Abstract
SMARCA4 (BRG1) and SMARCB1 (INI1) are tumor suppressor genes that are crucially involved in the formation of malignant rhabdoid tumors, such as atypical teratoid/rhabdoid tumor (AT/RT). AT/RTs typically affect infants and occur at various sites of the CNS with a particular frequency in the cerebellum. Here, granule neurons and their progenitors represent the most abundant cell type and are known to give rise to a subset of medulloblastoma, a histologically similar embryonal brain tumor. To test how Smarc proteins influence the development of granule neurons and whether this population may serve as cellular origin for AT/RTs, we specifically deleted Smarca4 and Smarcb1 in cerebellar granule cell precursors. Respective mutant mice displayed severe ataxia and motor coordination deficits, but did not develop any tumors. In fact, they suffered from a severely hypoplastic cerebellum due to a significant inhibition of granule neuron precursor proliferation. Molecularly, this was accompanied by an enhanced activity of Wnt/β-catenin signaling that, by itself, is known to cause a nearly identical phenotype. We further used an hGFAP-cre allele, which deleted Smarcb1 much earlier and in a wider neural precursor population, but we still did not detect any tumor formation in the CNS. In summary, our results emphasize cell-type-dependent roles of Smarc proteins and argue against cerebellar granule cells and other progeny of hGFAP-positive neural precursors as the cellular origin for AT/RTs.
Keywords: AT/RT, brain, cerebellum, development, Smarc, tumor
Introduction
Several core subunits of the chromatin remodeling complex SWI/SNF have been identified as classical tumor suppressor genes in a variety of human cancers (Kadoch et al., 2013). Biallelic inactivation of SMARCB1 (INI1) can occur in a number of tumors, but are regarded as entity-defining for malignant rhabdoid tumors when this inactivation is observed in small, blue, and round cell malignancies of infancy (Versteege et al., 1998; Sévenet et al., 1999; Klochendler-Yeivin et al., 2000). When occurring in the CNS, such tumors are called atypical teratoid/rhabdoid tumors (AT/RTs). AT/RTs are rare regarding absolute numbers, but make up approximately half of all malignant brain tumors in infancy and are associated with a particularly unfavorable outcome. Apart from SMARCB1 mutations, which are found in nearly all malignant rhabdoid tumors, no recurrent genomic alterations were found in the genome of AT/RTs so far (Kieran et al., 2012; Lee et al., 2012; Hasselblatt et al., 2013). Only a small proportion of AT/RTs were reported to carry SMARCA4 (BRG1) instead of SMARCB1 mutations (Schneppenheim et al., 2010; Hasselblatt et al., 2011). AT/RTs can be detected at diverse sites of the CNS, but predominantly occur in the cerebellum. Nevertheless, the exact cellular origin remains elusive, both in the cerebellum as well as in other parts of the CNS. In the cerebellum, granule neuron precursors not only give rise to the most abundant cell type, the granule neurons, but can also serve as the cell of origin for a subset of medulloblastoma, a histologically similar malignant embryonal brain tumor (Schüller et al., 2008). To investigate granule neuron precursors as a possible cell of origin for AT/RTs in the cerebellum, we conditionally deleted Smarca4 and Smarcb1 in these cells. However, none of the investigated mutants developed rhabdoid tumors. Instead, severe developmental defects were observed, pointing toward essential roles of Smarca4 and Smarcb1 for granule cell proliferation and strongly arguing against granule cells as the cellular origin for AT/RTs.
Materials and Methods
Transgenic mice.
The generation of mice carrying a loxP-flanked exon 1 of the Smarcb1 gene or a loxP-flanked Smarca4 gene has been previously described (Roberts et al., 2002; Indra et al., 2005). These mice were crossed to Math1-cre mice (Schüller et al., 2007) to generate Math1-cre::Smarcb1fl/fl and Math1-cre::Smarca4fl/fl mice. Mice were kept under standard conditions. Genotyping was performed by standard PCR using Cre-specific primers (5′-TCCGGGCTGCCACGACCAA-3′ and 5′-GGCGCGGCAACACCATTTT-3′), Smarcb1-specific primers (5′-TAGGCACTGGACATAAGGGC-3′ and 5′-GTAACTGTCAAGAATCAATGG-3′), and Smarca4-specific primers (5′-GCCTTGTCTCAAACTGATAAG-3′ and 5′-GTCATACTTATGTCATAGCC-3′ and 5′-GATCAGCTCATGCCCTAAGG-3′). Mice were intensely monitored twice a day including weight and size control.
Histology/immunostaining.
Paraffin-embedded tissue was sectioned, deparaffinized, and rehydrated before heat-induced antigen retrieval was conducted at 100°C for 20 min in 10 mm sodium citrate buffer for all antibodies. Immunohistochemical staining was done using primary antibodies (NeuN, Smarcb1, Calbindin, Cre, phospho-Histone H3) and the HRP/DAB Staining System (DAKO) according to the manufacturer's specifications. Hemalaun was used for nuclear counterstaining. For immunofluorescent double-stainings, sections were washed twice with PBS/0.1% Triton X-100 and then incubated in blocking buffer (I-Block protein-based blocking reagent; Applied Biosystems) for 30 min. Primary antibodies were diluted in blocking buffer and applied overnight at 4°C. Next, tissue was washed twice with PBS/0.1% Triton X-100 and incubated for another 60 min with a 1:500 dilution of fluorescence-labeled secondary antibodies (goat anti-mouse Alexa546; goat anti-rabbit Alexa488, Invitrogen) in blocking buffer. Sections were washed twice with PBS/0.1% Triton X-100, counterstained with 4′,6-diamidino-2-phenylindole (DAPI), and mounted in Fluorescent Mounting Medium (DAKO). All images of tissue sections were collected on an Olympus IX50 microscope in combination with the Color View Soft Imaging System.
Cell culture.
Cerebellar granule neuron precursor cultures were generated as previously described (Lorenz et al., 2011). Briefly, cerebella of postnatal day 5 (P5) pups were taken out and prepared in HBSS (Invitrogen) with added glucose. Meninges and plexus tissue were carefully removed. Dissociation of pooled cerebella was triggered by trypsin-EDTA-DNase. HBSS was replaced by culture medium containing DMEM-F12 (Invitrogen), 25 mm KCl, N2 supplement (Invitrogen), penicillin-streptomycin (Pen-strep), and 10% fetal calf serum (FCS) (Sigma). After centrifugation, cells were plated at a concentration of 3 million/ml in poly-l-ornithine (Sigma)-precoated wells and incubated at 37°C for 6–12 h to recover. Then, medium was changed to serum-free culture medium containing Shh-protein (R&D Systems) at a concentration of 3 μg/ml.
Production of IRES-GFP and Cre-IRES-GFP virus was performed as previously published (Lorenz et al., 2011). Briefly, 293 packaging cells (Invitrogen) were grown in DMEM-10% FCS-Pen-strep-300 μg/ml G418 and transfected with 10 μg of each virus construct, as well as vsv-g and gag-pol plasmids, using the Fugene 6 transfection reagent (Roche). Virus containing medium (without G418) was collected every 24 h for 3 consecutive days, pooled, filtered, and stored at −80°C until use.
For the experiments shown in Figure 2, cells were FACS-sorted based on GFP expression using a FACS ARIA III (BD Bioscience). After FACS-sorting, cells were replated at a density of 3 million/ml in poly-l-ornithine (Sigma)-precoated wells and incubated at 37°C for 6–12 h to recover. These cells were finally used for cell counting experiments [using the Neubauer chamber and automated cell counter TC10 (Bio-Rad)] and for RNA analyses.
Figure 2.
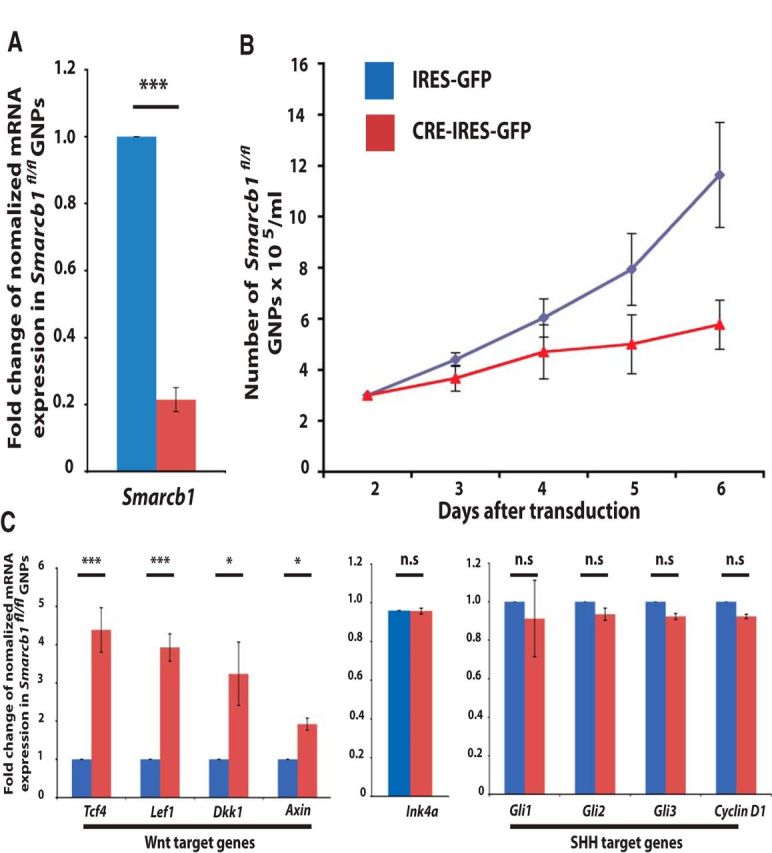
Loss of Smarcb1 impairs proliferation and activates Wnt signaling in cerebellar granule cell precursors. Cerebellar granule cell precursors were isolated from Smarcb1fl/fl mice on day P5, cultured, and transduced with IRES-GFP or Cre-IRES-GFP viruses. Thirty-six hours after transduction, cells were sorted for GFP-positive cells and experiments were performed. Cre-IRES-GFP-transduced cells displayed a severe reduction of Smarcb1 expression, as expected (A). Furthermore, proliferation was significantly decreased (B), and cells within the Cre-IRES-GFP condition were characterized by a massive upregulation of Wnt target genes Tcf4, Lef1, Dkk1, and Axin2. No significant changes were observed regarding the expression of Ink4a, Gli1, Gli2, Gli3 or Cyclin D1 (C).
Nucleic acid extraction and PCR analyses.
Total RNA from granule cell cultures was extracted using the RNAeasyMikro Kit (Qiagen). cDNA synthesis was performed using Prime Script RT reagent Kit (Takara) and applied according to the manufacturer's protocol. Quantitative reverse transcription-PCR (qRT-PCR) was performed using SYBR green (Promega) in the Step One Plus system (Applied Biosystems). Due to its consistent expression in murine granule neuron precursors, Actin was used as a housekeeping control gene. For each primer set, postamplification melting curves were analyzed with the Step One Plus software (Applied Biosystems) to verify the specific amplification of the expected PCR product. The following primers were used: mAxin2 forward: TGA-CTC-TCC-TTC-CAG-ATC-CCA; mAxin2 reverse: TGC-CCA-CAC-TAG-GCT-GAC-A; mLef1 forward: TGT-TTA-TCC-CAT-CAC-GGG-TGG; mLef1 reverse: CAT-GGA-AGT-GTC-GCC-TGA-CAG; mDkk1 forward: CTC-ATC-AAT-TCC-AAC-GCG-ATC-A; mDkk1 reverse: GCC-CTC-ATA-GAG-AAC-TCC-CG; mTcf4 forward: CGA-AAA-GTT-CCT-CCG-GGT-TTG; mTcf4 reverse: CGT-AGC-CGG-GCT-GAT-TCA-T; mSmarcb1 forward: GAG-GTG-GGA-AAC-TAC-CTG-CG; mSmarcb1 reverse: CGC-CAG-AGT-GAG-GGG-TAT-C; mβActin forward: GGC-TGT-ATT-CCC-CTC-CAT-CG; mβActin reverse: CCA-GTT-GGT-AAC-AAT-GCC-ATG-T; Ink4a forward: ACT-CTT-TCG-GTC-GTA-CCC-C; Ink4a reverse: GCG-TGC-TTG-AGC-TGA-AGC-TA; mGli1 forward: CCA-AGC-CAA-CTT-TAT-GTC-AGG-G; mGli1 reverse: AGC-CCG-CTT-CTT-TGT-TAA-TTT-GA; mGli2 forward: CAA-CGC-CTA-CTC-TCC-CAG-AC; mGli2 reverse: GAG-CCT-TGA-TGT-ACT-GTA-CCA-C; mGli3 forward: CAC-AGC-TCT-ACG-GCG-ACT-G; mGli3 reverse: CTG-CAT-AGT-GAT-TGC-GTT-TCT-TC; mCyclinD1 forward: GCG-TAC-CCT-GAC-ACC-AAT-CTC; mCyclin D1 reverse: CTC-CTC-TTC-GCA-CTT-CTG-CTC.
Quantifications and statistics.
Analysis of phenotypes from conditional knock-out mice was performed for at least three mice of each genotype and each age that were all taken from different litters.
For the experiment shown in Figures 1, 2, and 3, results were analyzed using the Fisher exact test, and p values of <0.05 were considered significant. All obtained results, including those from clinical testing of the mice, were analyzed using the Prism4 software program (GraphPad).
Figure 1.
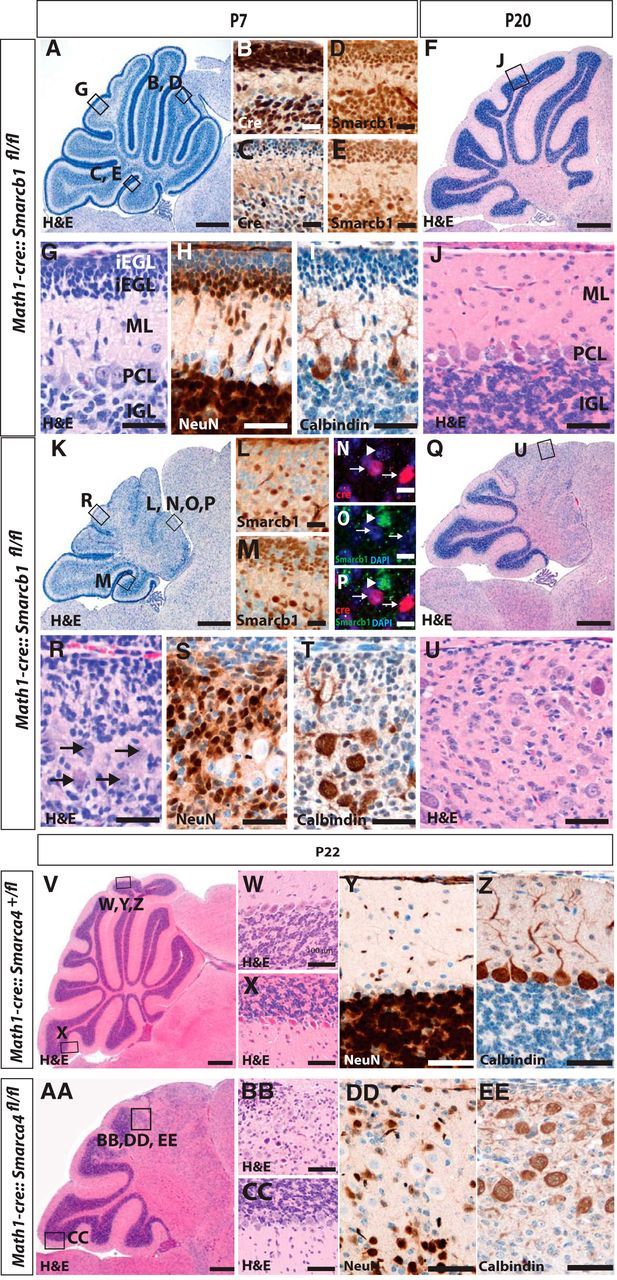
Loss of Smarcb1 in cerebellar granule cells results in a severe reduction of the cerebellar size. Cerebellar histopathology of control mice is normal (A, F) with the typical cortical layering (G, J) as visualized by H&E stains of P7 and P20 sagittal sections. Cre is strongly expressed in rostral parts (B), but less extensive in caudal lobes of control mice (C), and nuclear staining of Smarcb1 is retained in all control cells (D, E). In contrast, Math1-cre::Smarcb1fl/fl mice display a severe phenotype in the Cre-expressing rostral parts of the cerebellum. At P7 and P20, a loss of normal architecture and blurred cortical layers are apparent (K, Q and R, U, respectively), which is caused by a loss of Smarcb1 protein expression in rostral parts and, to a lesser extent, in caudal parts of the cerebellum (L, M). Smarcb1 is particularly lost in Cre-positive cells (N–P), the overall number of NeuN-positive cells is severely decreased in Math1-cre::Smarcb1fl/fl mice, and Purkinje cells are randomly distributed within the cerebellar cortex as compared with controls (H, I, S, T). With respect to histomorphology (V–X and AA–CC) and expression of NeuN and Calbindin (Y, Z, DD, EE), Math1-cre::Smarca4fl/fl mice display changes that are similar, if not identical, to Math1-cre::Smarcb1fl/fl mice. oEGL, Outer external granular layer; iEGL, inner external granular layer; ML, molecular layer; PCL, Purkinje cell layer; IGL, inner granular layer. Scale bars: A, K, 250 μm; B–E, L, M, 50 μm; F, Q, 500 μm; N–P, 10 μm; G–J, R–U, W–Z, BB, CC, DD, EE, 100 μm; V, AA,1 mm.
Figure 3.
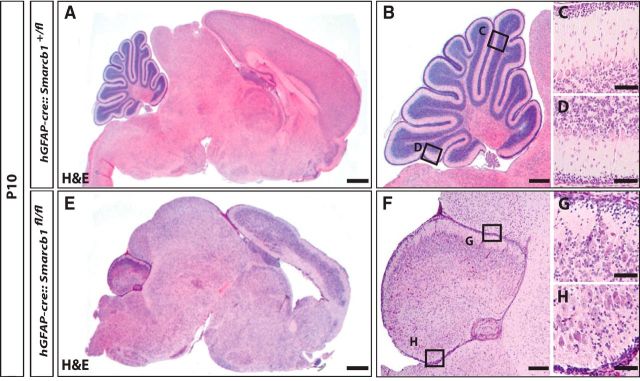
Loss of Smarcb1 in hGFAP-positive precursor cells similarly results in hypoplastic cerebella, but not in brain tumor formation. Cerebellar growth in hGFAP-cre::Smarcb1fl/fl was dramatically decreased compared with controls (H&E stains of sagitally cut brains; A, B vs E, F) with lack of foliation and cortical lamination (C, D vs G, H). Moreover, we detected severe lamination disturbances and a thinning of the cerebral cortex (A vs E), indicating that Smarcb1 is similarly important for the development of the forebrain. Scale bars: A, E, 2 mm; B, 500 μm; F, 300 μm; C, D, G, H, 100 μm.
Results
Loss of Smarcb1 or SmarcA4 in cerebellar granule neurons restricts their number and causes a severely hypoplastic cerebellum
To investigate the function of Smarcb1 in cerebellar development and to potentially identify cerebellar granule neurons as the cellular origin for AT/RTs within the cerebellum, we generated Math1-cre::Smarcb1fl/fl mice that are characterized by a deletion of Smarcb1 under the control of Math1 promoter sequences. Math1-cre::Smarcb1fl/fl mice were born with the expected Mendelian ratio, were fertile, and showed normal survival with 10/10 mice being alive after >6 months. No difference was observed between control mice (Math1-cre, Math1-cre::Smarcb1fl/+, Smarcb1fl/fl, or Smarcb1fll+) and Math1-cre::Smarcb1fl/fl mice concerning body weight, head size, or general condition as analyzed at postnatal day 0 (P0), P7, and P21 (data not shown). However, Smarcb1-deficient mice started to display severe signs of motor deficits and ataxia by day 14. To gain insights into the reasons for the observed motor deficits and ataxia, we investigated the brains by histomorphology and detected severe cerebellar hypoplasia using H&E stains of sagittally cut P7 and P20 cerebella (Fig. 1, A,F vs K,Q). As expected, Cre and Smarcb1 was strongly expressed in the cell nuclei of Math1-cre::Smarcb1fl/+ mice (Fig. 1B–E). Math1-cre::Smarcb1fl/fl mice similarly expressed Cre recombinase (Fig. 1N), but displayed a specific loss of Smarcb1 in Cre-positive granule cell precursors with a particular focus on rostral parts of the cerebellum (Fig. 1L–P), where Math1 promoter sequences are known to be strongly active (Schüller et al., 2008). As seen in high-power magnifications of H&E stains, the reduction of cerebellar size was explained by a dramatic reduction of cerebellar granule neuron precursors and mature granule neurons, especially in the rostral parts of the cerebellum. Moreover, Math1-cre::Smarcb1fl/fl mice did not display adequate cortical layering in these areas (Fig. 1, G,J vs R,U). Still, existing granule neurons of Math1-cre::Smarcb1fl/fl mice showed neuronal differentiation, as indicated by expression of NeuN, but a regular gradient from undifferentiated granule neuron precursors underneath the surface to fully differentiated granule cells in the internal granule cell layer was not detectable (Fig. 1, H vs S). Calbindin-positive Purkinje cells were still detected in Math1-cre::Smarcb1fl/fl mice, but were located in a disordered manner and without the typical dendritic tree (Fig. 1, I vs T).
Mutations in SMARCA4 are, albeit rare, also detectable in human AT/RTs, and we questioned whether a deletion of Smarca4 in cerebellar granule cell precursors resulted in tumor formation. As shown in Figure 1, V–EE, this was not the case, and Math1-cre::Smarca4fl/fl mice displayed a phenotype that was very similar to the changes seen in Math1-cre::Smarcb1fl/fl mice, both regarding histomorphological features as well as expression of NeuN and Calbindin.
Loss of Smarcb1 leads to severe proliferation deficits of granule neuron precursors and activation of Wnt signaling
To investigate why numbers of granule neuron precursors were severely diminished in Math1-cre::Smarcb1fl/fl mice, we analyzed the effects of Smarcb1 loss in granule neuron precursors in more detail using primary cell culture. Cultured cerebellar granule cell precursors of Smarcb1fl/fl were transduced either with the control viruses harboring IRES-GFP sequences or with Cre-IRES-GFP viruses to recombine the Smarcb1 allele in Smarcb1fl/fl cells. Twenty-four hours after transduction, cells were FACS-sorted and real time RT-PCR confirmed a strong reduction of Smarcb1 in the Cre-treated cells (Fig. 2A, p = 0.002). In parallel to the RT-PCR analyses, GFP-positive granule neuron precursors of the different conditions were recultured at equal cell numbers. Cell counts were then measured daily from day 2 until day 6 using a Neubauer chamber and an automated cell counter, TC10 (Bio-Rad). While the cell number of Smarcb1fl/fl cells treated with GFP viruses was highly increased after 6 d (12-fold on day 6 compared with day 2), Cre-treated Smarcb1fl/fl granule neuron precursors were significantly less increasing in numbers (Fig. 2B, p = 0.01). Hence, loss of Smarcb1 resulted in severely decreased proliferation of granule neuron precursors, explaining the hypoplastic cerebellum observed in vivo. As the phenotype caused by loss of Smarcb1 with a block of granule cell precursor proliferation and a severe hypoplastic cerebellum is similar to the cerebellar phenotype caused by constitutive activation of β-catenin (Lorenz et al., 2011; Pöschl et al., 2013), we questioned whether Smarcb1 deficiency resulted in deregulated Wnt/β-catenin signaling. Indeed, the expression of known Wnt-signaling target genes was significantly increased in Cre-IRES-GFP-treated Smarcb1fl/fl cells when compared with IRES-GFP-transduced Smarcb1fl/fl cells (Tcf4: p = 0.0097; Lef1: p = 0.0048; Dkk1: p = 0.043; Axin2: p = 0.0038; Fig. 2C). In contrast, the expression of Ink4a, Cyclin D1 and target genes of the Sonic hedgehog (SHH) signaling pathway (Gli1, Gli2, Gli3) was unchanged after the loss of Smarcb1. In summary, loss of Smarcb1 resulted in an upregulation of Wnt/β-Catenin target genes, which is compatible with a scenario where Smarcb1 maintains proliferation of granule neuron precursors via suppression of Wnt/β-catenin signaling.
No brain tumors after widespread loss of Smarcb1 in hGFAP-positive neural precursors
To investigate whether the loss of Smarcb1 resulted in the formation of AT/RTs outside the Math1-positive compartment, we used hGFAP promoter sequences to conditionally delete Smarcb1. Among other regions outside the CNS, the hGFAP promoter is active in very early neural progenitor cells as well as in mature astroglia. When breeding hGFAP-cre::Smarcb1fl/+ mice to Smarcb1fl/fl mice, mice with a hGFAP-cre::Smarcb1fl/fl genotype were observed at a significantly reduced frequency, indicating prenatal death. If these mice survived birth, they were generally smaller than control mice and had a reduced hair coat as well as a significantly decreased body weight and nose-to-tail length. Kaplan–Meier survival curves revealed that the mean survival of hGFAP-cre::Smarcb1fl/fl was 14.4 d with a maximum lifetime of 16 d of the generated animals. Analogous to Math1-cre::Smarcb1fl/fl mice, hGFAP-cre::Smarcb1fl/fl mice displayed severe motor deficits and ataxia (data not shown). We next assessed the overall brain morphology of hGFAP-cre::Smarcb1fl/fl mice. Cerebellar growth was dramatically decreased compared with controls (Fig. 3, A,B vs E,F) with a lack of foliation and cortical lamination (Fig. 3, C,D vs G,H). Moreover, we detected severe lamination disturbances and a thinning of the cerebral cortex (Fig. 1, A vs E), indicating that Smarcb1 is similarly important for the development of the forebrain. Interestingly, despite using the broadly active hGFAP-promoter, we did not detect any tumor formation within the brain up to 16 postnatal days. Hence, hGFAP-positive precursor cells and Math1-positive precursor cells are unlikely to serve as cells of origin for AT/RTs.
Discussion
SMARCA4 (BRG1) and SMARCB1 (INI1) are tumor suppressor genes that are crucially involved in the formation of malignant rhabdoid tumors. Specifically, loss-of-function mutations of these genes are detectable in the vast majority of AT/RTs, a subgroup of rhabdoid tumors occurring in the CNS. In the CNS, these tumors are particularly frequent in the cerebellum. In our study, we analyzed the role of Smarcb1 and Smarca4 during cerebellar development. Specific deletion of Smarca4 and Smarcb1 in cerebellar granule cell precursors, the most abundant cell type in the cerebellum, resulted in a significant inhibition of granule neuron precursor proliferation, but no tumor formation within the cerebellum. In general, granule neuron progenitors are known to be susceptible to oncogenic transformation as they can give rise to Sonic hedgehog-associated medulloblastoma (Schüller et al., 2008), a malignant brain tumor that shows histological similarities to AT/RTs. However, our results argue against cerebellar granule cells as the cellular origin for AT/RTs. We also did not detect tumor formation in hGFAP-cre::Smarcb1fl/fl mice, suggesting that hGFAP-positive progenitor cells are also unlikely to serve as a cell of origin of AT/RTs. Interestingly, SMARCB1 and SMARCA4 mutations are not only detectable in AT/RTs, but also in a small subset of medulloblastoma (Parsons et al., 2011; Pugh et al., 2012; Kool et al., 2014). In particular, a SMARCB1 N371Y mutation was identified in an adult SHH medulloblastoma (Kool et al., 2014), a tumor that is strongly believed to arise from Math1-positive granule cell precursors of the cerebellum (Schüller et al., 2008; Pöschl et al., 2014a). In this context, it appears worth mentioning that we not only failed to detect AT/RTs, but that a deletion of Smarcb1 or Smarca4 in mice is not sufficient to drive medulloblastoma tumorigenesis. One may speculate that either the timing or the type of mutation may not be modeled well enough by our systems or, alternatively, that a Smarcb1 or Smarca4 mutation alone is not sufficient to drive medulloblastoma from cerebellar granule neurons.
We show here that loss of Smarcb1 and Smarca4 leads to severe proliferation deficits of granule neuron precursors and a hypoplastic cerebellum. Hence, Smarcb1 and Smarca4 have pro-proliferative roles in cerebellar granule neuron precursors. Similar pro-proliferative functions of SMARCA4 have been detected in embryonic stem cells (Ho et al., 2011). In another study, Smarcb1inv/inv mice died due to severe bone marrow failure, which might be linked to a failure of hematopoietic stem cell proliferation (Roberts et al., 2002). Together, these results show that SMARCB1 and SMARCA4 clearly have diverse functions that may be cell-type and/or time-point-specific.
Apart from the involvement of Sonic hedgehog (Jagani et al., 2010) and Hippo signaling (Jeibmann et al., 2014), rhabdoid tumors have been demonstrated to display altered Wnt signaling (Mora-Blanco et al., 2014). Our results from granule cell precursors suggest that Smarcb1 leads to a deregulation of Wnt/β-catenin signaling, which appears context-independent. In contrast, a loss of Smarca4, which had effects comparable to those caused by a loss of Smarcb1, had previously been shown to inhibit the mitogenic effects of Sonic hedgehog signaling in normal cerebellar granule cells precursors (Zhan et al., 2011). At the same time, Wnt signaling has been described to impair the proliferation of granule neuron precursors and to counteract the formation of Shh-associated medulloblastoma (Pöschl et al., 2014b). Hence, our data are compatible with a scenario where a loss of Smarc proteins inhibits Shh-driven proliferation of cerebellar granule cell precursors via upregulation of Wnt-signaling.
Footnotes
This work was supported by the German Cancer Aid (Max-Eder-Programm), the Else-Kröner-Fresenius-Stiftung, and the Wilhelm Sander Stiftung (all to U.S.); and the IMF Münster, the IZKF Münster, and the Sonja Wasowicz Foundation (all to K.K.). We are indebted to M. Schmidt for expert technical assistance. We thank Charles Roberts (Dana Faber Cancer Institute, Boston) for providing Smarcb1fl/fl mice and Pierre Chambon (IGBMC, Illkirch-Graffenstaden, France) for providing the Smarca4fl/fl mice.
The authors declare no conflict of interest.
References
- Hasselblatt M, Gesk S, Oyen F, Rossi S, Viscardi E, Giangaspero F, Giannini C, Judkins AR, Frühwald MC, Obser T, Schneppenheim R, Siebert R, Paulus W. Nonsense mutation and inactivation of SMARCA4 (BRG1) in an atypical teratoid/rhabdoid tumor showing retained SMARCB1 (INI1) expression. Am J Surg Pathol. 2011;35:933–935. doi: 10.1097/PAS.0b013e3182196a39. [DOI] [PubMed] [Google Scholar]
- Hasselblatt M, Isken S, Linge A, Eikmeier K, Jeibmann A, Oyen F, Nagel I, Richter J, Bartelheim K, Kordes U, Schneppenheim R, Frühwald M, Siebert R, Paulus W. High-resolution genomic analysis suggests the absence of recurrent genomic alterations other than SMARCB1 aberrations in atypical teratoid/rhabdoid tumors. Genes Chromosomes Cancer. 2013;52:185–190. doi: 10.1002/gcc.22018. [DOI] [PubMed] [Google Scholar]
- Ho L, Miller EL, Ronan JL, Ho WQ, Jothi R, Crabtree GR. esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nat Cell Biol. 2011;13:903–913. doi: 10.1038/ncb2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indra AK, Dupé V, Bornert JM, Messaddeq N, Yaniv M, Mark M, Chambon P, Metzger D. Temporally controlled targeted somatic mutagenesis in embryonic surface ectoderm and fetal epidermal keratinocytes unveils two distinct developmental functions of BRG1 in limb morphogenesis and skin barrier formation. Development. 2005;132:4533–4544. doi: 10.1242/dev.02019. [DOI] [PubMed] [Google Scholar]
- Jagani Z, Mora-Blanco EL, Sansam CG, McKenna ES, Wilson B, Chen D, Klekota J, Tamayo P, Nguyen PT, Tolstorukov M, Park PJ, Cho YJ, Hsiao K, Buonamici S, Pomeroy SL, Mesirov JP, Ruffner H, Bouwmeester T, Luchansky SJ, Murtie J, et al. Loss of the tumor suppressor Snf5 leads to aberrant activation of the Hedgehog-Gli pathway. Nat Med. 2010;16:1429–1433. doi: 10.1038/nm.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeibmann A, Eikmeier K, Linge A, Kool M, Koos B, Schulz J, Albrecht S, Bartelheim K, Frühwald MC, Pfister SM, Paulus W, Hasselblatt M. Identification of genes involved in the biology of atypical teratoid/rhabdoid tumors using Drosophila melanogaster. Nat Commun. 2014;5:4005. doi: 10.1038/ncomms5005. [DOI] [PubMed] [Google Scholar]
- Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J, Crabtree GR. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet. 2013;45:592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieran MW, Roberts CW, Chi SN, Ligon KL, Rich BE, Macconaill LE, Garraway LA, Biegel JA. Absence of oncogenic canonical pathway mutations in aggressive pediatric rhabdoid tumors. Pediatr Blood Cancer. 2012;59:1155–1157. doi: 10.1002/pbc.24315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klochendler-Yeivin A, Fiette L, Barra J, Muchardt C, Babinet C, Yaniv M. The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO Rep. 2000;1:500–506. doi: 10.1093/embo-reports/kvd129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kool M, Jones DT, Jäger N, Northcott PA, Pugh TJ, Hovestadt V, Piro RM, Esparza LA, Markant SL, Remke M, Milde T, Bourdeaut F, Ryzhova M, Sturm D, Pfaff E, Stark S, Hutter S, Seker-Cin H, Johann P, Bender S, et al. Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened-inhibition. Cancer Cell. 2014;25:393–405. doi: 10.1016/j.ccr.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RS, Stewart C, Carter SL, Ambrogio L, Cibulskis K, Sougnez C, Lawrence MS, Auclair D, Mora J, Golub TR, Biegel JA, Getz G, Roberts CW. A remarkably simple genome underlies highly malignant pediatric rhabdoid cancers. J Clin Invest. 2012;122:2983–2988. doi: 10.1172/JCI64400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz A, Deutschmann M, Ahlfeld J, Prix C, Koch A, Smits R, Fodde R, Kretzschmar HA, Schüller U. Severe alterations of cerebellar cortical development after constitutive activation of Wnt signaling in granule neuron precursors. Mol Cell Biol. 2011;31:3326–3338. doi: 10.1128/MCB.05718-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora-Blanco EL, Mishina Y, Tillman EJ, Cho YJ, Thom CS, Pomeroy SL, Shao W, Roberts CW. Activation of beta-catenin/TCF targets following loss of the tumor suppressor SNF5. Oncogene. 2014;33:933–938. doi: 10.1038/onc.2013.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW, Li M, Zhang X, Jones S, Leary RJ, Lin JC, Boca SM, Carter H, Samayoa J, Bettegowda C, Gallia GL, Jallo GI, Binder ZA, Nikolsky Y, Hartigan J, Smith DR, Gerhard DS, Fults DW, VandenBerg S, Berger MS, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331:435–439. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöschl J, Grammel D, Dorostkar MM, Kretzschmar HA, Schüller U. Constitutive activation of beta-catenin in neural progenitors results in disrupted proliferation and migration of neurons within the central nervous system. Dev Biol. 2013;374:319–332. doi: 10.1016/j.ydbio.2012.12.001. [DOI] [PubMed] [Google Scholar]
- Pöschl J, Stark S, Neumann P, Gröbner S, Kawauchi D, Jones DT, Northcott PA, Lichter P, Pfister SM, Kool M, Schüller U. Genomic and transcriptomic analyses match medulloblastoma mouse models to their human counterparts. Acta Neuropathol. 2014a;128:123–136. doi: 10.1007/s00401-014-1297-8. [DOI] [PubMed] [Google Scholar]
- Pöschl J, Bartels M, Ohli J, Bianchi E, Kuteykin-Teplyakov K, Grammel D, Ahlfeld J, Schüller U. Wnt/beta-catenin signaling inhibits the Shh pathway and impairs tumor growth in Shh-dependent medulloblastoma. Acta Neuropathol. 2014b;127:605–607. doi: 10.1007/s00401-014-1258-2. [DOI] [PubMed] [Google Scholar]
- Pugh TJ, Weeraratne SD, Archer TC, Pomeranz Krummel DA, Auclair D, Bochicchio J, Carneiro MO, Carter SL, Cibulskis K, Erlich RL, Greulich H, Lawrence MS, Lennon NJ, McKenna A, Meldrim J, Ramos AH, Ross MG, Russ C, Shefler E, Sivachenko A, et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488:106–110. doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CW, Leroux MM, Fleming MD, Orkin SH. Highly penetrant, rapid tumorigenesis through conditional inversion of the tumor suppressor gene Snf5. Cancer Cell. 2002;2:415–425. doi: 10.1016/S1535-6108(02)00185-X. [DOI] [PubMed] [Google Scholar]
- Schneppenheim R, Frühwald MC, Gesk S, Hasselblatt M, Jeibmann A, Kordes U, Kreuz M, Leuschner I, Martin Subero JI, Obser T, Oyen F, Vater I, Siebert R. Germline nonsense mutation and somatic inactivation of SMARCA4/BRG1 in a family with rhabdoid tumor predisposition syndrome. Am J Hum Genet. 2010;86:279–284. doi: 10.1016/j.ajhg.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüller U, Zhao Q, Godinho SA, Heine VM, Medema RH, Pellman D, Rowitch DH. Forkhead transcription factor FoxM1 regulates mitotic entry and prevents spindle defects in cerebellar granule neuron precursors. Mol Cell Biol. 2007;27:8259–8270. doi: 10.1128/MCB.00707-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüller U, Heine VM, Mao J, Kho AT, Dillon AK, Han YG, Huillard E, Sun T, Ligon AH, Qian Y, Ma Q, Alvarez-Buylla A, McMahon AP, Rowitch DH, Ligon KL. Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer Cell. 2008;14:123–134. doi: 10.1016/j.ccr.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sévenet N, Sheridan E, Amram D, Schneider P, Handgretinger R, Delattre O. Constitutional mutations of the hSNF5/INI1 gene predispose to a variety of cancers. Am J Hum Genet. 1999;65:1342–1348. doi: 10.1086/302639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versteege I, Sévenet N, Lange J, Rousseau-Merk MF, Ambros P, Handgretinger R, Aurias A, Delattre O. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–206. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- Zhan X, Shi X, Zhang Z, Chen Y, Wu JI. Dual role of Brg chromatin remodeling factor in Sonic hedgehog signaling during neural development. Proc Natl Acad Sci U S A. 2011;108:12758–12763. doi: 10.1073/pnas.1018510108. [DOI] [PMC free article] [PubMed] [Google Scholar]