Abstract
The transcription factor encoded by Fez family zinc finger 2 (Fezf2) is necessary for normal development of the cerebral cortex. However, Fezf2 continues to be expressed in the mature brain, indicating that it might also be necessary for cortical function throughout life. Here, we show a unique identity of Fezf2-expressing intratelencephalic-projection neurons (IT-PNs) in layer 5 of the mature mouse motor cortex, using a Fezf2–Gfp reporter mouse, in vivo retrograde labeling, whole-cell electrophysiology with morphology reconstruction, and cluster analysis. Fezf2-expressing IT-PNs occupy layer 5A and display an apical dendritic tuft; functionally, they fire broad, adapting action potentials and exhibit an Ih-mediated voltage sag that influences their synaptic properties. In contrast, IT-PNs without Fezf2 expression mainly occupy layer 5B, do not display a tuft, and exhibit regular action potential firing and little sag. Both groups of IT-PNs demonstrated distinct frequency-selective synaptic responses to commissural inputs, indicating unique contributions within the cortical microcircuitry. Our findings establish a new, distinct physiological identity of Fezf2-expressing neurons within mature motor cortex.
Keywords: cluster analysis, Fezf2, intrinsic properties, layer 5, motor cortex, synaptic transmission
Introduction
Specific combinations of key transcription factors control neuron identity in the developing mammalian cerebral cortex and ultimately determine the complex diversity of neurons in the mature brain (Molyneaux et al., 2007). These cell type-specific transcription factors regulate the expression of downstream proteins necessary to acquire the unique position, morphology, electrical behavior, and synaptic connections, or functional identity, of each neuron type (Arlotta et al., 2005). Transcription factors important in development are often retained in the adult brain (Holmberg and Perlmann, 2012), raising the intriguing possibility that they also maintain neuron identity throughout life.
Fez family zinc finger 2 (Fezf2) encodes a zinc finger transcription factor that specifies layer 5 (L5) pyramidal tract-projection neurons (PT-PNs) during development (Chen et al., 2005a,b; Molyneaux et al., 2005). Nevertheless, a wider role for Fezf2 is emerging because deletion or ectopic expression of Fezf2 during development influences commissural connectivity (Chen et al., 2005a) and cortical microcircuits (De la Rossa et al., 2013), and deletion induces hyperactive motor behavior in mice (Hirata et al., 2004). Furthermore, although Fezf2 is essential in early neuron development and motor function, it continues to be expressed into adulthood (Chen et al., 2005b; Ozdinler et al., 2011). Therefore, we hypothesized that Fezf2 also influences the identity and function of other types of L5 projection neurons in the mature primary motor cortex (M1).
To test this possibility, we used a transgenic Fezf2–Gfp reporter mouse in combination with in vivo retrograde labeling, in vitro electrophysiological recordings, and morphological reconstructions of intratelencephalic projection neurons (IT-PNs). Our findings suggest that Fezf2 orchestrates the distinct physiological identity of a subpopulation of IT-PNs within the mature M1 microcircuitry.
Materials and Methods
Retrograde labeling, immunocytochemistry, and fluorescence in situ hybridization.
The University of Otago Animal Ethics Committee approved all procedures. We injected anesthetized hemizygous bacterial artificial chromosome (BAC) Fezf2–Gfp transgenic [(Fezf2–Gfp)CO61Gsat/Mmnc; Gong et al., 2003] or wild-type male Swiss-Webster mice (18–20 d old) with the retrograde tracer Alexa Fluor 647- or biotin-conjugated cholera toxin subunit B (CTB) into M1 (400 nl) or striatum (40 nl) (Oswald et al., 2013) to identify IT-PNs.
Immunocytochemistry (ICC; Oswald et al., 2013) used the following primary antibodies (diluted 1:1000, 70 h, 4°C): anti-special AT-rich sequence binding protein 2 (SATB2; Abcam), anti-COUP-TF interacting protein 2 (CTIP2; Abcam), anti-cut-like homeobox 1 (CUX1; Santa Cruz Biotechnology), and anti-vesicular glutamate transporter 2 (VGLUT2; Synaptic Systems), the latter to define layer boundaries (Fig. 1; Ueta et al., 2013), with 1:500 secondary antibody dilutions [Alexa Fluor 555-conjugated antibody or, for CTIP2, a biotin-conjugated antibody followed by Alexa Fluor 555-conjugated streptavidin (Invitrogen); absence of primary antibody was negative].
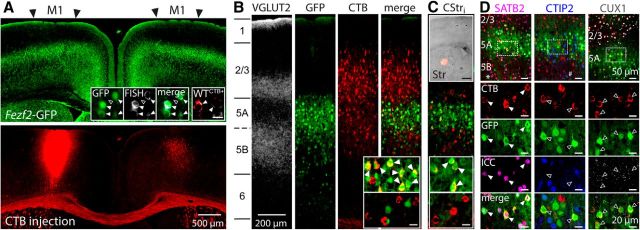
Figure 1.
Identification of Fezf2+ IT-PNs in mature M1. A, GFP fluorescence (top) in a coronal section (0.4 mm from bregma) that had received an injection of the retrograde tracer CTB into the left hemisphere M1 (bottom). Inset shows Fezf2 mRNA expression (bright gray spots) using FISH in GFP+ or CTB+ wild-type (WTCTB+, red) L5A neurons (arrowheads) but not in a GFP− neuron (open arrowhead). B, Layer distribution of Fezf2+CTB+ IT-PNs in the contralateral (right) hemisphere. Insets show enlarged views of L5A (top) and L5B (bottom). Arrowheads indicate Fezf2+CTB+ IT-PNs. We observed similar distributions of Fezf2+CTB+ and Fezf2−CTB+ IT-PNs in 6- to 8-week-old mice. C, CTB injection into the left striatum (top inset; 0.4 mm from bregma; scale bar, 500 μm) shows corticostriatal IT-PNs (CStri) in contralateral M1 (right hemisphere) that are Fezf2+ in L5A (arrowheads in inset) but not L5B (bottom). D, Confocal triple-labeled images of CTB, GFP with ICC for SATB2, CTIP2, or CUX1 shows that >90% of Fezf2+CTB+ IT-PNs in L5A are SATB2+ (arrowheads in enlarged panels), CTIP2− and CUX1− (open arrowheads in enlarged panels). Note that Fezf2−CTB+SATB2+ IT-PNs are in L5B (*); non-retrograde-labeled (CTB−) GFP-expressing L5B neurons are CTIP2+ (#), consistent with their PT-PN identity (Molyneaux et al., 2007). Scale bar, 20 μm unless otherwise stated. A–D from P28 mouse brain.
Fluorescence in situ hybridization (FISH) used 500–1000 nm Fezf2 mRNA probes (set of custom-designed 44 oligonucleotide Stellaris probes tagged with Quasar 570; Biosearch Technologies; Batish et al., 2012) incubated overnight with 12 μm frozen sections, according to the instructions of the manufacturer. Post hoc Alexa Fluor 405-conjugated streptavidin (1:250) detected CTB.
Image capture used standard epifluorescence (Olympus BX51) or confocal (Nikon A1R) microscopy with appropriate blue, green, red, and far red filter sets.
Electrophysiology.
After retrograde labeling (5–10 d or 6 weeks), we prepared acute coronal slices (300 μm thick) containing contralateral M1. Artificial CSF (ACSF) contained 126 mm NaCl, 3 mm KCl, 1 mm NaH2PO4, 2 mm MgSO4, 2 mm CaCl2, 25 mm NaHCO3, 15 glucose (equilibrated with 95% O2 and 5% CO2), 20 μm CNQX, 50 μm d-APV, and 50 μm picrotoxin to block fast synaptic input, except when studying synaptic physiology, when we used 10 μm picrotoxin. We used glass electrodes (containing biocytin) to target retrograde labeled (CTB+) GFP-positive (GFP+) or GFP-negative (GFP−) L5 IT-PNs, to probe their electrical properties with whole-cell current clamp and to post hoc recover their morphology (Oswald et al., 2013). Most recordings were at 26°C, but the differences between Fezf2+ (n = 9) and Fezf2− (n = 8) CTB+ IT-PNs remained at higher physiological temperature (36°C; data not shown).
A tungsten bipolar electrode (125 μm tip separation; MicroProbes) delivered pairs of stimuli to commissural axons (0.1 ms duration at 10 or 25 Hz; amplitude 1.1–1.15× the minimum required to evoke a response) before and after application of ACSF containing ZD7288 (4-ethylphenylamino-1,2-dimethyl-6-methylaminopyrimidinium chloride; 50 μm), the Ih antagonist. After recording and overnight fix, biocytin-filled IT-PNs were detected (Alexa Fluor 555-conjugated streptavidin) and reconstructed (NeuronJ; Meijering et al., 2004).
Data analysis and statistics.
As in the study by Oswald et al. (2013), we measured 19 parameters from single IT-PNs: (1) resting membrane potential; (2) input resistance; (3) membrane time constant; (4) sag; (5) action potential (AP) firing threshold; (6) AP peak amplitude; (7) AP half-width; (8) AP maximum rise slope; (9) AP maximum decay slope; (10) the amplitude of the depolarizing afterpotential; (11) instantaneous frequency of the first interspike interval; (12) adaptation index; (13) postburst afterhyperpolarization amplitude; (14) pial depth; (15) soma size; (16) tuft height; (17) tuft width; (18) apical dendrite height; and (19) shaft width. The EPSP2/EPSP1 ratio was the second EPSP amplitude measured from the trough of the first EPSP, normalized to the first EPSP amplitude. EPSP rise time was measured between 10 and 90% of the peak amplitude. The EPSP decay time constant was obtained by fitting a single exponential from the peak of the EPSP to baseline. We counted double-positive neurons from ICC and FISH as a percentage of IT-PNs retrograde labeled from contralateral M1 within L5. Statistical comparisons used an unsupervised cluster analysis (Statistica 10; Statsoft) with 32 L5 IT-PNs with all 19 parameters available (excludes synaptic properties) as in the study by Oswald et al. (2013), and Student's t test, nonparametric Mann–Whitney U test, or one-way ANOVA with Bonferroni's multiple comparisons (Prism; GraphPad Software).
Single-cell RT-PCR.
We harvested cytoplasm from individual L5 CTB+ IT-PNs for immediate RT (SuperScript III One-Step RT-PCR system; Invitrogen), followed by nested two-round PCR to amplify Fezf2 cDNA using two primer pairs: (1) one from the study by Wang et al. (2012); and (2) one custom designed to span at least one intron. The primers were as follows: outer pair 1, sense, 5′-AGACGCTGCTCAGTTACTCG and antisense, 5′-ATGACCTGCGGTTTGATGACC; inner pair 1, sense, 5′-CTCTACTGACAGCAAACCCAA and antisense, 5′-GCACACAAACGGTCTAGC; outer pair 2, sense, 5′-ACAGAAGAGACACGCTGCAT and antisense, 5′-ATGACCTGCGGTTTGATGACC; and inner pair 2, the same sense primers with antisense primer 5′-AGGCTGCTCTTCCAGAACTC. Amplified products were detected on a 2% agarose gel, and commercial sequencing confirmed their alignment with mouse Fezf2 mRNA (Applied Biosystems). We used two negative controls: (1) sample without RT; and (2) mock harvest with RT from close by neurons (no template control). Positive control was total forebrain RNA (Clontech) with RT, and all samples were processed in parallel.
Results
GFP-expressing neurons distributed throughout L5 and L6 of M1 (Fig. 1A, top). FISH reported Fezf2 mRNA expression in 93% of GFP+ neurons and 15% of GFP− L5 neurons (489 GFP+, 406 GFP−, n = 3 mice; Fig. 1A, inset), consistent with previously described adult Fezf2 mRNA expression (Chen et al., 2005b; Ozdinler et al., 2011) and indicating reliable reporting of Fezf2 mRNA by GFP in L5 of M1 in the BAC Fezf2–Gfp transgenic mouse. Injection of the retrograde tracer CTB into M1 or the striatum (Oswald et al., 2013; Sohur et al., 2014) identified CTB+ IT-PNs distributed across contralateral L2/3 to L6 homotopic to the injection site (Fig. 1A, bottom, B,C). Most CTB+ L5A IT-PNs were GFP+ (Fig. 1B,C, arrowheads in inset; 85%, n = 160, 7 mice) and Fezf2+ by FISH (94%, n = 32, 2 mice) and are hereafter called Fezf2+CTB+, whereas only 4% of L5B IT-PNs were GFP+ and Fezf2+ by FISH (14%, n = 28, 2 mice); indeed, most L5B IT-PNs were GFP− and Fezf2− (Fig. 1B,C; n = 136, 7 mice; also seen in the study by Sohur et al., 2014) and are hereafter called Fezf2−CTB+. A similar pattern existed in wild-type mice (Fig. 1A, inset); 97% (n = 95, 2 mice) of L5A CTB+ IT-PNs were Fezf2+ by FISH compared with 16% (n = 90, 2 mice) in L5B. We also conducted a broad assessment of the molecular identity of Fezf2+ L5A IT-PNs using ICC for known molecular markers (Fig. 1D); SATB2 defines IT-PN phenotype, CTIP2 defines PT-PN phenotype, and CUX1 defines L2/3 neurons (Sohur et al., 2014). L5A Fezf2+CTB+ IT-PNs expressed SATB2 but not CTIP2 or CUX1, confirming their L5 commissural projection phenotype; the same is true of L5B Fezf2−CTB+ IT-PNs (Fig. 1D, * symbol; see also Sohur et al., 2014). To further test for Fezf2 expression in L5A IT-PNs, we also sampled their cytosol for single-cell RT-PCR (Fig. 2B) and detected Fezf2 in GFP+CTB+ (n = 23) and wild-typeCTB+ (n = 3) L5A IT-PNs but not in GFP−CTB+ (n = 14) or wild-typeCTB+ (n = 9) L5B IT-PNs. The combination of single-cell RT-PCR and FISH provides verification of Fezf2 expression in Fezf2–Gfp transgenic and wild-type mice and validates the transgenic mouse as a useful model to study Fezf2-expressing L5A IT-PNs. Together with the ICC, our findings establish an exclusive subpopulation of Fezf2-expressing L5A IT-PNs in mature M1.
Figure 2.
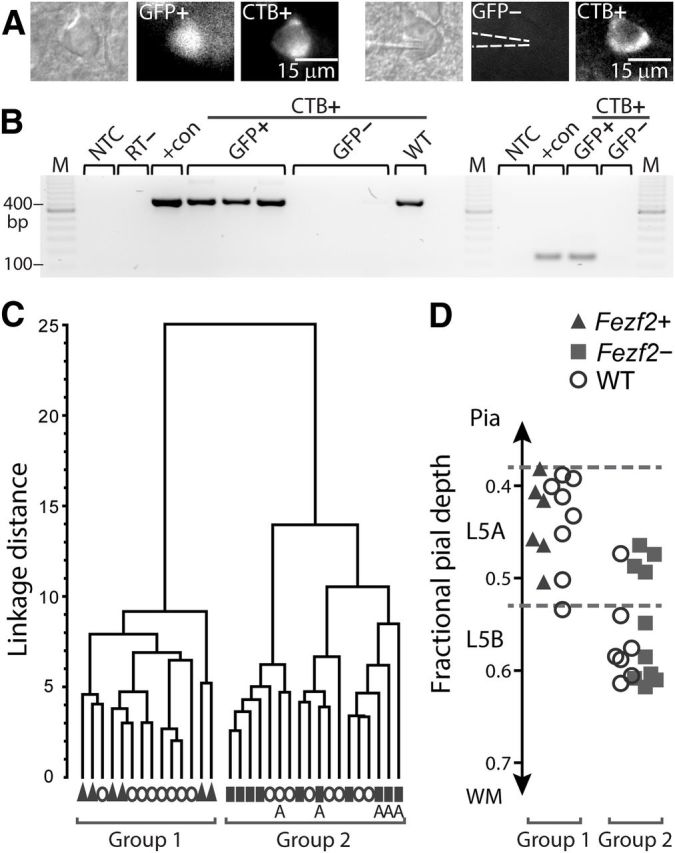
Specific identity of Fezf2+CTB+ IT-PNs in L5 of mature M1. A, We targeted identified Fezf2+/− CTB+ IT-PNs with patch-clamp recordings. Dashed lines indicate the recording electrode position on a GFP− neuron. B, Single-cell RT-PCR products for Fezf2 mRNA from four GFP+CTB+, four GFP−CTB+, and one wild-type (WT) IT-PNs. Amplification used two primer sets, either 1 or 2, to generate product lengths of 118 or 406 bp, respectively. C, Two broad groups of L5 IT-PNs were segregated in an unsupervised cluster analysis, calculated using 19 parameters measured for each Fezf2+CTB+, Fezf2−CTB+, or wild-typeCTB+ IT-PN. D, Pial depth position (as a fraction of cortical thickness) identified L5A neurons in C (A in Group 2). NTC, No template control; RT−, GFP+ single-cell sample without reverse transcription; +con, positive control; M, marker; WM, white matter.
To test whether Fezf2 expression in IT-PNs influences their identity and function, we sampled the electrophysiological and morphological properties of single Fezf2+CTB+ and Fezf2−CTB+ (and wild-typeCTB+) IT-PNs across L5A and L5B (Fig. 2A). We then used an unsupervised cluster analysis to unbiasedly compare multiple parameters in all neurons, independent of their Fezf2 expression status. (For a full description of selected parameters, see text below and Figure 3.) The cluster analysis accurately separated IT-PNs into two groups (Fig. 2C). Significantly, post hoc allocation of the Fezf2 expression status of each neuron revealed the groups as Fezf2+CTB+ (n = 6) and Fezf2−CTB+ (n = 10), strongly indicating a Fezf2-specific functional distinction in the IT-PN population. Allocation of wild-type information showed that wild-typeCTB+ IT-PNs (n = 16) intermixed within either Fezf2+CTB+ or Fezf2−CTB+ groups, indicating that similar cell types exist in both wild-type and transgenic mice. Furthermore, consistent with the sublayer arrangement reported in Figure 1, B and C, the Fezf2+ and wild-type IT-PNs of group 1 resided exclusively in L5A (Fig. 2C,D). In contrast, most Fezf2− and wild-type IT-PNs of group 2 occupied L5B. Note that the four Fezf2−CTB+ IT-PNs and one wild-typeCTB+ IT-PN found in L5A were also accurately distinguished from the Fezf2+CTB+ IT-PNs within the same sublayer (Figs. 2D, 3A for morphology). This indicates that the segregation of L5 IT-PNs was not solely based on sublayer position but rather on Fezf2 expression.
Figure 3.
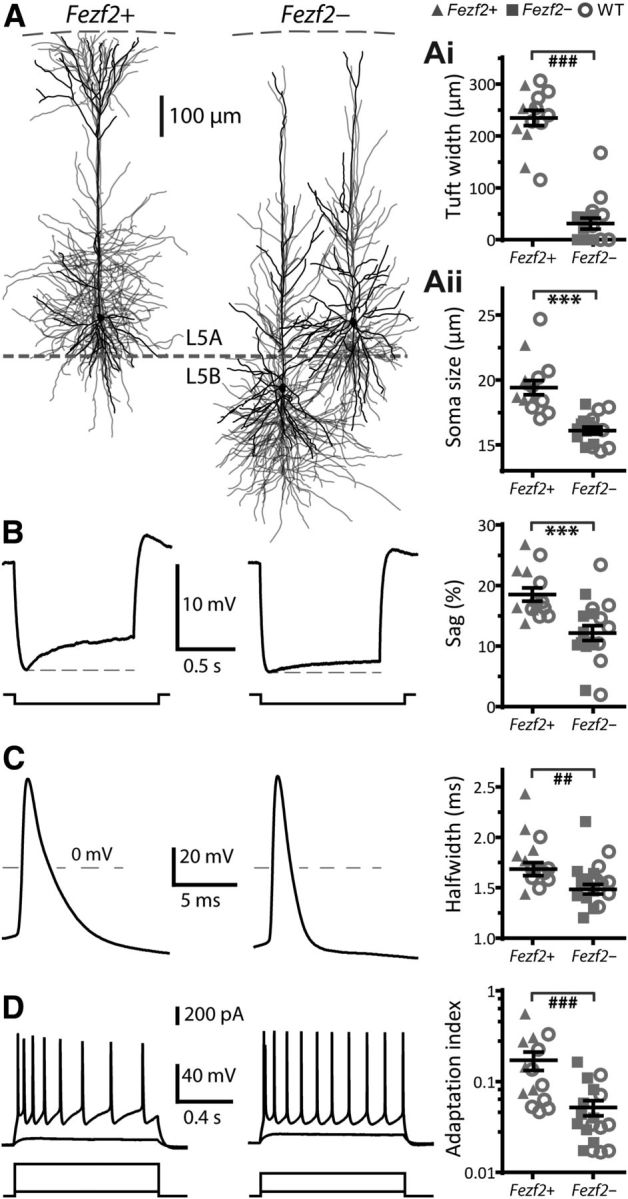
Unique electrophysiological and morphological properties of L5 Fezf2+CTB+ M1 IT-PNs. A, Morphology of Fezf2+CTB+ (n = 6) and Fezf2−CTB+ IT-PNs (separated as L5B, n = 6 and L5A, n = 4) aligned to pia and values of apical dendrite tuft width (Aii) and soma size (Aii). B, Representative voltage responses to hyperpolarizing current steps and values of Ih-mediated sag. C, Representative APs and half-widths. D, Repetitive AP firing in response to depolarizing current steps and adaptation index values. ##p < 0.01, ###p < 0.001 (Mann–Whitney U test), ***p < 0.001 (t test). We observed similar properties of Fezf2+CTB+ and Fezf2−CTB+ IT-PNs in 7- to 8-week-old mice.
Although the cluster analysis reveals two distinct types of L5 IT-PNs based on Fezf2 expression, it cannot reveal the differences. Therefore, we analyzed individual parameters for differences between Fezf2+CTB+ and Fezf2−CTB+ neuron groups, including wild-typeCTB+ IT-PNs, based on the segregation by the cluster analysis. Anatomically, the most striking feature of Fezf2+CTB+ IT-PNs was their large, tufted apical dendrite that reached L1 compared with the smaller, or absent, tuft in Fezf2−CTB+ IT-PNs (Fig. 3A,Ai). Fezf2+CTB+ IT-PNs also had larger somas (Fig. 3Aii) but similar dendritic shaft widths (Fezf2+, 2.7 ± 0.2 μm; Fezf2−, 2.5 ± 0.2 μm; p = 0.83, t test). Their intrinsic membrane properties also differed. The resting membrane potential was relatively depolarized in Fezf2+CTB+ IT-PNs (Fezf2+, −70 ± 1 mV, n = 14; Fezf2−, −77 ± 1 mV, n = 18; p < 0.001, t test). During hyperpolarizing steps, Fezf2+CTB+ IT-PNs showed a significantly larger voltage sag (Fig. 3B) indicative of Ih current (Sheets et al., 2011). Fezf2+CTB+ IT-PNs exhibited broader APs than Fezf2−CTB+ IT-PNs, seen as a longer half-width (Fig. 3C) and slower maximum decay rate (Fezf2+, −40 ± 2 mV/ms; Fezf2−, −48 ± 2 mV/ms; p < 0.01, t test). AP firing during sustained depolarization showed stronger frequency adaptation in Fezf2+CTB+ IT-PNs (Fig. 3D) seen as a larger adaptation index. Hence, we identified distinct electrical and morphological properties of Fezf2+CTB+ and Fezf2−CTB+ IT-PNs.
Next, we assessed whether Fezf2+CTB+ and Fezf2−CTB+ IT-PNs behaved differently within the M1 cortical microcircuit by testing their response to commissural inputs arising from the contralateral hemisphere (Hooks et al., 2013). A brief minimal electrical stimulation of the corpus callosum near the midline activated EPSPs in IT-PNs (Fig. 4A). Because electrical activity in M1 in vivo displays frequency shifts between 10 and 25 Hz during movements (Rickert et al., 2005; Isomura et al., 2009), we stimulated at these frequencies. At 10 Hz, a pair of stimuli evoked similarly sized EPSPs in Fezf2+CTB+ IT-PNs, whereas at 25 Hz, the second EPSP depressed (Fig. 4A,B; n = 10; paired t test, p < 0.001). In contrast, in Fezf2−CTB+ IT-PNs, the second EPSP depressed at 10 Hz but facilitated at 25 Hz (Fig. 4A,B; n = 9; paired t test, p < 0.05). At 25 Hz stimulation, the EPSP depression in Fezf2+CTB+ IT-PNs was significantly different from the EPSP facilitation in Fezf2−CTB+ IT-PNs (repeated-measures ANOVA, F(1,17) = 36.7; p < 0.0001; Fig. 4B). Together, these findings indicate that Fezf2+ and Fezf2− IT-PNs respond differently within the cortical microcircuitry.
Figure 4.
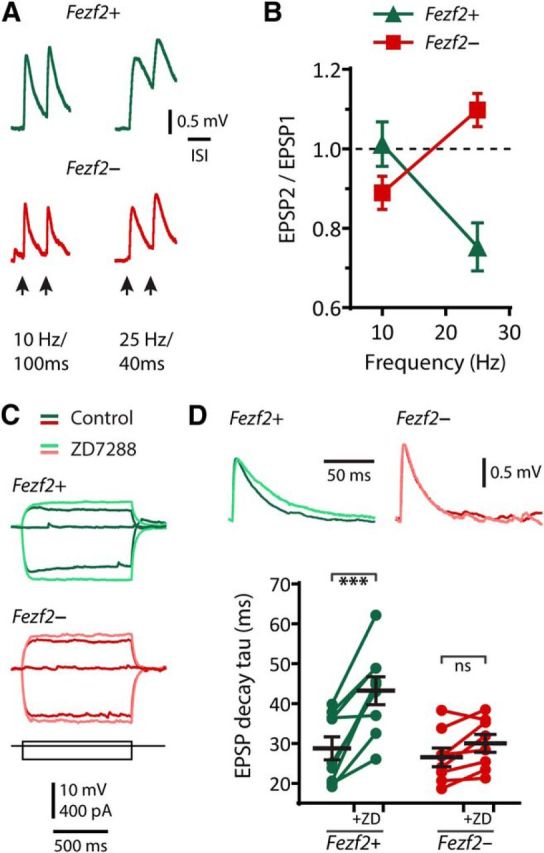
Motor-related frequency-selective synaptic responses of L5 Fezf2+CTB+ M1 IT-PNs. A, Representative EPSPs evoked by commissural stimulation at 10 and 25 Hz. B, Summary of the ratio of first and second EPSP amplitudes at 10 and 25 Hz stimulation. C, Response to current steps showing the membrane potential sag before (dark green and red) and after bath application of the Ih blocker ZD7288 (light green and red). D, Representative EPSP waveforms (top) before and after ZD7288 and changes in EPSP decay time constant, tau (bottom). ***p < 0.001 (paired t test).
The Ih current is neuron type-specific in L5 of M1 and influences synaptic behavior (Sheets et al., 2011). The differences we observed in voltage sag between Fezf2+CTB+ and Fezf2−CTB+ IT-PNs suggested that Ih might contribute differently to their synaptic properties. In both cell types, the Ih antagonist ZD7288 (50 μm) eliminated the sag (Fezf2+, sag(ZD7288) = 0.7 ± 0.1%, n = 7, p ≤ 0.01; Fezf2−, sag(ZD7288) = 0.8 ± 0.1%, n = 6, p < 0.01; paired t tests; Fig. 4C), increased their input resistance (Fezf2+, n = 8, p < 0.01; Fezf2−, n = 8, p < 0.01; paired t tests; data not shown), and slowed their membrane time constant (Fezf2+, p < 0.001; Fezf2−, p < 0.001; paired t tests; data not shown). Of greater significance was that ZD7288 selectively prolonged the EPSP in Fezf2+CTB+ IT-PNs, indicating that Ih specifically shapes synaptic events in Fezf2+ IT-PNs (Δrise time(ZD7288-control): Fezf2+, 1.1 ± 0.4 ms, p < 0.05; Fezf2−, 0.2 ± 0.2, p = 0.29; paired t tests; Δdecay tau shown in Fig. 4D).
Discussion
Here we show that expression of the transcription factor Fezf2 identifies a distinct IT-PN phenotype within L5A of mature M1. Although previous work shows that Fezf2 plays an important role during the development of L5B cortical PT-PNs (Chen et al., 2005a,b; Molyneaux et al., 2005), our findings now expand a role for Fezf2 to L5A IT-PNs. Interestingly, the distribution of Fezf2 expression in L5A neurons we report here (Fig. 1A) closely resembles that at postnatal day 4 (P4; Sohur et al., 2014, their Fig. 6E), P14 (Molyneaux et al., 2005, their Fig. 1D), and adult P120 (Ozdinler et al., 2011, their Fig. 3A). The idea that Fezf2 influences commissurally projecting IT-PNs is further supported by the aborted commissural axons in the corpus callosum of the P28 Fezf2 knock-out brain (Chen et al., 2005a; Molyneaux et al., 2005). Given the acknowledged diversity of IT-PNs (Le Be et al., 2007; Molyneaux et al., 2009; Otsuka and Kawaguchi, 2011; Ueta et al., 2013), it is remarkable that Fezf2 expression differentiates L5 IT-PNs based on their morphology, intrinsic electrophysiology, and synaptic responses even within the same layer. We propose that the distinct properties of Fezf2+ IT-PNs will be important for distinguishing their functional role within the cortical microcircuitry.
As a transcription factor, Fezf2 is likely to influence IT-PN function by coordinating multiple downstream partners within a wider gene network. Although our findings do not identify these partners, they do provide the resulting functional and morphological phenotypes of neurons with or without Fezf2 expression. Perhaps most striking was that only Fezf2+ IT-PNs displayed an apical dendritic tuft. Because some IT-PNs initially develop a tuft that is then retracted at maturity (Koester and O'Leary, 1992; Kasper et al., 1994), it is tempting to speculate that Fezf2 is central for both the specification of the tuft and to prevent its loss. This idea is also supported by the fact that Fezf2 deletion early in development results in fewer tuft branches in the apical dendrite of unidentified L5 neurons (Chen et al., 2005b), some of which exhibit the same type of adapting APs (Chen et al., 2008) seen here in Fezf2+ IT-PNs (Fig. 3D). More recently, ectopic expression of Fezf2 was reported to drive a tufted phenotype in cortical L4 stellate neurons (De la Rossa et al., 2013). The larger somatic Ih-mediated sag potential proved to be another key discriminator between Fezf2+ versus Fezf2− IT-PNs and is consistent with the appearance of Ih in L4 neurons during Fezf2-driven reprogramming of these neurons (De la Rossa et al., 2013). Furthermore, the greater presence of Ih in Fezf2+ IT-PNs provides an opportunity for selective neuromodulation of L5 output (Kalmbach et al., 2013).
From a wider functional perspective, the presence or absence of a tuft in Fezf2+ and Fezf2− L5 IT-PNs implies that they make different contributions during M1 processing, consistent with the recently shown sublayer-specific microcircuitry in M1 (Hooks et al., 2013; Ueta et al., 2013). The presence of the tuft may be essential for receiving L1 inputs and top-down influence specifically of Fezf2+ IT-PNs. Indeed, slender-tufted L5A neurons are known to actively propagate APs into their apical dendrite to support distal synaptic integration (Grewe et al., 2010), and the broad APs of Fezf2+ IT-PNs may also facilitate this function. In contrast, Fezf2− untufted IT-PNs with their deeper position within L5B and little opportunity for innervation from L1 may engage preferentially with deeper layers.
Finally, both Fezf2+ and Fezf2− IT-PNs are likely to be important during interhemispheric communication, so we stimulated their commissural inputs at frequencies known to occur during movement. The resulting Fezf2-specific frequency-selective facilitation or depression of EPSPs could serve to either boost or dampen M1 activity during different movement-related frequency oscillations (i.e., theta, 6–10 Hz; beta, 15–30 Hz; Rickert et al., 2005).
Together, our findings establish that expression of the developmental transcription factor Fezf2 functionally identifies a distinct subpopulation of mature L5 IT-PNs.
Footnotes
This work was supported by The Royal Society of New Zealand Marsden Fund and the Department of Physiology.
The authors declare no competing financial interests.
References
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Batish M, van den Bogaard P, Kramer FR, Tyagi S. Neuronal mRNAs travel singly into dendrites. Proc Natl Acad Sci U S A. 2012;109:4645–4650. doi: 10.1073/pnas.1111226109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Schaevitz LR, McConnell SK. Fezl regulates the differentiation and axon targeting of layer 5 subcortical projection neurons in cerebral cortex. Proc Natl Acad Sci U S A. 2005a;102:17184–17189. doi: 10.1073/pnas.0508732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Wang SS, Hattox AM, Rayburn H, Nelson SB, McConnell SK. The Fezf2-Ctip2 genetic pathway regulates the fate choice of subcortical projection neurons in the developing cerebral cortex. Proc Natl Acad Sci U S A. 2008;105:11382–11387. doi: 10.1073/pnas.0804918105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JG, Rasin MR, Kwan KY, Sestan N. Zfp312 is required for subcortical axonal projections and dendritic morphology of deep-layer pyramidal neurons of the cerebral cortex. Proc Natl Acad Sci U S A. 2005b;102:17792–17797. doi: 10.1073/pnas.0509032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Rossa A, Bellone C, Golding B, Vitali I, Moss J, Toni N, Lüscher C, Jabaudon D. In vivo reprogramming of circuit connectivity in postmitotic neocortical neurons. Nat Neurosci. 2013;16:193–200. doi: 10.1038/nn.3299. [DOI] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, Heintz N. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Grewe BF, Bonnan A, Frick A. Back-propagation of physiological action potential output in dendrites of slender-tufted L5A pyramidal neurons. Front Cell Neurosci. 2010;4:13. doi: 10.3389/fncel.2010.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata T, Suda Y, Nakao K, Narimatsu M, Hirano T, Hibi M. Zinc finger gene fez-like functions in the formation of subplate neurons and thalamocortical axons. Dev Dyn. 2004;230:546–556. doi: 10.1002/dvdy.20068. [DOI] [PubMed] [Google Scholar]
- Holmberg J, Perlmann T. Maintaining differentiated cellular identity. Nat Rev Genet. 2012;13:429–439. doi: 10.1038/nrg3209. [DOI] [PubMed] [Google Scholar]
- Hooks BM, Mao T, Gutnisky DA, Yamawaki N, Svoboda K, Shepherd GMG. Organization of cortical and thalamic input to pyramidal neurons in mouse motor cortex. J Neurosci. 2013;33:748–760. doi: 10.1523/JNEUROSCI.4338-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isomura Y, Harukuni R, Takekawa T, Aizawa H, Fukai T. Microcircuitry coordination of cortical motor information in self-initiation of voluntary movements. Nat Neurosci. 2009;12:1586–1593. doi: 10.1038/nn.2431. [DOI] [PubMed] [Google Scholar]
- Kalmbach BE, Chitwood RA, Dembrow NC, Johnston D. Dendritic generation of mGluR-mediated slow afterdepolarization in layer 5 neurons of prefrontal cortex. J Neurosci. 2013;33:13518–13532. doi: 10.1523/JNEUROSCI.2018-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasper EM, Lübke J, Larkman AU, Blakemore C. Pyramidal neurons in layer 5 of the rat visual cortex. III. Differential maturation of axon targeting, dendritic morphology, and electrophysiological properties. J Comp Neurol. 1994;339:495–518. doi: 10.1002/cne.903390404. [DOI] [PubMed] [Google Scholar]
- Koester SE, O'Leary DDM. Functional classes of cortical projection neurons develop dendritic distinctions by class-specific sculpting of an early common pattern. J Neurosci. 1992;12:1382–1393. doi: 10.1523/JNEUROSCI.12-04-01382.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bé JV, Silberberg G, Wang Y, Markram H. Morphological, electrophysiological, and synaptic properties of corticocallosal pyramidal cells in the neonatal rat neocortex. Cereb Cortex. 2007;17:2204–2213. doi: 10.1093/cercor/bhl127. [DOI] [PubMed] [Google Scholar]
- Meijering E, Jacob M, Sarria JCF, Steiner P, Hirling H, Unser M. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A. 2004;58A:167–176. doi: 10.1002/cyto.a.20022. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Hirata T, Hibi M, Macklis JD. Fezl is required for the birth and specification of corticospinal motor neurons. Neuron. 2005;47:817–831. doi: 10.1016/j.neuron.2005.08.030. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Menezes JRL, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Fame RM, MacDonald JL, MacQuarrie KL, Macklis JD. Novel subtype-specific genes identify distinct subpopulations of callosal projection neurons. J Neurosci. 2009;29:12343–12354. doi: 10.1523/JNEUROSCI.6108-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald MJ, Tantirigama ML, Sonntag I, Hughes SM, Empson RM. Diversity of layer 5 projection neurons in the mouse motor cortex. Front Cell Neurosci. 2013;7:174. doi: 10.3389/fncel.2013.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka T, Kawaguchi Y. Cell diversity and connection specificity between callosal projection neurons in the frontal cortex. J Neurosci. 2011;31:3862–3870. doi: 10.1523/JNEUROSCI.5795-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdinler PH, Benn S, Yamamoto TH, Güzel M, Brown RH, Jr, Macklis JD. Corticospinal motor neurons and related subcerebral projection neurons undergo early and specific neurodegeneration in hSOD1G93A transgenic ALS mice. J Neurosci. 2011;31:4166–4177. doi: 10.1523/JNEUROSCI.4184-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickert J, de Oliveira SC, Vaadia E, Aertsen A, Rotter S, Mehring C. Encoding of movement direction in different frequency ranges of motor cortical local field potentials. J Neurosci. 2005;25:8815–8824. doi: 10.1523/JNEUROSCI.0816-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheets PL, Suter BA, Kiritani T, Chan CS, Surmeier DJ, Shepherd GMG. Corticospinal-specific HCN expression in mouse motor cortex: Ih-dependent synaptic integration as a candidate microcircuit mechanism involved in motor control. J Neurophysiol. 2011;106:2216–2231. doi: 10.1152/jn.00232.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohur US, Padmanabhan HK, Kotchetkov IS, Menezes JRL, Macklis JD. Anatomic and molecular development of corticostriatal projection neurons in mice. Cereb Cortex. 2014;24:293–303. doi: 10.1093/cercor/bhs342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueta Y, Otsuka T, Morishima M, Ushimaru M, Kawaguchi Y. Multiple layer 5 pyramidal cell subtypes relay cortical feedback from secondary to primary motor areas in rats. Cereb Cortex. 2013 doi: 10.1093/cercor/bht088. doi: 10.1093/cercor/bht088. Advance online publication. Retrieved February 21, 2014. [DOI] [PubMed] [Google Scholar]
- Wang X, Spandidos A, Wang H, Seed B. PrimerBank: a PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Res. 2012;40:D1144–D1149. doi: 10.1093/nar/gkr1013. [DOI] [PMC free article] [PubMed] [Google Scholar]