Short abstract
Gr‐1 expression on γδ T cells helps differentiate IFNγ‐ and IL‐17A‐producing subsets involved in the early immune response to bacterial pneumonia.
Keywords: pneumonia, immune response, IFN‐γ, IL‐17
Abstract
T cell receptor γδ cells are known to be the primary effector T cells involved in the response to bacterial infections, yet their phenotypic characteristics are not as well established as other T cell subsets. In this study, we used cytometry by time‐of‐flight mass cytometry to better characterize the phenotypic response of T cell receptor γδ cells to Streptococcus pneumoniae lung infection. Mice were infected, and cells from lung washouts, spleen, and lymph nodes were stained to detect cell‐surface, intracellular, and signaling markers. We observed that infection caused a significant increase in T cell receptor γδ cells, which expressed high interferon‐γ and interleukin‐17A levels. Profiling T cell receptor γδ cells by cytometry by time‐of‐flight revealed that activated γδ T cells uniquely coexpressed cell‐surface Gr‐1, cluster of differentiation 14, and cluster of differentiation 274 (programmed death‐ligand 1). Further classification of Gr‐1 expression patterns on T cell receptor γδ cells demonstrated that Gr‐1+ T cell receptor γδ cells were the primary source of interferon‐γ, whereas Gr‐1− cells mostly expressed interleukin‐17A. Gr‐1+ T cell receptor γδ cells also showed higher ζ‐chain–associated protein kinase 70, p38, and 4eBP1 signaling in response to infection as compared with Gr‐1− T cell receptor γδ cells. Taken together, Gr‐1 expression patterns on γδ T cells in the lung provide a robust marker to differentiate interferon‐γ– and interleukin‐17A–producing subsets involved in the early immune response to bacterial pneumonia.
Abbreviations
- BAL
bronchoalveolar lavage
- CyTOF
cytometry by time‐of‐flight
- PD‐L1
programmed death‐ligand 1
- PFA
paraformaldehyde
- SPADE
spanning tree progression of density normalized events
- Vγ
variable‐γ
- ZAP70
ζ‐chain–associated protein kinase 70
Introduction
T cells that express the γδ TCR have a major role in immune surveillance in microbial‐exposed epithelia, such as the lung, the gastrointestinal tract, the reproductive tract, and the skin [1]. The localization of γδ T cells to these compartments suggests that they maintain antimicrobial immune function and homeostasis in microbial‐rich tissues. For example, the upper respiratory tract in humans is colonized by gram‐positive Streptococcus pneumoniae, and although colonization is asymptomatic in healthy individuals, S. pneumoniae can cause community‐acquired pneumonia [2, 3]. Although γδ T cells comprise a small percentage of immune cells in the lung, they have been shown to be required as a first line of defense against lung infections by producing chemotactic cytokines that contribute to the recruitment of neutrophils and macrophages to the sites of infection [4, 5]. In addition, γδ T cells are capable of secreting large amounts of IL‐17 and IFN‐γ, which activate neutrophils, macrophages, and cytotoxic lymphocytes to enhance antimicrobial immunity [6].
Traditionally, γδ T cells in mice were classified based on their tissue distribution and TCR Vγ chain expression patterns. As such, it was reported [7] that activated γδ T cells expressing Vγ1 secrete IFN‐γ, whereas cells expressing Vγ4 chains produce IL‐17. However, the results of another study [6] showed that γδ T cell phenotype and function are not solely determined by different Vγ chain expression profiles. Instead, γδ T cells expressing CD27 and NK1.1 produce IFN‐γ, whereas CD27− cells expressing CCR6 produced IL‐17 [8]. In a manner similar to describing helper CD4+ T cell subsets, for example, IFN‐γ–producing T cells as Th1 cells and IL‐17–producing cells as Th17 cells, these authors suggested using CD27 and CCR6 as markers to distinguish subsets of proinflammatory γδ T cells.
We recently discovered that injured mice showed defective γδ T cell reactivity to lung infections caused by S. pneumoniae (manuscript in preparation). Given the importance of γδ T cells in regulating the early response to infections, we wished to characterize γδ T cell subtypes involved in S. pneumoniae–induced lung infection. To accomplish this, we used CyTOF mass cytometry, which allowed us to perform simultaneous detection of multiple cell‐surface markers, transcription factors, and cytokines in BAL fluid, spleen, and lymph nodes prepared from S. pneumoniae–infected and uninfected groups of mice. CyTOF is a relatively new, single‐cell cytometry method that has several key advantages over conventional fluorescence‐based flow cytometry, including multiplex‐staining capabilities with low background and high sensitivity as reviewed by others [9] and Bendall et al [10]. Because CyTOF allows for simultaneous staining with multiple combinations of cell phenotyping markers, it can reveal new immune cell populations that would not otherwise be uncovered by conventional flow cytometry. In addition, studying the complexity of immune cell reactivity to lung infection is well‐suited to CyTOF because of the variety of cell types involved in this host response.
Here, we used CyTOF to identify γδ T cell responses to S. pneumoniae lung infection. Our findings demonstrate γδ T cell populations that are activated and involved in the acute response to S. pneumoniae lung infection in mice. Specifically, we show that γδ T cells expressing Gr‐1 represent the primary reactive T cell population in the lung airways of mice infected with bacteria. The Gr‐1+ γδ T cell subset was identified as a major source of IFN‐γ during active lung infection. By CyTOF analysis, we also determined that Gr‐1+ γδ T cells uniquely coexpress a high density of CD14 and CD274 (PD‐L1) but not CD27, a marker usually expressed on IFN‐γ–producing γδ T cells. Finally, we compared infection‐induced signaling response profiles between Gr‐1+ or Gr‐1− γδ T cells by CyTOF and found differences in ZAP70 and p38 MAPK pathway activation. Collectively, the data provide a comprehensive view of γδ T cell populations involved in the early response to lung infections and identify unique markers for γδ T cells involved in the host immune response to bacterial infections.
MATERIALS AND METHODS
Mice
Outbred, male CD‐1 mice (6–8 wk) were purchased from Charles River Laboratories (Wilmington, MA, USA). We intentionally used outbred mice for this study to generate robust population infection response data that cannot be collected using genetically identical inbred mice. All mice were maintained in our full‐barrier animal facility under controlled temperature and humidity, with a 12‐h light–dark regimen, and were provided with standard chow and water ad libitum. Mice were acclimatized for at least 1 wk before being used in the experiments. All animal protocols performed in this study were approved by the Harvard Medical School Standing Committee on Animal Research and were found to be in accordance with guidelines set by the U.S. Department of Agriculture (Washington, DC, USA) and the National Institutes of Health (Bethesda, MD, USA).
Streptococcus pneumoniae lung infection model
Streptococcus pneumoniae (strain 99.55, capsular subtype 6A) were maintained as frozen stocks at −80°C. Bacteria were grown for 16 h with gentle agitation in brain–heart infusion broth medium at 37°C. Bacteria was harvested at a midlog phase by centrifugation at 450 g and was washed twice by centrifugation with Dulbecco's PBS. Based on absorption spectroscopy measurements, bacteria were diluted to contain 3–5 × 107 CFUs/ml in PBS. This inoculum dose was found to cause significant mortality in normal, male CD‐1 mice during a 14‐d period, with deaths first occurring at d 4–5 after infection ( Fig. 1 ). To produce infection, mice were anesthetized by i.p. injection using Ketamine/Xylazine at 125/10 mg/kg. Mice were then held with their nares upright, and 40 μl of bacteria suspension was administered intranasal (1.2–2 × 106 CFUs). Control mice received anesthesia without being inoculated. Bacteria CFUs were quantified by drop‐plating of serial dilutions on tryptic soy agar plates, and colonies were counted the following day after incubating the tryptic soy agar plates at 37°C.
Figure 1.
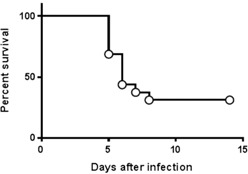
Survival curve for S. pneumoniae lung infection model. Male CD‐1 mice were infected intranasally with 1 × 106 CFUs of S. pneumoniae (n = 16). Mice were monitored daily for 14 d after infection to measure death from infection. The data shown indicate the first deaths from infection occurred on d 5, and the overall survival rate after 14 d was 31.25% by 8 d after infection.
Reagents
Cells were prepared in culture medium (C5); RPMI 1640 was supplemented with 5% heat‐inactivated FCS, 1 mM glutamine, 10 mM HEPES, 100× 2 M nonessential amino acids, penicillin/streptomycin/fungizone, and 2.5 × 10−5 M 2‐ME, all purchased from Gibco‐Invitrogen (Grand Island, NY, USA). Cells were fixed and permeabilized for conventional flow cytometry using PBS with PFA purchased from Alfa Aesar (Ward Hill, MA, USA) and HPLC‐grade methanol purchased from Sigma Chemical Company (St Louis, MO, USA). Fluorescent‐labeled antibodies specific for mouse CD3, CD27, CCR6, and TCRγδ for flow cytometry stains were purchased from BioLegend (San Diego, CA, USA). TruStain FcX Fc receptor blocking reagent from BioLegend was used in all stains to prevent nonspecific staining signals. For CyTOF mass cytometry, antibodies were strategically labeled with rare‐earth metal isotopes using MaxPar reagent kits from Fluidigm Sciences/DVS Sciences (Sunnyvale, CA, USA). The CyTOF staining panels used in this study are shown in Table 1 .
Table 1.
CyTOF antibody panels
| Cell‐surface and intracellular | Signaling | ||||
|---|---|---|---|---|---|
| Marker | Clone | Isotope label | Marker | Clone | Isotope label |
| CD115 | AFS98 | 142Nd | CD8α | 53‐6.7 | 142Nd |
| CD4 | RM4‐5 | 145Nd | CD4 | RM4‐5 | 145Nd |
| CD62L | MEL‐14 | 146Nd | CD120b | TR75‐54.7 | 147Sm |
| CD120b | TR75‐54.7 | 147Sm | CD274 | 10F.9G2 | 149Sm |
| TNF‐α | MP6‐XT22 | 148Nd | TCRβ | H57‐597 | 150Nd |
| CD274 | 10F.9G2 | 149Sm | CD27 | LG.3A10 | 151Eu |
| TCR‐β | H57‐597 | 150Nd | CD3ɛ | 145‐2C11 | 152Sm |
| CD27 | LG.3A10 | 151Eu | CD172a | P84 | 153Eu |
| CD3‐ɛ | 145‐2C11 | 152Sm | TCRγδ | GL3 | 154Sm |
| CD172a | P84 | 153Eu | NK1.1 | PK136 | 155Gd |
| IL‐17A | TC11‐18H10.1 | 154Sm | CD14 | Sa14‐2 | 156Gd |
| NK1.1 | PK136 | 155Gd | Tbet | 4B10 | 158Gd |
| CD14 | Sa14‐2 | 156Gd | CD23 | B3B4 | 159Tb |
| IL‐10 | JES5‐16E3 | 158Gd | ICOS | C398.A4 | 160Gd |
| IFN‐γ | XMG1.2 | 159Tb | FoxP3 | FJK‐16s | 162Dy |
| ICOS | C398.A4 | 160Gd | pZAP70 | (Y493/Y526) | 164Dy |
| FoxP3 | FJK‐16s | 162Dy | CD45 | 30‐F11 | 165Ho |
| CD8‐α | 53‐6.7 | 164Dy | p38 | T180/Y182 | 166Er |
| CD45 | 30‐F11 | 165Ho | pERK | T202/Y204 | 167Er |
| Gata‐3 | TWAJ | 166Er | CD64 | x54‐5/7.1 | 168Er |
| CD25 | 3C7 | 167Er | CCR6 | 29‐2L17 | 169Tm |
| CD64 | x54‐5/7.1 | 168Er | RoRγt | AFKJS‐9 | 170Er |
| CCR6 | 29‐2L17 | 169Tm | CD49b | DX5 | 171Yb |
| RoRγt | AFKJS‐9 | 170Er | 4eBP1 | 236B4 | 172Yb |
| CD49b | DX5 | 171Yb | I‐A/I‐E | M5/114.15.2 | 174Yb |
| TCRγδ | GL3 | 172Yb | pS6K | S235/236 | 175Lu |
| I‐A/I‐E | M5/114.15.2 | 174Yb | Gr‐1 | RB6‐8C5 | 176Yb |
| CCR4 | 2G12 | 175Lu | |||
| Gr‐1 | RB6‐8C5 | 176Yb | |||
Spleen, lymph node, and BAL cell preparations
Cells were prepared from CD‐1 mice 1 d after S. pneumoniae or sham infection. Mice were euthanized by CO2 asphyxiation. Spleen and lymph nodes (inguinal, axillary, and mediastinal) were harvested and immediately placed in ice‐cold C5 medium. To collect BAL fluid, the trachea was dissected and then cannulated with the outer sheath of a 22‐gauge intravenous catheter (Exel International, Los Angeles, CA, USA). The lung was rinsed repeatedly with a total of 3 ml of ice‐cold calcium/magnesium‐free PBS containing 2 mM EDTA. This method allowed us to measure immune cells that specifically entered the site of infection rather than the whole organ. To prepare single cell suspension from spleens and lymph nodes, tissues were minced through sterile 70‐μm cell strainers. Cells from BAL fluid were harvested and washed twice in C5 medium by centrifugation at 100 g for 10 min. RBCs in spleen cell preparations and BAL were lysed using our own formulated ammonium chloride–based mouse RBC lysis buffer (JL buffer) that was developed for gentle lysis to preserve fragile cells, such as neutrophils. Cell preparations were washed twice by centrifugation in C5 medium and then strained again in 70‐μm cell strainers. For flow cytometry and CyTOF stains, cells were plated in 96‐well round‐bottom plates at a density of 1 × 106 cells per well. For cytokine staining, cells were incubated for 4 h at 37°C with Brefeldin A to allow for intracellular accumulation of cytokines.
FACS
All FACS stains were performed at room temperature. For cell‐surface stains, cells were plated and Fc‐block reagent was added for 10 min to minimize nonspecific antibody staining before adding fluorochrome‐labeled antibodies: FITC‐labeled anti‐CD3, PE/Cy7‐labeled anti‐TCRγδ, APC‐labeled anti‐CD27, BV421‐labeled anti‐CCR6 (BioLegend). Stained samples were fixed in 0.6% PFA in PBS for storage, washed by centrifugation, then reconstituted in PBS for flow cytometry analysis on a MACS‐Quant Analyzer (Miltenyi Biotech, San Diego, CA, USA). Data analysis was performed using the FlowJo software program (Tree Star, Ashland, OR, USA).
CyTOF mass cytometry
All CyTOF staining was performed at room temperature. For all stains, rhodium viability staining reagent (Fluidigm Sciences) was added to plated cells for 10 min before staining. After washing by centrifugation, Fc‐Block reagent was added for 10 min to the cells before adding CyTOF antibody‐staining cocktails. Cells were stained for 30 min, then washed once in CyTOF staining buffer (cell staining buffer, calcium/magnesium‐free PBS, 0.2% BSA, 0.05% sodium azide). Cells were fixed and permeabilized using our own Fix/Perm Buffer (PBS with 1.6% PFA and 0.3% Saponin). Subsequently, intracellular antibodies were added for 30 min before the cells were washed and fixed with 1.6% PFA. For phospho‐mass cytometry stains, plated cells were fixed by adding PFA to a final concentration of 1.6%, centrifuged to pellet form, and permeabilized in ice‐cold methanol for 20 min incubation at −20 C. This optimized PFA/methanol fixation method preserves most cell markers, permeabilizes cells, and denatures proteins sufficiently for antiphospho‐peptide antibody staining. After washing cells twice by centrifugation in the cell staining buffer, Fc‐block reagent was added for 10 min preincubation followed by adding CyTOF antibody staining cocktail, which contained antiphospho–ZAP70, antiphospho–p38‐p38 MAPK, and antiphospho–4E‐BP1 antibodies (Table 1). Finally, cells were washed and fixed with 1.6% PFA. Iridium‐intercalator solution (MaxPar Intercalator‐Ir 500 μM; Fluidigm Sciences) was added to cells in Fix/Perm Buffer. After a final washing step, cells were reconstituted in MilliQ‐filtered distilled water (EMD Millipore, Billerica, MA, USA) at a concentration of 5 × 105 cells/ml containing EQ calibration beads (EQ Four Element Calibration Beads; Fluidigm Sciences), according to manufacturer's protocol. Cells were analyzed on a CyTOF 2 mass cytometer (Fluidigm Sciences). Data analysis was conducted using Cytobank (Cytobank, Mountain View, CA, USA) and the FlowJo software program. For the SPADE analysis, we used settings of 100 nodes and the default down‐sampled event target of 10%. To focus the presentation of SPADE analysis data to the cells of interest, the SPADE plots included the nodes in CD3+TCRγδ+‐gated cells.
Statistics
GraphPad Prism 6.0 software (GraphPad, San Diego, CA, USA) was used for statistical calculations. One‐way ANOVA with Tukey's multiple comparison test or 1‐tailed Student's t tests were used to analyze these data as indicated in the figure legends. For all data, P < 0.05 was considered statistically significant with a 95% confidence interval.
RESULTS
Comparison of conventional flow cytometry to CyTOF mass cytometry staining of TCRγδ T cell subsets
Because CyTOF is a relatively new method for single‐cell cytometry, one objective of this study was to validate that CyTOF generates cell‐staining data comparable to conventional flow cytometry. Because acute lung infection is a complex host response, it provides a good platform for this comparison. Thus, we prepared spleen cells from mice at 1 d after S. pneumoniae infection and performed matched FACS and CyTOF stains to identify TCRγδ T cell subsets. Cells were stained with fluorochrome‐coupled antibodies specific for CD3, TCRγδ, CD27, and CCR6 or were stained with metal isotope‐coupled CyTOF antibodies specific for the same cell markers. The data shown in Fig. 2 demonstrate that conventional flow cytometry and CyTOF staining showed no significant differences in staining profiles. This finding validates our finding that we were able to generate similar infection‐induced TCRγδ T cell phenotyping results by CyTOF and conventional fluorescence flow cytometry.
Figure 2.
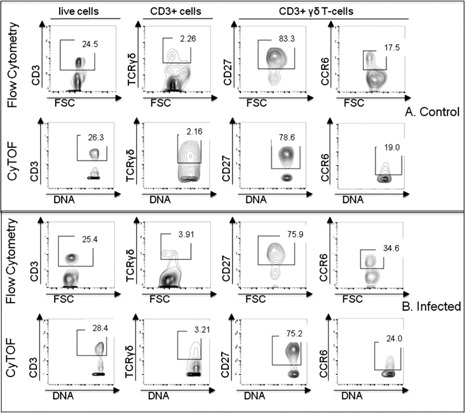
Comparison of conventional flow cytometry to CyTOF. Male mice were infected intranasally with 1 × 106 CFUs of S. pneumoniae (n = 4). Healthy, uninfected mice served as controls (n = 4). Spleen cells were harvested 1 d after infection and stained for either flow cytometry or CyTOF using anti‐CD3, anti‐TCRγδ, anti‐CD27, and anti‐CCR6 antibodies. Numbers in gates represent percentage of parent population as indicated in the row titles. (A) Biaxial plots of cells from control mice. (B) Plots of cells from mice at 1 d after infection. Representative plots demonstrate that conventional flow cytometry and CyTOF staining showed comparable staining profiles. Data are representative of 2 independent experiments. FSC, forward scatter.
Lung infection induces an increase in TCRγδ T cells in the spleen, lymph nodes, and BAL
Our initial experiments compared single‐cell events of CD3+TCRγδ T cells—referred to as γδ T cells from this point forward in this report—in the BAL, spleen, and lymph nodes in mice after lung infection with S. pneumoniae. CyTOF collects about 30% of the total input cell numbers as singlet cell DNA‐positive staining events; therefore, we analyzed data as singlet DNA staining events. We chose 1 d after infection as the study time point because we found the numbers of lung airway cells to be significantly increased at that point after lung infection. The relative percentages of immune cell types found in the BAL fluid at 1 d after infection were as follows: 90.5% neutrophils, 2.5% NK cells, 0.9% CD3+ T cells (46% CD8+, 38% CD4+, 4.3% NK1.1+, 3.7% γδ TCR+), and 0.8% macrophages. As shown in Fig. 3 , the DNA‐positive singlet‐cell event counts of γδ T cells were significantly increased in all these compartments at 1 d after infection. These results indicate that infection induces an acute increase in γδ T cells and agrees with findings in a prior report [11] demonstrating increased γδ T cells in lungs of mice early after infection with S. pneumoniae.
Figure 3.
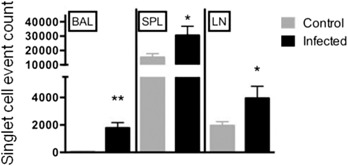
Infection with S. pneumoniae increases γδ T cells in the lungs, spleen, and lymph nodes of mice. Outbred mice received S. pneumoniae intranasally. Healthy mice served as control. Cells from the spleen (SPL), lymph node (LN), and BAL were collected at 1 d after infection. CyTOF staining was performed to detect changes in CD3+ TCRγδ+ T cells in the indicated tissue compartments in healthy control and S. pneumoniae–infected mice. CyTOF data were analyzed and plotted as DNA+ singlet staining events in gated CD45+ cells. Experiments were repeated 2 (SPL, LN) or 3 times (BAL) independently with n = 4 mice per group. Bars represent the means ± sem. *P < 0.05, **P < 0.01.
Cell‐surface and transcription factor expression among IL‐17A– and IFN‐γ–producing γδ T cells revealed by CyTOF mass cytometry
Activated γδ T cells are a major source of IL‐17A and IFN‐γ during bacterial infections [8, 12]. Moreover, γδ T cells have been classified into subsets based on expression of these cytokines and other cell surface markers [6]. Here, we used CyTOF to measure IL‐17A– and IFN‐γ–expressing γδ T cells in 3 tissue compartments at 1 d after lung infection with S. pneumoniae. CyTOF was also used to determine coexpression of multiple cell‐surface markers and transcription factors on IFN‐γ– and IL‐17A–producing γδ T cells. As shown in Fig. 4A , we detected IFN‐γ and IL‐17A expression in γδ T cells from the BAL, spleens, and lymph nodes of infected mice, whereas these cytokines were not detected in γδ T cells from healthy control mice (data not shown). γδ T cells from the lungs of infected mice expressed much higher levels of these cytokines than did those in the spleens or lymph nodes. To determine the distribution of cell‐subset identifying markers among IL‐17A– and IFN‐γ–expressing γδ T cells found in the BAL, we analyzed CyTOF data for coexpression of CCR6, CD27, CD8α, CD14, CD25, Gr‐1, ICOS, NK1.1, RoRγt, and Gata3 (Fig. 4B). We were particularly interested in CD27 and CCR6 because they were described as functional markers for cytokine‐producing γδ T cells [6]. As described previously, IFN‐γ+–, but not IL‐17A–producing γδ T cells expressed CD27. CCR6 was coexpressed on IL‐17A–producing γδ T cells only, but at low levels. We also confirmed that IFN‐γ–, but not IL‐17A–producing γδ T cells, expressed NK1.1. On the other hand, CD25 as well as the transcription factor RoRγt were expressed at higher levels in IL‐17A–expressing γδ T cells than they were in IFN‐γ–producing γδ T cells. The expression of CD25 on these activated γδ T cells is not surprising because CD25 is considered a T cell activation marker. Interestingly, the transcription factor Gata3 was expressed at low levels in IFN‐γ–producing γδ T cells only. This observation may be unique to γδ T cells and their reactivity to lung infection because Gata3 expression and IFN‐γ are generally not coexpressed in T cells. CD8α expression was slightly increased on IFN‐γ–expressing cells, without reaching statistical significance. No significant differences in expression levels of ICOS were detected on IL‐17A– or IFN‐γ–expressing γδ T cells. However, we discovered that high levels of Gr‐1 and CD14 were expressed on IFN‐γ–staining γδ T cells as compared with IL‐17A–staining γδ T cells in the BAL, although the difference was not statistically significant. These CyTOF findings indicate for the first time, to our knowledge, that Gr‐1 and CD14 might serve as novel markers to help differentiate IFN‐γ– from IL‐17A–producing γδ T cells during bacterial infections. These data also suggest that Gr‐1+ or CD14+ γδ T cells represent a primary reactive γδ T cell population during the acute phase of S. pneumoniae lung infections. Additionally, we were able to generate an overview of cytokine expression profiling data to identify the sources of TNF‐α, IFN‐γ, IL‐17A, and IL‐10 among immune cell subsets that arrive in the lung airways after infection. The heat map shown in Fig. 5 and the expression data in Table 2 show that NK cells were the major cell source of IFN‐γ and that γδ T cells were the primary source of IL‐17A. γδ T cells also expressed higher levels of IFN‐γ than CD4 or CD8 T cells, and γδ T cells appeared to be the primary source of TNFα. IL‐10 was not expressed at significant levels in any cell type.
Figure 4.
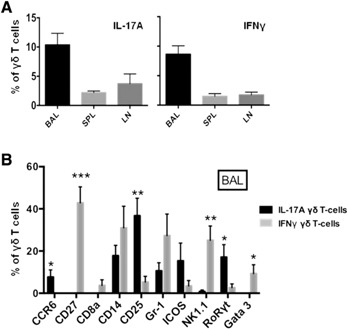
Distribution of cell subset identifying markers on IL‐17A– and IFN‐γ–expressing γδ T cells from the BAL of S. pneumoniae–infected mice. Groups of mice (n = 4) were challenged in the lung with S. pneumoniae. Cells from BAL were collected 1 d after infection and stained with a cell‐surface and intracellular CyTOF panel. (A) CyTOF data were analyzed for IL‐17A and IFN‐γ expression in gated CD3+/TCRβ−/TCRγδ+ single‐cell events. (B) IL‐17A– or IFN‐γ–expressing CD3+/TCRβ−/TCRγδ+ cells were analyzed for the indicated cell‐surface markers and transcription factors. Experiments were repeated 2 (spleen [SPL] and lymph node [LN]) or 3 times (BAL). Data are plotted as means ± sem. *P < 0.05, **P < 0.01, ***P < 0.001 for IL‐17A– vs. IFN‐γ–expressing γδ T cells.
Figure 5.
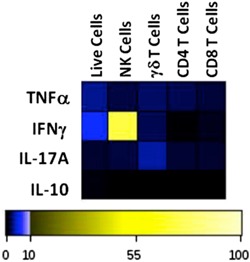
Heat map showing cytokine‐expressing cell types in the BAL from S. pneumoniae–infected mice. Groups of mice (n = 4) were challenged in the lung with S. pneumoniae. Cells from BAL were collected 1 d after infection into medium containing Brefeldin A and incubated for an additional 4 h at 37°C with Brefeldin A to allow for intracellular accumulation of cytokines. Cells were stained with a CyTOF antibody panel to identify the indicated cell subsets—NK cells (Gr‐1/CD14−, NK1.1+), γδ T cells (CD3+, TCR γδ+), CD4 T cells (Gr‐1/CD14−, CD3+, CD4+), and CD8 T cells (Gr‐1/CD14−, CD3+, CD8+). CyTOF data were concatenated to generate the heat map plot (see Table 2). The colored key indicates expression levels with low (0–10) and high (10–100) expression increments, and the table shows cytokine expression levels detected by CyTOF.
Table 2.
Cytokine expression levels in gated cell subsets
| Cytokine | Live cells | NK cells | γδ T cells | CD4 T cells | CD8 T cells |
|---|---|---|---|---|---|
| TNF‐α | 2.76 | 1.42 | 3.5 | 1.56 | 1.33 |
| IL‐17A | 0.98 | 1.22 | 2.22 | 0.96 | 0.94 |
| IFN‐γ | 5.41 | 70.2 | 7.18 | 2.41 | 0.67 |
| IL‐10 | 0.37 | 0 | 0.89 | 0 | 0 |
Characterization of Gr‐1+ γδ T cells in the spleen, lymph nodes, and BAL after S. pneumoniae infection
To determine the tissue distribution of Gr‐1+ γδ T cells after infection, we used the gating strategy displayed in Fig. 6A for cells prepared from the spleen, lymph nodes, and BAL fluid at 1 d after lung infection. In brief, we used rhodium viability and DNA intercalator staining reagents to gate on live, singlet cell events to exclude cell doublets and dead cells. We then used anti‐CD45 antibody staining to gate all immune cells. γδ T cells were identified with anti‐CD3 and anti‐γδ TCR‐specific antibodies. Finally, Gr‐1+ and Gr‐1− expression on γδ T cells prepared from the spleen, lymph nodes, or BAL of S. pneumoniae–infected and healthy control mice was identified by anti‐Gr‐1 antibody stains. As shown in Fig. 6C, lymph nodes from healthy, uninfected mice had very small subpopulations of Gr‐1+ γδ T cells. In the spleens of healthy mice, Gr‐1 was expressed on only 1.4% of γδ T cells, and in the BAL fluid, 5.6% of γδ T cells expressed Gr‐1. Thus, most γδ T cells in normal, uninfected mice do not express cell‐surface Gr‐1. However, we observed a significant increase in Gr‐1+ γδ T cells in the spleen (7.1% of γδ T cells) and in BAL fluid (79% of γδ T cells) from infected mice. Therefore, Gr‐1 expression on lung γδ T cells is induced by infection or, alternatively, γδ T cells that express Gr‐1 migrate to the infected lung airways. The substantial increase in Gr‐1+ γδ T cells in the lungs also suggests that Gr‐1 identifies an activated, effector γδ T cell subset. We used conventional flow cytometry to confirm and validate the existence of the Gr‐1+ γδ T cells in the BAL of S. pneumoniae–infected mice (Fig. 6B).
Figure 6.
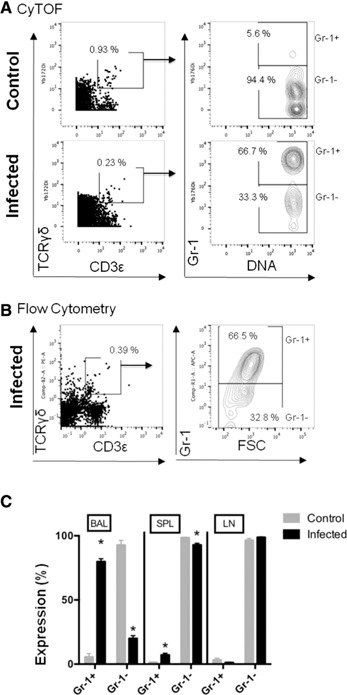
Gr‐1+ γδ T cells represent the primary, reactive γδ T cell population during the acute phase of S. pneumoniae lung infections. (A) BAL from healthy mice and infected mice was prepared at 1 d. Singlet, live cell events were identified by CyTOF viability and DNA intercalator staining reagents. Anti‐CD45 antibody staining was then used to gate all immune cells and γδ T cells were gated as CD3+, TCRγδ+ cell events. Expression of Gr‐1 on γδ T cells was then identified by anti‐Gr‐1 antibody staining events. The numbers shown on the plots indicate the percentage of gated events. (B) Conventional fluorescence flow cytometry used matched, fluorescent‐tagged antibodies to identify similar Gr‐1–expressing γδ T cell populations in the BAL from infected mice to compare with CyTOF staining patterns. (C) These data plots show Gr‐1 expression on γδ T cells prepared from the spleen (SPL), lymph node (LN), and BAL from healthy or S. pneumoniae–infected mice. Experiments were repeated 2 (fluorescence flow cytometry) or 3 times (CyTOF) using 4 mice per group. Data are plotted as means ± sem. *P < 0.001 for control vs. infected mice. FSC, forward scatter.
Characterization of differential cell‐surface marker expression on Gr‐1+ and Gr‐1− γδ T cells
Next, we used CyTOF to analyze Gr‐1+ and Gr‐1− γδ T cell subsets for differential expression levels of CCR4, CCR6, CD8α, CD14, CD25, CD27, CD274 (PD‐L1), I‐A/I‐E, and NK1.1 on BAL, spleen, and lymph node cells from uninfected and S. pneumoniae–infected mice. We observed that CD27, CD25, CD8α, and NK1.1 were expressed primarily on Gr‐1− γδ T cells ( Fig. 7A and B ) in the BAL at the site of infection, whereas, in the spleen, no differences were found in the expression of NK1.1 and CD25 on Gr‐1+ and Gr‐1− γδ T cells. This observation is best visualized in the SPADE plots shown in Fig. 7B. SPADE is a dimensional‐reduction analysis approach that allows for unsupervised analysis of CyTOF data by organizing staining data into nodes with similar staining patterns. The SPADE plots shown in Fig. 7B illustrate the differences in expression patterns of cell‐surface markers, cytokines, and RoRγt in Gr‐1+ and Gr‐1− γδ T cells. These SPADE plots show the expression of the indicated markers in the section of SPADE trees gated on CD3+TCRγδ+ staining cells. The size the circular nodes correlates with cell number, and the color represents expression intensity (blue, lower expression; red, higher expression). In the lymph nodes, Gr‐1− γδ T cells preferentially expressed CD27 and CD25, but not NK1.1 or CD8α. Of note, it was not possible to perform statistical analysis on the Gr‐1+ γδ T cell distribution in uninfected mice because of the low numbers in the BAL fluid, spleen, and lymph nodes. Nevertheless, we did observe that CD27, CD25, CD8α, and NK1.1 expression on Gr‐1− vs. Gr‐1+ γδ T cells largely correlated with what we measured in infected mice. As already described in this study, Gr‐1+ γδ T cells represent a distinct population of γδ T cells that are induced by S. pneumoniae infection (Fig. 6). Although most Gr‐1+ γδ T cells showed little or no expression of CD27, CD8α, and CD25, they did show high expression of CD274 (PD‐L1) and CD14 (Fig. 7A). CCR6 and CCR4 expression also increased significantly on spleen Gr‐1+ γδ T cells. In total, CyTOF staining helped identify differential expression patterns of multiple cell‐surface markers on Gr‐1+ and Gr‐1− γδ T cells. This information can now be used to better classify γδ T cell activation and subsets that arise following lung bacterial infections and possibly other types of infections or immune reactions.
Figure 7.
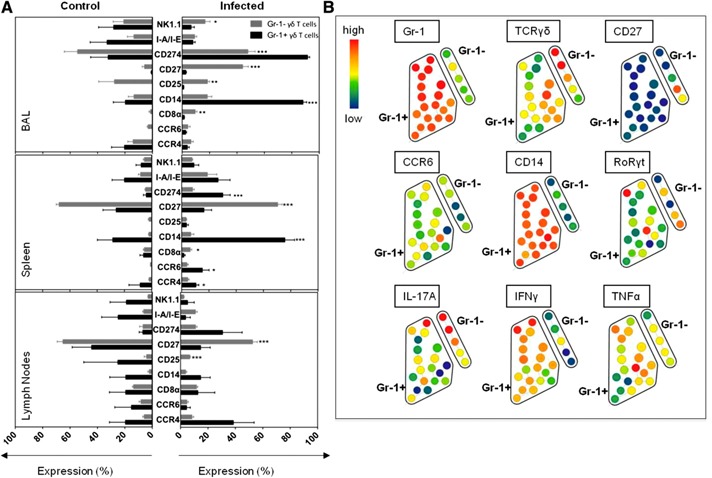
Characterization of cell‐surface marker expression on Gr‐1+ and Gr‐1‐ γδ T cells. (A) Cells from the spleen, lymph nodes, or BAL from uninfected and S. pneumoniae–infected mice were stained for the indicated cell‐surface markers on gated Gr‐1+ and Gr‐1− γδ T cells. The data are presented as means ± sem percentage of expression on γδ T cells with staining results from control mice on the left side and from infected mice on the right side. The experiments were repeated 3 times (n = 4 mice per group). *P < 0.05, **P < 0.01, ***P < 0.001 for Gr‐1+ vs. Gr‐1− γδ T cells. (B) Representative SPADE plots of BAL cells prepared from S. pneumoniae–infected mice stained with the cytokine CyTOF antibody panel (Table 1). The SPADE plots show gated CD3+TCRγδ+ nodes with Gr‐1+ and Gr‐1− γδ T cells accentuated to point out the differences in expression patterns of cell‐surface markers, cytokines, and RoRγt. Node size correlates with cell number, color represents expression intensity (blue, lower expression; red, higher expression). Experiments were repeated 3 (BAL) or 2 (SPL and LN) times with n = 4 mice per group.
Cytokine production profiles of Gr‐1+ and Gr‐1− γδ T cell subsets after infection with S. pneumoniae
We compared infection‐induced cytokine expression in subsets of Gr‐1+ and Gr‐1− γδ T cells by CyTOF. To accomplish this, cells prepared from spleens, lymph nodes, or BAL from infected or healthy control mice were incubated at 37°C for 4 h with Brefeldin A to prevent cytokine secretion. Cells were then stained with CyTOF antibodies to detect intracellular cytokines, including IFN‐γ, IL‐10, IL‐17A, and TNFα, in γδ T cell subsets. We also stained for RoRγt transcription factor expression. Although no production of cytokines could be detected in cells from uninfected mice, we observed significant levels of these cytokines in cells prepared from infected mice ( Fig. 8 ). First, we observed that Gr‐1+ γδ T cells produced significantly higher levels of IFN‐γ compared with Gr‐1− γδ T cells in the BAL and spleen. In addition, Gr‐1+ γδ T cells produced significantly higher amounts of TNFα than Gr‐1− γδ T cells did at the site of infection, but not in the spleen or lymph nodes. In contrast, Gr‐1− γδ T cells were the almost exclusive source of IL‐17A in cells prepared from the BAL, spleen, and lymph nodes. Finally, we found that Gr‐1+ γδ T cells were a source of IL‐10 in the BAL and spleen following lung infection.
Figure 8.
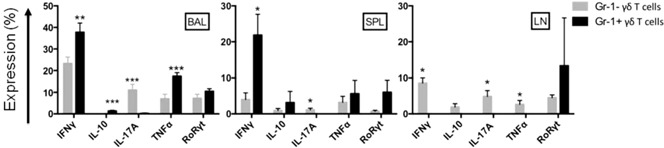
Cytokine production profiles of Gr‐1+ and Gr‐1− γδ T cells after lung infection. Cells prepared from BAL, spleen (SPL), or lymph node (LN) from infected mice were incubated for 4 h at 37°C with Brefeldin A to allow for intracellular accumulation of cytokines. Cells were stained for CyTOF analysis to detect the indicated cytokines and RoRγt in gated Gr‐1+ or Gr‐1−, CD3+/TCRγδ + cell populations. The results of 3 (BAL) or 2 (SPL, LN) experiments each are plotted as means ± sem (n = 4 mice per group). *P < 0.05, **P < 0.01, ***P < 0.001 for Gr‐1− vs. Gr‐1+ γδ T cells.
Differences in infection‐induced signaling between Gr‐1+ and Gr‐1− γδ T cell subsets
As shown, Gr‐1 expression can be used to describe phenotypically different subtypes of γδ T cells that arise during acute lung infection. Therefore, we decided to study whether there might be differences in infection‐induced cell signaling responses between Gr‐1+ and Gr‐1− γδ T cell subsets. Using CyTOF, we measured phosphorylated ZAP70, p38 MAPK (p38), and 4E‐BP1 levels in spleen and BAL cells prepared from infected or healthy control mice. As illustrated in the SPADE plots in Fig. 9A and plotted in Fig. 9B, Gr‐1+ γδ T cells in the spleen showed significantly higher levels of infection‐induced phosphorylated ZAP70, whereas, in the BAL, both Gr‐1+ and Gr‐1− γδ T cells showed increased phosphorylated ZAP70. The SPADE plots shown in Fig. 9A are the SPADE tree sections gated on CD3+TCRγδ+ cells. In addition, Gr‐1+ γδ T cells in the BAL showed high infection‐induced p38 phosphorylation. However, phosphorylated p38 was markedly decreased in splenic Gr‐1− γδ T cells. The phosphorylation of 4E‐BP1 was increased in both Gr‐1+ and Gr‐1− γδ T cells in the spleen, whereas only Gr‐1+ γδ T cells showed increased phosphorylation of 4E‐BP1 in the BAL. These results suggest that Gr‐1+ and Gr‐1− γδ T cells demonstrate different signaling responses to S. pneumoniae infection. Increased, combined phosphorylation of p38 and ZAP70 in Gr‐1+ γδ T cells suggests that both innate and TCR signaling pathways may be involved in their activation, whereas the predominant ZAP70 phosphorylation in Gr‐1− γδ T cells suggests their activation may be more TCR‐dependent. These observations provide more support for the idea that γδ T cells show diverse functional phenotypes in response to infection.
Figure 9.
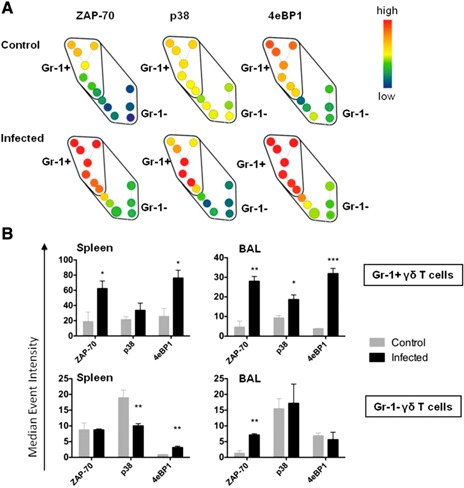
Infection‐induced differences in signaling between Gr‐1+ and Gr‐1− γδ T cells. Spleen, lymph node, and BAL cells prepared from uninfected and S. pneumoniae–infected mice at 1 d were stained to detect phosphorylated ZAP70, p38 MAPK (p38), and 4E‐BP1 in Gr‐1+ and Gr‐1− γδ T cells by CyTOF. (A) Representative SPADE analysis plots showing differences in expression intensity of signaling molecules in gated CD3+ TCRγδ+ spleen cells from control (top row) or infected mice (bottom row). Nodes represent clustered cell events, and node color indicates signal intensity (blue, lower; and red, higher event intensity). (B) Means ± sem (n = 4 mice per group) for median‐intensity staining of the indicated phosphorylated signaling molecules in Gr‐1+ vs. Gr‐1− γδ T cells in the spleen or BAL at 1 d after infection as analyzed using FlowJo software. *P < 0.05, **P < 0.01, ***P < 0.001 for control vs. infected mice.
DISCUSSION
The first‐line of defense against infections is a critical determinant of both clearance and resolution of infections. Although innate immune cells, such as neutrophils and macrophages, have potent antimicrobial activity, other immune and nonimmune cells are needed to orchestrate the infection process by producing factors to alert the immune system. One of these cell types includes γδ T cells, which are known to be involved in the initial immune response to pathogens, especially at mucosal surfaces and skin [12, 13]. We developed a specific interest in studying and more‐precisely characterizing γδ T cells as immune cell types that are reactive to acute lung infection for several reasons. First, we discovered that γδ T cell responses in the lung were significantly different in groups of injured mice that fail to survive lung S. pneumoniae infection. In addition, other experiments performed with γδ T cell knockout mice showed that they have a protective effect in mouse injury and infection models [11, 14]. Second, review of the literature suggested to us that γδ T cells are a diverse immune cell type that may act as innate or adaptive immune cells to regulate immune responses to infections or injury by producing regulatory cytokines, such as IL‐17A or IFN‐γ. For these reasons, we focused our efforts on studying the phenotypic features of infection‐induced γδ T cells to better understand how γδ T cells might influence antimicrobial immunity.
γδ T cells were shown to be involved in viral immune responses [15] and bacterial infections [6, 14, 16]. In addition, γδ T cells were shown to have the ability to either promote [17] or inhibit [18] autoimmune disease development in experimental mouse models of autoimmune encephalomyelitis and inflammatory bowel disease. It is well established that γδ T cells are required for effective antimicrobial immune function because γδ T cell–deficient mice were shown to be highly susceptible to infections caused by Klebsiella pneumoniae, Escherichia coli, Listeria monocytogenes, S. pneumoniae, and Toxoplasma gondii [6, 11, 19]. Currently, it is unclear whether the functional diversity of γδ T cells is due to the existence of distinct γδ T cell subsets or their activation status. However, their capacity to release high levels of proinflammatory cytokines, such as IL‐17A and IFN‐γ, during bacterial infections is well documented [6, 13, 20]. Some studies suggest that this rapid response to pathogens is attributable to the functional maturity that γδ T cells develop in the thymus before migration to peripheral sites [21]. In contrast, recent studies have shown that CD27− γδ T cells, previously considered to be functionally “predetermined” for IL‐17 production, can acquire the ability to produce IFN‐γ under certain circumstances, thereby providing strong evidence for the possible functional plasticity of γδ T cells [6].
Cytokine production by γδ T cells appears to be central to their function during bacterial infections. For example, the rapid production of IL‐17A by γδ T cells after bacterial infection has been shown to be required for neutrophil recruitment [22]. Similarly, early γδ T cell production of IFN‐γ after bacterial infection was shown to contribute to the initial recruitment and activation of macrophages and cytotoxic lymphocytes in several mouse infection models [19, 23]. In these studies, depletion of IFN‐γ–producing γδ T cells resulted in an increased bacterial growth after L. monocytogenes or S. pneumoniae infection. Furthermore, during the resolution phase, at 6 d after S. pneumoniae infection, it was shown that IFN‐γ–producing γδ T cells regulated macrophage and dendritic cell numbers in the lung [4]. Their data also suggested that S. pneumoniae infection caused expansion of resident, lung‐specific γδ T cells rather than migration of γδ T cells into the site of infection. The results of our studies fully support that lung infection induces the appearance of IFN‐γ–producing γδ T cells in the lung; but, for the first time, to our knowledge, we identified that they coexpress Gr‐1, CD14, and CD274 and were distinct from previously described CD27+/NK1.1+ IFN‐γ–producing γδ T cell populations. Furthermore, Gr‐1+/CD14+ IFN‐γ–producing γδ T cells were mostly restricted to the site of infection because only low numbers were found in the spleen, and relatively few were detected in the lymph nodes. These observations support the idea that Gr‐1+/CD14+ γδ T cells represent an infection‐induced expansion of lung‐resident γδ T cells.
The CD14 expression found on Gr1+ γδ T cells may indicate that these cells could respond to bacterial antigens, such as peptidoglycan, lipoteichoic acid, bacterial lipopeptides, or lipopolysaccharides, from gram‐negative bacteria [24, 25]. Thus, CD14 expression on γδ T cells may be involved in their antimicrobial function or in activating TCR‐independent cytokines. Accordingly, we observed increased expression of TNF‐α and IL‐10 in Gr‐1+/CD14+ γδ T cells—cytokines that are known to be induced by bacterial antigens via TLR signaling. In support of this idea, Gr‐1+ γδ T cells showed increased phosphorylated p38 MAPK in BAL cells prepared from S. pneumoniae–infected mice. Another novel finding was that CD274 (PD‐L1) expression increased significantly on Gr‐1+ γδ T cells at the site of infection. CD274 is an inhibitory receptor that is most commonly found on antigen‐presenting cells and is involved in suppressing immune responses [26]. We think the unique identification of its expression on γδ T cells suggests it may be acting as a feedback mechanism for the excessive inflammation that occurs during severe infections [27].
Now that we identified that infection induces an increase in Gr‐1+ γδ T cells in the lung airway, we would like to explore how they might affect infection outcome. However, at this time, it is not feasible to deplete these cells in vivo because they express cell‐surface markers shared by multiple immune cell subsets, including neutrophils (Gr‐1) and other innate immune cell types (CD14). Instead, future experiments will focus on how immune response–modifying drugs modulate these cells and affect S. pneumoniae lung infection outcomes. In addition, experiments will be done to identify unique transcription factors or markers by molecular approaches, such as RNA sequencing, so that how Gr‐1+ γδ T cells influence the immune response to infections can be more‐precisely determine.
CyTOF mass cytometry is a new flow cytometry–like technology that allows for multiplex, single‐cell staining of cells with combinations of antibodies that cannot be efficiently achieved by conventional fluorescence‐based flow cytometry. The technology uses rare‐earth element isotope–tagged antibodies, rather than fluorescently‐labeled antibodies, to detect cell‐surface or intracellular markers. Because rare‐earth element isotopes are not naturally found in mammalian cells, there is virtually no background signal. The multiplex staining capacity of CyTOF, as well as the low background staining, made the work presented here possible. What made CyTOF so effective for this work, however, was that we were able to stain lung, spleen, and lymph node cells simultaneously with cell markers that are not normally used in combination. This led to the discovery that γδ T cells express significant levels of markers normally associated with mouse neutrophils and macrophages (e.g., Gr‐1 and CD14). We were then able to further characterize expression levels of cytokines and other regulatory cell markers on infection‐induced γδ T cell populations.
As already stated, CyTOF multiplex staining generates high‐dimensional staining data that allows for characterization of cell subsets or phenotypes. CyTOF data are best visualized by analytic methods that reduce multidimensional staining results into 2‐dimensional plots. In this report, CyTOF data were analyzed by SPADE in Cytobank, a cloud‐based data sharing and analysis platform. SPADE allowed us to first visualize data to identify groupings of cells showing changes between normal, healthy mice vs. S. pneumoniae–infected mice. This analysis included cell‐surface marker, transcription factor, and cytokine expression patterns. CyTOF data were uploaded into Cytobank for initial analysis, which included gating on single‐cell events as detected by DNA intercalator staining. This was important to exclude doublet cell events that may occur among activated cell types found in the lungs after infection. After analysis in Cytobank, we then decided to analyze CyTOF staining data using FlowJo software because it allowed us to create specific gating strategies for statistical analysis. This data analysis workflow proved to be particularly important, once we identified that lung infection caused a significant increase in Gr‐1+ γδ T cells, because cells staining for Gr‐1 include Ly6G and Ly6C populations. The Gr‐1+ γδ T cells defined in this report were found to express high levels of Gr‐1, which correlates with the expression of Ly6G, as shown by us (data not shown) and others [28]. Unfortunately, the functions for many of the Ly6 protein family members, including Ly6C and Ly6G, are not well known because they were specifically developed as hematopoietic cell‐differentiation markers. In relation to this study, Ly6C has been shown to be expressed on a population of memory CD8 T cells, and Ly6G has been shown to associate with LFA‐1 (CD11a/CD18) and Mac‐1 (CD11b/CD18) [29, 30, 31, 32–33]. Because LFA‐1 and Mac‐1 are integrins, it may be that the Gr‐1 expression on γδ T cells has a role in cell‐to‐cell contact or adhesion during lung bacterial infections. Mac‐1 is a complement receptor (CR3) and responds to complement activation on bacteria. Thus, the expression of Gr‐1 on γδ T cells during acute bacterial infections may be involved in directly activating γδ T cells by a TCR‐independent pathway.
In summary, our results identify that S. pneumoniae infection induces an IFN‐γ–producing γδ T cell population in the lung, characterized by the coexpression of cell‐surface Gr‐1, CD14, and CD274 (PD‐L1). Moreover, we found that Gr‐1+ γδ T cells represent the dominant γδ T cell subset in the lungs after S. pneumoniae infection, suggesting that they are the primary reactive subset. Finally, we demonstrate that CyTOF mass cytometry is an efficient and effective way to characterize complex cellular immune responses to infection or other pathologic reactions and will likely prove to be a valuable way to study the effects of therapeutics on pneumonia.
AUTHORSHIP
L.W.J. and J.A.L. planned the study and wrote the manuscript. L.W.J., J.W.K., and J.W.D. performed the experiments.
DISCLOSURES
All authors declare no conflicts of interest.
ACKNOWLEDGMENTS
This study was supported by grant funding from the U.S. National Institutes of Health National Institute of Allergy and Infectious Diseases (Grants AI092905‐04 and AI107360‐02), the Deutsche Forschungsgemeinschaft Research (Grant WA 3426/1‐1), the Brigham and Women's Hospital Biomedical Research Institute, and the Harvard Medical Area CyTOF Consortium. We wish to thank Nicole Paul and John Daley of the Dana Farber Cancer Institute Flow Cytometry Core Facility for running the CyTOF2 instrument.
REFERENCES
- 1. Chodaczek, G. , Papanna, V. , Zal, M.A. , Zal, T. (2012) Body‐barrier surveillance by epidermal γδ TCRs. Nat. Immunol. 13, 272–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Koppe, U. , Suttorp, N. , Opitz, B. (2012) Recognition of Streptococcus pneumoniae by the innate immune system. Cell. Microbiol. 14, 460–466. [DOI] [PubMed] [Google Scholar]
- 3. De Stoppelaar, S.F. , Van't Veer, C. , van den Boogaard, F.E. , Nieuwland, R. , Hoogendijk, A.J. , de Boer, O.J. , Roelofs, J.J. , van der Poll, T. (2013) Protease activated receptor 4 limits bacterial growth and lung pathology during late stage Streptococcus pneumoniae induced pneumonia in mice. Thromb. Haemost. 110, 582–592. [DOI] [PubMed] [Google Scholar]
- 4. Kirby, A.C. , Newton, D.J. , Carding, S.R. , Kaye, P.M. (2007) Evidence for the involvement of lung‐specific γδ T cell subsets in local responses to Streptococcus pneumoniae infection. Eur. J. Immunol. 37, 3404–3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Weaver, C.T. , Hatton, R.D. , Mangan, P.R. , Harrington, L.E. (2007) IL‐17 family cytokines and the expanding diversity of effector T cell lineages. Annu. Rev. Immunol. 25, 821–852. [DOI] [PubMed] [Google Scholar]
- 6. Serre, K. , Silva‐Santos, B. (2013) Molecular mechanisms of differentiation of murine pro‐inflammatory γδ T cell subsets. Front. Immunol. 4, 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ribot, J.C. , de Barros, A. , Pang, D.J. , Neves, J.F. , Peperzak, V. , Roberts, S.J. , Girardi, M. , Borst, J. , Hayday, A.C. , Pennington, D.J. , Silva‐Santos, B. (2009) CD27 is a thymic determinant of the balance between interferon‐γ‐ and interleukin 17‐producing γδ T cell subsets. Nat. Immunol. 10, 427–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Haas, J.D. , González, F.H. M. , Schmitz, S. , Chennupati, V. , Föhse, L. , Kremmer, E. , Förster, R. , Prinz, I. (2009) CCR6 and NK1.1 distinguish between IL‐17A and IFN‐γ‐producing γδ effector T cells. Eur. J. Immunol. 39, 3488–3497. [DOI] [PubMed] [Google Scholar]
- 9. Mitchell, A.J. , Pradel, L.C. , Chasson, L. , Van Rooijen, N. , Grau, G.E. , Hunt, N.H. , Chimini, G. (2010) Technical advance: autofluorescence as a tool for myeloid cell analysis. J. Leukoc. Biol. 88, 597–603. [DOI] [PubMed] [Google Scholar]
- 10. Bendall, S.C. , Nolan, G.P. , Roederer, M. , Chattopadhyay, P.K. (2012) A deep profiler's guide to cytometry. Trends Immunol. 33, 323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nakasone, C. , Yamamoto, N. , Nakamatsu, M. , Kinjo, T. , Miyagi, K. , Uezu, K. , Nakamura, K. , Higa, F. , Ishikawa, H. , O'Brien, R.L. , Ikuta, K. , Kaku, M. , Fujita, J. , Kawakami, K. (2007) Accumulation of γδ T cells in the lungs and their roles in neutrophil‐mediated host defense against pneumococcal infection. Microb. Infect. 9, 251–258. [DOI] [PubMed] [Google Scholar]
- 12. Chien, Y. , Zeng, X. , Prinz, I. (2013) The natural and the inducible: interleukin (IL)‐17‐producing γδ T cells. Trends. Immunol. 34, 151–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sutton, C.E. , Mielke, L.A. , Mills, K.H. (2012) IL‐17‐producing γδ T cells and innate lymphoid cells. Eur. J. Immunol. 42, 2221–2231. [DOI] [PubMed] [Google Scholar]
- 14. Cheng, P. , Liu, T. , Zhou, W.‐Y. , Zhuang, Y. , Peng, L. , Zhang, J.Y. , Yin, Z.N. , Mao, X.H. , Guo, G. , Shi, Y. , Zou, Q.M. (2012) Role of γ‐δ T cells in host response against Staphylococcus aureus‐induced pneumonia. BMC Immunol. 13, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mukasa, A. , Lahn, M. , Pflum, E.K. , Born, W. , O'Brien, R.L. (1997) Evidence that the same αδ T cells respond during infection‐induced and autoimmune inflammation. J. Immunol. 159, 5787–5794. [PubMed] [Google Scholar]
- 16. Martin, B. , Hirota, K. , Cua, D.J. , Stockinger, B. , Veldhoen, M. (2009) Interleukin‐17‐producing γ‐δ T cells selectively expand in response to pathogen products and environmental signals. Immunity 31, 321–330. [DOI] [PubMed] [Google Scholar]
- 17. Spahn, T.W. , Issazadah, S. , Salvin, A.J. , Weiner, H.L. (1999) Decreased severity of myelin oligodendrocyte glycoprotein peptide 33 ‐ 35‐induced experimental autoimmune encephalomyelitis in mice with a disrupted TCR δ chain gene. Eur. J. Immunol. 29, 4060–4071. [DOI] [PubMed] [Google Scholar]
- 18. Tsuchiya, T. , Fukuda, S. , Hamada, H. , Nakamura, A. , Kohama, Y. , Ishikawa, H. , Tsujikawa, K. , Yamamoto, H. (2003) Role of γδ T cells in the inflammatory response of experimental colitis mice. J. Immunol. 171, 5507–5513. [DOI] [PubMed] [Google Scholar]
- 19. Hiromatsu, K. , Yoshikai, Y. , Matsuzaki, G. , Ohga, S. , Muramori, K. , Matsumoto, K. , Bluestone, J.A. , Nomoto, K. (1992) A protective role of γ/δ T cells in primary infection with Listeria monocytogenes in mice. J. Exp. Med. 175, 49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hayday, A.C. (2009) γδ T cells and the lymphoid stress‐surveillance response. Immunity 31, 184–196. [DOI] [PubMed] [Google Scholar]
- 21. Prinz, I. , Silva‐Santos, B. , Pennington, D.J. (2013) Functional development of γδ T cells. Eur. J. Immunol. 43, 1988–1994. [DOI] [PubMed] [Google Scholar]
- 22. Toth, B. , Alexander, M. , Daniel, T. , Chaudry, I.H. , Hubbard, W.J. , Schwacha, M.G. (2004) The role of γδ T cells in the regulation of neutrophil‐mediated tissue damage after thermal injury. J. Leukoc. Biol. 76, 545–552. [DOI] [PubMed] [Google Scholar]
- 23. Berguer, R. , Ferrick, D.A. (1995) Differential production of intracellular γ interferon in α β and γ δ T‐cell subpopulations in response to peritonitis. Infect. Immun. 63, 4957–4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schröder, N.W. J. , Morath, S. , Alexander, C. , Hamann, L. , Hartung, T. , Zähringer, U. , Göbel, U.B. , Weber, J.R. , Schumann, R.R. (2003) Lipoteichoic acid (LTA) of Streptococcus pneumoniae and Staphylococcus aureus activates immune cells via Toll‐like receptor (TLR)‐2, lipopolysaccharide‐binding protein (LBP), and CD14, whereas TLR‐4 and MD‐2 are not involved. J. Biol. Chem. 278, 15587–15594. [DOI] [PubMed] [Google Scholar]
- 25. Ranoa, D.R. E. , Kelley, S.L. , Tapping, R.I. (2013) Human lipopolysaccharide‐binding protein (LBP) and CD14 independently deliver triacylated lipoproteins to Toll‐like receptor 1 (TLR1) and TLR2 and enhance formation of the ternary signaling complex. J. Biol. Chem. 288, 9729–9741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Francisco, L.M. , Sage, P.T. , Sharpe, A.H. (2010) The PD‐1 pathway in tolerance and autoimmunity. Immunol. Rev. 236, 219–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Boomer, J.S. , To, K. , Chang, K.C. , Takasu, O. , Osborne, D.F. , Walton, A.H. , Bricker, T.L. , Jarman, S.D. , II, Kreisel, D. , Krupnick, A.S. , Srivastava, A. , Swanson, P.E. , Green, J.M. , Hotchkiss, R.S. (2011) Immunosuppression in patients who die of sepsis and multiple organ failure. JAMA 306, 2594–2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Daley, J.M. , Thomay, A.A. , Connolly, M.D. , Reichner, J.S. , Albina, J.E. (2008) Use of Ly6G‐specific monoclonal antibody to deplete neutrophils in mice. J. Leukoc. Biol. 83, 64–70. [DOI] [PubMed] [Google Scholar]
- 29. Mueller, S.N. , Gebhardt, T. , Carbone, F.R. , Heath, W.R. (2013) Memory T cell subsets, migration patterns, and tissue residence. Annu. Rev. Immunol. 31, 137–161. [DOI] [PubMed] [Google Scholar]
- 30. Slaney, C.Y. , Toker, A. , La Flamme, A. , Bäckström, B.T. , Harper, J.L. (2011) Naïve blood monocytes suppress T‐cell function. A possible mechanism for protection from autoimmunity. Immunol. Cell Biol. 89, 7–13. [DOI] [PubMed] [Google Scholar]
- 31. Miyamoto, M. , Emoto, M. , Emoto, Y. , Brinkmann, V. , Yoshizawa, I. , Seiler, P. , Aichele, P. , Kita, E. , Kaufmann, S.H. (2003) Neutrophilia in LFA‐1‐deficient mice confers resistance to listeriosis: possible contribution of granulocyte‐colony‐stimulating factor and IL‐17. J. Immunol. 170, 5228–5234. [DOI] [PubMed] [Google Scholar]
- 32. DuMont, A.L. , Yoong, P. , Day, C.J. , Alonzo, F., III , McDonald, W.H. , Jennings, M.P. , Torres, V.J. (2013) Staphylococcus aureus LukAB cytotoxin kills human neutrophils by targeting the CD11b subunit of the integrin Mac‐1. Proc. Natl. Acad. Sci. U. S. A. 110, 10794–10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Murali‐Krishna, K. , Lau, L.L. , Sambhara, S. , Lemonnier, F. , Altman, J. , Ahmed, R. (1999) Persistence of memory CD8 T cells in MHC class I‐deficient mice. Science 286, 1377–1381. [DOI] [PubMed] [Google Scholar]


