Abstract
The mammalian target of rapamycin (mTOR) pathway integrates multiple signals and regulates crucial cell functions via the molecular complexes mTORC1 and mTORC2. These complexes are functionally dependent on their raptor (mTORC1) or rictor (mTORC2) subunits. mTOR has been associated with oligodendrocyte differentiation and myelination downstream of the PI3K/Akt pathway, but the functional contributions of individual complexes are largely unknown. We show, by oligodendrocyte-specific genetic deletion of Rptor and/or Rictor in the mouse, that CNS myelination is mainly dependent on mTORC1 function, with minor mTORC2 contributions. Myelin-associated lipogenesis and protein gene regulation are strongly reliant on mTORC1. We found that also oligodendrocyte-specific overactivation of mTORC1, via ablation of tuberous sclerosis complex 1 (TSC1), causes hypomyelination characterized by downregulation of Akt signaling and lipogenic pathways. Our data demonstrate that a delicately balanced regulation of mTORC1 activation and action in oligodendrocytes is essential for CNS myelination, which has practical overtones for understanding CNS myelin disorders.
Keywords: mTOR, myelin, oligodendrocytes, TSC1, TSC2, tuberous sclerosis
Introduction
CNS myelination requires multiple-level coordination from controlling the proliferation and differentiation of oligodendrocyte precursor cells (OPCs) to wrapping of myelin sheaths around axons by mature oligodendrocytes as the prerequisite of rapid saltatory nerve conduction. Unraveling the mechanisms linking external signals to intracellular signaling and myelination is required for our understanding of oligodendrocyte biology and neuron–glia interactions, which has practical implications for treatments of demyelinating and dysmyelinating disorders and enhancing repair.
The kinase Akt is critically involved in CNS myelination. Expression of constitutively active Akt in oligodendrocytes induces hypermyelination (Flores et al., 2008) and the underlying mechanism has been linked to the mammalian target of rapamycin (mTOR, also referred to as mechanistic target of rapamycin) pathway (Narayanan et al., 2009), a crucial mediator of PI3K/Akt signaling (Laplante and Sabatini, 2012). The two mTOR-containing complexes mTORC1 and mTORC2 contain the functionally required proteins raptor (regulatory associated protein of mTOR) and rictor (rapamycin-insensitive companion of mTOR), respectively. Akt–mTOR interactions are complex because mTOR acts downstream and upstream of Akt. Full Akt activation requires phosphorylation of threonine 308 (T308) and serine 473 (S473). PI3K signaling leads to phosphorylation of Akt at T308 by PDK1, whereas mTORC2 activates Akt directly by phosphorylating S473. Activated Akt can phosphorylate the tuberous sclerosis complex 1/2 (consisting of TSC1 and TSC2, also called hamartin and tuberin, and TBC1D7; Dibble et al., 2012), which then releases its inhibitory effect on Rheb (Ras homolog enriched in brain), a GTPase that activates mTORC1 (Huang and Manning, 2009). Inactivating mutations in either TSC1/2 result in the autosomal-dominant multisystem disorder tuberous sclerosis complex, which is associated with multiple CNS manifestations (Crino et al., 2006).
Experiments using rapamycin, an inhibitor of mTORC1 and partial inhibitor of mTORC2 signaling, suggested that mTOR regulates oligodendrocyte differentiation (Norrmén and Suter, 2013; Wood et al., 2013). In addition, widespread conditional Rheb1 inactivation in neural precursors causes CNS myelination deficits (Zou et al., 2011). Here, we examined the specific roles of mTORC1 and mTORC2 in oligodendrocytes and myelination in vivo. To this end, we analyzed mice lacking raptor (called raptor mutants hereafter) or rictor (called rictor mutants hereafter) individually or raptor and rictor together (called raptor/rictor mutants hereafter) to assess potential compensatory effects and possible cooperative functions. The mTORC1 inhibitor TSC1 (called TSC1 mutants hereafter) was also deleted to analyze consequences of putative mTORC1 hyperactivation. We used conditional gene ablation via Cre-mediated recombination under the control of cyclic nucleotide phosphodiesterase (CNP) gene regulatory elements (Genoud et al., 2002; Lappe-Siefke et al., 2003), together with conditional LoxP-based raptor, rictor, and Tsc1-null alleles (Kwiatkowski et al., 2002; Bentzinger et al., 2008; Polak et al., 2008). In parallel, we analyzed consequences of raptor and/or rictor ablation in myelin maintenance (called induced mutants thereafter) using inducible Cre-mediated recombination in adult oligodendrocytes (Plp1-CreERT2 (Leone et al., 2003). Our results identify mTORC1 as crucial regulator of CNS myelination, whereas mTORC2 plays subtle functional roles in oligodendrocyte biology. Precisely controlled mTORC1 activation levels and actions in oligodendrocytes are critical for proper CNS myelination, with implications for the etiology of tuberous sclerosis complex.
Materials and Methods
Generation of conditional knock-out mice.
Mice homozygous for floxed alleles of Tsc1 (Tsc1lox/lox; JAX stock #005680; The Jackson Laboratory), Rictor (Rictorloxp/loxp; Bentzinger et al., 2008), and/or Rptor (Rptorloxp/loxp; Polak et al., 2008) were crossed with mice expressing the Cre recombinase under the control of the CNP promoter (CNP-Cre; Lappe-Siefke et al., 2003) or the Plp1 promoter (Plp1-CreERT2; Leone et al., 2003). Tissue from S6K1;S6K2 double-null mice were a generous gift Dr. Mario Pende (INSERM, Paris, France; Shima et al., 1998; Pende et al., 2004). Tissue from Eif4ebp1;Eif4ebp2 double-null mice were a generous gift from Dr. Nahum Sonenberg (McGill University, Montreal, Canada; Le Bacquer et al., 2007). Animals of either sex were used for the study and animals not carrying the Cre transgene were used as controls unless otherwise indicated. Genotypes were determined by PCR using genomic DNA. Primer sequences are listed in Table 1. Animal experiments were approved by the veterinary office of the Canton of Zurich, Switzerland.
Table 1.
List of PCR primers
| Target | Forward primer | Reverse primer | Product (in bp) |
|---|---|---|---|
| Primers for genotyping | |||
| Raptor | 5′-ATGGTAGCAGGCACACTCTTCATG-3′ | 5′-GGGAGATGTGACCTTAACCAGCTTC-3′ | WT : 141 |
| Flox : 228 | |||
| Rictor | 5′-TTATTAACTGTGTGTGGGTTG-3′ | 5′-CGTCTTAGTGTTGCTGTCTAG-3′ | WT : 197 |
| Flox : 295 | |||
| TSC1 | 5′-GTCACGACCGTAGGAGAAGC-3′ | 5′-GAATCAACCCCACAGAGCAT-3′ | WT : 193 |
| Flox : 230 | |||
| Cre | 5′-ACCAGGTTCGTTCACTCATGG-3′ | 5′-AGGCTAAGTGCCTTCTCTACA-3′ | KI : 200 |
| CNPcre | 5′-GATGGGGCTTACTCTTGC-3′ | 5′-CATAGCCTGAAGAACGAGA-3′ | KI : 900 |
| Primers for RT-PCR | |||
| GAPDH | 5′-CGTCCCGTAGACAAAATGGT-3′ | 5′-TTGATGGCAACAATCTCCAC-3′ | |
| PLP | 5′-GTTCCAGAGGCCAACATCAAGCTC-3′ | 5′-AGCCATACAACAGTCAGGGCATAG-3′ | |
| MOG | 5′-TGCACCGAAGACTGGCAGGAC-3′ | 5′-AAGGACCTGCTGGGCTCTCCT-3′ | |
| MAG | 5′-TGCCTTCAACCTGTCTGTG-3′ | 5′-CGGGTTGGATTTTACCACAC-3′ | |
| MBP | 5′-GGCCTCAGAGGACAGTGATG-3′ | 5′-TCTGCTGTGTGCTTGGAGTC-3′ | |
| MRF | 5′-GTATGACGCTAACTACAAGGAGCTGCC-3′ | 5′-CGGTCACCTGGAAGTGGTTCTTCTTC-3′ | |
| HMGCR | 5′-CACAATAACTTCCCAGGGGT-3 | 5′-GGCCTCCATTGAGATCCG-3′ | |
| SCAP | 5′-CTGCTACCCGCTGCTGAA-3′ | 5′-AGAATTCCACAGGTCCCGTTC-3′ | |
| IDI1 | 5′-TCAACTTCATGTTCACCCCA-3′ | 5′-GAGTTGGGAATACCCTTGGA-3′ | |
| SCD1 | 5′-CAGCCGAGCCTTGTAAGTTC-3′ | 5′-GCTCTACACCTGCCTCTTCG-3′ | |
| S1P | 5′-GGTGCTGGAGTGCGGGGTTC-3′ | 5′-ACCCCAGGAAGTCTCCGGGC-3′ | |
| S2P | 5′-TCACCAGTCCAGCAGCTAAGGA-3′ | 5′-GCCTCTGGGTCCAATGGCAGG-3′ | |
| Lipin-1 | 5′-GCTTTTGGGAACCGTGC-3′ | 5′-ACCACTTCGCAGAGCCGCAC-3′ | |
| PGC-1α | 5′-ACACACCGCAATTCTCCCTT-3′ | 5′-TTTGGCCCTTTCAGACTCCC-3′ | |
| PPARγ | 5′-ATTGAGTGCCGAGTCTGTGG-3′ | 5′-GCAAGGCACTTCTGAAACCG-3′ | |
| Sox10 | 5′-CATGTCAGATGGGAACCCAGAGCACC-3′ | 5′-TTGCCGAAGTCGATGTGGGGCTTC-3′ | |
| YY1 | 5′-AAGACCCTGGAGGGCGAGTT-3 | 5′-TCCAGGAGGGAGTTTCTTGCCT-3′ | |
| Olig2 | 5′-CGCAGCGAGCACCTCAAATC-3′ | 5′-TCATCGGGTTCTGGGGACGA-3′ | |
| Srebp2 | 5′-CCC ACT CAG AAC ACC AAG CA-3′ | 5′-AGTAGCTCGCTCTCGTTGGC-3′ | |
| Srebp1c | 5′-GCCATGGATTGCACATTTGA-3′ | 5′-TGGTTGTTGATGAGCTGGAGC-3′ | |
| Srebp1a | 5′-GAACAGACACTGGCCGAGATG T-3′ | 5′-GGTTGTTGATGAGCTGGAGCA T-3′ | |
| FASN | 5′-CAGCAGAGTCTACAGCTACCT-3′ | 5′-ACCACCAGAGACCGTTATGC-3′ | |
| Rxr α | 5′-GTTGGAGAGTTGAGGGACGA-3′ | 5′-GGGCATGAGTTAGTCGCAGA-3′ | |
| Rxr β | 5′-GTCCACAGGCATCTCCTCAG-3′ | 5′-ACTGGCATGAAAAGGGAGG-3′ | |
| Rxr γ | 5′-CAAGGCTACTGAAGGGCTCA-3′ | 5′-GCAGCCAACATGTATGGAAA-3′ | |
| Rar α | 5′-GGGAGGGCTGGGTACTATCT-3′ | 5′-AGCACCAGCTTCCAGTCAGT-3′ | |
| Rar β | 5′-CTCTGTGCATTCCTGCTTTG-3′ | 5′-AAGTGCTTTGAAGTGGGCAT-3′ | |
| Rar γ | 5′-CGAGCTGGTGCTCTGTGTC-3 | 5′-ACCATTTGAGATGCTGAGCC-3′ | |
| Lxr α | 5′-GCCCTGCACGCCTACGT-3′ | 5′-TAGCATCCGTGGGAACATCA-3′ | |
| Lxr β | 5′-GCTGATGATCCAGCAGTTAG-3′ | 5′-CGGAGAAAGATCGTTTGTTG-3′ | |
| Insig2a | 5′-CCCTCAATGAATGTACTGAAGGATT-3′ | 5′-TGTGAAGTGAAGCAGACCAATGT-3′ | |
| Insig2 | 5′-TGGGTACCACCATGCTGCGG-3′ | 5′-TACCCACGAACACCGCCACG-3′ | |
| Insig1 | 5′-TCGTTGGCATCAACCACGCCA-3′ | 5′-GGCCGCTTCGGGAACGATCA-3′ |
Electron microscopy.
Tissue processing was performed as described previously (Pereira et al., 2010).
Morphometric analysis.
Transverse sections of spinal cords were analyzed at the junction level of the thoracic and lumbar vertebrae (T13–L1). The ratio of the axon diameter/fiber diameter (g-ratio) was measured in pixels on EM photographs (ultrathin sections, 65 nm) using Adobe Photoshop CS5 software and converted to micrometers. Axonal diameter was measured in toluidine-blue-stained semithin sections (0.65 μm) using a light microscope (Axioplan 2; Carl Zeiss). To achieve this, all axons were measured in a determined area (45 × 45 μm) of the spinal cord ventricular funiculus. Sections were taken at the junction of the thoracic and lumbar vertebrae.
Immunohistochemistry.
Mice were killed by intraperitoneal injection of pentobarbital (150 mg/kg, Nembutal; Abbott Laboratories) and perfused with 4% paraformaldehyde (PFA). Spinal cords were dissected, postfixed for 2 h in 4% PFA, washed in PBS, dehydrated in 30% sucrose overnight, embedded in optimal cutting temperature medium, and stored at −80°C. Cryosections (10 μm thick, T13–L1) were permeabilized and blocked for 1 h at room temperature (RT) in blocking buffer (0.3% Triton X-100 and 5% bovine serum albumin in PBS) and incubated with primary antibodies overnight at 4°C in blocking buffer. For enhanced Tcf-4 and Ki67 immunostaining, slides were preincubated in 10 mm citrate buffer, pH 6.5, for 5 min at 95°C and then washed with PBS before blocking. All primary antibodies are listed in Table 2. After washing with PBS, sections were incubated with secondary antibodies coupled to Alexa Fluor 488 (1:200; Invitrogen) or Cy3 (1:250; Jackson ImmunoResearch Laboratories) for 1 h at RT, washed in PBS, incubated for 5 min in DAPI, and mounted with IMMU-MOUNT (Thermo Scientific). Staining was observed using a fluorescence microscope (Axioplan2 Imaging; Carl Zeiss). Images were digitized with with a Powershot G5 camera (Canon) and acquired with Axio Vision 4.5 software (Carl Zeiss). Images were processed (levels adjusted) using Photoshop CS5 (Adobe). To assess double-positive phospho-S6 (P-S6)/CC1 or FASN/Olig2 cells, three random areas were chosen from each of three different transverse spinal cord sections derived from the junction of the thoracic and lumbar vertebrae (three different animals per condition) and the number of strongly P-S6- or FASN-positive (above a given threshold) cells of total CC1- or Olig2-positive cells was established. For confocal analyses, images were acquired using a Leica SP2-AOBS confocal laser-scanning microscope equipped with an argon laser using the HCX PL APO CS 63× oil-immersion objective. After optimizing image parameters, a series of 2D images was collected to form 3D spatial data. Processed images were obtained with Imaris software.
Table 2.
List of antibodies
| Target | Host | Dilution | Source |
|---|---|---|---|
| Antibodies for IHC | |||
| APC (CC1) | Mouse | 1:200 | Abcam |
| FASN | Rabbit | 1:200 | |
| Ki67 | Rat | 1:25 | DakoCytomation |
| P-S6 | Rabbit | 1:200 | Cell Signaling Technology |
| Lipin-1 | Rabbit | 1:50 | |
| MBP | Rat | 1:200 | AbD Serotec |
| NF-160 | Mouse | 1:200 | Sigma |
| Olig2 | Rabbit | 1:500 | Millipore |
| PDGFRα | Rat | 1:100 | BD Pharmingen |
| Tcf-4 | Mouse | 1:100 | Millipore |
| Antibodies for WB | |||
| P-Akt S473 | Rabbit | 1:1000 | Cell Signaling Technology |
| P-Akt T308 | Rabbit | 1:1000 | |
| Akt | Rabbit | 1:1000 | |
| P-4-EBP1 T37/46 | Rabbit | 1:1000 | |
| P-4-EBP1 S65 | Rabbit | 1:1000 | |
| 4EBP1 | Rabbit | 1:1000 | |
| P-Erk1/2 T202/Y204 | Rabbit | 1:1000 | |
| Erk1/2 | Mouse | 1:1000 | |
| Lipin-1 | Rabbit | 1:1000 | |
| mTOR | Rabbit | 1:1000 | |
| P-S6 S235/236 | Rabbit | 1:1000 | |
| S6 | Rabbit | 1:1000 | |
| P-p70 S6K T389 | Rabbit | 1:1000 | |
| p70 S6K | Rabbit | 1:1000 | |
| TSC1 | Rabbit | 1:1000 | |
| TSC2 | Rabbit | 1:1000 | |
| Raptor | Rabbit | 1:200 | |
| Rictor | Rabbit | 1:200 | |
| MBP | Rat | 1:1000 | AbD Serotec |
| α-tubulin | Mouse | 1:10000 | Sigma |
| PLP | Rabbit | 1:1000 | Abcam |
| FASN | Rabbit | 1:1000 | |
| Scap isoform 4 | Rabbit | 1:500 | |
| SREBP2 | Rabbit | 1:200 | |
| MOG | Mouse | 1:500 | Santa Cruz Biotechnology |
| SREBP1 K-10 | Rabbit | 1:50 | |
| Scap isoform 1 | Goat | 1:200 | |
Antibodies to MAG were a generous gift from Dr Steven Scherer (University of Philadelphia, Philadelphia, PA); antibodies to HMGCR and IDI1 were a generous gift from Dr. Werner Kovacs (ETH Zürich, Switzerland).
Western blot.
Analyses were performed with spinal cord lysates of postnatal day 10 (P10) mutant mice and their littermate controls unless otherwise indicated. Mice were killed and spinal cords were isolated. Whole spinal cord protein extracts were used unless otherwise indicated. Spinal cords were normalized by weight and homogenized with a chilled mortar and pestle in RIPA lysis buffer containing the following: 50 mm Tris-HCl, 1% NP-40, 0.25% Na-deoxycholate, 150 mm NaCl, and protease inhibitor (Sigma-Aldrich); phosphatase inhibitor (Roche); urea lysis buffer containing 8 m urea, 1% SDS, 5 mm DTT, 1 mm Tris-HCl, 10% glycerol, and protease inhibitor (Sigma-Aldrich); and phosphatase inhibitor (Roche). Extracts were processed using standard SDS-PAGE and Western blotting procedures using precast Mini-PROTEAN TGX gels (Bio-Rad). Equal volumes were loaded on the gels. Primary antibodies are listed in Table 2. Secondary antibodies were obtained from Southern Biotech and JacksonImmunoresearch Laboratories. Quantity One software (Bio-Rad) was used for quantification.
qRT-PCR.
Analyses have been conducted on RNA extracts from spinal cords of P10 mutant mice and their littermate controls. Total RNA was extracted using the Quiazol (QIAGEN) protocol. cDNA was produced using Superscript III Reverse Transcriptase (Invitrogen). qRT-PCR analysis was performed on a Light Cycler 480 II (Roche) using LightCycler SYBR Green I Master Mix. Primer sequences are listed in Table 1.
Lipid analysis.
Chemicals were purchased either from Merck KGaA or Sigma-Aldrich. Lipid standards were obtained from Avanti Polar Lipids. Spinal cords from P60 animals were isolated. Lipid extraction was performed as described previously (Matyash et al., 2008). Internal standardization and data acquisition by HPLC coupled to an FT-ICR-MS hybrid mass spectrometer (LTQ-FT; Thermo Scientific) has been described previously (Fauland et al., 2011). Data processing was performed using Lipid Data Analyzer software as described previously (Hartler et al., 2011), a process that relies on exact mass and retention time. Annotation of lipid species was according to the shorthand nomenclature of the International Lipid Classification and Nomenclature Committee (Liebisch et al., 2013).
Processing of lipid extracts.
Lipid extracts were evaporated and resuspended in 1 ml of chloroform/methanol (1/1; v/v). Each lipid extract was then split for analysis of total fatty acids (FAs; 350 μl), free FAs (200 μl), positive ESI LC-MS/MS (18 μl), and negative ESI LC-MS/MS (18 μl). Lipid extracts for LC-MS/MS analysis were evaporated, spiked with a set of internal standards (Table 3), and resuspended in 90 μl of chloroform/methanol (1/1; v/v).
Table 3.
List of internal standards used in LC-MS
| Product no. | Shorthand nomenclature | Amount/sample (pmol) |
|---|---|---|
| LM-1100 | PE 12:0/13:0 | 160 |
| LM-1102 | PE 17:0/20:4 | 160 |
| LM-1103 | PE 21:0/22:6 | 160 |
| LM-1104 | PE 17:0/14:1 | 160 |
| LM-1302 | PS 17:0/20:4 | 240 |
| LM-1300 | PS 12:0/13:0 | 240 |
| LM-1304 | PS 17:0/14:1 | 240 |
| LM-1000 | PC 12:0/13:0 | 200 |
| LM-1002 | PC 17:0/20:4 | 200 |
| LM-1003 | PC 21:0/22:6 | 200 |
| LM-1004 | PC 17:0/14:1 | 200 |
| LM-1601 | LPC 17:1 | 80 |
| LM-4100 | Cholesterol (D7) | 6400 |
| LM-6002 | Sphingolipid mix I | 120 |
| LM-1500 | PI 12:0/13:0 | 320 |
| LM-1502 | PI 17:0/20:4 | 320 |
| LM-1504 | PI 17:0/14:1 | 320 |
Chromatography with electron impact mass spectrometry of total FAs (free + esterified).
Aliquots of lipid extracts were dried and suspended in 1 ml of methanolic NaOH. After 10 min of incubation at 80°C, samples were cooled for 5 min on ice, 1 ml of BF3 was added, and the mixture was incubated for 10 min at 80°C. FA methyl esters were extracted with 1 ml of saturated NaCl and 2 ml of hexane. The hexane phase was dried and methyl esters dissolved in 1.5 ml of hexane.
A Trace-DSQ GC-MS (Thermo Scientific) equipped with a TR-FAME 30 m column was used for analysis. Helium was used as carrier gas at a flow of 1.3 ml/min, in split mode, at 250°C injector temperature. The initial oven temperature of 150°C was held for 0.5 min and then the temperature was increased to 180°C at a rate of 10°C/min. This was followed by a further increase to 190°C at a rate of 0.5°C/min and then increased to 250°C at a rate of 40°C/min and kept for 3 min. The mass spectrometer was run in electron impact mode and FAs were detected in full scan of m/z 80–400. Source temperature was set to 250°C and the transfer line temperature to 200°C.
Rapamycin treatment.
Rapamycin was obtained from Calbiochem. Tween 80% and polyethylene glycol PEG-400 were obtained from Sigma-Aldrich. Rapamycin was dissolved in a vehicle solution of 4% ethanol, 5% PEG-400, and 5% Tween 80. Mice were administered daily intraperitoneal injections of rapamycin (5 mg/kg body weight) from P3 to P10. At P10, mice were killed, perfused, and analyzed by electron microscopy.
Statistical analysis.
Prism 5 software (GraphPad) was used for statistical analyses. Data are shown as the mean ± SEM. Statistical significance was determined using an ANOVA test for groups of more than two and a two-tailed Student's t test for groups of two. Significance was set at *p < 0.05, **p < 0.01, and ***p < 0.001. n refers to the number of independent animals analyzed unless otherwise indicated.
Results
Conditional deletion of Rptor in the oligodendrocyte lineage affects the timing of myelination and leads to persistent hypomyelination
To address the functions of raptor, rictor, and raptor/rictor in oligodendrocytes during CNS myelination, we generated mice lacking these proteins in the oligodendrocyte lineage using Cre-loxP technology (Fig. 1A). As expected, Western blot analysis of microdissected spinal cord white matter fragments revealed strong reduction of the target protein(s) in the corresponding mutants compared with controls (Fig. 1B). To obtain a first overview of the consequences of raptor/rictor loss, we used immunohistochemistry on spinal cord sections of raptor/rictor mutants. Staining for myelin basic protein (MBP) at P10 exposed an overall much reduced myelinated area and more sparse myelin labeling in mutants compared with littermate controls (Fig. 1C). Subsequently, we started a systematic investigation by carrying out Western blot analysis of P10 spinal cord homogenates of the different mutants compared with controls. MBP, myelin oligodendrocyte glycoprotein (MOG), myelin-associated glycoprotein (MAG), and proteolipid protein (PLP) levels were all reduced in P10 raptor and raptor/rictor mutants, whereas no significant changes (but a consistent trend to reductions) were detected in rictor mutants (Fig. 1D). To corroborate these data at the mRNA level, we performed qRT-PCR analysis for the same targets and found strong reductions of MOG, MAG, and PLP in raptor and raptor/rictor mutants compared with controls (Fig. 1E). Small but significant reductions were also detectable for the mRNAs of MOG and PLP in rictor mutants. In contrast, MBP mRNA levels were unchanged in all mutants, consistent with the interpretation that altered posttranscriptional regulation of MBP causes the reductions observed at the protein level in raptor and raptor/rictor mutants.
Figure 1.
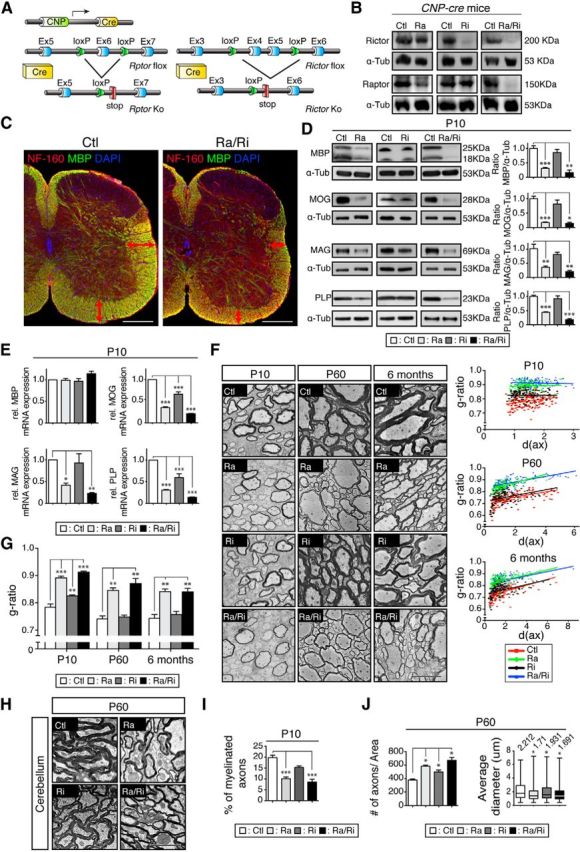
Myelination deficiencies due to mTORC1 or mTORC2 inactivation. A, Regulatory elements of the CNP promoter drive the expression of Cre in oligodendrocytes. Schematics map of the Rptor and Rictor allele depicts the location of the loxP sequences. B, Microdissected fragments of the spinal cord white matter were analyzed by Western blot to confirm the loss of raptor and/or rictor in the conditional knock-out tissue. C, Transverse sections of the spinal cord from P10 raptor/rictor mutants and littermate controls were immunolabeled for MBP (green), neurofilament 160 (red), and DAPI (blue). White matter area is reduced in raptor/rictor mutant with lower density of MBP staining (red double-sided arrows). Scale bar, 200 μm D, Western blot analysis of spinal cord lysates shows a striking reduction of the myelin proteins MBP, MOG, MAG, and PLP in raptor and raptor/rictor mutants when normalized to α-tubulin. E, Levels of transcripts encoding myelin proteins analyzed by qRT-PCR reveal significant reductions of MOG and PLP in all mutants, reduced MAG expression in raptor and raptor/rictor mutants, and no difference for MBP in any mutants. F, EM micrographs of the ventral funiculus of spinal cords showing thinner myelin in the raptor/rictor and raptor mutants at all ages examined (P10, P60, 6 months). Shown are scatter plots of the g-ratios of individual fibers in relation to respective axon diameters (in micrometers) quantified at P10, P60, and 6 months from raptor/rictor (blue triangles), raptor (green diamonds), rictor (black circles), and littermate controls (red squares). Scale bars, 5 μm. G, Average g-ratios at P10, P60, and 6 months. Rictor mutants displayed a minor deficit in myelin thickness (higher g-ratio) at P10, which recovered at P60 and 6 months. H, EM micrographs of the cerebellum show thinner myelin in raptor/rictor and raptor mutants at P60. Scale bar, 2.5 μm. I, Percentage of myelinated fibers in the ventral spinal cord funiculus was reduced in raptor and raptor/rictor mutants. Error bars represent SEM. n = 3; *p < 0.05; **p < 0.01; ***p < 0.001. J, Quantification of axon number and axonal diameter per defined area (45 × 45 μm) of the spinal cord ventral funiculus on cross-sections stained with toluidine blue. Axons with diameter <0.4 μm were excluded.
Next, we analyzed morphological changes in our mutants. Consistent with the lower amount of myelin proteins present, ultrastructural analysis of P10 spinal cord ventral funiculi revealed strongly reduced myelin thickness in raptor and raptor/rictor mutants compared with controls (Fig. 1F). These alterations persisted into adulthood (P60, 6 months old). Upon qualitative inspection, changes in myelin thickness were less obvious in rictor mutants. Morphometric quantification by g-ratio analysis (axon diameter/axon plus myelin diameter), however, revealed a slightly but significantly increased g-ratio (indicating thinner myelin) for P10 rictor mutants compared with controls (Fig. 1F,G). This rictor-dependent hypomyelination was transient and fully recovered at later time points analyzed (P60, 6 months old). In raptor and raptor/rictor mutants, the g-ratios remained abnormally high throughout development and in adulthood (Fig. 1F,G). CNS hypomyelination was not restricted to the spinal cord, because cerebella of P60 raptor and raptor/rictor mutants were also similarly affected (Fig. 1H). In addition, we quantified the fraction of myelinated axons and found a lower percentage in P10 raptor and raptor/rictor mutants compared with controls (Fig. 1I), indicating that raptor/mTORC1 regulates myelination initiation. Furthermore, quantification of axonal diameter showed a lower average in P60 mutants versus controls, including the rictor mutants, albeit to a lesser extent (Fig. 1H,J). These findings draw a parallel to previous studies in the PNS showing that lack of mTOR protein in Schwann cells affects axonal diameter (Sherman et al., 2012).
In summary, altered initiation of myelination and hypomyelination were associated with reduced myelin protein expression in our mutants. The effects were strong in raptor mutants, indicating that raptor/mTORC1 is the main crucial determinant of mTOR signaling in oligodendrocytes. In rictor mutants, the effects were small, often showing a tendency not reaching statistical significance. In raptor/rictor mutants, there was a general trend toward additive effects. Therefore, it appears likely that also rictor/mTORC2 in oligodendrocytes is critically involved in CNS myelination, although to a much lesser extent compared with raptor/mTORC1.
Combined loss of raptor and rictor is required to impair OPC differentiation and maturation
The mTORC1 inhibitor rapamycin can block progression of OPC differentiation at the late progenitor stage in vitro (Tyler et al., 2009). Therefore, our findings of defects in the timing of CNS myelination and the observed hypomyelination in early development in the mutants might be at least partially explained by oligodendrocyte differentiation deficiencies. Therefore, we addressed the functional role of mTORC1 and mTORC2 in OPC survival, proliferation, and differentiation in vivo by analyzing spinal cord sections of mutants and controls with established oligodendrocyte lineage markers. Proliferation of cells of the oligodendrocyte lineage (Ki67+/Olig2+ cells) at P0 did not differ among mutants and controls (Fig. 2A,B). Quantification of Olig2+ cells through postnatal development (P0, P10, P60; Fig. 2C) showed that oligodendrocyte-specific ablation of raptor, rictor, or both had no significant effects on OPC/oligodendrocyte cell numbers, which is consistent with the results of in vitro studies (Tyler et al., 2009). Next, we quantified OPCs specifically by counting Olig2+/PDGFRα+ cells. Consistently, from P0 to P60, only raptor/rictor double mutants contained significantly higher numbers of OPCs compared with controls and single mutants (Fig. 2D). Using Tcf-4 as marker of late progenitors to premyelinating oligodendrocytes (Fancy et al., 2011), we found a significant reduction of Olig2+/Tcf-4+ cells in P10 raptor/rictor mutants only, with tendencies toward decreases at P0 and P60 as well (Fig. 2E). Quantification of CC1+/Olig2+ mature oligodendrocytes revealed lower numbers in raptor/rictor mutants from birth to adulthood (Fig. 2F). Together, these results suggest synergistic contributions of mTORC1 and mTORC2 to oligodendrocyte maturation, because only the raptor/rictor double-mutant mice were affected by significantly impaired oligodendrocyte differentiation. To follow up on the underlying molecular mechanism, we used qRT-PCR to analyze the expression of transcription factors crucially involved in oligodendrocyte maturation. Analysis of P10 spinal cords revealed strongly decreased levels of SOX10, MRF, and Olig2, not only in raptor/rictor mutants, but also in raptor mutants (Fig. 2G). YY1 levels remained unchanged. We interpret these data as meaning that the examined transcription factors not only regulate oligodendrocyte differentiation (significantly changed in raptor/rictor mutants only), but also myelination per se, which is strongly altered in both raptor/rictor and raptor mutants at P10.
Figure 2.
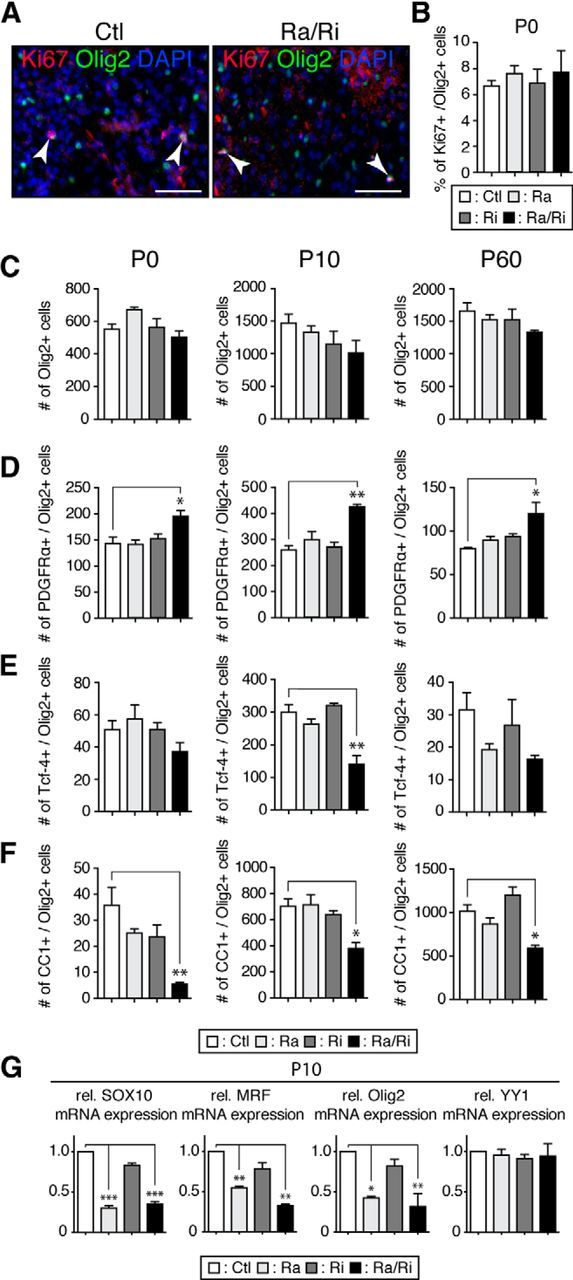
Ablation of raptor/rictor affects OPC maturation. A, Immunostaining for the cell proliferation marker Ki67 combined with Olig2 staining on spinal cord transverse sections of P0 raptor/rictor mutants showed no difference compared with controls. Arrowheads indicate double-labeled cells. Scale bars, 50 μm B, Percentage of Ki67-positive cells/Olig-2 positive cells did not significantly differ among mutants and controls. C–F, Transverse spinal cord sections at the junction of thoracic and lumbar vertebrae (T13–L1) were used to count total numbers of cells of the oligodendrocyte lineage (Olig2 staining; C), OPCs (PDGFRα/Olig2 staining; D), premyelinating oligodendrocytes (Tcf-4/Olig2 staining; E), and mature oligodendrocytes (CC1/Olig2 staining; F) at P0, P10, and P60. All mutants showed the same number of oligodendrocyte lineage cells. Only double mutants (raptor/rictor) exhibited negative effects on OPC maturation (higher number of PDGFR+ cells, lower number of CC1+ cells at all time points). Three sections each from three mice of each genotype were analyzed. G, qRT-PCR for the transcription factors SOX 10, MRF, and Olig2 showed a striking reduction in spinal cords of raptor/rictor and raptor mutant. YY1 mRNA did not change in any mutants. Error bars represent SEM. n = 3; *p < 0.05; **p < 0.01; ***p < 0.001.
mTORC1 function in oligodendrocytes is required for the maintenance of myelinated fibers
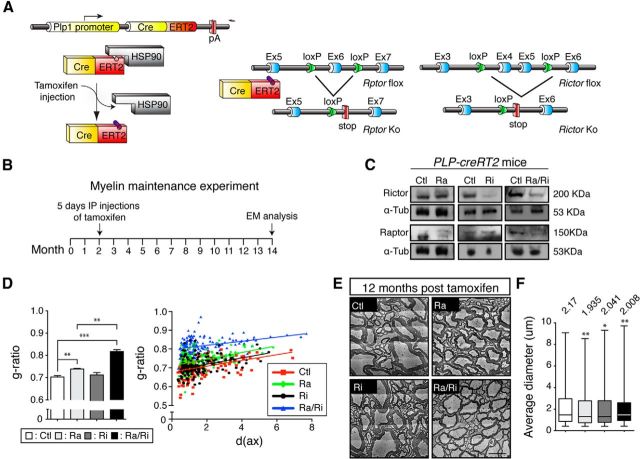
A dynamic turnover of myelin membranes throughout the lifespan has been suggested (Ando et al., 2003) and was supported by recent evidence of CNS myelin remodeling in the adult (Young et al., 2013). Furthermore, the major CNS myelin regulator MRF (the expression of which is reduced in raptor and raptor/rictor mutants; Fig. 2G), is required to maintain adult myelinating oligodendrocytes, providing further evidence for ongoing crucial dynamic renewal processes (Koenning et al., 2012). Therefore, we assessed the overall requirements for mTOR signaling in adult oligodendrocytes. To achieve this goal, we used tamoxifen-inducible PLP1-CreRT2 mice (Fig. 3A,B) combined with conditional Rptor, Rictor, and Rptor/Rictor double-null alleles to disrupt mTOR signaling and components thereof in oligodendrocytes after established myelination. After induction of recombination in 2-month-old mice, we confirmed loss of the corresponding proteins (Fig. 3C) and analyzed the animals 12 months later by spinal cord ventral funiculus morphometry (Fig. 3D,E). Although the g-ratio in raptor mutants was slightly but significantly higher than in controls and rictor mutants, raptor/rictor double mutants showed a more pronounced hypomyelination. In addition, analysis of the average axonal diameters revealed significant reductions in all induced mutants (Fig. 3F), comparable to our findings in development. We conclude that mTOR signaling in oligodendrocytes is required to mantain the full integrity of myelinated fibers. Axons depend on healthy oligodendrocytes in various ways, including metabolic support (Nave, 2010; Morrison et al., 2013). Which specific aspects are disturbed by defective mTOR signaling in oligodendrocytes remains to be determined.
Figure 3.
Role of mTORC1 and mTORC2 in myelin maintenance. A, PLP1 gene regulatory elements drive expression of a tamoxifen-activatable Cre fusion protein in oligodendrocytes. Schematic map of raptor and rictor alleles depicts the location of loxP sequences. Upon Cre-mediated recombination after tamoxifen injection, the genomic region located between loxP sites is excised, thereby inactivating the conditional raptor or rictor alleles. B, Outline of the experimental procedure: raptor, raptor/rictor, and rictor mutants carrying the PLP1-CreRT2 allele and their littermate controls were injected with tamoxifen for 5 consecutive days at 2 months of age and killed for analysis 12 months later. C, Microdissected fragments of the spinal cord white matter were analyzed by Western blot to confirm loss of targeted raptor and/or rictor in the conditional knock-out mice. D, g-ratio values for raptor mutants showed a small but significant increase compared with control. The raptor/rictor mutant display a strikingly higher average g-ratio compared with both controls and single raptor mutants. Scatter plot graphics of single fiber g-ratio measurements versus axonal diameter (in micrometers) detailing the myelin thickness among mutants and controls. Shown are raptor/rictor (blue triangles), raptor (green diamonds), rictor (black circles), and their littermate controls (red squares). E, Representative electron micrographs of the spinal cord ventral funiculus showing thinner myelin of raptor/rictor mutants and the minor effect in raptor mutants. Scale bars, 2.5 μm. Error bars indicate SEM. n = 3; **p < 0.01; ***p < 0.001. F, Quantification of axonal diameters per defined area of the spinal cord ventral funiculus on cross-sections stained with toluidine blue. Axons with diameter <0.4 μm were excluded. The average axonal diameter was lower in all mutants. Error bars indicate SEM. n = 3; *p < 0.05; **p < 0.01; ***p < 0.001.
mTOR signaling in raptor, rictor, and raptor/rictor mutants
To unravel the molecular mechanisms that underlie the observed phenotypes along mTOR signaling, we analyzed spinal cord homogenates of P10 mice using Western blot analysis. Loss of raptor, rictor, or raptor/rictor did not affect total mTOR protein levels in the corresponding animals (Fig. 4A). Next, we examined phosphorylation of mTOR downstream targets as reliable measures of mTOR activity regarding stimulation of protein translation (Roux and Topisirovic, 2012). Phosphorylation levels of the main targets of mTORC1, including the ribosomal protein S6 kinase (S6K), its substrate ribosomal protein S6, and the initiation factor 4E-binding protein 1 (4E-BP1), were drastically reduced in raptor and raptor/rictor mutants, but not in rictor mutants (Fig. 4A). To corroborate our data and to show cell-type specificity, we used P-S6 immunohistochemistry on P10 spinal cord sections. CC1+ oligodendrocytes showed strongly reduced P-S6 levels in raptor and raptor/rictor mutants compared with rictor mutants and controls (Fig. 4B). These findings were confirmed by quantifications (strongly P-S6+/CC1+ cells: 63.4 ± 2.1% for controls compared with 4.8 ± 2.2% for raptor mutants, p < 0.001; 54.5 ± 3.1% for rictor mutants, not significant; and 4.9 ± 1.0% for raptor/rictor mutants, p < 0.001). Our results indicate that raptor ablation is sufficient to downregulate the mTORC1 pathway in developing oligodendrocytes, whereas ablation of rictor/mTORC2 does not affect mTORC1 signaling detectably in this setting. Next, we hypothesized that the observed reduced S6K activation in raptor mutants could be responsible for the observed hypomyelination on its own via impaired protein synthesis. However, morphological g-ratio analysis of 2-month-old S6K1/2 double-null mice (Shima et al., 1998; Pende et al., 2004) did not reveal detectable differences compared with littermate controls (Fig. 4C), indicating that S6K alone is not responsible for this phenotype. Following a related rational, we performed a similar analysis using 2-month-old 4E-BP1/2 double-null mice (Le Bacquer et al., 2007). 4E-BP1 binds to the translation initiation factor eIF4E, preventing its assembly into the EIF4F initiation complex and so inhibiting cap-dependent translation (Morita et al., 2013). Once phosphorylated by mTORC1, 4E-BP1 dissociates from eIF4E and translation is activated. In the raptor and raptor/rictor mutants, 4E-BP1 phosphorylation levels were strongly reduced, presumably associated with diminished translation. In contrast, translation might be aberrantly regulated, possibly enhanced, in 4E-BP1/2 double-null mice, potentially causing CNS myelination changes. However, we could not detect myelin alterations in such mice (Fig. 4D).
Figure 4.
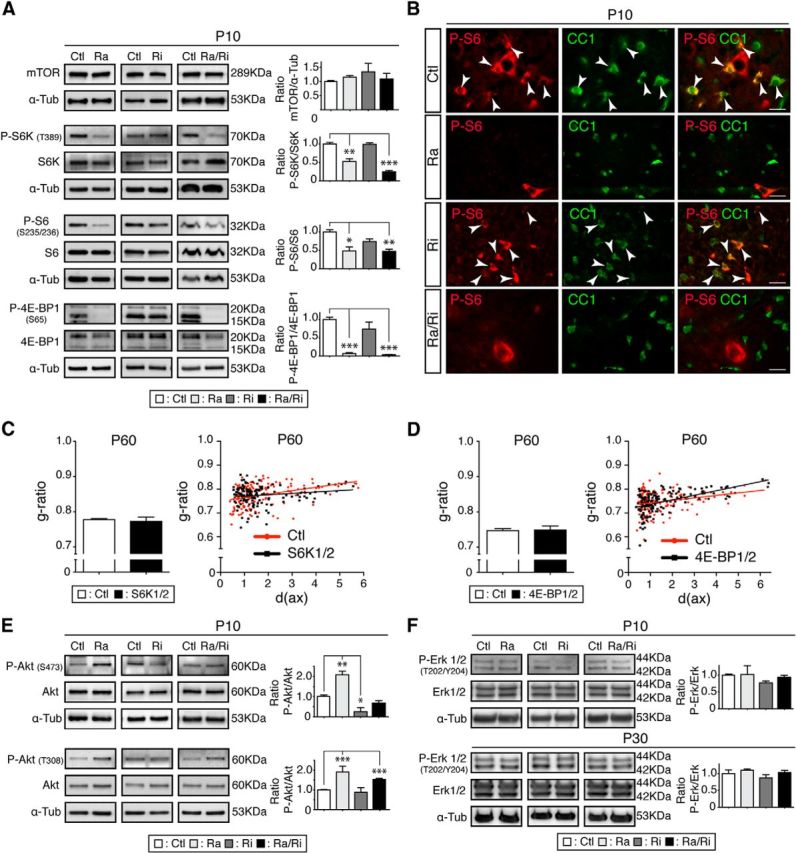
mTOR signaling pathway in raptor, rictor, and raptor/rictor mutants. A, Spinal cord lysates of raptor, rictor, and raptor/rictor mutants and littermate controls were analyzed by immunoblotting to detect mTOR protein and phosphorylation levels of the main mTORC1 targets: P-S6K (T389), P-S6 (S235/236), and P-4E-BP1 (S65). B, Immunostaining on spinal cord transverse sections revealed loss of P-S6 (red) in oligodendrocytes (marked by CC1; green) in raptor and raptor/rictor mutants, but not in rictor mutants and controls. Arrowheads indicate double-labeled cells. Scale bars, 20 μm. C, g-ratio values did not differ in S6K1/2 double-mutant animals compared with their littermate controls. Scatter plot graphics showing individual measurements of g-ratios versus axonal diameter (in micrometers) for S6K1/2 double mutants (black squares) and their littermate controls (red circles). D, Analogous analysis and outcome for 4E-BP1/2 double mutants as in C. Black squares, 4E-BP1/2; red circles, controls. Error bars indicate SEM. n = 3; *p < 0.05. E, Western blot analysis of the phosphorylation state of Akt at T308 or S473. F, Western blot analysis revealed no change of phosphorylation levels of Erk1/2 at P10 or P30, indicating no shift toward the Erk1/2-mitogen-activated protein kinase (MAPK) pathway. Error bars indicate SEM. n = 3; *p < 0.05; **p < 0.01; ***p < 0.001.
The mTOR pathway is intimately linked with Akt signaling, both upstream of mTORC1 and downstream of mTORC2 (Laplante and Sabatini, 2012). We found that total protein levels of Akt were not changed in raptor, rictor, and raptor/rictor mutants compared with controls (Fig. 4E). As expected, however, Akt phosphorylation at S473 was strongly impaired in rictor mutants because this residue is phophorylated directly by mTORC2 (Fig. 4E), further confirming inactivation of the mTORC2 complex by rictor deletion in the mutant. Raptor mutants showed a twofold increase of P-Akt at S473, consistent with previous reports demonstrating that specific inhibition of mTORC1 increases Akt phosphorylation at S473 in a rictor-dependent manner (Breuleux et al., 2009). Upstream of mTOR, Akt can be phosphorylated at T308 through the action of insulin and various growth factors via the PI3K pathway, often referred to as the PI3K/Akt/mTOR pathway. Both raptor and raptor/rictor mutants showed an increase in T308 phosphorylation. Such hyperphosphorylation has been attributed to the failure of raptor-deficient cells to activate inhibitory feedback loops (Bentzinger et al., 2008). A significant crosstalk exists between PI3K/Akt/mTOR and Ras/MAPK pathways, which can influence each other both negatively and positively (Aksamitiene et al., 2012). Because the Erk1/2 MAPK pathway is required to achieve proper myelin thickness (Ishii et al., 2012), we measured both total protein levels and phosphorylation of Erk1/2 in raptor, rictor, and raptor/rictor mutants compared with controls. No alterations could be detected at either P10 or P30 (Fig. 4F).
mTORC1 regulates proper lipid biosynthesis in myelinated fibers
Myelin is dependent on both its precise protein and characteristic lipid contents. Lipids provide the basic building blocks of this multilayered cell membrane structure and proteins contribute the critical connections and regulators. Therefore, assessing the role of mTOR in myelinating oligodendrocytes must include the regulation of lipogenesis, a major function attributed to mTOR signaling (Laplante and Sabatini, 2010; Lewis et al., 2011; Soliman, 2011; Lamming and Sabatini, 2013). Endogenous lipid synthesis is mainly controlled by the SREBP family of transcription factors (Horton et al., 2002), which regulate the expression of genes encoding sets of enzymes required for lipid metabolism. SREBP-1c mainly controls genes involved in FA synthesis. SREBP-2 mainly regulates genes involved in cholesterol synthesis. SREBP-1a targets both sets of genes. We started our analysis by examining expression of SREBPs and their main targets in raptor, rictor, and raptor/rictor mutants (Fig. 5A,B). No changes for SREBP1 proteins and the two transcripts encoding SREBP1a and SREBP1c could be detected between mutants and controls. However, Western blot and qRT-PCR analyses aimed at determining FA synthase (FASN) and stearoyl-CoA desaturase-1 (SCD1) levels, two main targets of SREBP1, revealed significant reductions in raptor and raptor/rictor mutants. In the same mutants, SREBP2 expression was also diminished at the protein and mRNA levels, accompanied by reductions of the SREBP2 targets HMG-CoA reductase (HMGCR) and isopentenyl-diphosphate delta isomerase 1 (IDI1). In rictor mutants, we detected a small but significant diminution of FASN and HMGCR mRNA levels. Note that we used whole spinal cord extracts for these analyses; therefore, contributions from cell types other than oligodendrocytes may have partially masked the degree of expression attenuation at the cellular level in these assays. To address this issue with respect to FASN reduction, we performed immunohistochemistry on P10 spinal cord sections. In raptor and raptor/rictor mutants, Olig2+ oligodendrocytes showed strongly reduced FASN staining compared with controls, rictor mutants, and non-Olig2+ cells present in the sections (Fig. 5C), corroborating and extending the Western blot data. These findings were further confirmed by quantifications (strongly FASN+/Olig2+ cells: 85.3 ± 3.0% for controls compared with 11.6 ± 0.5% for raptor mutants, p < 0.001; 87.9 ± 0.8% for rictor mutants, not significant; and 7.5 ± 2.0% for raptor/rictor mutants (p < 0.001)).
Figure 5.
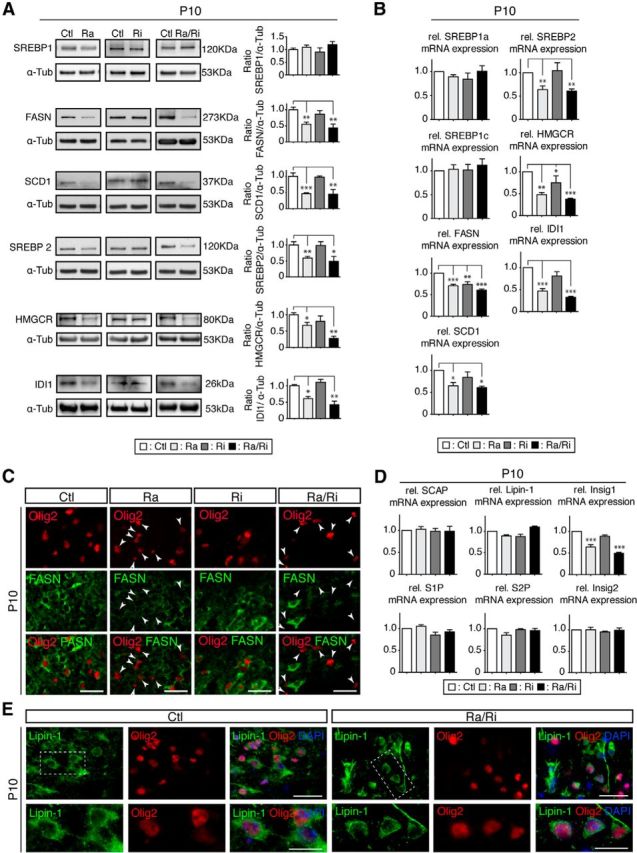
mTORC1 regulates lipogenesis via SREBP transcription factors. A, Western blot analysis of protein levels of SREBPs and their main targets in raptor, rictor, and raptor/rictor mutants. SREBP1 protein levels showed no change, whereas while its targets, FASN and SCD1, were reduced in raptor and raptor/rictor mutants. The same mutants showed a reduction at the protein level for SREBP2 and its targets HMGCR and IDI1. B, qRT-PCR of SREBPS and their main targets. C, Transverse sections of spinal cords revealed reduced staining for FASN (green) in oligodendrocyte lineage cells (Olig2+, red) in raptor and raptor/rictor mutants. Arrowheads indicate cells positive for Olig2 and negative for FASN. Scale bars, 25 μm. Error bars indicate SEM. n = 3; *p < 0.05; **p < 0.01; ***p < 0.001. D, Quantification of mRNA levels of key elements responsible for the maturation of the SREBPs did not display significant changes compared with controls, except for Insig1, a target of SREBPs, in raptor and raptor/rictor mutants. E, Transverse sections of spinal cords double-labeled for Lipin-1 (green) and the oligodendrocyte lineage marker Olig2 (red) shows no detectable nuclear localization of Lipin-1 in raptor/rictor mutants and no difference in the subcellular localization of Lipin-1 compared with controls. Scale bars, 50 μm.
Next, we approached how SREBP1 levels can be unchanged in raptor and raptor/rictor mutants, whereas the main targets of SREBP1 are reduced. Therefore, we examined major players involved in the maturation and activation of SREBPs in mutants and controls. This group included the SREBP cleavage-activating protein (SCAP; a SREBP activator), insulin-induced gene 1 (Insig1), insulin-induced gene 2 (Insig2), site-1 protease (S1P), site-2 protease (S2P), and Lipin-1. No expression alterations were found, except for the known SREBP1 target Insig1 in raptor and raptor/rictor mutants. However, this finding cannot explain the downregulation of targets, because Insig1 inhibits translocation of the SCAP/SREBP complex from the ER to the Golgi apparatus (Fig. 5D; Yang et al., 2002). mTORC1 can control the SREBP pathway by regulating the cellular localization of Lipin-1 via phosphorylation (Peterson et al., 2011). Therefore, to gain further insights into the connection between mTOR signaling and SREBP regulation in oligodendrocytes, we analyzed Lipin-1 localization by immunohistochemistry on spinal cord sections in raptor and raptor/rictor mutants (Fig. 5E). No changes were detected compared with controls. We conclude that raptor/mTORC1 function regulates lipid biogenesis in myelinated oligodendrocytes in a complex manner that remains partially unclear.
Disarray of lipid composition in raptor mutants
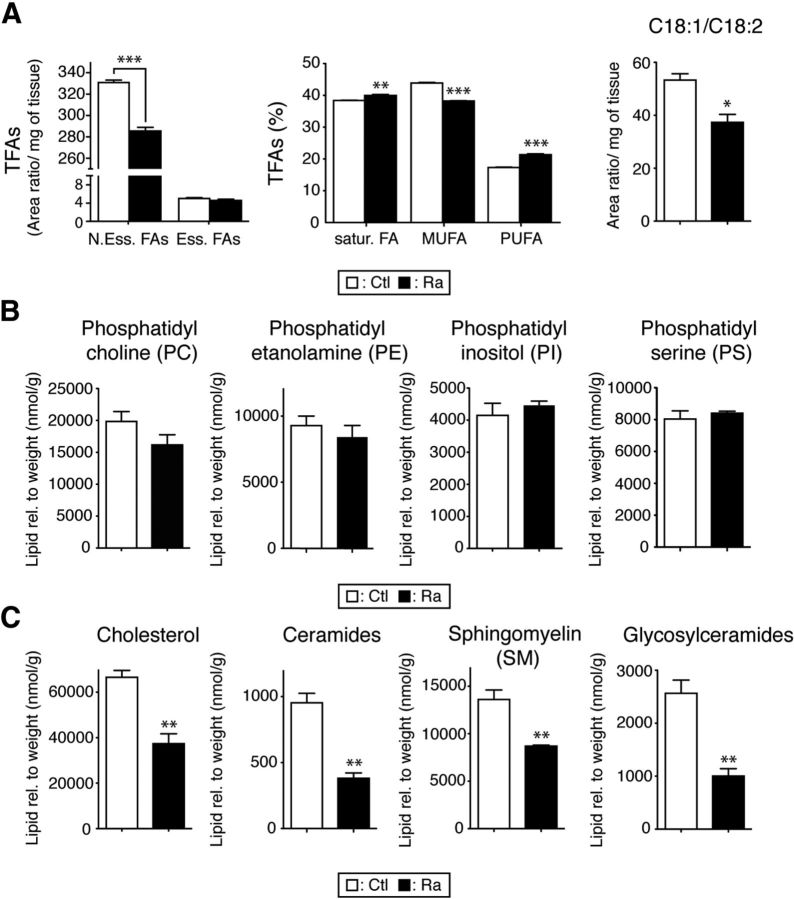
Because the biosynthesis of lipids in myelinated fibers is regulated by mTORC1 signaling, differences in raptor mutants would be expected. In indirect support of this hypothesis, Schwann cells lacking SCAP showed PNS hypomyelination that was characterized by lipid composition abnormalities (Verheijen et al., 2009). Therefore, we performed quantitative lipid profiling of spinal cords derived from raptor mutants. Robust changes were found compared with controls (Figs. 6, 7). Essential FAs cannot be synthesized by cells, so, as expected, their levels were unchanged in raptor mutants. However, nonessential FAs, which can be produced by the organism itself, were reduced significantly. Analyzing the levels of total FAs revealed a small increase in saturated FAs and a shift from monounsaturated FAs to polyunsaturated FAs. In addition, the C18:1/C18:2 ratio, a marker for progression of myelination (Huether et al., 1986), was decreased in raptor mutants (Fig. 6A). Total levels of phosphatidyl choline, phosphatidyl ethanolamine, phosphatidyl inositol, and phosphatidyl serine lipids were not changed (Fig. 6B), although we detected significant variations between individual lipid species (Fig. 7B). Finally, cholesterol, ceramide, sphingomyelin, and glycosylceramide levels were severely reduced in raptor mutant spinal cords (Figs. 6C, 7C).
Figure 6.
Abnormal lipid composition in raptor mutant spinal cords. A, Quantification of total FAs (TFAs) showed a significant reduction of nonessential FAs (N.Ess.FAs) in raptor mutants, whereas the level of essential FAs (Ess.FAs) did not change. Percentage of saturated, monounsaturated, and polyunsaturated FAs indicated a shift from monounsaturated to polyunsaturated FAs. The ratio of 18:1/18:2, relatively high in normal myelin, was reduced in mutants. B, Phosphatidyl choline, phosphatidyl ethanolamine, phosphatidyl inositol, and phosphatidyl serine levels showed no change between controls and mutants. C, Cholesterol, ceramides, sphingomyelin, and glycosylceramides were strongly reduced in mutant spinal cords. Lipid species were normalized to the weight of the sample. Error bars indicate SEM. n = 4; *p < 0.05; **p < 0.01; ***p < 0.001.
Figure 7.
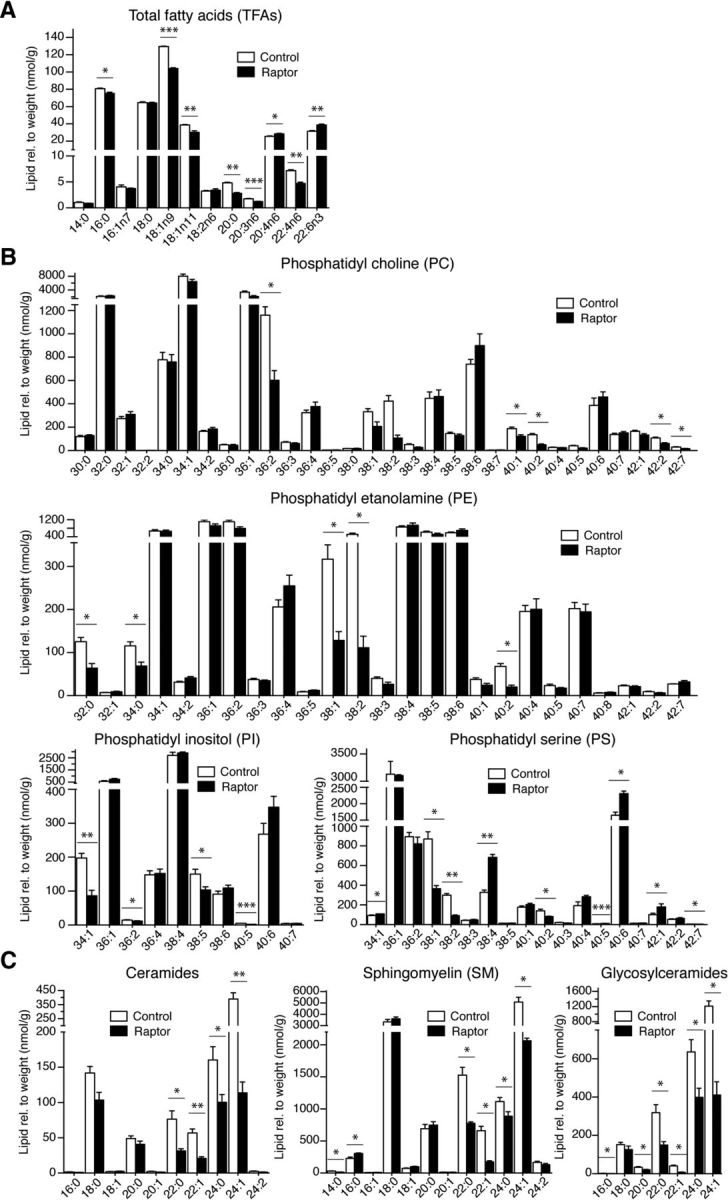
Lipid species analysis of the raptor mutant spinal cords. A, Quantification of the total FA (TFA) species. B, Quantification of the individual lipid species phosphatidyl choline, phosphatidyl ethanolamine, phosphatidyl inositol, and phosphatidyl serine of raptor mutants compared with littermate controls. C, Quantification of the individual lipid species ceramides, sphingomyelin, and glycosylceramides in raptor mutants. Lipid species were normalized to sample weight. Error bars indicate SEM. n = 3; *p < 0.05; **p < 0.01; ***p < 0.001.
Hyperactivation of mTORC1 via TSC1 ablation causes hypomyelination
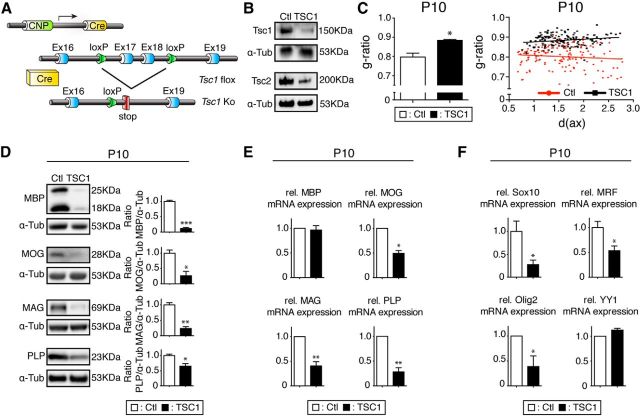
We had shown that inhibiting mTORC1 by raptor ablation in oligodendrocytes leads to hypomyelination. To evaluate the opposite situation, hyperactivation of mTORC1 specifically in oligodendrocytes, we crossed CNP-cre mice with mice carrying floxed alleles of the Tsc1 gene encoding the tuberous sclerosis 1 protein, an inhibitor of mTORC1 (TSC1 mutants; Fig. 8A). TSC1/2 stabilize each other critically as a physical and functional complex (van Slegtenhorst et al., 1998). Because disruption of the complex by mutations in Tsc1 or Tsc2 cause tuberous sclerosis complex, our experimental design was also expected to aid in the evaluation of oligodendrocyte-specific contributions to this syndrome. Western blot analysis of microdissected fragments of spinal cord white matter revealed loss of both TSC1 and TSC2 in TSC1 mutants, as anticipated (Fig. 8B). Morphological analysis showed that mice with oligodendrocytes lacking TSC1 were affected by hypomyelination comparable to raptor mutants (Fig. 8C). In addition, we found similar decreases in the levels of myelin proteins (MBP, MOG, MAG, and PLP) in TSC1 mutants as in raptor mutants, shown by immunoblotting (Fig. 8D). qRT-PCR analysis revealed reduced MOG, MAG, PLP, but not MBP, mRNA levels, as well as reduced transcript levels of the key transcription factors Sox10, MRF, and Olig2, but not YY1, a pattern shared between TSC1 mutants and raptor mutants (Fig. 8E,F). In contrast to raptor mutants, however, the percentage of myelinated axons was not altered in TSC1 mutants (18.1 ± 4%) compared with controls at P10 (19.8 ± 1.1%). These findings indicate that TSC1 function in oligodendrocytes is required for proper myelination and point to a crucial contribution of loss of TSC1 in oligodendrocytes to the CNS myelination defects and their potential functional consequences in tuberous sclerosis complex.
Figure 8.
Overactivation of mTORC1 caused by TSC1 ablation leads to hypomyelination. A, Regulatory elements of the CNP promoter drive the expression of Cre in oligodendrocytes. Schematics map of the TSC1 allele depicts the location of loxP sequences. B, Microdissected fragments of the spinal cord white matter were analyzed by Western blot to confirm TSC1 loss in conditional knock-outs. C, Higher g-ratio values of TSC1 mutants in the spinal cord ventral funiculi indicate hypomyelination compared with controls. Shown are scatter plot graphics of single fiber measurements detailing the thinner myelin in TSC1 mutants (black squares) compared with littermate controls (red circles). D, Spinal cord lysates of TSC1 mutants revealed strongly reduced levels of the myelin proteins MBP, MOG, MAG, and PLP. E, qRT-PCR revealed reduced levels of the myelin proteins MOG, MAG, and PLP in the TSC1 mutants compared with controls, whereas MBP was unchanged. F, qRT-PCR of transcripts encoding SOX 10, MRF, and Olig2 showed significant reductions in the spinal cord of TSC1 mutants. YY1 mRNA levels did not change.
Next, we confirmed that TSC1 ablation leads to mTORC1 overactivation. In support of these expectations, S6K and its substrate S6 were hyperphosphorylated in TSC1 mutants (Fig. 9A). Surprisingly, the level of P-4E-BP1 was reduced, similar to raptor mutants. Phosphorylation of Akt at T308 was also diminished, consistent with high activation of S6K triggering negative feedback loops (Huang and Manning, 2009; Song et al., 2012). In addition, phosphorylation of Akt at S473 was decreased in TSC1 mutants compared with controls (Fig. 9B). This finding is in agreement with in vitro data demonstrating that TSC1/2 is required for proper activation of mTORC2, the complex responsible for phosphorylation of Akt at S473 (Huang et al., 2008). Altered feedback signaling may also contribute (Bentzinger et al., 2013; Castets et al., 2013). Our results on mTOR signaling in TSC1 mutants are in agreement with earlier studies showing that a lack of the TSC1/2 complex leads to mTORC1 activation and defective PI3K-Akt signaling (Zhang et al., 2003). Next, we investigated the effects of mTORC1 overactivation on the lipid biosynthesis pathway. Comparable to raptor mutants, SREBP2 and SREBP targets were reduced at the protein and mRNA levels in TSC1 mutants (Fig. 9C,D). Therefore, precisely balanced activation of mTORC1 appears to be essential for lipogenesis. Offering a potential explanation for our results, Yecies et al. (2011) have shown that Akt can directly negatively regulate transcription of Insig2a, an inhibitor of SREBP maturation, whereas this Akt-mediated transcriptional suppression is missing in TSC1-ablated cells (Yecies et al., 2011). However, we found that Insig2a is a liver-specific transcript (Yabe et al., 2003) that is not expressed in the spinal cord (Fig. 9E). Furthermore, expression of the major isoform, Insig2, was not altered in the spinal cords of all our mutants (Figs. 5D, 9E,F).
Figure 9.
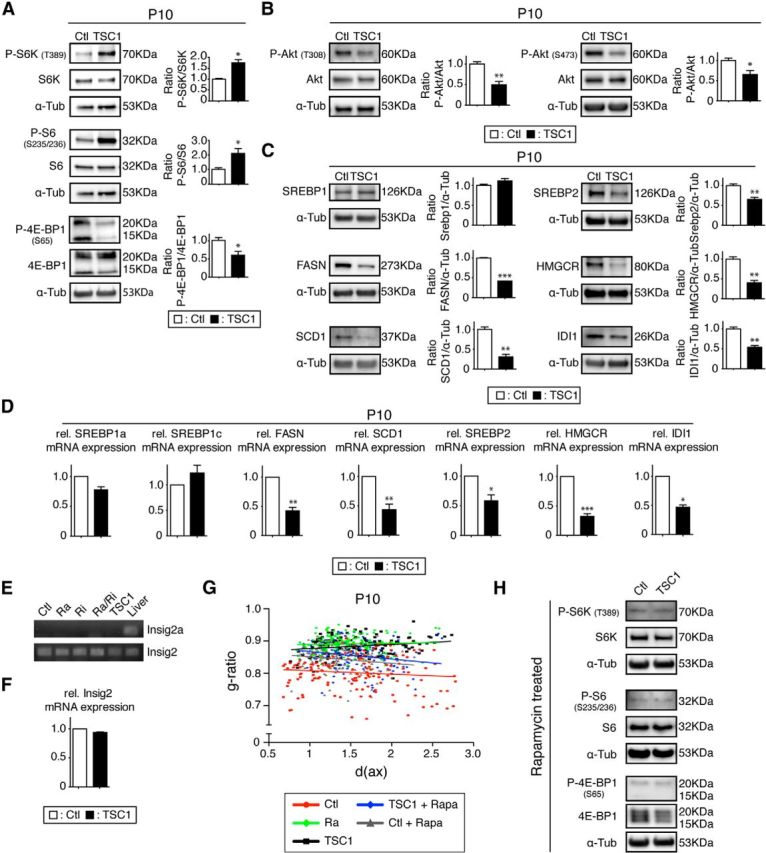
TSC1 mutant analysis. A, Phosphorylation levels of mTORC1 downstream targets P-S6K (T389) and P-S6K (S235/236) were strongly upregulated, except for P-4E-BP1 (S65), which was reduced. B, Phosphorylation of Akt was decreased on both T308 and S473. C, As in the raptor mutants, protein levels of main targets of SREBP and SREBP2 itself were reduced in mutants. D, qRT-PCR of the SREBPs and their main targets. E, RT-PCR for Insig2a and Insig2 in raptor, rictor, raptor/rictor, TSC1 mutant, and control spinal cords compared with liver positive control (ethidium-bromide-stained agarose gel). F, qRT-PCR of Insig2 in TSC1 mutants and control spinal cords. Error bars indicate SEM. n = 3; *p < 0.05; **p < 0.01; ***p < 0.001. G, Scatter plot graphics of g-ratios versus diameter of axons (in micrometers) of animal treated with rapamycin from P3 to P10. Shown are controls (gray triangles) and TSC1 mutants (blue diamonds) compared with control animals (red circles), raptor mutants (green diamonds), and TSC1 mutants (black squares) without treatment. Error bars indicate SEM. n = 3; *p < 0.05; **p < 0.01; ***p < 0.001. H, Spinal cord samples of rapamycin-treated TSC1 mutants and littermate controls showed the same downregulation of the main mTORC1 downstream targets.
We reasoned that mechanisms independent of mTORC1 hyperactivation but dependent on Akt for the activation of the SREBPs may explain the observed CNS hypomyelination in oligodendrocyte-specific TSC1 mutants. Therefore, we treated TSC1 mutant pups and their littermate controls from P3 to P10 with rapamycin, an inhibitor of mTORC1, to decrease mTORC1 activity. Analysis of g-ratios in spinal cords revealed that rapamycin treatment led to a partial rescue of the phenotype, mainly increasing myelination of larger axons (Fig. 9G). Rapamycin-treated controls showed comparable g-ratio profiles to treated TSC1 mutants, consistent with modulation of mTORC1 causing the effects. Western blot analysis on spinal cord samples confirmed the ability of rapamycin injection to reduce the overactivation of the mTORC1 downstream targets in the TSC1 mutants, reducing the phosphorylation to the same level as the littermate controls treated with rapamycin. We conclude that overactivation of mTORC1 mediated by TSC1 ablation in oligodendrocytes is the main contributor to the observed CNS hypomyelination.
Discussion
In the present study, we show that mice lacking raptor in oligodendrocytes exhibit robust CNS hypomyelination, likely due to defects in lipogenesis and alterations in the expression of myelin proteins and critical regulators. In contrast, analogous rictor mutants were only mildly and transiently affected in the early phase of myelination. To cause significantly delayed OPC differentiation, combined loss of raptor and rictor was required. In addition, our data suggest essential roles of the mTOR pathway in myelin maintenance. Finally, overactivation of mTORC1, caused by TSC1 ablation in oligodendrocytes, also resulted in thinner myelin and defective lipogenesis, with implications for our understanding of tuberous sclerosis complex.
Studies with rapamycin have shown that mTOR is a key factor regulating oligodendrocyte differentiation in vitro. They have not led to a consensus, however, about which stage(s) of oligodendrocyte differentiation is mainly dependent on mTOR signaling (Tyler et al., 2009; Guardiola-Diaz et al., 2012). Because rapamycin inhibits many (but not all) mTORC1 activities and mTORC2 is known to be insensitive to short-term rapamycin applications but inhibited by prolonged treatments (Sarbassov et al., 2006), the specific individual functional roles of mTORC1 and mTORC2 in oligodendrocytes remained unclear. Here, we show in vivo that mTOR signaling is indeed crucial for oligodendrocyte differentiation. Initial analyses revealed that proliferation and numbers of oligodendrocyte lineage cells were not changed in raptor, rictor, and raptor/rictor mutants in the ventral spinal cord funiculus at birth, consistent with the available in vitro results (Tyler et al., 2009). Subsequently, we examined oligodendrocyte differentiation using marker proteins. We found significant alterations only if raptor and rictor were lacking in combination, even though deletion of raptor alone led to comparable decreases in transcription factors involved in oligodendrocyte differentiation such as SOX10, MRF, and Olig2. The presence of detectable maturation defects exclusively in raptor/rictor mutants is somewhat in contrast to the reduced number of myelinated axons and the hypomyelination observed already in single raptor mutants and the transient hypomyelination in rictor mutants early in life. Therefore, our findings suggest complex and partially intermingled roles of mTORC1 and mTORC2 in the different steps of oligodendrocyte differentiation, maturation, myelination initiation, and myelin growth. It is clear, however, that both complexes together regulate oligodendrocyte maturation, whereas mTORC1 signaling is the main regulator of myelin sheath growth.
Akt is a major player in the regulation of CNS myelination, presumably acting through activation of the mTOR pathway (Norrmén and Suter, 2013). Myelinating-glia-specific conditional knock-out of PTEN (phosphatase and tensin homolog deleted on chromosome 10), an upstream inhibitor of PI3K/Akt signaling, induced hypermyelination. This phenotype was likely mediated by mTOR, because phosphorylation of S6K and S6 downstream of mTORC1 were increased and partial rescue with rapamycin treatment was observed (Goebbels et al., 2010; Harrington et al., 2010). In addition, mice overexpressing constitutively active Akt exhibited CNS hypermyelination associated with mTOR pathway activation (Narayanan et al., 2009). Here, we have targeted and inactived raptor/mTORC1 as a major signaling hub downstream of Akt. In indirect agreement with these previous studies, raptor mutants were affected by CNS hypomyelination. TSC1 mutants were also hypomyelinated despite hyperactivated mTORC1 signaling through S6K. Another common consequence of both mTORC1 deficiency or overactivation was dowregulation of the SREBP pathway. The observed dysregulation of SREBPs in raptor mutants is consistent with the positive regulatory functions of mTORC1 in lipid biosynthesis (Porstmann et al., 2008; Lewis et al., 2011). Furthermore, mTOR has been shown to induce the expression of proteins involved in lipogenesis during oligodendrocyte differentiation in vitro (Tyler et al., 2011). However, we were puzzled by the downregulation of SREBPs in the TSC1 mutant. Hyperactivation of downstream targets of mTOR are characteristic in tuberous sclerosis complex (Napolioni et al., 2009), but mTORC1 activation also induces negative feedback loops acting on the PI3K/Akt pathway and leading to defective Akt signaling (Song et al., 2012). A recent study showed that SREBPs can be activated in TSC1-deficient cells by a mechanism independent of mTORC1 but dependent on Akt and mediated by Insig2a (Yecies et al., 2011). Although we did not find evidence that this mechanism acts in TSC1-null oligodendrocytes, those results provided the proof-of-principle that such mTOR-independent/Akt-dependent mechanisms exist. Similar mechanisms may explain the difference in phenotypes observed between models in which the PI3K/Akt pathway is activated directly (mainly hypermyelination) and models in which mTORC1 is activated by TSC1 ablation (mainly hypomyelination). Elucidating the specific mechanisms of SREBP activation and its specific relation to PI3K/Akt/mTOR signaling in oligodendrocytes will be an exciting challenge for the future.
Dysfunction of the SREBP pathway in both TSC1- and raptor-ablated mutants had direct and similar repercussions on the levels of their targets. Most previous studies have focused on SREBP1 activation by mTOR. In raptor mutants and TSC1-mutant oligodendrocytes, we found that SREBP2 and its targets were strongly downregulated. SREBP2 tightly regulates cholesterol levels, consistent with the reduced levels of cholesterol present in raptor mutant spinal cords. Cholesterol is essential for membrane growth (Brown and Goldstein, 1999; Saher et al., 2005) and the myelin sheath tolerates only a moderate variation in cholesterol-protein ratio (Saher and Simons, 2010). However, despite robust reductions in the levels of enzymes responsible for cholesterol and lipid synthesis, the mutant oligodendrocytes were still able to produce myelin, albeit with reduced thickness. It remains to be shown whether uptake of external lipids may have helped to compensate for the deficient lipid synthesis, as was previously suggested in Schwann cells (Verheijen et al., 2009).
To model tuberous sclerosis complex due to loss of TSC1, appropriate animal models have been generated (Meikle et al., 2007; Goto et al., 2011; Carson et al., 2012; Mietzsch et al., 2013). Invariably, myelin deficiencies were observed in these studies, similar to the hypomyelination described in tuberous sclerosis complex patients. None of these studies, however, addressed whether lack of TSC1 specifically in oligodendrocytes contributes to the pathology. This led to speculations that signals from other TSC1-deleted cells in the mutant CNS may impair myelination (Wood et al., 2013). We show here that TSC1 expression by oligodendrocytes is required for proper myelination, because oligodendrocyte-specific TSC1 deletion leads to robust mTORC1-dependent hypomyelination. Minor contributions to this phenotype due to reduced mTORC2 activity are conceivable, because P-Akt S473 was diminished in TSC1 mutants and rictor mutants showed transient hypomyelination. However, mTORC2 is unlikely to be majorly involved, because the hypomyelination in TSC1 mutants was as robust as in raptor mutants and much more extensive than in rictor mutants.
In conclusion, we identified here a critical role of mTORC1 in governing correct protein and lipid synthesis, which is required for proper CNS myelin formation and maintenance. mTORC2-mediated signaling is a minor but important contributor. Balanced mTOR signaling and strictly regulated activation of mTORC1 in oligodendrocytes is required for myelination and correct lipogenesis. Although this study has focused mainly on development, it will be an important future task to evaluate the different roles of mTORC1 and mTORC2 in the regulation of demyelination and remyelination in diseases such as multiple sclerosis, its experimental models, or after injury to assess potential therapeutic efficacies of treatments along the mTOR signaling pathway.
During the time when this manuscript was under review, two related studies were published (Bercury et al., 2014; Wahl et al., 2014) supporting that mTOR signaling, and in particular mTORC1 signaling (Bercury et al., 2014), are critical for CNS myelination.
Footnotes
Research by F.L.J. was supported by a Marie Curie International Incoming Fellowship within the 7th European Community framework and a Swiss Federal Institute of Technology (ETH) fellowship. C.N. was supported by a Marie Curie Intra European Fellowship within the 7th European Community framework. Work in the laboratory of U.S. is funded by the Swiss National Science Foundation and the National Center of Competence in Research “Neural Plasticity and Repair.” We thank Klaus-Armin Nave for providing CNP-cre mice, Mario Pende and Karim Nadra for providing processed tissues of S6K1;S6K2 double-null mice, Nahum Sonenberg and Arkady Khoutorsky for providing processed tissue of Eif4ebp1;Eif4ebp2 double-null mice, Steven Scherer and Werner Kovacs for antibodies, the Electron Microscopy and Light Microscopy Centers of the ETH Zürich, members of the Suter laboratory for fruitful discussions, Ned Mantei for critically reading the manuscript, and Jorge Pereira for support and input.
The authors declare no competing financial interests.
References
- Aksamitiene E, Kiyatkin A, Kholodenko BN. Cross-talk between mitogenic Ras/MAPK and survival PI3K/Akt pathways: a fine balance. Biochem Soc Trans. 2012;40:139–146. doi: 10.1042/BST20110609. [DOI] [PubMed] [Google Scholar]
- Ando S, Tanaka Y, Toyoda Y, Kon K. Turnover of myelin lipids in aging brain. Neurochem Res. 2003;28:5–13. doi: 10.1023/A:1021635826032. [DOI] [PubMed] [Google Scholar]
- Bentzinger CF, Romanino K, Cloëtta D, Lin S, Mascarenhas JB, Oliveri F, Xia J, Casanova E, Costa CF, Brink M, Zorzato F, Hall MN, Rüegg MA. Skeletal muscle-specific ablation of raptor, but not of rictor, causes metabolic changes and results in muscle dystrophy. Cell Metab. 2008;8:411–424. doi: 10.1016/j.cmet.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Bentzinger CF, Lin S, Romanino K, Castets P, Guridi M, Summermatter S, Handschin C, Tintignac LA, Hall MN, Rüegg MA. Differential response of skeletal muscles to mTORC1 signaling during atrophy and hypertrophy. Skelet Muscle. 2013;3:6. doi: 10.1186/2044-5040-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bercury KK, Dai J, Sachs HH, Ahrendsen JT, Wood TL, Macklin WB. Conditional ablation of raptor or rictor has differential impact on oligodendrocyte differentiation and CNS myelination. J Neurosci. 2014;34:4466–4480. doi: 10.1523/JNEUROSCI.4314-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuleux M, Klopfenstein M, Stephan C, Doughty CA, Barys L, Maira SM, Kwiatkowski D, Lane HA. Increased AKT S473 phosphorylation after mTORC1 inhibition is rictor dependent and does not predict tumor cell response to PI3K/mTOR inhibition. Mol Cancer Ther. 2009;8:742–753. doi: 10.1158/1535-7163.MCT-08-0668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc Natl Acad Sci U S A. 1999;96:11041–11048. doi: 10.1073/pnas.96.20.11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson RP, Van Nielen DL, Winzenburger PA, Ess KC. Neuronal and glia abnormalities in Tsc1-deficient forebrain and partial rescue by rapamycin. Neurobiol Dis. 2012;45:369–380. doi: 10.1016/j.nbd.2011.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castets P, Lin S, Rion N, Di Fulvio S, Romanino K, Guridi M, Frank S, Tintignac LA, Sinnreich M, Rüegg MA. Sustained activation of mTORC1 in skeletal muscle inhibits constitutive and starvation-induced autophagy and causes a severe, late-onset myopathy. Cell Metab. 2013;17:731–744. doi: 10.1016/j.cmet.2013.03.015. [DOI] [PubMed] [Google Scholar]
- Crino PB, Nathanson KL, Henske EP. The tuberous sclerosis complex. N Engl J Med. 2006;355:1345–1356. doi: 10.1056/NEJMra055323. [DOI] [PubMed] [Google Scholar]
- Dibble CC, Elis W, Menon S, Qin W, Klekota J, Asara JM, Finan PM, Kwiatkowski DJ, Murphy LO, Manning BD. TBC1D7 is a third subunit of the TSC1-TSC2 complex upstream of mTORC1. Mol Cell. 2012;47:535–546. doi: 10.1016/j.molcel.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fancy SP, Chan JR, Baranzini SE, Franklin RJ, Rowitch DH. Myelin regeneration: a recapitulation of development? Annu Rev Neurosci. 2011;34:21–43. doi: 10.1146/annurev-neuro-061010-113629. [DOI] [PubMed] [Google Scholar]
- Fauland A, Köfeler H, Trötzmüller, Knopf A, Hartler J, Eberl A, Chitraju C, Lankmayr E, Spener F. A comprehensive method for lipid profiling by liquid chromatography-ion cyclotron resonance mass spectrometry. J Lipid Res. 2011;52:2314–2322. doi: 10.1194/jlr.D016550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores AI, Narayanan SP, Morse EN, Shick HE, Yin X, Kidd G, Avila RL, Kirschner DA, Macklin WB. Constitutively active Akt induces enhanced myelination in the CNS. J Neurosci. 2008;28:7174–7183. doi: 10.1523/JNEUROSCI.0150-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genoud S, Lappe-Siefke C, Goebbels S, Radtke F, Aguet M, Scherer SS, Suter U, Nave KA, Mantei N. Notch1 control of oligodendrocyte differentiation in the spinal cord. J Cell Biol. 2002;158:709–718. doi: 10.1083/jcb.200202002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebbels S, Oltrogge JH, Kemper R, Heilmann I, Bormuth I, Wolfer S, Wichert SP, Möbius W, Liu X, Lappe-Siefke C, Rossner MJ, Groszer M, Suter U, Frahm J, Boretius S, Nave KA. Elevated phosphatidylinositol 3,4,5-trisphosphate in glia triggers cell-autonomous membrane wrapping and myelination. J Neurosci. 2010;30:8953–8964. doi: 10.1523/JNEUROSCI.0219-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto J, Talos DM, Klein P, Qin W, Chekaluk YI, Anderl S, Malinowska IA, Di Nardo A, Bronson RT, Chan JA, Vinters HV, Kernie SG, Jensen FE, Sahin M, Kwiatkowski DJ. Regulable neural progenitor-specific Tsc1 loss yields giant cells with organellar dysfunction in a model of tuberous sclerosis complex. Proc Natl Acad Sci U S A. 2011;108:E1070–E1079. doi: 10.1073/pnas.1106454108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guardiola-Diaz HM, Ishii A, Bansal R. Erk1/2 MAPK and mTOR signaling sequentially regulates progression through distinct stages of oligodendrocyte differentiation. Glia. 2012;60:476–486. doi: 10.1002/glia.22281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington EP, Zhao C, Fancy SP, Kaing S, Franklin RJ, Rowitch DH. Oligodendrocyte PTEN is required for myelin and axonal integrity, not remyelination. Ann Neurol. 2010;68:703–716. doi: 10.1002/ana.22090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartler J, Trötzmüller M, Chitraju C, Spener F, Köfeler HC, Thallinger GG. Lipid Data Analyzer: unattended identification and quantitation of lipids in LC-MS data. Bioinformatics. 2011;27:572–577. doi: 10.1093/bioinformatics/btq699. [DOI] [PubMed] [Google Scholar]
- Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Manning BD. A complex interplay between Akt, TSC2 and the two mTOR complexes. Biochem Soc Trans. 2009;37:217–222. doi: 10.1042/BST0370217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Dibble CC, Matsuzaki M, Manning BD. The TSC1-TSC2 complex is required for proper activation of mTOR complex 2. Mol Cell Biol. 2008;28:4104–4115. doi: 10.1128/MCB.00289-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huether G, Klapproth M, Neuhoff V. Fatty acid composition of myelin lipids from developing rat forebrain and spinal cord: influence of experimental hyperphenylalaninaemia. Neurochem Res. 1986;11:1303–1311. doi: 10.1007/BF00966124. [DOI] [PubMed] [Google Scholar]
- Ishii A, Fyffe-Maricich SL, Furusho M, Miller RH, Bansal R. ERK1/ERK2 MAPK signaling is required to increase myelin thickness independent of oligodendrocyte differentiation and initiation of myelination. J Neurosci. 2012;32:8855–8864. doi: 10.1523/JNEUROSCI.0137-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenning M, Jackson S, Hay CM, Faux C, Kilpatrick TJ, Willingham M, Emery B. Myelin gene regulatory factor is required for maintenance of myelin and mature oligodendrocyte identity in the adult CNS. J Neurosci. 2012;32:12528–12542. doi: 10.1523/JNEUROSCI.1069-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski DJ, Zhang H, Bandura JL, Heiberger KM, Glogauer M, el-Hashemite N, Onda H. A mouse model of TSC1 reveals sex-dependent lethality from liver hemangiomas, and up-regulation of p70S6 kinase activity in Tsc1 null cells. Hum Mol Genet. 2002;11:525–534. doi: 10.1093/hmg/11.5.525. [DOI] [PubMed] [Google Scholar]
- Lamming DW, Sabatini DM. A central role for mTOR in lipid homeostasis. Cell Metab. 2013;18:465–469. doi: 10.1016/j.cmet.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTORC1 activates SREBP-1c and uncouples lipogenesis from gluconeogenesis. Proc Natl Acad Sci U S A. 2010;107:3281–3282. doi: 10.1073/pnas.1000323107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling. Cold Spring Harb Perspect Biol. 2012;4:a011593. doi: 10.1101/cshperspect.a011593. pii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappe-Siefke C, Goebbels S, Gravel M, Nicksch E, Lee J, Braun PE, Griffiths IR, Nave KA. Disruption of Cnp1 uncouples oligodendroglial functions in axonal support and myelination. Nat Genet. 2003;33:366–374. doi: 10.1038/ng1095. [DOI] [PubMed] [Google Scholar]
- Le Bacquer O, Petroulakis E, Paglialunga S, Poulin F, Richard D, Cianflone K, Sonenberg N. Elevated sensitivity to diet-induced obesity and insulin resistance in mice lacking 4E-BP1 and 4E-BP2. J Clin Invest. 2007;117:387–396. doi: 10.1172/JCI29528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone DP, Genoud S, Atanasoski S, Grausenburger R, Berger P, Metzger D, Macklin WB, Chambon P, Suter U. Tamoxifen-inducible glia-specific Cre mice for somatic mutagenesis in oligodendrocytes and Schwann cells. Mol Cell Neurosci. 2003;22:430–440. doi: 10.1016/S1044-7431(03)00029-0. [DOI] [PubMed] [Google Scholar]
- Lewis CA, Griffiths B, Santos CR, Pende M, Schulze A. Regulation of the SREBP transcription factors by mTORC1. Biochem Soc Trans. 2011;39:495–499. doi: 10.1042/BST0390495. [DOI] [PubMed] [Google Scholar]
- Liebisch G, Vizcaíno JA, Köfeler H, Trötzmüller M, Griffiths WJ, Schmitz G, Spener F, Wakelam MJ. Shorthand notation for lipid structures derived from mass spectrometry. J Lipid Res. 2013;54:1523–1530. doi: 10.1194/jlr.M033506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyash V, Liebisch G, Kurzchalia TV, Shevchenko A, Schwudke D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J Lipid Res. 2008;49:1137–1146. doi: 10.1194/jlr.D700041-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meikle L, Talos DM, Onda H, Pollizzi K, Rotenberg A, Sahin M, Jensen FE, Kwiatkowski DJ. A mouse model of tuberous sclerosis: neuronal loss of Tsc1 causes dysplastic and ectopic neurons, reduced myelination, seizure activity, and limited survival. J Neurosci. 2007;27:5546–5558. doi: 10.1523/JNEUROSCI.5540-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mietzsch U, McKenna J, 3rd, Reith RM, Way SW, Gambello MJ. Comparative analysis of Tsc1 and Tsc2 single and double radial glial cell mutants. J Comp Neurol. 2013;521:3817–3831. doi: 10.1002/cne.23380. [DOI] [PubMed] [Google Scholar]
- Morita M, Gravel SP, Chénard V, Sikström K, Zheng L, Alain T, Gandin V, Avizonis D, Arguello M, Zakaria C, McLaughlan S, Nouet Y, Pause A, Pollak M, Gottlieb E, Larsson O, St-Pierre J, Topisirovic I, Sonenberg N. mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab. 2013;18:698–711. doi: 10.1016/j.cmet.2013.10.001. [DOI] [PubMed] [Google Scholar]
- Morrison BM, Lee Y, Rothstein JD. Oligodendroglia: metabolic supporters of axons. Trends Cell Biol. 2013;23:644–651. doi: 10.1016/j.tcb.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napolioni V, Moavero R, Curatolo P. Recent advances in neurobiology of Tuberous Sclerosis Complex. Brain Dev. 2009;31:104–113. doi: 10.1016/j.braindev.2008.09.013. [DOI] [PubMed] [Google Scholar]
- Narayanan SP, Flores AI, Wang F, Macklin WB. Akt signals through the mammalian target of rapamycin pathway to regulate CNS myelination. J Neurosci. 2009;29:6860–6870. doi: 10.1523/JNEUROSCI.0232-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nave KA. Myelination and support of axonal integrity by glia. Nature. 2010;468:244–252. doi: 10.1038/nature09614. [DOI] [PubMed] [Google Scholar]
- Norrmén C, Suter U. Akt/mTOR signalling in myelination. Biochem Soc Trans. 2013;41:944–950. doi: 10.1042/BST20130046. [DOI] [PubMed] [Google Scholar]
- Pende M, Um SH, Mieulet V, Sticker M, Goss VL, Mestan J, Mueller M, Fumagalli S, Kozma SC, Thomas G. S6K1(-/-)/S6K2(-/-) mice exhibit perinatal lethality and rapamycin-sensitive 5′-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol Cell Biol. 2004;24:3112–3124. doi: 10.1128/MCB.24.8.3112-3124.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira JA, Baumann R, Norrmén C, Somandin C, Miehe M, Jacob C, Lühmann T, Hall-Bozic H, Mantei N, Meijer D, Suter U. Dicer in Schwann cells is required for myelination and axonal integrity. J Neurosci. 2010;30:6763–6775. doi: 10.1523/JNEUROSCI.0801-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson TR, Sengupta SS, Harris TE, Carmack AE, Kang SA, Balderas E, Guertin DA, Madden KL, Carpenter AE, Finck BN, Sabatini DM. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 2011;146:408–420. doi: 10.1016/j.cell.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak P, Cybulski N, Feige JN, Auwerx J, Rüegg MA, Hall MN. Adipose-specific knockout of raptor results in lean mice with enhanced mitochondrial respiration. Cell Metab. 2008;8:399–410. doi: 10.1016/j.cmet.2008.09.003. [DOI] [PubMed] [Google Scholar]
- Porstmann T, Santos CR, Griffiths B, Cully M, Wu M, Leevers S, Griffiths JR, Chung YL, Schulze A. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 2008;8:224–236. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux PP, Topisirovic I. Regulation of mRNA translation by signaling pathways. Cold Spring Harb Perspect Biol. 2012:4. doi: 10.1101/cshperspect.a012252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saher G, Simons M. Cholesterol and myelin biogenesis. Subcell Biochem. 2010;51:489–508. doi: 10.1007/978-90-481-8622-8_18. [DOI] [PubMed] [Google Scholar]
- Saher G, Brügger B, Lappe-Siefke C, Möbius W, Tozawa R, Wehr MC, Wieland F, Ishibashi S, Nave KA. High cholesterol level is essential for myelin membrane growth. Nat Neurosci. 2005;8:468–475. doi: 10.1038/nn1426. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- Sherman DL, Krols M, Wu LM, Grove M, Nave KA, Gangloff YG, Brophy PJ. Arrest of myelination and reduced axon growth when Schwann cells lack mTOR. J Neurosci. 2012;32:1817–1825. doi: 10.1523/JNEUROSCI.4814-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima H, Pende M, Chen Y, Fumagalli S, Thomas G, Kozma SC. Disruption of the p70(s6k)/p85(s6k) gene reveals a small mouse phenotype and a new functional S6 kinase. EMBO J. 1998;17:6649–6659. doi: 10.1093/emboj/17.22.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soliman GA. The integral role of mTOR in lipid metabolism. Cell Cycle. 2011;10:861–862. doi: 10.4161/cc.10.6.14930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol. 2012;13:283–296. doi: 10.1038/nrm3330. [DOI] [PubMed] [Google Scholar]
- Tyler WA, Gangoli N, Gokina P, Kim HA, Covey M, Levison SW, Wood TL. Activation of the mammalian target of rapamycin (mTOR) is essential for oligodendrocyte differentiation. J Neurosci. 2009;29:6367–6378. doi: 10.1523/JNEUROSCI.0234-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler WA, Jain MR, Cifelli SE, Li Q, Ku L, Feng Y, Li H, Wood TL. Proteomic identification of novel targets regulated by the mammalian target of rapamycin pathway during oligodendrocyte differentiation. Glia. 2011;59:1754–1769. doi: 10.1002/glia.21221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Slegtenhorst M, Nellist M, Nagelkerken B, Cheadle J, Snell R, van den Ouweland A, Reuser A, Sampson J, Halley D, van der Sluijs P. Interaction between hamartin and tuberin, the TSC1 and TSC2 gene products. Hum Mol Genet. 1998;7:1053–1057. doi: 10.1093/hmg/7.6.1053. [DOI] [PubMed] [Google Scholar]
- Verheijen MH, Camargo N, Verdier V, Nadra K, de Preux Charles AS, Médard JJ, Luoma A, Crowther M, Inouye H, Shimano H, Chen S, Brouwers JF, Helms JB, Feltri ML, Wrabetz L, Kirschner D, Chrast R, Smit AB. SCAP is required for timely and proper myelin membrane synthesis. Proc Natl Acad Sci U S A. 2009;106:21383–21388. doi: 10.1073/pnas.0905633106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl SE, McLane LE, Bercury KK, Macklin WB, Wood TL. Mammalian target of rapamycin promotes oligodendrocyte differentiation, initiation and extent of CNS myelination. J Neurosci. 2014;34:4453–4465. doi: 10.1523/JNEUROSCI.4311-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood TL, Bercury KK, Cifelli SE, Mursch LE, Min J, Dai J, Macklin WB. mTOR: a link from the extracellular milieu to transcriptional regulation of oligodendrocyte development. ASN Neuro. 2013;5:e00108. doi: 10.1042/AN20120092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe D, Komuro R, Liang G, Goldstein JL, Brown MS. Liver-specific mRNA for Insig-2 down-regulated by insulin: implications for fatty acid synthesis. Proc Natl Acad Sci U S A. 2003;100:3155–3160. doi: 10.1073/pnas.0130116100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Espenshade PJ, Wright ME, Yabe D, Gong Y, Aebersold R, Goldstein JL, Brown MS. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell. 2002;110:489–500. doi: 10.1016/S0092-8674(02)00872-3. [DOI] [PubMed] [Google Scholar]
- Yecies JL, Zhang HH, Menon S, Liu S, Yecies D, Lipovsky AI, Gorgun C, Kwiatkowski DJ, Hotamisligil GS, Lee CH, Manning BD. Akt stimulates hepatic SREBP1c and lipogenesis through parallel mTORC1-dependent and independent pathways. Cell Metab. 2011;14:21–32. doi: 10.1016/j.cmet.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young KM, Psachoulia K, Tripathi RB, Dunn SJ, Cossell L, Attwell D, Tohyama K, Richardson WD. Oligodendrocyte dynamics in the healthy adult CNS: evidence for myelin remodeling. Neuron. 2013;77:873–885. doi: 10.1016/j.neuron.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Cicchetti G, Onda H, Koon HB, Asrican K, Bajraszewski N, Vazquez F, Carpenter CL, Kwiatkowski DJ. Loss of Tsc1/Tsc2 activates mTOR and disrupts PI3K-Akt signaling through downregulation of PDGFR. J Clin Invest. 2003;112:1223–1233. doi: 10.1172/JCI17222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Zhou L, Du XX, Ji Y, Xu J, Tian J, Jiang W, Zou Y, Yu S, Gan L, Luo M, Yang Q, Cui Y, Yang W, Xia X, Chen M, Zhao X, Shen Y, Chen PY, Worley PF, Xiao B. Rheb1 is required for mTORC1 and myelination in postnatal brain development. Dev Cell. 2011;20:97–108. doi: 10.1016/j.devcel.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]