Abstract
Guanylate‐binding proteins (GBPs) are essential components of cell‐autonomous immunity. In response to IFN signaling, GBPs are expressed in the cytoplasm of immune and nonimmune cells, where they unleash their antimicrobial activity toward intracellular bacteria, viruses, and parasites. Recent studies have revealed that GBPs are essential for mediating activation of the caspase‐1 inflammasome in response to the gram‐negative bacteria Salmonella enterica serovar Typhimurium, Francisella novicida, Chlamydia muridarum, Chlamydia trachomatis, Legionella pneumophila, Vibrio cholerae, Enterobacter cloacae, and Citrobacter koseri. During infection with vacuolar‐restricted gram‐negative bacteria, GBPs disrupt the vacuolar membrane to ensure liberation of LPS for cytoplasmic detection by caspase‐11 and the noncanonical NLRP3 inflammasome. In response to certain cytosolic bacteria, GBPs liberate microbial DNA for activation of the DNA‐sensing AIM2 inflammasome. GBPs also promote the recruitment of antimicrobial proteins, including NADPH oxidase subunits and autophagy‐associated proteins to the Mycobacterium‐containing vacuole to mediate intracellular bacterial killing. Here, we provide an overview on the emerging relationship between GBPs and activation of the inflammasome in innate immunity to microbial pathogens.
Keywords: caspase‐1, caspase‐11, pyroptosis, GBPs, bacteria, viruses
Short abstract
Review of how GBPs attack pathogens and activate inflammasomes, cell death, and innate immunity.
Abbreviations
- BMDM
bone marrow‐derived macrophage
- CARD
caspase recruitment domain
- CSFV
classical swine fever virus
- EMCV
encephalomyocarditis virus
- GBP
guanylate‐binding protein
- HCV
hepatitis C virus
- IAV
influenza virus
- IRG
immunity‐related GTPases
- NLR
nucleotide‐binding domain–leucine‐rich repeat containing proteins
- VSV
vesicular stomatitis virus
Introduction
Cell‐autonomous immunity is a part of the innate immune system specialized in host defense against intracellular pathogens. Production of type I IFNs (including IFN‐α subtypes and IFN‐β) and type II IFN (IFN‐γ) upon recognition of pathogens engages cell‐autonomous immunity, which is required for restriction of microbial replication [1]. IFN signaling induced by pathogens results in the expression of hundreds of genes, which contribute to innate and adaptive immune responses in both immune and nonimmune cells [2, 3]. Many of the genes induced by IFNs encode potent effectors of cell‐autonomous immunity, including the 65–67 kDa proteins called GBPs. IFN‐γ is a more potent inducer of GBPs than type I IFNs, and most of the work investigating cell‐autonomous effects of GBPs to pathogens involves preactivation of host cells with IFN‐γ [4].
GBPs comprise a family of highly conserved IFN‐inducible GTPases. They were originally identified as 67‐kDa proteins in human and mouse cell lines treated with IFNs [5]. Two of these proteins were later isolated from IFN‐treated human diploid fibroblastic cells and were found to have guanylate‐binding activity [6]. The GBP family now consists of 7 human GBPs, 11 murine GBPs, and 2 mouse pseudogenes encoding GBPs ( Fig. 1 ) [4, 7, 8, 9–10]. Although all human GBPs are located in a single cluster on chromosome 1, murine GBPs have been mapped to 2 chromosomal clusters, 1 on chromosome 3 and the other on chromosome 5 (Fig. 1) [11, 12]. Genomic analysis of murine GBPs revealed the presence of Gbp1, Gbp2, Gbp3, Gbp5, Gbp7, and 1 pseudogene on chromosome 3, whereas Gbp4, Gbp6, Gbp8, Gbp9, Gbp10, Gbp11, and the second pseudogene are located on chromosome 5 [12].
Figure 1.
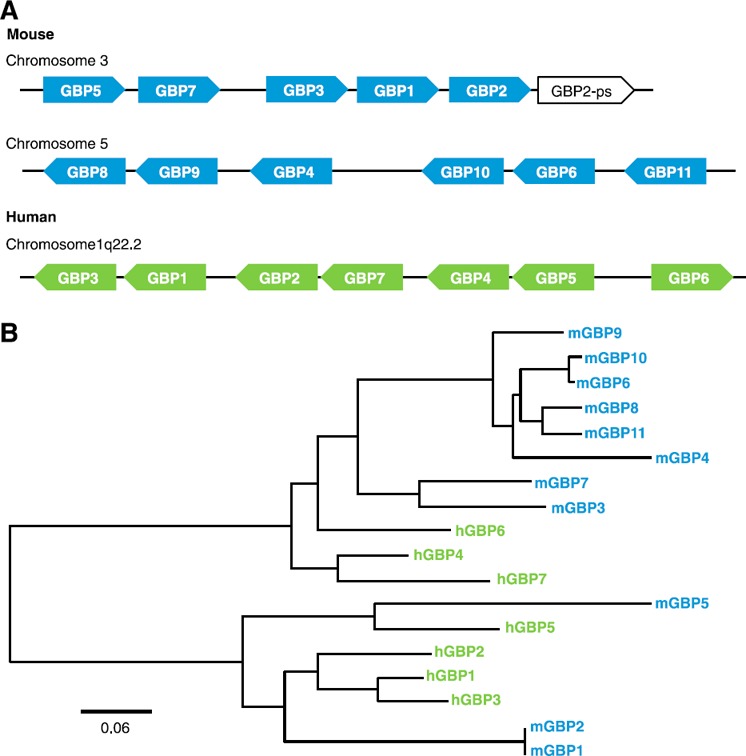
Genomic and phylogenetic organization of GBPs. (A) Mouse and human GBP open‐reading frames organized by chromosome location based on National Center for Biotechnology Information (Bethesda, MD, USA) annotations. (B) A phylogenetic tree was generated by amino acid multiple sequence alignments using the MUSCLE (multiple sequence alignment software) algorithm followed by the PhyML program (phylogeny.fr) with downstream visualization using FigTree version 1.4.2 (http://tree.bio.ed.ac.uk/software/figtree). Scale bar indicates the number of substitutions per site.
GBPs are classified under the dynamin superfamily of GTPases based on their structural and biochemical properties [4, 13]. Similar to other dynamin family members, GBPs undergo nucleotide‐dependent oligomerization and can hydrolyze GTP to GDP and GMP [14, 15, 16–17]. Structural analysis of GBPs revealed the presence of an N‐terminal globular GTP‐binding domain (also known as the large G or LG domain) and a C‐terminal α‐helical GTPase effector domain [18]. These 2 domains are interconnected by a short middle region consisting of an α‐helical domain and 2 β sheets [18]. The amphipathic α‐helices in the C‐terminal region facilitate protein–protein and protein–lipid interactions [13]. In addition, the CaaX sequence (“aa” indicates 2 aliphatic residues and “X” indicates any amino acid) at the C‐terminus of murine and human GBP1, GBP2, and GBP5 promotes isoprenylation to mediate both protein–protein interactions and targeting of proteins to membranous compartments [4, 7, 19, 20–21]. Regardless of protein modifications, GBPs are predominantly cytoplasmic (mouse GBP5) or within vesicle‐like structures (mouse GBP1, GBP2, GBP3, and GBP6) in the absence of an infection [20, 21–22]. The ability of GBPs to associate with other proteins and to anchor to membranes contributes to their role in pathogen restriction [3, 13, 23, 24]. For example, mouse GBP7 can recruit components of the NADPH oxidase subunits and the autophagy‐associated protein ATG4B to the Mycobacterium‐containing vacuole to promote bacterial killing [25].
Although characterized in the early 1980s, the function of GBPs in host defense has remained relatively obscure until now. Recent studies have unraveled a requirement for GBPs in cell‐autonomous immunity against bacterial, viral, and protozoan pathogens [13, 23, 26]. Moreover, emerging studies elucidated a specific role for GBPs in the activation of the inflammasome, an innate immune caspase‐1–containing protein complex, which drives the release of the proinflammatory cytokines IL‐1β and IL‐18 and the induction of pyroptosis [27]. In this review, we provide an overview of the role of GBPs at the intersection of cell‐autonomous immunity and activation of the inflammasome.
EMERGING ROLES FOR GBPs IN THE ACTIVATION OF THE INFLAMMASOME
The inflammasome is a multiprotein innate immune complex composed of an inflammasome‐initiating sensor, the adaptor protein apoptosis‐associated speck‐like protein containing a CARD and the cysteine protease caspase‐1 [27]. Inflammasome‐initiating sensors include members of the NLRs (e.g., NLRP1, NLRP3, and NLRC4), the pyrin and HIN domain‐containing (also known as PYHIN, AIM2‐like receptors, or ALRs; e.g., AIM2), or the TRIM (e.g., pyrin) family [28]. Previous studies in the inflammasome field have identified a crucial role for type I IFN signaling in the activation of the AIM2 and NLRP3 inflammasomes in response to bacterial infection [29, 30, 31, 32, 33–34]. How type I IFN signaling regulated activation of these inflammasomes was unknown. Recent studies have now identified GBPs as a missing link between IFN signaling and activation of the inflammasome.
In 2012, Shenoy et al. [35] searched protein databases and found GBP‐like proteins in lower organisms Danio and Branchiostoma, which contain a CARD resembling those of inflammasome proteins. Based on this observation, they hypothesized that GBP proteins could have a role in activation of the inflammasome. They generated a mouse strain lacking GBP5 and demonstrated that this GBP was required for activation of the NLRP3 inflammasome in IFN‐γ‐primed mouse BMDMs treated with LPS and the NLRP3 activators ATP or nigericin ( Fig. 2 ) [35]. IFN‐γ priming was not necessary but potentiated activation of the NLRP3 inflammasome by LPS plus ATP [35].
Figure 2.
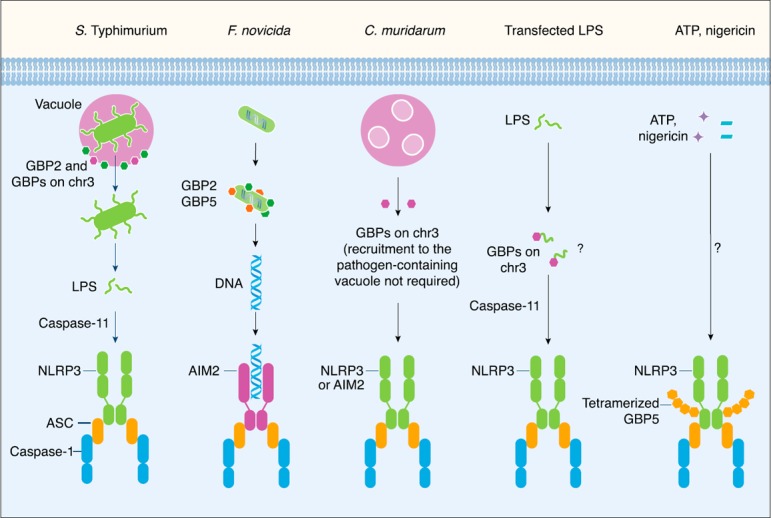
The role of GBPs in the activation of inflammasomes. The gram‐negative bacterium Salmonella Typhimurium normally resides in a vacuole. Mouse GBP2 and other GBP members within the chromosome 3 cluster (GBPs on chr3) are recruited to the vacuole where they induce vacuolar rupture. This process allows the bacteria to become cytosolic, where LPS is sensed by caspase‐11 to drive activation of the noncanonical NLRP3 inflammasome [ 37 ]. However, the gram‐negative bacterium Francisella novicida is a cytosolic pathogen that directly escapes the vacuole and does not activate caspase‐11. Mouse GBP2 and GBP5 directly target the bacteria to induce bacteriolysis and liberate bacterial DNA for sensing by the AIM2 inflammasome [ 38 , 44 ]. Mouse GBPs encoded on chromosome 3 can partially promote pyroptosis in IFN‐γ–primed BMDMs infected with the gram‐negative bacteria Chlamydia trachomatis or Chlamydia muridarum [ 42 ]. Recruitment of GBPs to the pathogen‐containing vacuole is not necessary to drive inflammasome activation in response to C. muridarum infection. There is some evidence to suggest that GBPs encoded on chromosome 3 might also induce pyroptosis in IFN‐γ‐primed BMDMs transfected with LPS [ 41 ]. However, the mechanism regulating this process remains unclear. Human and mouse GBP5 have been shown to undergo tetramerization and to interact with the pyrin domain of NLRP3. Tetramerization of GBP5 might potentiate activation of the NLRP3 inflammasome in response to the soluble NLRP3 activators ATP and nigericin, but not to crystalline NLRP3 activators [ 35 ].
The GBP5‐dependent response appeared to be specific to ATP and nigericin because GBP5 was dispensable for inflammasome activation induced by the NLRP3 activators alum and monosodium urate and the activator of the AIM2 inflammasome poly(dA:dT) dsDNA or the activator of the NLRC4 inflammasome Salmonella flagellin [35]. Experiments using small hairpin RNAs and small interfering RNAs to knock down the expression of GBP5 in a human macrophage cell line further validated the requirement for GBP5 in activation of the NLRP3 inflammasome in response to LPS plus ATP [35]. Using an overexpression system, the authors [35] found that tetramerization of GBP5 promoted assembly of the NLRP3 inflammasome. Tetramerized GBP5 mediated interaction between the GTPase domain of GBP5 and the pyrin domain of NLRP3 via an unknown mechanism [35].
Although this study demonstrated that GBP5 is involved in the activation of the NLRP3 inflammasome induced by soluble (ATP) but not crystalline (alum and monosodium urate) NLRP3 inflammasome activators, it is somewhat surprising that GBP5 is exclusively involved in the activation of the NLRP3 inflammasome induced by a subset of NLRP3 inflammasome activators. Phagosomal rupture and destabilization is a molecular hallmark by which crystalline substances engage activation of the NLRP3 inflammasome [36]. Owing to its ability to target cellular membranes, it is reasonable to speculate that GBP5 may also be involved in mediating rupture of phagosomes containing crystals. Nevertheless, the study by Shenoy et al. [35] unveiled that the function of GBPs and inflammasomes could intersect in higher species to mediate innate immune responses.
Following the initial study by Shenoy et al. [35], Meunier et al. [37] generated 2 independent lines of GBP5‐deficient mouse strains and found that the involvement of GBP5 in the activation of the NLRP3 inflammasome is not always apparent. They found that GBP5 was not required for activation of the NLRP3 inflammasome in LPS‐primed BMDMs stimulated with ATP or nigericin. Our laboratory was also unable to observe a requirement for GBP5 for the activation of the NLRP3 inflammasome in LPS‐primed BMDMs stimulated with ATP [38]. Furthermore, analysis of inflammasome activation performed by Meunier et al. [37] using a mouse strain lacking GBPs encoded on chromosome 3 (GBP1, GBP2, GBP3, GBP5, and GBP7) also revealed that these GBPs are dispensable for the activation of the NLRP3 inflammasome induced by soluble or crystalline activators. These observations are in line with a previous study [29] showing no role for type I IFN signaling in the secretion of IL‐1β in BMDMs treated with LPS plus ATP, suggesting that GBPs—whose expression depends on type I IFN signaling—are dispensable in this process. Why the role for GBP5 in the activation of the canonical NLRP3 inflammasome induced by soluble activators is not consistently apparent has remained unclear. It is possible that prolonged priming by type I or type II IFNs or LPS to induce robust levels of GBP5 is required to fully reveal its role for activation of the NLRP3 inflammasome by soluble NLRP3 inflammasome activators. A more likely explanation is offered by Pilla‐Moffett et al. [24], who highlighted that the Gbp5 −/− mouse line used by Shenoy et al. [35], was generated using 129 embryonic stem cells, resulting in a backcrossed C57BL/6 Gbp5 −/− mouse line carrying the 129 alleles of the adjacent Gbp1, Gbp2, Gbp3, and Gbp7 genes. The C57BL/6 Gbp5 −/− mouse lines used by Man et al. [38] and Meunier et al. [37] were generated using C57BL/6 embryonic stem cells, which carry the C57BL/6 alleles of Gbp1, Gbp2, Gbp3, and Gbp7 genes [24]. Further studies are required to untangle the function of GBP5 in the activation of the canonical NLRP3 inflammasome.
GBPs AND ACTIVATION OF THE INFLAMMASOME BY BACTERIA
In addition to GBP5, a role for other GBPs has also been identified in the activation of inflammasomes by intracellular bacteria. Intracellular bacteria have distinct lifestyles; some are vacuole‐restricted, whereas others disrupt the vacuole and enter the cytoplasm for replication. Type I IFN signaling is essential for activation of the inflammasome by certain vacuolar‐restricted and cytosolic bacteria [29, 32, 33]. Emerging studies have identified GBPs as a missing link between type I IFN signaling and activation of inflammasomes to certain pathogenic bacteria.
Vacuolar bacteria
In 2014, a study by Meunier et al. [37] demonstrated that GBPs encoded on chromosome 3 are essential for activation of the caspase‐11 noncanonical NLRP3 inflammasome in mouse BMDMs infected with the vacuolar‐restricted gram‐negative bacteria Salmonella serovar Typhimurium mutant lacking the Salmonella pathogenicity island‐2 type III secretion system (which preferentially activates caspase‐11 rather than the NLRC4 inflammasome [39, 40]), Vibrio cholerae, Enterobacter cloacae, and Citrobacter koseri [37]. Specifically, GBP2 is required for activation of the caspase‐11–dependent NLRP3 inflammasome in response to these gram‐negative bacteria (Fig. 2) [37]. In addition, Pillar et al. [41] demonstrated the contribution from GBPs encoded on chromosome 3 in caspase‐11–dependent pyroptosis in response to infection by the gram‐negative bacterium Legionella pneumophila. Silencing of the expression of GBPs encoded on chromosome 3 by small hairpin RNAs revealed a partial role for GBP1, GBP2, GBP3 and GBP5 in driving pyroptosis induced by L. pneumophila [41]. A role for GBP2 was further confirmed in mouse BMDMs lacking GBP2 [41].
Further studies have found that GBPs encoded on chromosome 3 can partially promote pyroptosis in IFN‐γ–primed BMDMs infected with the gram‐negative bacteria Chlamydia trachomatis or Chlamydia muridarum (Fig. 2) [42]. GBPs encoded on chromosome 3 can also partially contribute to the secretion of IL‐1β and IL‐18 in LPS‐primed BMDMs infected with C. trachomatis, a pathogen that activates both the AIM2 and NLRP3 inflammasomes [42].
The mechanism by which GBPs contribute to activation of the inflammasomes by vacuolar‐restricted bacteria—at least for the Salmonella Typhimurium serovar—involves the ability of GBP2, and to a smaller extent other GBPs on the chromosome 3 cluster, to target the pathogen‐containing vacuole (Fig. 2). BMDMs lacking GBPs encoded on chromosome 3 harbor less Salmonella Typhimurium associated with galectin‐8, a marker for lysed vacuoles, suggesting that GBP‐mediated expulsion of the bacteria from the vacuole into the cytoplasm is the molecular basis for LPS release and sensing [37].
It is noteworthy that the autophagy marker LC3 was also recruited to Salmonella Typhimurium in a GBP‐dependent manner; however, pharmacological inhibition of autophagy or genetic deletion of ATG5 in BMDMs did not impair activation of the noncanonical NLRP3 inflammasome [37]. This finding suggests that autophagy is a downstream process independent of inflammasome signaling. This study by Meunier et al. [37] provided the first mechanistic link to explain how GBPs govern activation of the noncanonical NLRP3 inflammasome by gram‐negative bacteria that are normally sequestered in the pathogen‐containing vacuole.
Further development in the field has shown that inflammasome activation can occur independent of GBP recruitment to the pathogen. Chlamydia muridarum is resistant to cell‐autonomous immunity mediated by GBPs and does not induce recruitment of GBPs to the pathogen‐containing vacuole to drive activation of the inflammasomes (Fig. 2) [42, 43]. How GBPs regulate inflammasome activation in this context requires further investigation.
In addition to GBP‐mediated liberation of gram‐negative bacteria from the pathogen‐containing vacuole, LPS can be delivered directly into the cytoplasm by transfection techniques or following pathogen‐mediated disruption of phagosomes, resulting in activation of caspase‐11. It is less clear whether GBPs are involved in mediating caspase‐11 sensing of LPS in these scenarios. Meunier et al. [37] found no requirement for GBPs encoded on chromosome 3 in the secretion of IL‐1β and induction of cell death in Pam3CSK4‐primed BMDMs transfected with Escherichia coli or Salmonella minnesota LPS. In contrast, Pillar et al. [41] identified a partial requirement for GBPs encoded on chromosome 3 in the induction of pyroptosis in IFN‐γ‐primed BMDMs transfected with LPS from L. pneumophila, E. coli, or S. minnesota (Fig. 2). It is possible that the subtle effect of GBPs can only be revealed when IFN‐γ priming is used to induce abundant levels of GBPs in the cell.
Cytosolic bacteria
Unlike bacteria that reside within vacuolar compartments, certain bacteria escape from the vacuole into the cytoplasm for replication. GBPs are also essential for driving inflammasome responses to some of these cytosolic pathogens. The cytosolic gram‐negative bacterium Francisella novicida induces type I IFN production, triggering expression of GBPs via the transcription factor IRF1 [38]. Of the GBPs expressed during F. novicida infection, GBP2 and GBP5 are both recruited to the bacteria, where they contribute to the lysis of cytosolic bacteria and the release of bacterial DNA [38, 44]. DNA is detected by AIM2, a cytoplasmic DNA sensor that activates the inflammasome (reviewed in [45, 46]). It is noteworthy that both these GBPs are required for controlling F. novicida burden in BMDMs and in mice [38, 44]. The presence of both GBP2 and GBP5 is required to fully activate the AIM2 inflammasome; however, whether these 2 GBPs are recruited to the bacteria sequentially or are recruited simultaneously has remained unclear. Further studies are required to unravel the mechanisms by which GBPs induce bacteriolysis.
Although GBPs are essential for inflammasome activation during infection with some cytosolic bacteria, the requirement for GBPs in the activation of inflammasomes in response to cytosolic bacteria is not universal. GBPs encoded on chromosome 3 did not appear to have a role for activation of the inflammasome in unprimed BMDMs infected with Burkholderia thailandensis, a cytosolic gram‐negative bacterium known to activate caspase‐11 [37]. In addition, GBPs encoded on chromosome 3 are dispensable for the activation of the noncanonical caspase‐11 NLRP3 inflammasome in response to a genetically engineered strain of Salmonella Typhimurium, which aberrantly enters the cytoplasm because of a lack of the Salmonella vacuole‐maintenance protein, sifA [37]. These studies suggest that, although GBPs encoded on chromosome 3 contribute to activation of the noncanonical caspase‐11 NLRP3 inflammasome in response to vacuolar‐restricted, gram‐negative bacteria, they are not necessary for inflammasome activation during infection with certain cytosolic gram‐negative bacteria that disrupt the vacuole and expose their LPS in the cytoplasm.
It is noteworthy that, when BMDMs were primed with LPS or IFN‐γ to induce robust expression of GBPs, a partial role for GBPs encoded on chromosome 3 for caspase‐11–dependent pyroptosis in response to Salmonella Typhimurium lacking sifA was revealed [37, 41]. In this case, whether GBPs contribute to the destabilization of the Salmonella‐containing vacuole or enhance liberation of LPS by bacteriolysis for sensing by caspase‐11 is not known. Overall, these studies have clearly demonstrated a requirement for GBPs in the activation of inflammasomes in response to certain vacuolar and cytosolic bacteria. However, evidence for the existence of this functional relationship during viral and parasitic infections has remained elusive.
IS THERE A ROLE FOR GBPs IN INFLAMMASOME ACTIVATION DURING VIRAL INFECTION?
Although the induction of an antiviral state is regarded as the hallmark of an IFN response, the importance of IFN‐inducible GBPs in antiviral effector mechanisms has not been thoroughly explored. Earlier studies reported a role for human GBP1 in restricting viral replication and mediating cytopathic effects during infection with VSV and EMCV [47]. Indeed, HeLa cells constitutively expressing human GBP1 produced a lower yield of progeny virions compared with cells which did not constitutively express GBP1 [47]. The antiviral effects of GBP1 were also observed in cells infected with HCV, dengue virus, IAV, and CSFV [48, 49, 50–51]. GBP1 suppresses HCV replication in a GTPase‐dependent manner [48]. However, the NS5B protein of HCV directly binds to GBP1 and inhibits its GTPase activity to counteract the antiviral effects of GBP1 [48]. Similar to the NS5B of HCV, the nonstructural protein 1 of IAV as well as the NS5A protein of CSFV directly interacted with and inhibited the GTPase activity of GBP1 [50, 51]. The presence of these viral antagonists of GBP1 underscores the importance of this GTPase in antiviral host defense.
In addition to human GBP1, mouse GBP2 has antiviral activities against VSV and EMCV [52]. The enzymatic activity of mouse GBP2 is required for restriction of EMCV, whereas VSV inhibition does not rely on the GTP‐binding motif of GBP2 [52]. Anti‐IAV activity was also demonstrated for human GBP3 [53]. A splice variant of human GBP3 with a modified C‐terminal α‐helical domain, called GBP3‐ΔC, confers substantial antiviral activity in epithelial cells by inhibiting the IAV RNA polymerase complex [53]. The antiviral activity of human GBP3‐ΔC was dependent on GTP binding, but GTP hydrolysis was dispensable [53]. Unlike other GBPs with antiviral functions, mouse GBP4 disrupts TRAF6 and IRF7 interaction and negatively regulates type I IFN production and antiviral responses in cells infected with Sendai virus [54].
In addition to various RNA viruses, the antiviral activity of GBPs against retroviruses has also been demonstrated [55, 56]. Overexpression of human GBP5 in HEK293T cells reduces the yield of infectious HIV‐1 NL4‐3 [55]. Moreover, cotransfection of GBP5 with HIV and SIV proviral constructs reduces the yield of infectious virions [56]. GBP5 interferes with virion incorporation of mature envelope protein gp120, and thereby, impairs the ability of virus to infect target cells [56].
Despite various reports demonstrating the importance of GBPs in inflammasome activation in response to canonical inflammasome activators and during bacterial infections, the role of GBPs in mediating inflammasome activation in response to virus infection is not known. A study by Krapp et al. [56] found that silencing of GBP5 in human macrophages does did not impair IL‐1β production in response to HIV. This could be due to an increase in virus replication in the absence of GBP5 that amplifies the signals leading to inflammasome activation. Because GBPs are also important in restricting replication of other viruses, including VSV, EMCV, and IAV, which are known to activate inflammasome, further studies are warranted to investigate whether GBPs positively or negatively regulate inflammasome activation in response to viruses.
IS THERE A ROLE FOR GBPs IN INFLAMMASOME ACTIVATION DURING TOXOPLASMA GONDII INFECTION?
Even though inflammasome activation is studied during parasitic infections, the link between inflammasome activation and GBPs has not been reported for any protozoan parasites. Toxoplasma gondii is one of the model pathogens commonly used to explore cell‐autonomous immunity and the antimicrobial effects of GBPs. Toxoplasma gondii is an obligate, intracellular protozoan and the etiologic agent of toxoplasmosis. The Toxoplasma‐containing vacuole is targeted by multiple GBPs, including mouse GBP1, GBP2, GBP3, GBP5, GBP6, GBP7, and GBP9 [7, 57, 58–59]. However, recruitment of human GBP1 to the Toxoplasma‐containing vacuole is not required for restriction of the parasite in human epithelial cells [60]. Notably, members from a family of IFN‐inducible GTPases closely related to the GBPs called IRGs are also effectively recruited to the Toxoplasma‐containing vacuole [61, 62, 63, 64, 65, 66–67].
Both GBPs and IRGs mediate the rupture of the Toxoplasma‐containing vacuole [61, 68], allowing T. gondii to become cytosolic for subsequent attack by GBP2 [22]. Liberation of the T. gondii into the cytoplasm ultimately results in the death of the parasite and causes pyronecrosis of the host cell, which is characterized by host cell‐membrane permeabilization and release of the alarmin HMGB1 [68]. Based on these observations, it is very likely that GBP‐mediated cell‐autonomous immunity to T. gondii is inherently linked to activation of inflammasomes.
It is known that T. gondii infection can induce IL‐1β secretion and cell death in macrophages by activating the mouse NLRPB1 and NLRP3 or rat NLRP1 inflammasomes [69, 70–71]. Mice lacking the 3 Nlrp1 genes—Nlrp1a, Nlrp1b, and Nlrp1c—or the Nlrp3 gene are more susceptible to T. gondii infection, show increased parasitic load, and reduced serum IL‐18 levels compared with infected wild‐type mice [71], indicating an involvement for multiple inflammasomes in host defense against T. gondii. However, secretion of IL‐1β in mouse BMDMs infected with T. gondii is predominantly dependent on the NLRP3 inflammasome [71]. No published reports, to our knowledge, have examined the direct link between any IFN‐inducible GTPases and activation of these various inflammasomes during T. gondii infection. Future studies are required to unravel any functional relationships that likely exist between GBPs and inflammasomes in the context of T. gondii and other parasitic infections.
CONCLUSIONS
GBPs are major effectors of the IFN‐mediated host defense to a whole range of intracellular pathogens. Recent studies have highlighted divergent roles of GBPs in cell‐autonomous immunity as well as inflammasome activation, although these 2 functions are not mutually exclusive for some pathogens. The function of GBPs usually involves structural modifications of membranes that destroy pathogen‐containing vacuoles or bacterial cell walls and membrane to release pathogen‐associated molecular patterns for detection by innate immune sensors to initiate an immune response, including activation of inflammasomes. Despite being an active area of research, the molecular mechanisms regulating activation of these GTPases and how these proteins combat against compartmentalized vs. cytosolic pathogens is still unclear.
A number of important questions that could be a focus of future studies include (1) Do GBPs target and destroy membranes of pathogen‐containing vacuoles and those of the bacteria, and which components of the membrane are targets for GBPs? (2) What determines the selectivity of GBPs, specifically, why do certain GBPs target bacteria and others do not? (3) GBPs are known to confer protection against viruses and protozoan parasites, but are they involved in mediating inflammasome activation in response to those pathogens? (4) How do GBPs coordinate with one another? (5) Do GBPs guide other unique cellular functions driving inflammasome activation? Unveiling the novel regulatory functions and processes of GBPs will strengthen the tie between this important family of IFN‐inducible GTPases and innate immune responses.
AUTHORSHIP
S.M. Man, D.E. Place, T. Kuriakose, and T.‐D. Kanneganti contributed to the writing of this review.
DISCLOSURES
The authors declare no conflicts of interest.
ACKNOWLEDGMENTS
This work was supported by the U.S. National Institutes of Health (NIH) National Institute of Allergy and Infectious Diseases Grants AI101935, AI124346; NIH National Institute of Arthritis and Musculoskeletal and Skin Diseases Grant AR056296; and NIH National Cancer Institute Grant CA163507 (to T.D.K.); the American Lebanese Syrian Associated Charities (to T.D.K.); and the R.G. Menzies Early Career Fellowship from the National Health and Medical Research Council of Australia (to S.M.M.).
REFERENCES
- 1. McNab, F. , Mayer‐Barber, K. , Sher, A. , Wack, A. , O'Garra, A. (2015) Type I interferons in infectious disease. Nat. Rev. Immunol. 15, 87–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Howard, J. C. (2007) Introduction: cell‐autonomous immunity. Microbes Infect. 9, 1633–1635. [DOI] [PubMed] [Google Scholar]
- 3. MacMicking, J. D. (2012) Interferon‐inducible effector mechanisms in cell‐autonomous immunity. Nat. Rev. Immunol. 12, 367–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vestal, D. J. , Jeyaratnam, J. A. (2011) The guanylate‐binding proteins: emerging insights into the biochemical properties and functions of this family of large interferon‐induced guanosine triphosphatase. J. Interferon Cytokine Res. 31, 89–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gupta, S. L. , Rubin, B. Y. , Holmes, S. L. (1979) Interferon action: induction of specific proteins in mouse and human cells by homologous interferons. Proc. Natl. Acad. Sci. U.S.A. 76, 4817–4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cheng, Y. S. , Patterson, C. E. , Staeheli, P. (1991) Interferon‐induced guanylate‐binding proteins lack an N(T)KXD consensus motif and bind GMP in addition to GDP and GTP. Mol. Cell. Biol. 11, 4717–4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Degrandi, D. , Konermann, C. , Beuter‐Gunia, C. , Kresse, A. , Würthner, J. , Kurig, S. , Beer, S. , Pfeffer, K. (2007) Extensive characterization of IFN‐induced GTPases mGBP1 to mGBP10 involved in host defense. J. Immunol. 179, 7729–7740. [DOI] [PubMed] [Google Scholar]
- 8. Nguyen, T. T. , Hu, Y. , Widney, D. P. , Mar, R. A. , Smith, J. B. (2002) Murine GBP‐5, a new member of the murine guanylate‐binding protein family, is coordinately regulated with other GBPs in vivo and in vitro. J. Interferon Cytokine Res. 22, 899–909. [DOI] [PubMed] [Google Scholar]
- 9. Staeheli, P. , Colonno, R. J. , Cheng, Y. S. (1983) Different mRNAs induced by interferon in cells from inbred mouse strains A/J and A2G. J. Virol. 47, 563–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vestal, D. J. , Buss, J. E. , McKercher, S. R. , Jenkins, N. A. , Copeland, N. G. , Kelner, G. S. , Asundi, V. K. , Maki, R. A. (1998) Murine GBP‐2: a new IFN‐γ–induced member of the GBP family of GTPases isolated from macrophages. J. Interferon Cytokine Res. 18, 977–985. [DOI] [PubMed] [Google Scholar]
- 11. Olszewski, M. A. , Gray, J. , Vestal, D. J. (2006) In silico genomic analysis of the human and murine guanylate‐binding protein (GBP) gene clusters. J. Interferon Cytokine Res. 26, 328–52. [DOI] [PubMed] [Google Scholar]
- 12. Kresse, A. , Konermann, C. , Degrandi, D. , Beuter‐Gunia, C. , Wuerthner, J. , Pfeffer, K. , Beer, S. (2008) Analyses of murine GBP homology clusters based on in silico, in vitro and in vivo studies. BMC Genomics 9, 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim, B. H. , Shenoy, A. R. , Kumar, P. , Bradfield, C. J. , MacMicking, J. D. (2012) IFN‐inducible GTPases in host cell defense. Cell Host Microbe 12, 432–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Praefcke, G. J. , Geyer, M. , Schwemmle, M. , Robert Kalbitzer, H. , Herrmann, C. (1999) Nucleotide‐binding characteristics of human guanylate‐binding protein 1 (hGBP1) and identification of the third GTP‐binding motif. J. Mol. Biol. 292, 321–332. [DOI] [PubMed] [Google Scholar]
- 15. Prakash, B. , Renault, L. , Praefcke, G. J. , Herrmann, C. , Wittinghofer, A. (2000) Triphosphate structure of guanylate‐binding protein 1 and implications for nucleotide binding and GTPase mechanism. EMBO J. 19, 4555–4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vestal, D. J. (2005) The guanylate‐binding proteins (GBPs): proinflammatory cytokine‐induced members of the dynamin superfamily with unique GTPase activity. J. Interferon Cytokine Res. 25, 435–443. [DOI] [PubMed] [Google Scholar]
- 17. Schwemmle, M. , Staeheli, P. (1994) The interferon‐induced 67‐kDa guanylate‐binding protein (hGBP1) is a GTPase that converts GTP to GMP. J. Biol. Chem. 269, 11299–11305. [PubMed] [Google Scholar]
- 18. Prakash, B. , Praefcke, G. J. , Renault, L. , Wittinghofer, A. , Herrmann, C. (2000) Structure of human guanylate‐binding protein 1 representing a unique class of GTP‐binding proteins. Nature 403, 567–571. [DOI] [PubMed] [Google Scholar]
- 19. Britzen‐Laurent, N. , Bauer, M. , Berton, V. , Fischer, N. , Syguda, A. , Reipschläger, S. , Naschberger, E. , Herrmann, C. , Stürzl, M. (2010) Intracellular trafficking of guanylate‐binding proteins is regulated by heterodimerization in a hierarchical manner. PLoS One 5, e14246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vestal, D. J. , Gorbacheva, V. Y. , Sen, G. C. (2000) Different subcellular localizations for the related interferon‐induced GTPases, MuGBP‐1 and MuGBP‐2: implications for different functions? J. Interferon Cytokine Res. 20, 991–1000. [DOI] [PubMed] [Google Scholar]
- 21. Nantais, D. E. , Schwemmle, M. , Stickney, J. T. , Vestal, D. J. , Buss, J. E. (1996) Prenylation of an interferon‐gamma‐induced GTP‐binding protein: the human guanylate binding protein, huGBP1. J. Leukoc. Biol. 60, 423–431. [DOI] [PubMed] [Google Scholar]
- 22. Kravets, E. , Degrandi, D. , Ma, Q. , Peulen, T. O. , Klumpers, V. , Felekyan, S. , Kuhnemuth, R. , Weidtkamp‐Peters, S. , Seidel, C. A. , Pfeffer, K. (2016) Guanylate binding proteins (GBPs) directly attack via supramolecular complexes. Elife 5, e11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Meunier, E. , Broz, P. (2016) Interferon‐inducible GTPases in cell autonomous and innate immunity. Cell. Microbiol. 18, 168–180. [DOI] [PubMed] [Google Scholar]
- 24. Pilla‐Moffett, D. , Barber, M. F. , Taylor, G. A. , Coers, J. (2016) Interferon‐inducible GTPases in host resistance, inflammation and disease. J. Mol. Biol. doi: 10.1016/j.jmb.2016.04.032 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim, B. H. , Shenoy, A. R. , Kumar, P. , Das, R. , Tiwari, S. , MacMicking, J. D. (2011) A family of IFN‐γ‐inducible 65‐kD GTPases protects against bacterial infection. Science 332, 717–721. [DOI] [PubMed] [Google Scholar]
- 26. Kim, B. H. , Chee, J. D. , Bradfield, C. J. , Park, E. S. , Kumar, P. , MacMicking, J. D. (2016) Interferon‐induced guanylate‐binding proteins in inflammasome activation and host defense. Nat. Immunol. 17, 481–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Man, S. M. , Kanneganti, T. D. (2016) Converging roles of caspases in inflammasome activation, cell death and innate immunity. Nat. Rev. Immunol. 16, 7–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Man, S. M. , Kanneganti, T. D. (2015) Regulation of inflammasome activation. Immunol. Rev. 265, 6–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Henry, T. , Brotcke, A. , Weiss, D. S. , Thompson, L. J. , Monack, D. M. (2007) Type I interferon signaling is required for activation of the inflammasome during Francisella infection. J. Exp. Med. 204, 987–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sander, L. E. , Davis, M. J. , Boekschoten, M. V. , Amsen, D. , Dascher, C. C. , Ryffel, B. , Swanson, J. A. , Müller, M. , Blander, J. M. (2011) Detection of prokaryotic mRNA signifies microbial viability and promotes immunity. Nature 474, 385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cole, L. E. , Santiago, A. , Barry, E. , Kang, T. J. , Shirey, K. A. , Roberts, Z. J. , Elkins, K. L. , Cross, A. S. , Vogel, S. N. (2008) Macrophage proinflammatory response to Francisella tularensis live vaccine strain requires coordination of multiple signaling pathways. J. Immunol. 180, 6885–6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Broz, P. , Ruby, T. , Belhocine, K. , Bouley, D. M. , Kayagaki, N. , Dixit, V. M. , Monack, D. M. (2012) Caspase‐11 increases susceptibility to Salmonella infection in the absence of caspase‐1. Nature 490, 288–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rathinam, V. A. , Vanaja, S. K. , Waggoner, L. , Sokolovska, A. , Becker, C. , Stuart, L. M. , Leong, J. M. , Fitzgerald, K. A. (2012) TRIF licenses caspase‐11‐dependent NLRP3 inflammasome activation by gram‐negative bacteria. Cell 150, 606–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gurung, P. , Malireddi, R. K. , Anand, P. K. , Demon, D. , Vande Walle, L. , Liu, Z. , Vogel, P. , Lamkanfi, M. , Kanneganti, T. D. (2012) Toll or interleukin‐1 receptor (TIR) domain‐containing adaptor inducing interferon‐β (TRIF)‐mediated caspase‐11 protease production integrates Toll‐like receptor 4 (TLR4) protein‐ and Nlrp3 inflammasome‐mediated host defense against enteropathogens. J. Biol. Chem. 287, 34474–34483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shenoy, A. R. , Wellington, D. A. , Kumar, P. , Kassa, H. , Booth, C. J. , Cresswell, P. , MacMicking, J. D. (2012) GBP5 promotes NLRP3 inflammasome assembly and immunity in mammals. Science 336, 481–485. [DOI] [PubMed] [Google Scholar]
- 36. Hornung, V. , Bauernfeind, F. , Halle, A. , Samstad, E. O. , Kono, H. , Rock, K. L. , Fitzgerald, K. A. , Latz, E. (2008) Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat. Immunol. 9, 847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Meunier, E. , Dick, M. S. , Dreier, R. F. , Schürmann, N. , Kenzelmann Broz, D. , Warming, S. , Roose‐Girma, M. , Bumann, D. , Kayagaki, N. , Takeda, K. , Yamamoto, M. , Broz, P. (2014) Caspase‐11 activation requires lysis of pathogen‐containing vacuoles by IFN‐induced GTPases. Nature 509, 366–370. [DOI] [PubMed] [Google Scholar]
- 38. Man, S. M. , Karki, R. , Malireddi, R. K. , Neale, G. , Vogel, P. , Yamamoto, M. , Lamkanfi, M. , Kanneganti, T. D. (2015) The transcription factor IRF1 and guanylate‐binding proteins target activation of the AIM2 inflammasome by Francisella infection. Nat. Immunol. 16, 467–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Broz, P. , Newton, K. , Lamkanfi, M. , Mariathasan, S. , Dixit, V. M. , Monack, D. M. (2010) Redundant roles for inflammasome receptors NLRP3 and NLRC4 in host defense against Salmonella . J. Exp. Med. 207, 1745–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Man, S. M. , Hopkins, L. J. , Nugent, E. , Cox, S. , Glück, I. M. , Tourlomousis, P. , Wright, J. A. , Cicuta, P. , Monie, T. P. , Bryant, C. E. (2014) Inflammasome activation causes dual recruitment of NLRC4 and NLRP3 to the same macromolecular complex. Proc. Natl. Acad. Sci. U.S.A. 111, 7403–7408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pilla, D. M. , Hagar, J. A. , Haldar, A. K. , Mason, A. K. , Degrandi, D. , Pfeffer, K. , Ernst, R. K. , Yamamoto, M. , Miao, E. A. , Coers, J. (2014) Guanylate binding proteins promote caspase‐11‐dependent pyroptosis in response to cytoplasmic LPS. Proc. Natl. Acad. Sci. U.S.A. 111, 6046–6051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Finethy, R. , Jorgensen, I. , Haldar, A. K. , de Zoete, M. R. , Strowig, T. , Flavell, R. A. , Yamamoto, M. , Nagarajan, U. M. , Miao, E. A. , Coers, J. (2015) Guanylate binding proteins enable rapid activation of canonical and noncanonical inflammasomes in Chlamydia‐infected macrophages. Infect. Immun. 83, 4740–4749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Coers, J. , Bernstein‐Hanley, I. , Grotsky, D. , Parvanova, I. , Howard, J. C. , Taylor, G. A. , Dietrich, W. F. , Starnbach, M. N. (2008) Chlamydia muridarum evades growth restriction by the IFN‐γ–inducible host resistance factor Irgb10. J. Immunol. 180, 6237–6245. [DOI] [PubMed] [Google Scholar]
- 44. Meunier, E. , Wallet, P. , Dreier, R. F. , Costanzo, S. , Anton, L. , Rühl, S. , Dussurgey, S. , Dick, M. S. , Kistner, A. , Rigard, M. , Degrandi, D. , Pfeffer, K. , Yamamoto, M. , Henry, T. , Broz, P. (2015) Guanylate‐binding proteins promote activation of the AIM2 inflammasome during infection with Francisella novicida . Nat. Immunol. 16, 476–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Man, S. M. , Karki, R. , Kanneganti, T. D. (2016) AIM2 inflammasome in infection, cancer, and autoimmunity: Role in DNA sensing, inflammation, and innate immunity. Eur. J. Immunol. 46, 269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Man, S. M. , Karki, R. , Kanneganti, T. D. (2016) DNA‐sensing inflammasomes: regulation of bacterial host defense and the gut microbiota. Pathog. Dis. 74, ftw028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Anderson, S. L. , Carton, J. M. , Lou, J. , Xing, L. , Rubin, B. Y. (1999) Interferon‐induced guanylate binding protein‐1 (GBP‐1) mediates an antiviral effect against vesicular stomatitis virus and encephalomyocarditis virus. Virology 256, 8–14. [DOI] [PubMed] [Google Scholar]
- 48. Itsui, Y. , Sakamoto, N. , Kakinuma, S. , Nakagawa, M. , Sekine‐Osajima, Y. , Tasaka‐Fujita, M. , Nishimura‐Sakurai, Y. , Suda, G. , Karakama, Y. , Mishima, K. , Yamamoto, M. , Watanabe, T. , Ueyama, M. , Funaoka, Y. , Azuma, S. , Watanabe, M. (2009) Antiviral effects of the interferon‐induced protein guanylate binding protein 1 and its interaction with the hepatitis C virus NS5B protein. Hepatology 50, 1727–1737. [DOI] [PubMed] [Google Scholar]
- 49. Pan, W. , Zuo, X. , Feng, T. , Shi, X. , Dai, J. (2012) Guanylate‐binding protein 1 participates in cellular antiviral response to dengue virus. Virol. J. 9, 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhu, Z. , Shi, Z. , Yan, W. , Wei, J. , Shao, D. , Deng, X. , Wang, S. , Li, B. , Tong, G. , Ma, Z. (2013) Nonstructural protein 1 of influenza A virus interacts with human guanylate‐binding protein 1 to antagonize antiviral activity. PLoS One 8, e55920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li, L. F. , Yu, J. , Li, Y. , Wang, J. , Li, S. , Zhang, L. , Xia, S. L. , Yang, Q. , Wang, X. , Yu, S. , Luo, Y. , Sun, Y. , Zhu, Y. , Munir, M. , Qiu, H. J. (2016) Guanylate‐binding protein 1, an interferon‐induced GTPase, exerts an antiviral activity against classical swine fever virus depending on its GTPase activity. J. Virol. 90, 4412–4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Carter, C. C. , Gorbacheva, V. Y. , Vestal, D. J. (2005) Inhibition of VSV and EMCV replication by the interferon‐induced GTPase, mGBP‐2: differential requirement for wild‐type GTP binding domain. Arch. Virol. 150, 1213–1220. [DOI] [PubMed] [Google Scholar]
- 53. Nordmann, A. , Wixler, L. , Boergeling, Y. , Wixler, V. , Ludwig, S. (2012) A new splice variant of the human guanylate‐binding protein 3 mediates anti‐influenza activity through inhibition of viral transcription and replication. FASEB J. 26, 1290–1300. [DOI] [PubMed] [Google Scholar]
- 54. Hu, Y. , Wang, J. , Yang, B. , Zheng, N. , Qin, M. , Ji, Y. , Lin, G. , Tian, L. , Wu, X. , Wu, L. , Sun, B. (2011) Guanylate binding protein 4 negatively regulates virus‐induced type I IFN and antiviral response by targeting IFN regulatory factor 7. J. Immunol. 187, 6456–6462. [DOI] [PubMed] [Google Scholar]
- 55. McLaren, P. J. , Gawanbacht, A. , Pyndiah, N. , Krapp, C. , Hotter, D. , Kluge, S. F. , Götz, N. , Heilmann, J. , Mack, K. , Sauter, D. , Thompson, D. , Perreaud, J. , Rausell, A. , Munoz, M. , Ciuffi, A. , Kirchhoff, F. , Telenti, A. (2015) Identification of potential HIV restriction factors by combining evolutionary genomic signatures with functional analyses. Retrovirology 12, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Krapp, C. , Hotter, D. , Gawanbacht, A. , McLaren, P. J. , Kluge, S. F. , Sturzel, C. M. , Mack, K. , Reith, E. , Engelhart, S. , Ciuffi, A. , Hornung, V. , Sauter, D. , Telenti, A. , Kirchhoff, F. (2016) Guanylate binding protein (GBP) 5 is an interferon‐inducible inhibitor of HIV‐1 infectivity. Cell Host Microbe. 19, 504–514. [DOI] [PubMed] [Google Scholar]
- 57. Virreira Winter, S. , Niedelman, W. , Jensen, K. D. , Rosowski, E. E. , Julien, L. , Spooner, E. , Caradonna, K. , Burleigh, B. A. , Saeij, J. P. , Ploegh, H. L. , Frickel, E. M. (2011) Determinants of GBP recruitment to Toxoplasma gondii vacuoles and the parasitic factors that control it. PLoS One 6, e24434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yamamoto, M. , Okuyama, M. , Ma, J. S. , Kimura, T. , Kamiyama, N. , Saiga, H. , Ohshima, J. , Sasai, M. , Kayama, H. , Okamoto, T. , Huang, D. C. , Soldati‐Favre, D. , Horie, K. , Takeda, J. , Takeda, K. (2012) A cluster of interferon‐γ–inducible p65 GTPases plays a critical role in host defense against Toxoplasma gondii. Immunity 37, 302–313. [DOI] [PubMed] [Google Scholar]
- 59. Degrandi, D. , Kravets, E. , Konermann, C. , Beuter‐Gunia, C. , Klümpers, V. , Lahme, S. , Wischmann, E. , Mausberg, A. K. , Beer‐Hammer, S. , Pfeffer, K. (2013) Murine guanylate binding protein 2 (mGBP2) controls Toxoplasma gondii replication. Proc. Natl. Acad. Sci. U.S.A. 110, 294–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Johnston, A. C. , Piro, A. , Clough, B. , Siew, M. , Virreira Winter, S. , Coers, J. , Frickel, E. M. (2016) Human GBP1 does not localise to pathogen vacuoles but restricts Toxoplasma gondii . Cell. Microbiol. doi: 10.1111/cmi.12579 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Martens, S. , Parvanova, I. , Zerrahn, J. , Griffiths, G. , Schell, G. , Reichmann, G. , Howard, J. C. (2005) Disruption of Toxoplasma gondii parasitophorous vacuoles by the mouse p47‐resistance GTPases. PLoS Pathog. 1, e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Haldar, A. K. , Saka, H. A. , Piro, A. S. , Dunn, J. D. , Henry, S. C. , Taylor, G. A. , Frickel, E. M. , Valdivia, R. H. , Coers, J. (2013) IRG and GBP host resistance factors target aberrant, “non‐self” vacuoles characterized by the missing of “self” IRGM proteins. PLoS Pathog. 9, e1003414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Butcher, B. A. , Greene, R. I. , Henry, S. C. , Annecharico, K. L. , Weinberg, J. B. , Denkers, E. Y. , Sher, A. , Taylor, G. A. (2005) p47 GTPases regulate Toxoplasma gondii survival in activated macrophages. Infect. Immun. 73, 3278–3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hunn, J. P. , Koenen‐Waisman, S. , Papic, N. , Schroeder, N. , Pawlowski, N. , Lange, R. , Kaiser, F. , Zerrahn, J. , Martens, S. , Howard, J. C. (2008) Regulatory interactions between IRG resistance GTPases in the cellular response to Toxoplasma gondii . EMBO J. 27, 2495–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Henry, S. C. , Daniell, X. G. , Burroughs, A. R. , Indaram, M. , Howell, D. N. , Coers, J. , Starnbach, M. N. , Hunn, J. P. , Howard, J. C. , Feng, C. G. , Sher, A. , Taylor, G. A. (2009) Balance of Irgm protein activities determines IFN‐γ–induced host defense. J. Leukoc. Biol. 85, 877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhao, Y. , Ferguson, D. J. , Wilson, D. C. , Howard, J. C. , Sibley, L. D. , Yap, G. S. (2009) Virulent Toxoplasma gondii evade immunity‐related GTPase‐mediated parasite vacuole disruption within primed macrophages. J. Immunol. 182, 3775–3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Khaminets, A. , Hunn, J. P. , Könen‐Waisman, S. , Zhao, Y. O. , Preukschat, D. , Coers, J. , Boyle, J. P. , Ong, Y. C. , Boothroyd, J. C. , Reichmann, G. , Howard, J. C. (2010) Coordinated loading of IRG resistance GTPases on to the Toxoplasma gondii parasitophorous vacuole. Cell. Microbiol. 12, 939–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhao, Y. O. , Khaminets, A. , Hunn, J. P. , Howard, J. C. (2009) Disruption of the Toxoplasma gondii parasitophorous vacuole by IFNgamma‐inducible immunity‐related GTPases (IRG proteins) triggers necrotic cell death. PLoS Pathog. 5, e1000288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ewald, S. E. , Chavarria‐Smith, J. , Boothroyd, J. C. (2014) NLRP1 is an inflammasome sensor for Toxoplasma gondii . Infect. Immun. 82, 460–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cirelli, K. M. , Gorfu, G. , Hassan, M. A. , Printz, M. , Crown, D. , Leppla, S. H. , Grigg, M. E. , Saeij, J. P. , Moayeri, M. (2014) Inflammasome sensor NLRP1 controls rat macrophage susceptibility to Toxoplasma gondii . PLoS Pathog. 10, e1003927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Gorfu, G. , Cirelli, K. M. , Melo, M. B. , Mayer‐Barber, K. , Crown, D. , Koller, B. H. , Masters, S. , Sher, A. , Leppla, S. H. , Moayeri, M. , Saeij, J. P. , Grigg, M. E. (2014) Dual role for inflammasome sensors NLRP1 and NLRP3 in murine resistance to Toxoplasma gondii . MBio 5, e01117–13. [DOI] [PMC free article] [PubMed] [Google Scholar]


