acts on regulatory T cells to maintain expression of the transcription factor Foxp3 and suppressive function in mice with colitis. Nat. Immunol. 10, 1178–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fiorentino, D. F. , Zlotnik, A. , Mosmann, T. R. , Howard, M. , O'Garra, A. (1991) IL‐10 inhibits cytokine production by activated macrophages. J. Immunol. 147, 3815–3822. [PubMed] [Google Scholar]
- 19. Fiorentino, D. F. , Zlotnik, A. , Vieira, P. , Mosmann, T. R. , Howard, M. , Moore, K. W. , O'Garra, A (1991) IL‐10 acts on the antigen‐presenting cell to inhibit cytokine production by Th1 cells. J. Immunol. 146, 3444–3451. [PubMed] [Google Scholar]
- 20. Huber, S. , Gagliani, N. , Esplugues, E. , O'Connor Jr., W. , Huber, F. J. , Chaudhry, A. , Kamanaka, M. , Kobayashi, Y. , Booth, C. J. , Rudensky, A. Y. , Roncarolo, M. G. , Battaglia, M. , Flavell, R. A. (2011) Th17 cells express interleukin‐10 receptor and are controlled by Foxp3 and Foxp3+ regulatory CD4+ T cells in an interleukin‐10‐dependent manner. Immunity 34, 554–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kamanaka, M. , Huber, S. , Zenewicz, L. A. , Gagliani, N. , Rathinam, C. , O'Connor, W., Jr. , Wan, Y. Y. , Nakae, S. , Iwakura, Y. , Hao, L. , Flavell, R. A. (2011) Memory/effector (CD45RB(lo)) CD4 T cells are controlled directly by IL‐10 and cause IL‐22‐dependent intestinal pathology. J. Exp. Med. 208, 1027–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Roncarolo, M. G. , Gregori, S. , Battaglia, M. , Bacchetta, R. , Fleischhauer, K. , Levings, M. K. (2006) Interleukin‐10‐secreting type 1 regulatory T cells in rodents and humans. Immunol. Rev. 212, 28–50. [DOI] [PubMed] [Google Scholar]
- 23. Shoemaker, J. , Saraiva, M. , O'Garra, A. (2006) GATA‐3 directly remodels the IL‐10 locus independently of IL‐4 in CD4+ T cells. J. Immunol. 176, 3470–3479. [DOI] [PubMed] [Google Scholar]
- 24. Cretney, E. , Xin, A. , Shi, W. , Minnich, M. , Masson, F. , Miasari, M. , Belz, G. T. , Smyth, G. K. , Busslinger, M. , Nutt, S. L. , Kallies, A. (2011) The transcription factors Blimp‐1 and IRF4 jointly control the differentiation and function of effector regulatory T cells. Nat. Immunol. 12, 304–311. [DOI] [PubMed] [Google Scholar]
- 25. Cao, S. , Liu, J. , Song, L. , Ma, X. (2005) The protooncogene c‐Maf is an essential transcription factor for IL‐10 gene expression in macrophages. J Immunol. 174, 3484–3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. VanDeusen, J. B. , Shah, M. H. , Becknell, B. , Blaser, B. W. , Ferketich, A. K. , Nuovo, G. J. , Ahmer, B. M. , Durbin, J. , Caligiuri, M. A. (2006) STAT‐1‐mediated repression of monocyte interleukin‐10 gene expression in vivo. Eur. J. Immunol. 36, 623–630. [DOI] [PubMed] [Google Scholar]
- 27. Chang, E. Y. , Guo, B. , Doyle, S. E. , Cheng, G. (2007) Cutting edge: involvement of the type I IFN production and signaling pathway in lipopolysaccharide‐induced IL‐10 production. J. Immunol. 178, 6705–6709. [DOI] [PubMed] [Google Scholar]
- 28. Tone, M. , Powell, M. J. , Tone, Y. , Thompson, S. A. , Waldmann, H. (2000) IL‐10 gene expression is controlled by the transcription factors Sp1 and Sp3. J. Immunol. 165, 286–291. [DOI] [PubMed] [Google Scholar]
- 29. Kim, W. Y. , Sharpless, N. E. (2006) The regulation of INK4/ARF in cancer and aging. Cell 127, 265–275. [DOI] [PubMed] [Google Scholar]
- 30. He, X. S. , Su, Q. , Chen, Z. C. , He, X. T. , Long, Z. F. , Ling, H. , Zhang, L. R. (2001) Expression, deletion [was deleton] and mutation of p16 gene in human gastric cancer. World. J. Gastroenterol. 7, 515–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ohtsubo, K. , Watanabe, H. , Yamaguchi, Y. , Hu, Y. X. , Motoo, Y. , Okai, T. , Sawabu, N. (2003) Abnormalities of tumor suppressor gene p16 in pancreatic carcinoma: immunohistochemical and genetic findings compared with clinicopathological parameters. J. Gastroenterol. 38, 663–671. [DOI] [PubMed] [Google Scholar]
- 32. Serrano, M. , Lee, H. , Chin, L. , Cordon‐Cardo, C. , Beach, D. , DePinho, R. A. (1996) Role of the INK4a locus in tumor suppression and cell mortality. Cell 85, 27–37. [DOI] [PubMed] [Google Scholar]
- 33. Elson, C. O. , Beagley, K. W. , Sharmanov, A. T. , Fujihashi, K. , Kiyono, H. , Tennyson, G. S. , Cong, Y. , Black, C. A. , Ridwan, B. W. , McGhee, J. R. (1996) Hapten‐induced model of murine inflammatory bowel disease. J. Immunol. 157, 2174–2185. [PubMed] [Google Scholar]
- 34. Fuss, I. J. , Boirivant, M. , Lacy, B. , Strober, W. (2002) The interrelated roles of TGF‐β and IL‐10 in the regulation of experimental colitis. J. Immunol. 168, 900–908. [DOI] [PubMed] [Google Scholar]
- 35. Maynard, C. L. , Harrington, L. E. , Janowski, K. M. , Oliver, J. R. , Zindl, C. L. , Rudensky, A. Y. , Weaver, C. T. (2007) Regulatory T cells expressing interleukin 10 develop from Foxp3+ and Foxp3‐recursor cells in the absence of interleukin 10. Nat. Immunol. 8, 931–941. [DOI] [PubMed] [Google Scholar]
- 36. Duan, X. , Yarmush, D. , Leeder, A. , Yarmush, M. L. , Mitchell, R. N. (2008) Burn‐induced immunosuppression: attenuated T cell signaling independent of IFN‐γ‐ and nitric oxide‐mediated pathways. J. Leukoc. Biol. 83, 305–313. [DOI] [PubMed] [Google Scholar]
- 37. Chen, W. , Jin, W. , Hardegen, N. , Lei, K. J. , Li, L. , Marinos, N. , McGrady, G. , Wahl, S. M. (2003) Conversion of peripheral CD4 + CD25‐naive T cells to CD4 + CD25+ regulatory T cells by TGF‐β induction of transcription factor Foxp3. J. Exp. Med. 198, 1875–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bai, A. , Lu, N. , Guo, Y. , Liu, Z. , Chen, J. , Peng, Z. (2009) All‐trans retinoic acid down‐regulates inflammatory responses by shifting the Treg/Th17 profile in human ulcerative and murine colitis. J. Leukoc Biol. 86, 959–969. [DOI] [PubMed] [Google Scholar]
- 39. Ito, R. , Shin‐Ya, M. , Kishida, T. , Urano, A. , Takada, R. , Sakagami, J. , Imanishi, J. , Kita, M. , Ueda, Y. , Iwakura, Y. , Kataoka, K. , Okanoue, T. , Mazda, O. (2006) Interferon‐γ is causatively involved in experimental inflammatory bowel disease in mice. Clin. Exp. Immunol. 146, 330–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cox, M. A. , Kahan, S. M. , Zajac, A. J. (2013) Anti‐viral CD8 T cells and the cytokines that they love. Virology 435, 157–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shanker, A. , Verdeil, G. , Buferne, M. , Inderberg‐Suso, E. M. , Puthier, D. , Joly, F. , Nguyen, C. , Leserman, L. , Auphan‐Anezin, N. , Schmitt‐Verhulst, A. M. (2007) CD8 T cell help for innate antitumor immunity. J. Immunol. 179, 6651–6662. [DOI] [PubMed] [Google Scholar]
- 42. Harty, J. T. , Schreiber, R. D. , Bevan, M. J. (1992) CD8 T cells can protect against an intracellular bacterium in an interferon γ‐independent fashion. Proc. Natl. Acad. Sci. USA 89, 11612–11616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hamada, H. , Garcia‐Hernandez Mde, L. , Reome, J. B. , Misra, S. K. , Strutt, T. M. , McKinstry, K. K. , Cooper, A. M. , Swain, S. L. , Dutton, R. W. (2009) Tc17, a unique subset of CD8 T cells that can protect against lethal influenza challenge. J. Immunol. 182, 3469–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mosmann, T. R. , Li, L. , Sad, S. (1997) Functions of CD8 T‐cell subsets secreting different cytokine patterns. Semin. Immunol. 9, 87–92. [DOI] [PubMed] [Google Scholar]
- 45. Zhu, J. , Yamane, H. , Paul, W. E. (2010) Differentiation of effector CD4 T cell populations. Annu. Rev. Immunol. 28, 445–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chen, L. , Grabowski, K. A. , Xin, J. P. , Coleman, J. , Huang, Z. , Espiritu, B. , Alkan, S. , Xie, H. B. , Zhu, Y. , White, F. A. , Clancy, J. Jr. , Huang, H. (2004) IL‐4 induces differentiation and expansion of Th2 cytokine‐producing eosinophils. J. Immunol. 172, 2059–2066. [DOI] [PubMed] [Google Scholar]
- 47. Noble, A. , Giorgini, A. , Leggat, J. A. (2006) Cytokine‐induced IL‐10‐secreting CD8 T cells represent a phenotypically distinct suppressor T‐cell lineage. Blood 107, 4475–4483. [DOI] [PubMed] [Google Scholar]
- 48. Kim, H. J. , Verbinnen, B. , Tang, X. , Lu, L. , Cantor, H. (2010) Inhibition of follicular T‐helper cells by CD8(+) regulatory T cells is essential for self tolerance. Nature 467, 328–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wilson, E. H. , Wille‐Reece, U. , Dzierszinski, F. , Hunter, C. A. (2005) A critical role for IL‐10 in limiting inflammation during toxoplasmic encephalitis. J. Neuroimmunol. 165, 63–74. [DOI] [PubMed] [Google Scholar]
- 50. Kullberg, M. C. , Jankovic, D. , Gorelick, P. L. , Caspar, P. , Letterio, J. J. , Cheever, A. W. , Sher, A. (2002) Bacteria‐triggered CD4(+) T regulatory cells suppress Helicobacter hepaticus‐mduced colitis. J. Exp. Med. 196, 505–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kossodo, S. , Monso, C. , Juillard, P. , Velu, T. , Goldman, M. , Grau, G. E. (1997) Interleukin‐10 modulates susceptibility in experimental cerebral malaria. Immunology 91, 536–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Redpath, S. , Angulo, A. , Gascoigne, N. R. , Ghazal, P. (1999) Murine cytomegalovirus infection down‐regulates MHC class II expression on macrophages by induction of IL‐10. J. Immunol. 162, 6701–6707. [PubMed] [Google Scholar]
- 53. Fichtner‐Feigl, S. , Fuss, I. J. , Preiss, J. C. , Strober, W. , Kitani, A. (2005) Treatment of murine Th1‐ and Th2‐mediated inflammatory bowel disease with NF‐κ B decoy oligonucleotides. J. Clin. Invest. 115, 3057–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data


