Short abstract
CD84 is a modulator of the amplitude and the quality of the TLR‐induced response in murine macrophages.
Keywords: RAW‐264.7, SLAM family, mutant, tyrosine, TLR4
Abstract
CD84 is 1 of the 9 SLAM family cell‐surface receptors involved in leukocyte activation. The CD84 ectodomain is highly glycosylated, and its cytoplasmic tail contains 2 copies of an ITSM, which can be phosphorylated. Here, we report that although mouse CD84 was present on all BM HSCs, its expression declined in developing thymic and BM lymphocytes. However, CD84 expression levels did increase significantly during the later maturation stages and were expressed abundantly on mature B and T cells. Among lymphocyte subsets, the highest expression was found on innate‐like lymphocytes; specifically, on NKT and marginal zone B cells. Splenic CD4+ TFH cells exhibited higher levels of CD84 compared with the other CD4+ T cell subsets. CD84 was expressed abundantly on monocytes, macrophages, granulocytes, and DCs. Moreover, as the function of CD84 in myeloid cells remains unknown, we focused on the role this receptor plays in mouse macrophage activation. Transfection of CD84 in RAW‐264.7 macrophages led to an increase in MAPK phosphorylation and NF‐κB activation upon LPS stimulation. Concomitantly, the presence of CD84 increased the LPS‐induced secretion of TNF‐α and MCP‐1 but lowered IL‐10 and IL‐6 production significantly. This modulatory effect was mediated by Y300 within the second ITSM of CD84. Additionally, CD84 knock‐down decreased TNF‐α and IL‐6 production in LPS‐activated BMDMs. Taken together, these results show that mouse CD84 is a pan‐leukocyte receptor, able to modulate signaling pathways downstream of TLR4, and regulates macrophage cell‐fate decisions and effector functions.
Abbreviations
- APC
allophycocyanin
- BMDC
bone marrow‐derived DC
- BMDM
bone marrow‐derived macrophage
- |gDcyt
lack of cytoplasmic tail
- EAT‐2
EWS/FLI1‐activated transcript‐2
- F
phenylalanine
- FL
full‐length
- H1‐M2
human domain 1–mouse domain 2
- HSC
hematopoietic stem cells
- ITSM
immunoreceptor tyrosine‐based switch motif
- M1‐H2
mouse domain 1–human domain 2
- mCD84.7
anti‐mouse CD84
- MFI
mean fluorescence intensity
- MZ
marginal zone
- POD
peroxidase
- RT
room temperature
- SAP
signaling lymphocytic activation molecule‐associated protein
- Sca‐1
stem cell antigen 1
- SEAP
secreted embryonic alkaline phosphatase
- SH2
Src homology 2
- siRNA
small interfering RNA
- SLAMF5
signaling lymphocytic activation molecule family member 5
- TFH
T follicular helper
- Treg
regulatory T cell
- Y
tyrosine
Introduction
Human and mouse CD84 (SLAMF5) are type I transmembrane glycoproteins that contain two extracellular Ig domains [1, 2]. Human CD84 is not only expressed on T and B cells but also on a variety of other leukocytes, including monocytes/macrophages, granulocytes, DCs, and mast cells [3, 4]. Expression of human CD84 is up‐regulated in distinct lymphocyte subsets, e.g., memory T and B cells, as well as TFH cells, suggesting that CD84 functionality may be restricted to specific stages of lymphocyte development [5, –, 7]. Moreover, CD84 has been found to be expressed on HSCs [8, 9].
CD84 belongs to the SLAM family of cell‐surface receptors, a structural subgroup of the Ig superfamily, most of which bind the cytoplasmic adaptors SAP (SH2D1A) and EAT‐2 (SH2D1B) [10, –, 12]. The genes encoding CD84 and the other members of the CD150 family are situated in a 400‐kb segment on human chromosome 1q23, and the homologous mouse genes are located on the syntenic region of mouse chromosome 1 [1, 2]. Members of the SLAM family are involved in the functional regulation of several immune cell types, including helper and cytotoxic T lymphocytes, NKs, and macrophages [11, 12]. Recent evidence indicates that members of this family of receptors and signaling intermediates are also involved in autoimmunity [13, –, 15].
These receptors share a common ectodomain organization consisting of a membrane‐proximal Ig constant domain and a membrane‐distal Ig variable domain. As has been shown with other SLAM members, the N‐terminal Ig domain of CD84 mediates homophilic adhesion in humans and mice [5, 16]. The crystal structure of human CD84 reveals a 2‐fold homophilic dimer similar to the reported homophilic dimer of the NK, T, and B cell antigen (NTB‐A), another member of the SLAM family [17].
The cytoplasmic domain of CD84, like several other members of the SLAM family, contains 2 copies of the ITSM (T‐I/V‐Y‐x‐x‐V/I), which is a docking site for SH2‐binding phosphatases and the adaptor molecules SAP and EAT‐2 [6, 18, –, 20]. The SH2D1A gene, which encodes SAP, is deleted or mutated in patients with X‐linked lymphoproliferative disease [21]. Human CD84 undergoes Y phosphorylation following CD84 ligation in lymphocytes and thereby, determines the specific recruitment of SAP and EAT‐2, an event that involves the Src kinase Lck [20]. Cross‐linking of human CD84 increases proliferative responses and IFN‐γ secretion of anti‐CD3 mAb‐stimulated human T cells, suggesting that CD84 regulates TCR‐mediated signaling [5, 20]. Recently, it has been shown that CD84 is required for optimal germinal center formation and T–B cell adhesion [22].
It has been demonstrated that some cell‐surface molecules are able to regulate TLR downstream signaling events, although the ability of CD84 to modulate these signals remains undetermined [23].
Thus, in the present study, we have characterized extensively the expression of mouse CD84 on hematopoietic cells. Moreover, as it is highly expressed on macrophages, we have also examined the hypothesis that CD84 can regulate LPS‐induced macrophage activation.
MATERIALS AND METHODS
Antibodies
The mCD84.7 mAb (IgG1 isotype) was generated in our laboratory by fusing NS1 myeloma cells with spleen cells obtained from Armenian hamsters, which were immunized 3 times with mouse CD84‐Fc fusion protein [16]. mAb mCD84.7 was purified using the Affi‐Gel Protein A MAPS II kit (Bio‐Rad, Hercules, CA, USA) from concentrated supernatants obtained from cultured hybridomas in INTEGRA CL 1000 flasks (Integra Biosciences AG, Switzerland). Purified mAb was biotinylated using biotinamidocaproate N‐hydroxysuccinimide ester (Sigma Chemical Co., St. Louis, MO, USA). Hamster IgG isotype was verified using an anti‐Armenian hamster IgG1 from BD PharMingen (San Diego, CA, USA) as a secondary antibody.
IgM FITC and IgD‐PE were obtained from Southern Biotechnology Associates, Ins. (Birmingham, AL, USA). The following anti‐mouse mAb were obtained from BD PharMingen: CD3‐FITC, CD4‐FITC, CD11c‐FITC, CD14‐FITC, CD21‐FITC, CD24‐FITC, CD41‐FITC, CD43‐FITC, CD45‐FITC, MHC‐II (I‐Ab)‐PE, CD3‐PE, CD5‐PE, CD8‐PE, CD11b‐PE, CD23‐PE, CD41‐PE, c‐kit‐PE, NK 1.1‐PE, Ter‐119‐PE, CD34‐FITC, GR‐1‐PE, GR‐1‐FITC, CD3‐APC, B220‐APC, Sca‐1‐APC, CXCR5‐biotinylated and Armenian hamster IgG1 as an isotype control. Streptavidin‐PE‐Cy5 was purchased from BD PharMingen. mAb c‐Kit‐FITC, CD19‐PE, and CD25‐PE were acquired from ImmunoTools (Friesoythe, Germany) and IL‐6R (CD126)‐PE from BioLegend (San Diego, CA, USA).
Mice
C57BL/6 mice were purchased from Charles River Laboratories (Saint‐Aubin‐lès‐Elbeuf, France) and used in experiments when they reached an age of 6–12 weeks. All procedures involving animals and their care were approved by the Ethics Committee of the University of Barcelona (Spain) and were conducted in compliance with institutional guidelines as well as with national (Generalitat de Catalunya decree 214/1997, DOGC 2450) and international (Guide for the Care and Use of Laboratory Animals, National Institutes of Health, 85‐23, 1985) laws and policies.
Cell culture
Thymocyte and splenocyte single‐cell suspensions were obtained by injecting culture medium in these tissues. BM cells were obtained by flushing culture media through femurs using a syringe with a 0.45‐mm‐diameter needle. Clusters within the marrow suspension were disaggregated by vigorous pipetting. Peritoneal cells were obtained by lavage of the peritoneum with 5 ml culture medium. All cells were resuspended in RPMI‐1640 (Gibco‐BRL, Paisley, UK) medium containing 10% FCS (Biological Industries, Kibbutz Beit Haemek, Israel) at 4°C until further processing. Typically, the tissues were processed within 1–3 h following surgical removal. Fresh blood was obtained by cardiac puncture using heparin as an anticoagulant. Blood lymphocyte subsets, granulocytes, and monocytes were identified by their forward‐ and side‐scatter characteristics, as well as by the expression of specific markers. BMDCs were obtained by culturing BM leukocytes (2×106) seeded in 100 mm bacteriological culture dishes (Falcon, Becton Dickinson, San Diego, CA, USA) in 10 ml RPMI‐1640 medium containing 10% FCS (Biological Industries) and 20 ng/ml mouse rGM‐CSF (R&D Systems, Wiesbaden, Germany). At Day 3, another 10 ml medium containing 20 ng/ml mouse rGM‐CSF was added to the cells. At Days 6 and 8, half of the culture supernatant was collected and centrifuged, and the cell pellet was resuspended in 10 ml fresh medium containing 20 ng/ml mouse rGM‐CSF. For complete maturation, Day 10 nonadherent cells were collected by gentle pipetting, centrifuged at 300 g for 5 min at RT, and resuspended in 10 ml fresh medium in a 100‐mm tissue‐culture dish containing 10 ng/ml mouse rGM‐CSF and 1 μg/ml LPS from Escherichia coli 0111:B4 (Sigma Chemical Co.). Cells were then cultured for 1 additional day. RBCs were washed twice in PBS (900 g for 5 min) at RT. At least 4 samples from different mice were analyzed for each cell population.
The mouse RAW‐264.7 macrophage cell line was maintained in DMEM (Gibco‐BRL), supplemented with 10% heat‐inactivated FCS (Biological Industries), plus 2 mM l‐glutamine and 100 U/ml penicillin. RAW‐Blue™ cells (Invivogen, Tolouse, France) were derived from RAW‐264.7 macrophages with chromosomal integration of a SEAP reporter construct inducible by NF‐κB and AP‐1. RAW‐Blue™ cells were maintained in Zeocin™ (Invivogen) selective medium. Mouse BMDMs were isolated and prepared by flushing the BM of tibias and femurs and then culturing them in DMEM supplemented with 20% FCS and 30% L929 supernatant containing a macrophage‐stimulating factor. After 6 days of culture, adherent macrophages were detached and resuspended in DMEM, supplemented with 10% FCS, 2 mM l‐glutamine, and 100 U/ml of penicillin.
Stable and transient transfections
COS‐7 cells were transiently transfected with mouse CD84 FL, CD84 chimeric H1‐M2, and CD84 chimeric M1‐H2 cDNAs [5] using a Nucleofector II device (Amaxa AG, Koeln, Germany). RAW‐264.7 cells (5–10×106 cells) were transfected by electroporation in a final volume of 600 μl at 250 V/950 μF (Gene Pulser II; Bio‐Rad) with 8 μg linearized mouse CD84 FL cDNA subcloned in pCI‐neo [2]. Twenty‐four hours after transfection, geneticin (1 mg/ml; Gibco‐BRL) was added to the cells for stable selection and maintenance. The resistant stable transfectant (CD84‐RAW‐264.7) was obtained as a polyclonal population after 2 cell‐sorting rounds using a FACSVantage (Becton Dickinson). RAW‐Blue™ cells were transiently transfected with mouse CD84 FL cDNA or pCI‐neo empty vector using the Nucleofector II device. FL mouse CD84 and its cytoplasmic mutants (Y at positions 265 and 300 were mutated to F: Y265F and Y300F, respectively), cloned in pCDNA‐3.1(+) vector, were generated by gene synthesis or site‐directed mutagenesis (GenScript, Piscataway, NJ, USA). A construct lacking mouse CD84 (Δcyt) was also synthesized by introducing a stop codon after aa 246, leaving only the first 4 cytoplasmic aa. CD84 mutants and pCDNA‐3.1(+) empty vector were transiently transfected in RAW‐264.7 cells using the Nucleofector as explained above. Mouse BMDMs were transfected with a 50‐pmol mixture of mouse CD84 siRNAs (AUGAGAAUAAGCAGAAAGAGCACGG and CCGUGCUCUUUCUGCUUAUUCUCAU) or scrambled siRNAs (AUGACAAGUAAACGGAAAGAGACGG and CCGUCUCUUUCCGUUUACUUGUCAU) using the Nucleofector II device and a mouse macrophage nucleofector kit (Amaxa AG). BMDMs were harvested 48 h after transfection.
Immunofluorescence analysis
Cells were stained using mAb followed by streptavidin‐PE‐Cy5. Fluorescence was analyzed using a FACSCalibur (Becton Dickinson) flow cytometer equipped with CellQuest™ software. Fluorescence intensity was plotted on a log scale. The number of cells acquired for each sample depended on the subpopulation studied, and at least 10,000 cells were analyzed. The fluorescence of the isotype‐matched negative controls (Becton Dickinson) was set at a MFI of 5.
Immunoprecipitation and immunoblotting
COS‐7 cells transfected with mouse CD84, thymocytes, splenocytes, and BMDCs were first biotinylated for 30 min at 4°C with sulfo‐normal human serum‐biotin (Sigma Chemical Co.), as described by the manufacturer. Cells were washed twice in PBS buffer. For sodium pervanadate stimulation, DCs were pretreated with 0.03% H2O2 and 100 μM Na3VO4 (Sigma Chemical Co.) for 10 min at 37°C. CD84‐RAW‐264.7 cells were incubated with LPS (100 ng/ml; Sigma Chemical Co.) for 1, 5, and 15 min at 37°C. Cells were then washed once in cold PBS buffer and lysed in 1 ml buffer containing 1% (v/v) Nonidet P‐40 (Sigma Chemical Co.) and protease inhibitors. Cell lysates were precleared once for 30 min using only Protein G‐Sepharose beads (Amersham Pharmacia Biotech, Sweden) and twice for 30 min using 40 μl (50% v/v) hamster IgG1‐coated beads. The precleared lysates were then incubated with 40 μl (7 μg) hamster mCD84.7‐coated beads or hamster IgG1‐coated beads with a constant rotation at 4°C for 6 h. For CD84 N‐deglycosylation, immunoprecipitates were digested for 1 h 15 min at 37°C with 6 U N‐glycosidase F (Roche Diagnostics GmbH, Mannheim, Germany), according to the manufacturerˈs instructions. Immunoprecipitates were washed and analyzed by SDS‐PAGE. The molecular mass was determined using prestained standard markers (Bio‐Rad or Fermentas, Burlington, ON, Canada). Proteins were transferred to PVDF membranes (Immobilon, Millipore, Boston, MA, USA). After 1 h of incubation at RT with blocking solution (3% nonfat dry milk in PBS or 5% BSA in PBS), the membrane was incubated for 45 min at RT with streptavidin‐POD conjugate (Roche Diagnostics GmbH) or with HRP‐conjugated antiphosphotyrosine Ab (Zymed Laboratories, San Francisco, CA, USA). The membrane was washed in TTBS, and the blot was developed using a SuperSignal West Pico chemiluminescent substrate (Pierce, Rockford, IL, USA).
Phosphorylation assays
Untransfected and mouse CD84‐transfected RAW‐264.7 cells (106/culture dish) were treated with LPS (100 ng/ml; Sigma Chemical Co.) for 5, 15, and 30 min. Afterwards, the protein concentration of these denatured cell lysate samples was measured using the BCA protein assay kit (Pierce). To determine the concentrations of phosphorylated MAPKs—ERK‐1/2, JNK‐1/2, and p38 molecules—in these lysates, the CBA cell signaling flex set system (Becton Dickinson) was used, according to the manufacturerˈs instructions. Briefly, 12 μg cell lysates were incubated with 50 μl capture beads for 3 h and with 50 μl PE mixed‐detection reagent for 1 h at RT and protected from light. The samples were then washed and analyzed using a FACSCanto II flow cytometer (Becton Dickinson). Data processing was carried out using the accompanying FACSDiVa™ and FCAP Array™ software.
NF‐κB activation
RAW‐Blue™ cells were transfected with pCI‐neo (mock) or mouse CD84‐pCI‐neo. Afterwards, cells were attached to a flat‐bottom 96‐well plate (5×104/well) and activated with soluble LPS (100 ng/ml; Sigma Chemical Co.). After 24 h, cell‐free supernatants were harvested. Upon TLR‐4 stimulation, RAW‐Blue™ cells activated NF‐κB and AP‐1, leading to the secretion of SEAP, which was easily detectable and measurable in a spectrophotometer at 650 nm with QUANTI‐Blue™ (Invivogen).
Cytokine determination
CD84‐transfected and untransfected RAW‐264.7 cells were seeded in a 24‐well plate (2×105/well) and activated with soluble LPS (100 ng/ml; Sigma Chemical Co.). After 1, 3, 6, and 18 h in culture, cell‐free supernatants were harvested. Alternatively, siRNA‐transfected BMDMs were seeded in a 96‐well plate (3×104/well) and activated with LPS (100 ng/ml; Sigma Chemical Co.) for 6 and 24 h. The concentrations of IL‐6, IL‐10, IL‐12p70, IFN‐γ, and MCP‐1 in these supernatants were measured by using the commercially available CBA mouse inflammation kit (Becton Dickinson). Briefly, 50 μl mixed capture beads were incubated with 50 μl diluted supernatant and 50 μl PE detection reagent for 2 h, carefully protected from light at RT. The beads were then washed and analyzed using a FACSCanto II flow cytometer (Becton Dickinson). Data processing was carried out using the accompanying FACSDiVa™ and FCAP Array™ software. Mouse TNF‐α, IL‐6, and IL‐10 levels were also measured by a sandwich ELISA (R&D Systems).
RESULTS
Molecular characterization of mouse CD84 using a novel mAb
The mAb mCD84.7 (IgG1), directed against mouse CD84, was generated by immunizing an Armenian hamster with mouse CD84‐Fc fusion protein. In FACS assays, mCD84.7 bound specifically to those transfectant COS‐7 cells that expressed a chimeric molecule comprised of the first Ig‐like domain of mouse CD84 and the second human CD84 domain (M1‐H2). By contrast, mCD84.7 did not stain COS‐7 cells expressing a chimeric molecule comprised of the first Ig‐like domain of human CD84 and the second Ig‐like domain of mouse CD84 (H1‐M2; Fig. 1 ). Thus, the mAb proved to be specific for the N‐terminal domain of mouse CD84. Next, mCD84.7 mAb was used to immunoprecipitate mouse CD84 from biotinylated COS‐7 cells that had been transfected previously with mouse CD84 full‐length cDNA. A single band of 48,000–59,000 kDa was observed ( Fig. 2 A). To determine the molecular mass of the protein in absence of N‐linked sugars, CD84 protein was digested with endoglycosylase F. A single band of 39,000 kDa was detected after deglycosylation. A shift in molecular mass of ∼9 kDa was consistent with the presence of 3 N‐linked oligosaccharides in positions 177, 122, and 126 in the predicted amino acid sequence [2]. Moreover, immunoprecipitation of CD84 showed a broad band of ∼55,500–63,200 kDa in thymocytes and of 55,500–69,100 kDa in BMDCs and splenocytes (Fig. 2B). Treatment of DCs with pervanadate induced the phosphorylation of CD84 (Fig. 2C). These data indicate that mouse CD84 undergoes extensive post‐translational modifications.
Figure 1.

Reactivity of mCD84.7 mAb (solid line) and isotype control (dotted line) on transiently transfected COS‐7 cells with the CD84 chimeric cDNAs M1‐H2 and H1‐M2.
Figure 2.

Immunoprecipitation, N‐deglycosylation, and phosphorylation of mouse CD84. (A) The biotinylated cell lysates from CD84 transiently transfected COS‐7 cells were immunoprecipitated (IP) with mCD84.7 mAb and treated with (+) or without (–) N‐glycosylase F (N‐Glyc. F). WB: Sv‐POD, Western blot streptavidin‐POD. Biotinylated thymocyte, splenocyte, and DC (B) lysates were immunoprecipitated with the mCD84.7 mAb or with an isotypic hamster IgG as a control. (C) Phosphorylation of mouse CD84 Y (pY) in immature DCs. Untreated (–) and pervanadate (PV)‐treated (+) cells were biotinylated and lysed and then immunoprecipitated with mCD84.7 mAb. (D) Phosphorylation of mouse CD84 Y in LPS‐stimulated CD84‐RAW‐264.7 cells. Molecular mass in kDa was determined by the migration of a known protein standard.
Expression of mouse CD84 on the surface of hematopoietic cells
HSCs.
We next analyzed the cell‐surface expression of CD84 on BM HSCs. CD84 was present on virtually all Sca‐1+c‐kit+Lineage– HSCs (95.8%±2.47%; MFI 71.59±6.12), indicating that it is expressed very early during hematopoietic development.
T lymphocytes and NK cells.
Consistent with its expression on HSCs, 89% of CD4–CD8– immature thymocytes were CD84‐positive ( Table 1 ). Whereas the highest expression was found on the single‐positive CD4 and CD8 thymocytes, CD84 was present only on 74% of CD4+CD8+ thymocytes (Table 1). CD84 was present on all mature blood and spleen T cells, and its expression levels were slightly higher than those found in the thymus (Table 1). Differences in these expression levels could be observed among T cells subsets (Table 1). For instance, the expression of CD84 was significantly higher in CD4+ cells compared with CD8+ cells. Treg (CD4+CD25+), however expressed CD84 levels similar to those detected in other CD4+ T cells (Table 1). Interestingly, CD4+ TFH cells (CD4+CXCR5+) presented significantly higher levels of CD84 as compared with other CD4+ T cells ( Fig. 3 ). NK cells also expressed CD84, albeit to a lesser extent than did NKT cells (Table 1).
Table 1.
CD84 Expression on T, NK, and NKT Cells
| T and NK cell subset | CD84 expression a | Source | |
| % | MFI | ||
| DN (CD4–CD8–) | 89 ± 7 | 70 ± 7 | Thymus |
| DP (CD4+CD8+) | 74 ± 7 | 39 ± 4 | Thymus |
| CD4+ | 99 ± 1 | 94 ± 8 | Thymus |
| CD8+ | 95 ± 3 | 72 ± 7 | Thymus |
| CD4+ | 96 ± 3 | 101 ± 11 | Spleen |
| CD8+ | 93 ± 6 | 84 ± 3 | Spleen |
| Treg (CD4+CD25+) | 99 ± 1 | 110 ± 5 | Spleen |
| TFH (CD4+CXCR5+) | 99 ± 1 | 154 ± 16 | Spleen |
| CD4+ | 99 ± 1 | 112 ± 10 | Blood |
| CD8+ | 99 ± 1 | 95 ± 11 | Blood |
| NK (NK1.1+CD3–) | 98 ± 2 | 140 ± 27 | Blood |
| NKT (NK1.1+CD3+) | 98 ± 4 | 174 ± 29 | Blood |
The values indicate the % and mean ± sd of at least 4 independent experiments. DN, Double‐negative; DP, double‐positive.
Figure 3.

Expression of mouse CD84. (A) Expression of CD84 on splenic CD4+ TFH and CD4+ TH cells. The plots were gated on B220– cells. (B) Differential expression levels of CD84 on peripheral blood NKT and T cells. (C) Expression of CD84 on follicular and MZ splenic B cell subsets. The plots were gated on B220+ cells. CD23 and CD21 staining was used to characterize follicular and MZ subsets. CD84 staining (solid line) is overlaid by a negative control (thin line). Percentage of positive cells and MFI is shown from 1 of the 4 experiments.
Precursor B cells and B cell subsets.
Between 53% and 75% of pro‐B, pre‐B, and immature B cells in the BM expressed CD84 ( Table 2 ). Interestingly, the CD84 expression on pro‐B proved to be significantly higher than that observed on pre‐B and immature B cells, and was identical to mature B cells in the BM. In contrast, virtually all mature B cells in the periphery expressed CD84. The highest levels were observed in MZ B lymphocytes (Table 2 and Fig. 3). No significant difference could be detected between B1 and B2 subsets in the blood and peritoneum.
Table 2.
CD84 Expression on B‐Cell Subsets
| B‐cell subset (B220+) | CD84 expression a | Source | |
| % | MFI | ||
| Pro‐B (cKit+CD24–) | 75 ± 10 | 118 ± 10 | BM |
| Pre‐B (CD25+CD43–) | 62 ± 8 | 32 ± 3 | BM |
| Pre‐B (CD25+CD43+) | 53 ± 3 | 74 ± 15 | BM |
| Immature (IgM+IgD–) | 73 ± 3 | 45 ± 7 | BM |
| Mature (IgM+IgD+) | 98 ± 1 | 116 ± 11 | BM |
| T1 (CD21–CD23–) | 92 ± 4 | 69 ± 20 | Spleen |
| Follicular (CD21+CD23+) | 99 ± 1 | 119 ± 30 | Spleen |
| MZ (CD21+CD23–low) | 99 ± 1 | 186 ± 38 | Spleen |
| B1a (CD5+IgM+) | 98 ± 1 | 83 ± 12 | Peritoneum |
| B1b/T1 (CD5–IgM–low) | 81 ± 10 | 79 ± 12 | Peritoneum |
| B2 (CD5–IgM+) | 97 ± 1 | 74 ± 7 | Peritoneum |
| B2 (CD5–IgM+) | 93 ± 4 | 51 ± 19 | Blood |
| B1a (CD5+IgM+) | 90 ± 8 | 62 ± 15 | Blood |
The values indicate the % and mean ± sd of at least 4 independent experiments. T1, Transitional 1.
Myeloid cells and platelets.
CD84 was present on myeloid cells such as monocytes, macrophages, granulocytes, and immature and mature BMDCs ( Table 3 ). Mature DCs presented significantly lower amounts of CD84 on their cell surfaces as compared with immature DCs (Table 3). To a lesser extent, CD84 could also be detected on plasmacytoid DCs, platelets, and megakaryocytes (Table 3). Mouse CD84 was not present on RBCs (data not shown).
Table 3.
CD84 Expression on Platelets and Myeloid Lineage Cells
| Myeloid cells and platelets | CD84 expression a | Source | |
| % | MFI | ||
| Monocytes (CD14+CD11b+) | 95 ± 2 | 120 ± 6 | Blood |
| Macrophages (CD14+CD11b+) | 87 ± 3 | 131 ± 1 | Spleen |
| Granulocytes (Gr‐1high) | 96 ± 6 | 57 ± 16 | Blood |
| Immature DC (CD11c+I‐Abhigh) | 99 ± 1 | 218 ± 8 | BMDC |
| Mature DC (CD11c+I‐Abhigh) | 99 ± 1 | 166 ± 3 | BMDC |
| Plasmacytoid DC (CD11c+B220+) | 63 ± 9 | 75 ± 19 | Spleen |
| Megakaryocytes (CD41+) | 40 ± 14 | 21 ± 5 | BM |
| Platelets (CD41+) | 49 ± 16 | 28 ± 4 | Blood |
The values indicate the % and mean ± sd of at least 4 independent experiments.
Functional role of mouse CD84 in macrophages
CD84 enhances LPS‐induced MAPK phosphorylation and NF‐κB activation in RAW‐264.7 macrophages.
The functional role of CD84 has not been analyzed in myeloid cells. To provide further insight into the biological role played by CD84 in the activation of myeloid cells, we stably transfected the mouse monocytic cell line RAW‐264.7, which did not express CD84 with mouse CD84 full‐length cDNA. This stably transfected polyclonal population used for these experiments expressed levels of CD84 similar to normal macrophages ( Fig. 4 and Table 3). Treatment of the CD84‐RAW‐264.7 cells with soluble LPS (100 ng/ml) induced rapid and markedly increased phosphorylation of ERK‐1/2, p38, and JNK‐1/2 MAPK compared with RAW‐264.7 cells ( Fig. 5 ). Importantly, the expression of CD14, a component of the LPS receptor complex, was identical in untransfected and CD84‐transfected cells (Fig. 4). Furthermore, upon TLR‐4 stimulation with LPS for 24 h, RAW‐Blue™ cells transiently transfected with mouse CD84 (37±3% of CD84‐positive cells) showed higher NF‐κB/AP‐1 activation levels compared with mock‐transfected cells ( Fig. 6 ). Thus, the LPS‐induced MAPK and NF‐κB/AP‐1 activation pathways were strikingly enhanced by CD84 signaling. Interestingly, CD84‐RAW‐264.7 cells presented constitutive Y phosphorylation, which transiently decreased following LPS activation (Fig. 2D).
Figure 4.

CD84 and CD14 expression levels on the RAW‐264.7 cell line. RAW‐264.7 cells and the CD84 stable transfectant (CD84‐RAW‐264.7) were screened by CD84 and CD14 (solid line) expression patterns, overlaid by a negative control (thin line) using flow cytometry. Percentage of positive cells and MFI is shown.
Figure 5.

MAPK phosphorylation by CD84‐RAW‐264.7 cells. RAW‐264.7 cells were treated with 100 ng/ml LPS (0111:B4) for 5, 15, and 30 min. A representative experiment is shown. p, Phosphorylated.
Figure 6.

Activation of NF‐|gkB/AP‐1 in RAW‐Blue™ cells. These cells were derived from RAW 264.7 macrophages with chromosomal integration of a SEAP reporter construct inducible by NF‐κB and AP‐1. CD84‐ and pCI‐neo‐transfected cells were treated with LPS (100 ng/ml) for 24 h. QUANTI‐Blue™ substrate was used to measure SEAP secretion. Measurements were performed in triplicate, and a representative experiment is shown.
CD84 modulates LPS‐induced cytokine secretion in RAW‐264.7 cells via cytoplasmic Y300.
As phosphorylation of MAPKs and NF‐κB activation was notably augmented in CD84‐RAW‐264.7 cells, we subsequently investigated whether cytokine release was also altered after LPS stimulation (100 ng/ml). We measured IL‐6, IL‐10, IL‐12p70, IFN‐γ, MCP‐1, and TNF‐α secretion after 1, 3, 6, and 18 h poststimulation. LPS‐stimulated CD84‐RAW‐264.7 cells secreted TNF‐α and MCP‐1 at levels that were considerably higher when compared with those of CD84‐negative cells ( Fig. 7 ). By contrast, the presence of CD84 markedly hindered the secretion of IL‐6 and IL‐10, as judged by a comparison with untransfected RAW‐264.7 cells (Fig. 7). Interestingly, the levels of IL‐6 at 18 h decreased in RAW‐264.7 and CD84‐RAW‐264.7 cells, probably as a result of the consumption of this cytokine. This may be a result of the high expression of the IL‐6R in these cells (61%±5% and 67%±2%, respectively). No significant levels of IL‐12p70 and IFN‐γ could be detected in the LPS‐stimulated cells. Furthermore, LPS‐induced secretion of IL‐6, IL‐10, and TNF‐α was also examined using RAW‐264.7 cells transfected with a Δcyt CD84 to test whether this modulatory effect was dependent on direct CD84‐mediated signaling. We chose IL‐6, IL‐10, and TNF‐α cytokines, as they had previously presented different secretion patterns. The Δcyt reversed the effects of CD84 by decreasing the levels of TNF‐α while increasing IL‐6 and IL‐10 production ( Fig. 8 ). Given that the cytoplasmic domain of mouse CD84 contains 2 Y residues (Y265 and Y300) embedded within the putative ITSM [2], we decided to identify which Y was involved in the modulation of LPS‐induced cytokine production by singly mutating them to F (Y265F and Y300F). Mutating Y265 had no significant effect on cytokine production compared with CD84 (FL) cells (Fig. 8). Nonetheless, when Y300 was mutated, IL‐6 and IL‐10 secretion increased, and TNF‐α levels diminished with a cytokine profile identical to that seen in mock and Δcyt transfectants.
Figure 7.
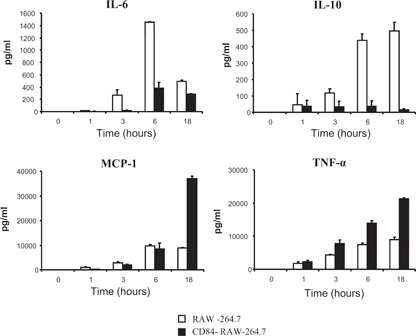
CD84‐RAW‐264.7 cytokine secretion after LPS‐induced activation. IL‐6, IL‐10, and MCP‐1 secretion levels at several time‐points (pg/ml) were measured using the CBA system and TNF‐α levels by ELISA. Measurements were performed in triplicate, and a representative experiment is shown.
Figure 8.

Cytokine profile of CD84 mutants after LPS activation. RAW‐264.7 cells were transiently transfected with mock, FL, Δcyt, Y265F, or Y300F CD84 constructs. CD84 transient transfectants expressed equivalent numbers of positive cells, as determined by flow cytometry (30.2%+2.7%). IL‐6, IL‐10, and TNF‐α secretion levels after 6 h of LPS activation were quantitatively detected by ELISA. Measurements were performed in triplicate, and a representative experiment is shown.
CD84 modulates LPS‐induced cytokine secretion in BMDMs.
As virtually all BMDMs expressed high levels of CD84, we knocked down mouse CD84 expression using siRNAs ( Fig. 9 A). Stimulation of CD84‐silenced BMDMs with LPS (100 ng/ml) resulted in remarkably reduced production of IL‐6 and TNF‐α, particularly after 24 h (Fig. 9B). No significant differences in IL‐10 and MCP‐1 levels were observed between CD84 and scrambled siRNA‐transfected cells (Fig. 9B). In addition, no detectable levels of IL‐12p70 and IFN‐γ could be observed in the supernatants (data not shown). Taken together, these results demonstrate that CD84 signaling modulates the activation of LPS‐induced BMDMs and the monocytic cell line RAW‐264.7.
Figure 9.

Cytokine production by CD84 knocked down BMDMs after LPS induction. (A) Percentage of CD84‐ and CD45‐positive, scrambled siRNA‐treated (white bars) and CD84 siRNA‐treated (black bars) cells is shown. (B) Secretion levels of CD84 knocked‐down macrophages (black bars; CD84 siRNA) and CD84‐positive macrophages (white bars; scrambled siRNA) at several time‐points (pg/ml) were measured using the CBA system and TNF‐α levels by ELISA. Measurements were performed in triplicate, and a representative experiment is shown.
DISCUSSION
Here, we present a comprehensive characterization of mouse CD84 using a new mCD84.7 mAb. Our results show that all of the leukocytes characterized in this study expressed the CD84 receptor. However, there are important differences in the expression levels among the various leukocytes and lymphocyte subsets. Mouse CD84 was expressed very early during hematopoiesis, as >90% of the HSCs expressed CD84, as was observed in human BM HSCs [9]. Although CD84 expression declined in developing lymphocytes in the thymus (70% of double‐positive thymocytes) and BM (only 62% of Pre‐B cells), its expression increased significantly during the later lymphocyte maturation stages, abundantly expressed on mature B and T lymphocytes.
CD84 levels were higher on CD4+ than on CD8+ T cells, which was true not only for peripheral blood and splenic lymphocytes but also for single‐positive thymocytes. In humans, however, expression was higher on CD8+ T cells [4]. Another discrepancy with human cells was observed in mice immature BMDCs, in which CD84 levels were higher than those seen on mature DCs, although just the opposite has been reported in humans [4]. Another significant divergence was the high expression of mouse CD84 present on NK cells, and human NK cells barely expressed this molecule [4, 6]. In contrast, the expression of CD84 on mouse megakaryocytes and platelets was strikingly lower than on their human counterparts [5].
Here, we also show the presence of CD84 on NKT cells and at levels higher than were noted on NK cells. In fact, the lymphocytes expressing the highest amounts of CD84 were the NKT cells and the MZ B cells. These two cell types are prototypes of the so‐called “innate‐like lymphocytes”, which are lymphocytes expressing rearranged antigen receptors that are invariant or semi‐invariant and recognize self‐antigens or simple molecular pathogens structures [24, 25]. When activated, they exhibit rapid and robust effector functions such as cytokine release, cytotoxicity, and/or antibody production. Thus, these cells are thought to serve as a bridge between the rapidly occurring innate immune response and the more slowly occurring adaptive immune responses [26]. NKT‐MZ B cell communication can lead to the rapid cytokine secretion [27, 28] and production of IgM and IgG autoantibodies in NZB/W mice [29]. CD84‐mediated adhesion could participate in this cognate interaction, determining their activation. Importantly, SLAM/SAP family molecules have been described recently as playing a pivotal role in the differentiation of innate‐like lymphocytes [30, –, 32, 33].
It was also noteworthy that splenic CD4+ TFH cells exhibited higher levels of CD84 compared with other CD4+ T cell subsets. In addition, we found that mouse CD84 was expressed on follicular splenic B cells to a notable degree. In the same way, human CD84 was similarly expressed preferentially on TFH cells, as was the adaptor protein SAP [7], which has been shown to be essential for the generation of humoral immunity [34]. Interestingly, a recent study shows that CD84 is required for prolonged T cell–B cell contact, TFH function, and germinal center formation [22].
In addition, our data show that mouse CD84 was highly expressed on myeloid cells, as has been observed similarly in humans [4]. We therefore analyzed the potential role of this receptor as a modulator of macrophage activation. The molecular mechanisms underlying LPS‐induced macrophage activation have been studied extensively, demonstrating the involvement of 2 major signaling pathways: NF‐κB and MAPK (ERK‐1/2, p38, and JNK‐1/2) activation, which govern the production of inflammatory cytokines and chemokines [35, –, 37, 38, 39]. Here, we show that the CD84‐RAW‐264.7 transfectant was hyper‐reactive to LPS stimulation, as was evident by the dramatic increase in ERK‐1/2, p38, and JNK‐1/2 phosphorylation levels and in NF‐κB/AP‐1 activity. The activating effect of CD84 on this monocytic cell line is reminiscent of that observed in T and B cells, in which receptor ligation induced by an agonistic mAb and CD84‐CD84 homophilic interactions resulted in IFN‐γ secretion and enhanced proliferation [5, 6, 20]. With this in mind, we decided to analyze whether the observed differences in MAPK phosphorylation and NF‐κB/AP‐1 activation in CD84‐transfected cells could affect cytokine production. As has been shown previously [40], LPS induced RAW‐264.7 to secrete proinflammatory (TNF‐α, IL‐6, and MCP‐1) and anti‐inflammatory (IL‐10) cytokines. The presence of CD84 on RAW‐264.7 cells altered cytokine production, not only increasing the release of TNF‐α and MCP‐1 but also lowering the secretion of IL‐10 significantly. Interestingly, LPS‐induced production of IL‐6 was impaired by the presence of CD84. Human CD84 undergoes Y phosphorylation following CD84 ligation in lymphocytes, thereby determining the specific recruitment of adaptors and phosphatases containing SH2 domains [6]. Interestingly, mouse CD84 was already found constitutively phosphorylated in CD84‐RAW‐264.7 cells, probably as a result of CD84 homophilic interactions, as has been reported previously in other CD84‐tranfected cells [6, 41].
Analysis of human CD84‐SAP/EAT‐2 interactions suggests that Y262 is critical for SAP recruitment, whereas EAT‐2 is capable of binding Y262 and Y298 [20]. In contrast, the cytosolic phosphatase SH2 domain‐containing Y phosphatase 2 binds to Y298 specifically [41]. Mouse CD84 cytoplasmic tail also contains two conserved ITSMs (TVY265 AVV and TIY300 SSV) [2]. Although SAP and EAT‐2 are not expressed in RAW‐264.7 cells, we decided to explore whether these two ITSM motifs are required for the effect of CD84 on cytokine production. When mouse CD84 lacked its cytoplasmic domain or when Y300 was mutated, its cytokine modulatory ability was abolished, whereas mutating Y265 had no significant effects on cytokine production, as compared with CD84 (FL). The absence of SAP and EAT‐2 in these cells may well allow other adaptor molecules or enzymes to bind to the Y300, thereby determining its functionality. To demonstrate that the observed regulatory effects mediated by CD84 in the monocytic cell line RAW‐264.7 were relevant to normal cells, we carried out a series of experiments silencing CD84 expression in BMDMs. As expected, our results showed that abrogating the expression of CD84 in these cells caused a reduction in LPS‐induced TNF‐α secretion. Surprisingly, the knock‐down of CD84 in BMDMs also resulted in a decrease of IL‐6 without considerably affecting IL‐10 production. Although we do not have an explanation presently for this phenomenon, a possible reason for this different signaling behavior could be the relative abundance of certain intracellular molecules in different cell subsets or maturation stages, as has been observed for other SLAM family receptors [42].
In summary, here, we present a detailed description of mouse CD84 expression and propose a model in which CD84 molecules are able to modulate signaling pathways downstream of TLR4. This cross‐talk between TLRs and CD84 could regulate macrophage cell‐fate decisions and effector functions. This study will help to develop new hypotheses about CD84 functionality in innate and adaptive immune responses.
AUTHORSHIP
J.S. designed and performed experiments, analyzed data, and wrote the manuscript. X.R. and J.d.S. performed experiments and analyzed data. C.T. contributed to the design and manuscript writing. P.E. planned, designed experiments, and wrote the manuscript.
DISCLOSURE
The authors have no potential conflicts of interest.
ACKNOWLEDGMENTS
This publication was made possible by a Senior Research Award from the Crohnˈs and Colitis Foundation of America (CCFA) and by grants from the Ministerio de Ciencia e Innovación (SAF 2006‐00490 and SAF 2009‐07071). We thank Isabel Crespo for her expert technical assistance in cell sorting and Pilar Muñoz for her help in performing immunoprecipitation studies.
REFERENCES
- 1. De la Fuente, M. A. , Pizcueta, P. , Nadal, M. , Bosch, J. , Engel, P. (1997) CD84 leukocyte antigen is a new member of the Ig superfamily. Blood 90, 2398–2405. [PubMed] [Google Scholar]
- 2. De la Fuente, M. A. , Tovar, V. , Pizcueta, P. , Nadal, M. , Bosch, J. , Engel, P. (1999) Molecular cloning, characterization, and chromosomal localization of the mouse homologue of CD84, a member of the CD2 family of cell surface molecules. Immunogenetics 49, 249–255. [DOI] [PubMed] [Google Scholar]
- 3. Krause, S. W. , Rehli, M. , Heinz, S. , Ebner, R. , Andreesen, R. (2000) Characterization of MAX.3 antigen, a glycoprotein expressed on mature macrophages, dendritic cells and blood platelets: identity with CD84. Biochem. J. 346, 729–736. [PMC free article] [PubMed] [Google Scholar]
- 4. Romero, X. , Benítez, D. , March, S. , Vilella, R. , Miralpeix, M. , Engel, P. (2004) Differential expression of SAP and EAT‐2‐binding leukocyte cell‐surface molecules CD84, CD150 (SLAM), CD229 (Ly9) and CD244 (2B4). Tissue Antigens 64, 132–144. [DOI] [PubMed] [Google Scholar]
- 5. Martin, M. , Romero, X. , de la Fuente, M. A. , Tovar, V. , Zapater, N. , Esplugues, E. , Pizcueta, P. , Bosch, J. , Engel, P. (2001) CD84 functions as a homophilic adhesion molecule and enhances IFN‐γ secretion: adhesion is mediated by Ig‐like domain 1. J. Immunol. 167, 3668–3676. [DOI] [PubMed] [Google Scholar]
- 6. Tangye, S. G. , van de Weerdt, B. C. , Avery, D. T. , Hodgkin, P. D. (2002) CD84 is up‐regulated on a major population of human memory B cells and recruits the SH2 domain containing proteins SAP and EAT‐2. Eur. J. Immunol. 32, 1640–1649. [DOI] [PubMed] [Google Scholar]
- 7. Chtanova, T. , Tangye, S. G. , Newton, R. , Frank, N. , Hodge, M. R. , Rolph, M. S. , Mackay, C. R. (2004) T follicular helper cells express a distinctive transcriptional profile, reflecting their role as non‐Th1/Th2 effector cells that provide help for B cells. J. Immunol. 173, 68–78. [DOI] [PubMed] [Google Scholar]
- 8. Zaiss, M. , Hirtreiter, C. , Rehli, M. , Rehm, A. , Kunz‐Schughart, L. A. , Andreesen, R. , Hennemann, B. (2003) CD84 expression on human hematopoietic progenitor cells. Exp. Hematol. 31, 798–805. [DOI] [PubMed] [Google Scholar]
- 9. Sintes, J. , Romero, X. , Marin, P. , Terhorst, C. , Engel, P. (2008) Differential expression of CD150 (SLAM) family receptors by human hematopoietic stem and progenitor cells. Exp. Hematol. 36, 1199–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Engel, P. , Eck, M. J. , Terhorst, C. (2003) The SAP and SLAM families in immune responses and X‐linked lymphoproliferative disease. Nat. Rev. Immunol. 3, 813–821. [DOI] [PubMed] [Google Scholar]
- 11. Ma, C. S. , Nichols, K. E. , Tangye, S. G. (2007) Regulation of cellular and humoral immune responses by the SLAM and SAP families of molecules. Annu. Rev. Immunol. 25, 337–379. [DOI] [PubMed] [Google Scholar]
- 12. Calpe, S. , Wang, N. , Romero, X. , Berger, S. B. , Lanyi, A. , Engel, P. , Terhorst, C. (2008) The SLAM and SAP gene families control innate and adaptive immune responses. Adv. Immunol. 97, 177–250. [DOI] [PubMed] [Google Scholar]
- 13. Wandstrat, A. E. , Nguyen, C. , Limaye, N. , Chan, A. Y. , Subramanian, S. , Tian, X. H. , Yim, Y. S. , Pertsemlidis, A. , Garner Jr., H. R. , Morel, L. , Wakeland, E. K. (2004) Association of extensive polymorphisms in the SLAM/CD2 gene cluster with murine lupus. Immunity 21, 769–780. [DOI] [PubMed] [Google Scholar]
- 14. Limaye, N. , Belobrajdic, K. A. , Wandstrat, A. E. , Bonhomme, F. , Edwards, S. V. , Wakeland, E. K. (2008) Prevalence and evolutionary origins of autoimmune susceptibility alleles in natural mouse populations. Genes Immun. 9, 61–68. [DOI] [PubMed] [Google Scholar]
- 15. Chan, A. Y. , Westcott, J. M. , Mooney, J. M. , Wakeland, E. K. , Schatzle, J. D. (2006) The role of SAP and the SLAM family in autoimmunity. Curr. Opin. Immunol. 18, 656–664. [DOI] [PubMed] [Google Scholar]
- 16. Romero, X. , Zapater, N. , Calvo, M. , Kalko, S. G. , de la Fuente, M. A. , Tovar, V. , Ockeloen, C. , Pizcueta, P. , Engel, P. (2005) CD229 (Ly9) lymphocyte cell surface receptor interacts homophilically through its N‐terminal domain and relocalizes to the immunological synapse. J. Immunol. 174, 7033–7042. [DOI] [PubMed] [Google Scholar]
- 17. Yan, Q. , Malashkevich, V. N. , Fedorov, A. , Fedorov, E. , Cao, E. , Lary, J. W. , Cole, J. L. , Nathenson, S. G. , Almo, S. C. (2007) Structure of CD84 provides insight into SLAM family function. Proc. Natl. Acad. Sci. USA 104, 10583–10588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sayos, J. , Martin, M. , Chen, A. , Simarro, M. , Howie, D. , Morra, M. , Engel, P. , Terhorst, C. (2001) Cell surface receptors Ly‐9 and CD84 recruit the X‐linked lymphoproliferative disease gene product SAP. Blood 97, 3867–3874. [DOI] [PubMed] [Google Scholar]
- 19. Morra, M. , Lu, J. , Poy, F. , Martin, M. , Sayos, J. , Calpe, S. , Gullo, C. , Howie, D. , Rietdijk, S. , Thompson, A. , Coyle, A. J. , Denny, C. , Yaffe, M. B. , Engel, P. , Eck, M. J. , Terhorst, C. (2001) Structural basis for the interaction of the free SH2 domain EAT‐2 with SLAM receptors in hematopoietic cells. EMBO J. 20, 5840–5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tangye, S. G. , Nichols, K. E. , Hare, N. J. , van de Weerdt, B. C. (2003) Functional requirements for interactions between CD84 and Src homology 2 domain containing proteins and their contribution to human T cell activation. J. Immunol. 171, 2485–2495. [DOI] [PubMed] [Google Scholar]
- 21. Morra, M. , Howie, D. , Grande, M. S. , Sayos, J. , Wang, N. , Wu, C. , Engel, P. , Terhorst, C. (2001) X‐linked lymphoproliferative disease: a progressive immunodeficiency. Annu. Rev. Immunol. 19, 657–682. [DOI] [PubMed] [Google Scholar]
- 22. Cannons, J. L. , Qi, H. , Lu, K. T. , Dutta, M. , Gomez‐Rodriguez, J. , Cheng, J. , Wakeland, E. K. , Germain, R. N. , Schwartzberg, P. L. (2010) Optimal germinal center responses require a multistage T cell:B cell adhesion process involving integrins, SLAM‐associated protein, and CD84. Immunity 32, 253–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ivashkiv, L. B. (2008) A signal‐switch hypothesis for cross‐regulation of cytokine and TLR signaling pathways. Nat. Rev. Immunol. 8, 816–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lopes‐Carvalho, T. , Foote, J. , Kearney, J. F. (2005) Marginal zone B cells in lymphocyte activation and regulation. Curr. Opin. Immunol. 17, 244–250. [DOI] [PubMed] [Google Scholar]
- 25. Bendelac, A. , Savage, P. B. , Teyton, L. (2007) The biology of NKT cells. Annu. Rev. Immunol. 25, 297–336. [DOI] [PubMed] [Google Scholar]
- 26. Kearney, J. F. (2005) Innate‐like B cells. Springer Semin. Immunopathol. 26, 377–383. [DOI] [PubMed] [Google Scholar]
- 27. Sonoda, K. H. , Stein‐Streilein, J. (2002) CD1d on antigen‐transporting APC and splenic marginal zone B cells promotes NKT cell dependent tolerance. Eur. J. Immunol. 32, 848–857. [DOI] [PubMed] [Google Scholar]
- 28. Colgan, S. P. , Hershberg, R. M. , Furuta, G. T. , Blumberg, R. S. (1999) Ligation of intestinal epithelial CD1d induces bioactive IL‐10: critical role of the cytoplasmic tail in autocrine signaling. Proc. Natl. Acad. Sci. USA 96, 13938–13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Takahashi, T. , Strober, S. (2008) Natural killer T cells and innate immune B cells from lupus‐prone NZB/W mice interact to generate IgM and IgG autoantibodies. Eur. J. Immunol. 38, 156–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nichols, K. E. , Hom, J. , Gong, S. Y. , Ganguly, A. , Ma, C. S. , Cannons, J. L. , Tangye, S. G. , Schwartzberg, P. L. , Koretzky, G. A. , Stein, P. L. (2005) Regulation of NKT cell development by SAP, the protein defective in XLP. Nat. Med. 11, 340–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li, W. , Sofi, M. H. , Rietdijk, S. , Wang, N. , Terhorst, C. , Chang, C. H. (2007) The SLAM‐associated protein signaling pathway is required for development of CD4+ T cells selected by homotypic thymocyte interaction. Immunity 27, 763–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Griewank, K. , Borowski, C. , Rietdijk, S. , Wang, N. , Julien, A. , Wei, D. G. , Mamchak, A. A. , Terhorst, C. , Bendelac, A. (2007) Homotypic interactions mediated by Slamf1 and Slamf6 receptors control NKT cell lineage development. Immunity 27, 751–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Veillette, A. , Dong, Z. , Latour, S. (2007) Consequence of the SLAM‐SAP signaling pathway in innate‐like and conventional lymphocytes. Immunity 27, 698–710. [DOI] [PubMed] [Google Scholar]
- 34. Crotty, S. , Kersh, E. N. , Cannons, J. , Schwartzberg, P. L. , Ahmed, R. (2003) SAP is required for generating long‐term humoral immunity. Nature 421, 282–287. [DOI] [PubMed] [Google Scholar]
- 35. Fujihara, M. , Muroi, M. , Tanamoto, K. , Suzuki, T. , Azuma, H. , Ikeda, H. (2003) Molecular mechanisms of macrophage activation and deactivation by lipopolysaccharide: roles of the receptor complex. Pharmacol. Ther. 100, 171–194. [DOI] [PubMed] [Google Scholar]
- 36. Weinstein, S. L. , Sanghera, J. S. , Lemke, K. , DeFranco, A. L. , Pelech, S. L. (1992) Bacterial lipopolysaccharide induces tyrosine phosphorylation and activation of mitogen‐activated protein kinases in macrophages. J. Biol. Chem. 267, 14955–14962. [PubMed] [Google Scholar]
- 37. Han, J. , Lee, J. D. , Bibbs, L. , Ulevitch, R. J. (1994) A MAP kinase targeted by endotoxin and hyperosmolarity in mammalian cells. Science 265, 808–811. [DOI] [PubMed] [Google Scholar]
- 38. Hambleton, J. , Weinstein, S. L. , Lem, L. , DeFranco, A. L. (1996) Activation of c‐Jun N‐terminal kinase in bacterial lipopolysaccharide‐stimulated macrophages. Proc. Natl. Acad. Sci. USA 93, 2774–2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kawai, T. , Akira, S. (2007) TLR signaling. Semin. Immunol. 19, 24–32. [DOI] [PubMed] [Google Scholar]
- 40. Pradervand, S. , Maurya, M. R. , Subramaniam, S. (2006) Identification of signaling components required for the prediction of cytokine release in RAW 264.7 macrophages. Genome Biol. 7, R11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Oliver‐Vila, I. , Saborit‐Villarroya, I. , Engel, P. , Martin, M. (2008) The leukocyte receptor CD84 inhibits Fc ε RI‐mediated signaling through homophilic interaction in transfected RBL‐2H3 cells. Mol. Immunol. 45, 2138–2149. [DOI] [PubMed] [Google Scholar]
- 42. Chlewicki, L. K. , Velikovsky, C. A. , Balakrishnan, V. , Mariuzza, R. A. , Kumar, V. (2008) Molecular basis of the dual functions of 2B4 (CD244). J. Immunol. 180, 8159–8167. [DOI] [PubMed] [Google Scholar]


