Abstract
The clock network of Drosophila melanogaster expresses various neuropeptides, but a function in clock-mediated behavioral control was so far only found for the neuropeptide pigment dispersing factor (PDF). Here, we propose a role in the control of behavioral rhythms for the ion transport peptide (ITP), which is expressed in the fifth small ventral lateral neuron, one dorsal lateral neuron, and in only a few nonclock cells in the brain. Immunocytochemical analyses revealed that ITP, like PDF, is most probably released in a rhythmic manner at projection terminals in the dorsal protocerebrum. This rhythm continues under constant dark conditions, indicating that ITP release is clock controlled. ITP expression is reduced in the hypomorph mutant ClkAR, suggesting that ITP expression is regulated by CLOCK. Using a genetically encoded RNAi construct, we knocked down ITP in the two clock cells and found that these flies show reduced evening activity and increased nocturnal activity. Overexpression of ITP with two independent timeless-GAL4 lines completely disrupted behavioral rhythms, but only slightly dampened PER cycling in important pacemaker neurons, suggesting a role for ITP in clock output pathways rather than in the communication within the clock network. Simultaneous knockdown (KD) of ITP and PDF made the flies hyperactive and almost completely arrhythmic under constant conditions. Under light-dark conditions, the double-KD combined the behavioral characteristics of the single-KD flies. In addition, it reduced the flies' sleep. We conclude that ITP and PDF are the clock's main output signals that cooperate in controlling the flies' activity rhythms.
Keywords: circadian rhythms, Drosophila melanogaster, ITP, locomotor activity, neuropeptides, PDF
Introduction
The fruit fly Drosophila melanogaster has served as model organism for the investigation of biological rhythms for decades. The master clock in the central brain of the fly consists of ∼150 clock neurons, which can be divided into several subgroups: the small ventral lateral neurons (sLNv), fifth sLNv, large ventral lateral neurons (lLNv), dorsal lateral neurons (LNd), and lateral posterior neurons (LPN) in the lateral protocerebrum and the dorsal neurons DN1, DN2, and DN3 in the dorsal brain (Helfrich-Förster et al., 2007). These neurons are characterized by cell autonomous molecular oscillations of different clock proteins, which constitute the core clock mechanism (for review, see Peschel and Helfrich-Förster, 2011). The most prominent circadian output in the fly is the rhythm in daily locomotor activity, which consists of a morning (M) and an evening (E) activity bout. Previous studies showed that the M activity is mainly controlled by the sLNv, whereas the fifth sLNv and three of the LNd's constitute the E oscillator cells (Grima et al., 2004; Stoleru et al., 2004; Rieger et al., 2006; Picot et al., 2007; reviewed by Yoshii et al., 2012).
M and E oscillator cells express different neuropeptides that seem to be involved in communication pathways within the clock network, as well as in output signaling pathways. (for review, see Peschel and Helfrich-Förster, 2011). The neuropeptide pigment dispersing factor (PDF), which is expressed in the sLNv and lLNv, was shown to act as a synchronizing signal between different clock neurons (Shafer et al., 2008; Yoshii et al., 2009) and is important for the maintenance of rhythmicity in constant darkness (DD; Renn et al., 1999). In light-dark (LD) cycles, PDF was further shown to promote M activity, suggesting that it is the main output factor of the M oscillator cells (Renn et al., 1999; Shafer and Taghert, 2009).
The E oscillator cells are more heterogeneous with respect to their neuropeptide expression. Some contain the long form of neuropeptide F (NPF), others its short form (sNPF) and few neurons express the ion transport peptide (ITP) (Johard et al., 2009; Hermann et al., 2012). So far, only few clock-related functions of these neuropeptides have been demonstrated (Hermann et al., 2012; Damulewicz et al., 2013) and it is not clear which of them is the main output factor of the E cells to control rhythmic behavior. Here, we have investigated the role of ITP, which is expressed in the fifth sLNv and one LNd (Johard et al., 2009) and has so far received the most attention for its antidiuretic functions in the insect gut (Dircksen, 2009). Through RNA interference (RNAi) and overexpression experiments, we show for the first time that ITP participates in the control of locomotor rhythms. As part of the E oscillator neurons, ITP promotes E activity and acts as a weak period shortening component in DD. We further demonstrate that its clock-related functions may be mediated by rhythmic ITP release from the two clock cells into the pars intercerebralis (PI) and that this occurs at different times than the PDF release.
Materials and Methods
Fly stocks.
All fly stocks were reared on Drosophila food (0.8% agar, 2.2% sugar-beet syrup, 8.0% malt extract, 1.8% yeast, 1.0% soy flour, 8.0% corn flour, and 0.3% hydroxybenzoic acid) in a humidity-controlled climate chamber in an LD 12:12 cycle at 25°C. As wild-type, we used the laboratory strain Canton S (CS). w1118 was crossed to GAL4-and UAS-lines to obtain heterozygous control flies. We also used the mutants per01 and ClkAR (M. Rosbash, Brandeis University, Waltham, MA) and, for the RNAi experiments, w1118;UAS-dicer2;+;+ (#60012), w1118;+;UAS-itp-RNAi (#43848), and w1118;+;UAS-pdf-RNAi (#4380), which were all obtained from the Vienna Drosophila RNAi Center (VDRC). The used driver lines were the following: yw;+;pdf-GAL4, w;tim-GAL4/CyO and yw;per-GAL4 (all from J. C. Hall and M Kaneko, Brandeis University, Waltham, MA), w;tim(UAS)-GAL4 (M. W. Young, Rockefeller University, New York, NY), w;clk856-GAL4 (O. T. Shafer, University of Michigan, Ann Arbor, MI; Gummadova et al., 2009), w;cry-GAL4#39 (F. Rouyer, CNRS, Gif-sur-Yvette, France), w;elav-GAL4/CyO (Bloomington Stock Center, #8765), and 386y(amon)-GAL4 (C. Wegener, University of Würzburg, Würzburg, Germany).
Generation of UAS-ITP flies.
RNA was extracted from D. melanogaster CS heads and was subsequently reversely transcribed into cDNA. The cDNA of the short ITP isoform (ITP-PE; DrmITP in Dircksen et al., 2008) was then amplified in its full length using a primer pair, which created EcoRI and XbaI restriction sites. Forward primer from 5′ to 3′: ACG-AAT-TCG-TTT-CTG-CCC-CAC-AAC-AAC-AC; reverse primer from 5′ to 3′: TCC-TCT-AGA-ATC-GCA-CTT-TAC-TTG-CGA-CC. The amplicon was ligated into the EcoRI-XbaI-digested pUAST vector (containing genes encoding ampicillin resistance and mini-white; kindly donated by A. Fiala, University of Göttingen, Göttingen, Germany) and NEB 10-β competent E. coli bacteria (New England BioLabs) were used for transformation with the ITP-pUAST vector. Positive clones were selected on ampicillin-containing agar plates and one clone was chosen for vector amplification, sequencing, and injection into w1118 flies by BestGene Drosophila Embryo Injection Services. Ten different red-eyed UAS-ITP lines, in which the construct was inserted either on the second or on the third chromosome, were obtained.
Antibodies and immunocytochemistry.
Immunohistochemical analysis was performed to investigate the ITP expression pattern in the brain of wild-type and overexpressing flies to confirm RNAi efficiency and to quantify clock protein cycling and ITP staining intensity.
The monoclonal mouse anti-PDF-C7 antibody was purchased from the Developmental Studies Hybridoma Bank (DSHB) at the University of Iowa (investigator: Justin Blau, New York University, New York, NY). To counterstain all clock neurons, we used a polyclonal guinea pig antiserum against the clock protein Vrille (anti-VRI), which was described by Glossop et al. (2003) and kindly provided by Paul E. Hardin (Texas A&M University, College Station, TX). For the quantification of the PERIOD (PER) protein cycling, we used a polyclonal rabbit anti-PER antibody (Stanewsky et al., 1997), which was a gift from R. Stanewsky (University College London, London, UK).
The polyclonal rabbit anti-ITP antibody was commercially generated against the C-terminal fragment of D. melanogaster ITP, CEMDKYNEWRDTL-NH2, coupled to bovine thyroglobulin via maleimide-coupling methodology. Rabbits were repeatedly injected subcutaneously and were terminally bled after 110 d. Immunocytochemistry, antisera titrations, and analyses of specificity were performed as described previously (Dircksen et al. (2008)); that is, via dilution series, preabsorption controls, Western blots, and combined HPLC-ELISA analysis.
The staining protocol for D. melanogaster adult whole-mount brains has been described previously (Hermann et al., 2012, 2013). We used only male 3- to 5-d-old flies, which were entrained for at least 4 d in LD 12:12, before they were collected at various zeitgeber times (ZTs) in LD or circadian times (CTs) on the first or third day in DD. Brains were embedded and confocal images were obtained using a Leica TCS SPE confocal microscope. Z-stack images were visualized and edited with the ImageJ distribution Fiji (http://fiji.sc/wiki/index.php/Fiji or http://rsb.info.nih.gov/ij/). Stacks were cropped and compiled as maximum projections. Brightness and contrast were adjusted, but no other manipulations were performed on the images if not explicitly stated otherwise.
For intensity quantification, samples were processed in exactly the same way during the staining protocol and were scanned with identical laser settings. The quantifications were conducted in ImageJ (Fiji). For quantification of PER or ITP in cell bodies, a square-shaped area of 9 pixels (3 × 3 pixels) was placed on each cell of interest and the average pixel intensity was measured in the brightest focal plane. Cells of at least five different hemispheres were analyzed and the intensity values were first background corrected and then averaged for each neuronal group and genotypes. For quantification of ITP and PDF in the terminals, we compiled maximum projections containing the PI and the pars lateralis (PL) and removed all staining in addition to the ITP and PDF terminals in this area (see Fig. 2A,B). All resulting images were therefore of the exact same size and contained only a defined part of the staining in the dorsal terminals. We then set the background of each image to zero and measured the total intensity of the whole image, which then reflected the staining intensity in the dorsal projection terminals. These manipulations were done without knowing the ZT or CT at which the samples were taken to avoid any subjective influence of the investigator. We quantified at least 10 brains for each time point and ITP and PDF were analyzed in the same specimens.
Figure 2.
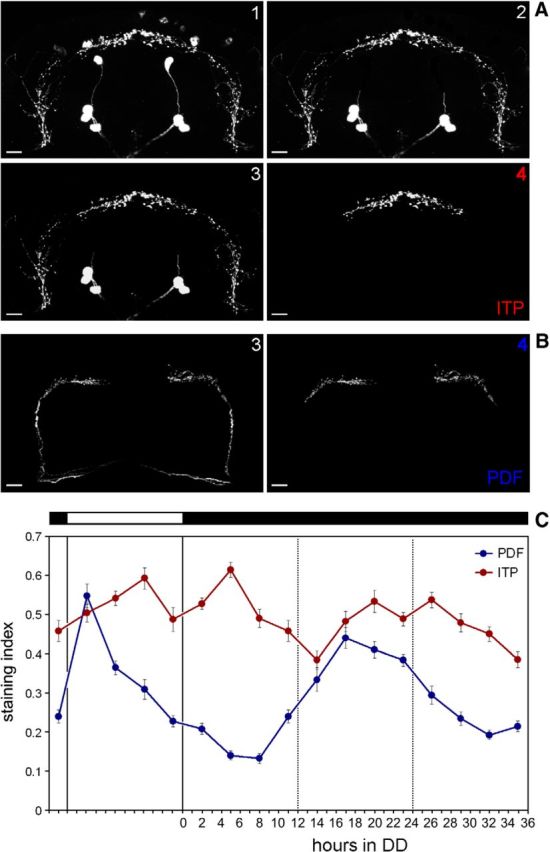
Exact manipulation of the pictures to determine staining intensity and ITP/PDF cycling in DD. A, Manipulation of the ITP fibers for quantification of staining intensity in the terminals is indicated in pictures 1–4. Picture 1 represents the original picture obtained from overlaying 20 confocal sections (of 2 μm thickness). In Picture 2, the cell bodies of the ipc-1, ipc-2, and ipc-4 neurons that were close to the ITP terminals in the PI were removed manually with the erase tool of Corel Photopaint. In few cases, single ipc-1 or ipc-4 neurons were located directly on the varicosities of the ITP terminals (left brain hemisphere). Their removal left a black spot in the varicosities, but because this happened at the same frequency in all samples, we assume that the overall results were not affected. Next (picture 3), the background was set to zero by adjusting brightness of the picture (in Corel Photopaint). In picture 4, the ITP fibers were cut so that only the ITP fibers in the median brain were left (see also Fig. 1C). This picture was used to calculate average pixel intensity of the entire picture. B, Manipulation of the PDF fibers for quantification. As for ITP 20, confocal sections were combined, the background was set to zero (picture 3), and finally the PDF fibers were cut at their characteristic first bend toward the median dorsal protocerebrum (picture 4). C, Quantification of ITP and PDF terminals for the first day under DD. For details, see text. n ≥ 10 per time point. Error bars indicate SEM. Scale bars, 10 μm.
Behavioral assay.
For analysis of daily locomotor rhythms, we used 3- to 5-d-old male flies, which were recorded using the commercially available Drosophila Activity Monitoring system by TriKinetics. The exact procedure was described in Hermann et al. (2012). Experiments were performed in light-proof boxes, which were equipped with a computer-controlled white light LED system. The whole setup was located in a climate chamber with controlled humidity and constant 20°C. Light intensity during light phases was set to 100 lux. We recorded the flies in LD 12:12 for 7 d, followed by at least 14 d of DD. Experimental genotypes were always recorded together with their respective control genotypes in the same box and at the same time.
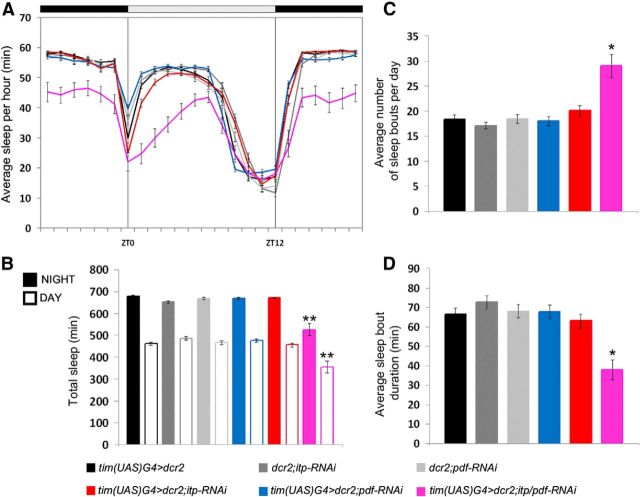
Analysis of LD behavioral data was performed using Microsoft Excel and the procedure of calculating normalized average activity profiles was described in Hermann et al. (2012). Free-running period lengths in DD were determined using χ2-periodogram analysis and actograms were depicted using ElTemps (Diez-Noguera, http://www.el-temps.com; upper limit 5) and the ImageJ plugin ActogramJ (Schmid et al., 2011). Average activity levels were calculated from mean activity counts of single flies during daytime (ZT0 to ZT12) or nighttime (ZT12 to ZT24) relative to the average of activity counts over the whole day. We further calculated the average number of beam crosses during the evening (ZT06 to ZT18) relative to the average activity during the morning (ZT18 to ZT06). Sleep amount was defined as the sum of time in which the flies did not cross the infrared light beam within 5 consecutive minutes. We calculated average sleep profiles in 1 h bins over the whole day, quantified total sleep during the light phase and the dark phase, and calculated the number of sleep bouts per day and the average duration of one sleep bout.
Statistics.
Data were tested for normal distribution applying a one-sample Kolmogorov–Smirnov test. To test for significant differences in normally distributed datasets, we then applied a one-way ANOVA followed by a post hoc pairwise comparison with Bonferroni's correction. Data that were not distributed normally were tested for significant differences with a Kruskal–Wallis test followed by pairwise comparison with Wilcoxon analysis. Percentages of rhythmicity were compared by a χ2 test. Data were considered as significantly different at *p < 0.05 and as highly significant at **p < 0.001. Significances are either indicated by asterisks or by a letter code within all graphical charts.
Results
ITP peptide levels cycle in dorsal projection terminals
ITP is expressed only in few brain neurons in the adult fly. The whole pattern was described in detail in Dircksen et al. (2008) and the original nomenclature of ITP-positive (ITP+) cells was mostly adopted into this work. We will, however, refer to the two ITP+ clock neurons as fifth sLNv and LNd, which were originally included in the ipc-3 neuronal group (Fig. 1A). According to this partly new nomenclature, the ITP+ cells in the brain can be divided into five groups: the two clock cells in the lateral brain, the ipc-1 in a posterior dorsal or medial position, the ipc-2 and the remaining ipc-3 cell(s) in the dorsal medial brain, and the ipc-4 in the dorsal central brain (Fig. 1A).
Figure 1.
ITP staining intensity in clock neuron cell bodies and projection terminals in LD 12:12. A, Anti-ITP staining on male CS brains at different ZTs in LD 12:12 (20°C). B, Quantification of the ITP staining intensity at different ZTs in the fifth sLNv (top) and the LNd (bottom). We found no significant oscillation in staining intensity in the ITP+ cell bodies. (n = 12 per time point), C, Terminals of the ITP and PDF clock neurons in the dorsal protocerebrum. The ipc-1, ipc-2, and ipc-3 neurons were removed for better clarity (see Fig. 2A for the detailed methodology). The two ITP neurons (LNd and fifth sLNv, magenta) terminate predominantly in the PI, whereas the PDF-expressing sLNv (blue) terminate in the PL. The PDF terminals were maximally stained at ZT2 and the ITP terminals at ZT20. Only staining intensity of the terminals in between the yellow bars was quantified, as indicated for PDF in the top and for ITP in the bottom picture. D, ITP and PDF staining intensities in the terminals depicted in C (n ≥ 10 per time point). PDF staining intensity significantly peaks at ZT2, decreases during the rest of the light phase, and remains low during the night. Quantification of the ITP staining intensity revealed two statistically significant peaks: one around noon and one around midnight. The troughs occurred at the time of lights-on and lights-off. Error bars indicate SEM; lowercase letters indicate significant differences between time points (different letters indicate p < 0.05); black and white bars indicate light regime. Scale bars, 10 μm.
To investigate ITP peptide levels over the day, we immunostained brains of adult male CS flies every 4 h in LD 12:12 with anti-ITP (Fig. 1A) and quantified the staining intensity in the cell bodies of the fifth sLNv and in the LNd (Fig. 1B). We did not see a significant cycling in staining intensity of the two cell bodies (fifth sLNv ANOVA F(6,65) = 0.685; p = 0.663; LNd ANOVA F(6,65) = 0.484; p = 0.818). Because the amount of PDF is also not cycling within the PDF+ cell bodies, but rather within the axon terminals in the dorsal protocerebrum, indicating a rhythm in peptide release (Park et al., 2000), we pursued similar investigations concerning ITP. Male CS brains were immunostained every 3 h in LD 12:12 and ITP staining intensity was quantified in the clock neuron terminals in the dorsal protocerebrum, which are close to the PI (Fig. 1C). We costained the same brains with anti-PDF and also quantified the PDF staining in the dorsal projection terminals of the sLNv. Consistent with Park et al. (2000), PDF immunostaining peaked at the beginning of the light phase, decreased during the rest of the day, and was quite low during the night (ANOVA F(7,111) = 25.64; p < 0.0001; Fig. 1D). ITP immunostaining in the projection terminals also showed significant differences during the LD cycle (ANOVA F(7,111) = 8,86; p < 0.0001; Fig. 1D). The quantification revealed a peak in the middle of the light phase and a second peak around ZT20 during the night. Staining levels were minimal at around lights-on and lights-off. A decrease of immunostaining in the projection terminals may indicate a loss of peptide that is possibly mediated by peptide release from large dense core vesicles. Therefore, our results suggest that PDF is released during the light phase, whereas ITP might be released in the end of the dark phase and the end of the light phase.
ITP levels in the dorsal brain terminals continue to cycle during the first day in constant darkness
The next question was whether ITP cycling is circadian and thus controlled by the clock. To answer this question, we repeated the above described staining experiment, but this time extended it into DD (Fig. 2C). In this second experiment, staining intensity was slightly higher than in the first experiment, and this was especially true for PDF. Therefore, the PDF peak staining index reached the ITP staining index during the day (Fig. 2C), but otherwise the staining curves of PDF and ITP were similar to the first experiment, with ITP exhibiting two staining peaks and PDF only one. As expected, we found PDF cycling to continue into DD (ANOVA F(7,89) = 19,21; p < 0.0001). PDF levels had already increased before the beginning of the first subjective day and reached the maximum in its first half. The PDF peak was considerably broader than under LD conditions, but PDF levels dropped continuously in the second half of the subjective day reaching the next minimum in the following subjective night. ITP also continued to cycle into DD (ANOVA F(7,89) = 7,05; p < 0.0001). The two peaks in ITP staining intensity were still marginally visible under DD conditions, but they moved much closer together (Fig. 2C). The first ITP peak occurred in the second half of the subjective day, as it did under LD conditions, but the second peak had already occurred at the beginning of the subjective night instead of in the middle of the night as under LD conditions. These changes in staining pattern may correspond to changes in the activity pattern under DD conditions: already during the first day in DD, M and E peaks come closer together often merging into one activity bout (see Fig. 6).
Figure 6.
Representative individual double-plotted actograms of ITP-KD, PDF-KD, and ITP/PDF-KD flies and controls in LD 12:12 followed by DD. ITP-KD flies (tim(UAS)G4>dcr2;itp-RNAi) (B) have significantly longer free-running periods in DD compared with the relevant controls (A). Many of the PDF-KD flies (tim(UAS)G4>dcr2;pdf-RNAi) were arrhythmic in DD (D). The still rhythmic individuals free ran with a short period (B) that was significantly different from the relevant controls (A). The majority of the ITP/PDF-double-KD flies (tim(UAS)G4>dcr2;itp-RNAi/pdf-RNAi) were arrhythmic (C, right actogram, D). The remaining flies showed several free-running components in DD (C, left actogram), the period of which was impossible to determine (F). Furthermore, all ITP/PDF-double-KD flies had a high activity level that was significantly different from all other genotypes (E). Black and white bars indicate the light regime in LD 12:12 (100 lux, 20°C). The control strains in D and E (dark gray bars) are in the following order from left to right: tim(UAS)G4>dcr2, dc2;itp-RNAi, and dcr2;pdf-RNAi. Error bars indicate SEM. Lowercase letters indicate statistical significances (different letters indicate p < 0.05). F, Characteristic periodograms for PDF-KD and ITP/PDF-KD flies. When only PDF was knocked down (top left periodogram), a significant period around 23 h was visible. In the ITP/PDF-double-KD flies, either no significant period was detectable (bottom left periodogram) or multiple (>3) periods appeared (right periodogram). The top right periodogram stems from the left actogram shown in C. A–C and F show individual actograms and periodograms, respectively. n ≥ 21 in D and E.
ITP levels are reduced in clock neurons of the hypomorph ClkAR mutants
After we showed that ITP levels are clock controlled, we wanted to know whether the clock genes influence overall ITP expression in the clock neurons. To answer this, we analyzed ITP staining intensity in the clock cell bodies in different clock-impaired mutants (Fig. 3A). We found that there is no difference in staining intensity in per01 flies compared with wild-type CS (Fig. 3B). However, per01 is thought to retain residual clock function because only one of the two molecular feedback loops is impaired (Helfrich and Engelmann, 1987; Helfrich-Förster, 2001; Kempinger et al., 2009; Goda et al., 2011; Bywalez et al., 2012; Menegazzi et al., 2012; Vanin et al., 2012). In ClkJrk mutants, clock function seems to be completely abolished (Allada et al., 1998); however, in addition to its deficits in clock functionality, ClkJrk flies show strong developmental defects that also affect the presence of certain clock neurons (Park et al., 2000). We therefore decided to investigate ITP staining in the hypomorph clock mutant ClkAR. Interestingly, we found that the ITP staining intensity in the two clock cells was significantly reduced compared with wild-type and per01 (fifth sLNv ANOVA F(2,31) = 30,469; p < 0.001; LNd ANOVA F(2,33) = 37,900; p < 0.001), suggesting that ITP expression is under CLK regulation (Fig. 3B; note that both ITP+ clock cells are only faintly stained, but clearly present in ClkAR; Fig. 3A). When searching through the upstream region of the itp gene, we did not find any indications for the presence of E-boxes, indicating that ITP abundance is probably regulated indirectly by CLK.
Figure 3.
ITP staining intensity in clock neuron cell bodies in CS compared with the clock mutants per01 and ClkAR. A, Anti-ITP staining on male adult brains of CS, per01, and ClkAR at ZT02 in LD 12:12. B, Quantification of the ITP staining intensity in the fifth sLNv (left) and the LNd (right) in the different genotypes (n = 12). Anti-ITP staining intensity was significantly reduced in both cells in ClkAR mutants compared with CS and per01. Error bars indicate SEM. Scale bars, 10 μm. **p < 0.001 in pairwise comparisons.
ITP knock-down affects LD locomotor activity, especially the activity level during the night and during the evening
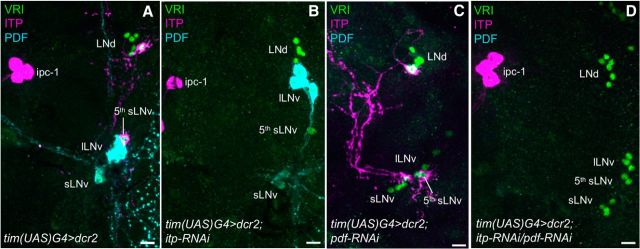
To investigate the function of ITP for locomotor rhythms in the fly, we expressed a genetically encoded itp-RNAi construct with the help of the GAL4/UAS system. We chose the very strong tim(UAS)-GAL4 line (tim(UAS)G4; described in Blau and Young, 1999) to express both UAS-dicer2 (dcr2) and the RNAi-construct, to knock down ITP (itp-RNAi) only in the ITP+ clock cells (ITP-KD). The tim(UAS)-GAL4 line is not entirely specific to the clock network, but does not include ITP+ nonclock cells (data not shown). Therefore, ITP levels should not be affected in these cells, but only in the two clock neurons per hemisphere. We used the same driver line to also manipulate PDF levels via pdf-RNAi, as described previously (Shafer and Taghert, 2009; Hermann et al., 2012; PDF-KD). To verify the RNAi efficiency, we stained adult male brains of the respective genotypes with anti-ITP and anti-PDF and counterstained them with anti-VRI (Fig. 4). PDF and ITP staining was wild-type-like in tim(UAS)G4>dcr2 control flies (Fig. 4A) and in heterozygous RNAi-construct controls (dcr2;itp-RNAi and dcr2;pdf-RNAi; data not shown). ITP was, however, undetectable in both clock neurons in ITP-KD flies in all investigated specimens, but remained present in the ITP+ nonclock cells (Fig. 4B). PDF immunostaining was also completely lost in a consistent way when pdf-RNAi was expressed in the clock neurons (Fig. 4C). When itp-RNAi and pdf-RNAi were expressed together, neither PDF nor ITP was detectable in the clock cells (ITP/PDF-double-KD; Fig. 4D). Therefore, both RNAi constructs worked very efficiently, even when expressed in conjuncture inside the clock neurons. It is worth mentioning that the complete KD of ITP in all ITP+ neurons is lethal (data not shown), whereas ITP-KD only in the clock neurons did not seem to affect viability.
Figure 4.
Immunohistochemistry on RNAi-expressing flies to validate RNAi efficiency. RNAi constructs were expressed with tim(UAS)G4. Repeatedly, at least 10 male adult brains of each genotype were stained with anti-ITP (magenta), anti-VRI (green), and anti-PDF (cyan). A, Control flies (tim(UAS)G4>dcr2) always showed wild-type-like expression pattern of ITP and PDF in the clock neurons. B, ITP was undetectable in the fifth sLNv and the LNd in ITP-KD flies (tim(UAS)G4>dcr2;itp-RNAi) in all investigated brains, whereas it was still present in the ITP+ nonclock neurons (ipc-1). C, PDF was consistently undetectable in sLNv and lLNv in PDF-KD flies (tim(UAS)G4>dcr2;pdf-RNAi). D, Both ITP and PDF were undetectable in the clock neurons in ITP/PDF-double-KD flies (tim(UAS)G4>dcr2;itp-RNAi/pdf-RNAi). Scale bars, 10 μm.
After ensuring that the RNAi was working efficiently, we tested the locomotor rhythms of ITP-KD flies and corresponding controls in LD 12:12 cycles. We calculated normalized average activity profiles to better depict the general shape of the daily activity pattern. The ITP-KD did not seem to have any severe effect on the shape of the bimodal activity profile (Fig. 5A). In particular, the phasing of the activity peaks seemed to be normal. We also recorded these flies under longer and shorter photoperiods because changes in phase of the M peak or E peak become more apparent when the activity peaks do not occur at the exact time of the light transitions (Majercak et al., 1999, Rieger et al., 2003). These experiments also did not reveal any impairment in activity peak timing (data not shown). However, the KD of ITP had effects on relative activity levels. We calculated daytime and nighttime activity as the average number of beam crosses during the light phase and the dark phase relative to the average activity during the whole day (Fig. 5B). We found that ITP-KD flies show a significantly reduced relative daytime activity (Kruskal–Wallis H(2) = 37.637; p < 0.001) and a significantly enhanced nighttime activity (Kruskal–Wallis H(2) = 37.637; p < 0.001). Furthermore, they seem to reduce E activity. When calculating the relation between average E activity (average beam crosses from ZT06 to ZT18) and average M activity (average beam crosses from ZT18 to ZT06), ITP-KD flies revealed a significant reduction in E activity relative to M activity (Kruskal–Wallis H(2) = 30.345; p < 0.001; Fig. 5C).
Figure 5.
Locomotor activity of ITP-KD flies and controls in LD 12:12. A, Average activity profiles were calculated for each genotype and light condition and were normalized to the highest activity value to better visualize the shape of the profile. No obvious differences in the shape of the bimodal activity pattern of ITP-KD flies (tim(UAS)G4>dcr2;itp-RNAi) were visible compared with controls. n = number of investigated flies; black areas indicate darkness, gray areas indicate light of 100 lux; black line = mean, gray lines = SEM. B, Relative activity levels for day (left) or night (right) were calculated as mean beam crosses per minute during the light phase or the dark phase relative to the average of beam crosses during the whole day. ITP-KD flies (light gray) showed significantly less daytime activity compared with both controls (darker grays) and significantly higher nighttime activity. C, When calculating mean E activity (ZT06 to ZT18) relative to mean M activity (ZT18 to ZT06), ITP-KD flies show a reduction in relative E amplitude compared with both controls. T = 20°C. Error bars indicate SEM. *p < 0.05 and **p < 0.001 in pairwise comparisons. n.s., Not significant. n = 32 for tim(UAS)G4>dcr2, n = 31 for dcr2;itp-RNAi, n = 32 for tim(UAS)G4>dcr2;itp-RNAi in all panels.
ITP KD slightly prolongs the free-running period in DD
To determine the effect of the ITP-KD on the free-running rhythm, we recorded ITP-KD flies together with their respective controls in LD 12:12 cycles, followed by at least 2 weeks of DD. The ITP-KD did not affect general rhythmicity of the flies, but slightly lengthened period (Kruskal–Wallis H(2) = 15.447; p < 0.001; pairwise comparisons: ITP-KD to timG4>dcr2 p < 0.001; ITP-KD to dcr2;itp-RNAi p < 0.05; Fig. 6, Table 1). We conclude that the presence of ITP is not necessary for maintaining rhythmicity under DD, but that ITP has a slight period-shortening effect on the free-running period.
Table 1.
Rhythmicity data of ITP-KD flies, PDF-KD flies, and ITP/PDF-double-KD flies and controls in DD
| Genotype | Period in hours (SEM) (n rhythmic flies) | Power (SEM) | Rhythmic flies |
|---|---|---|---|
| tim(UAS)G4>dcr2 | 23.7 (0.05) (32) | 22.8 (0.68) | 100% |
| dcr2;itp-RNAi | 23.9 (0.05) (28) | 35.1 (2.00) | 100% |
| tim(UAS)G4>dcr2;itp-RNAi | 24.1 (0.08) (30)* | 34.1 (1.96) | 94% |
| dcr2;pdf-RNAi | 23.8 (0.06) (32) | 36.1 (2.29) | 100% |
| tim(UAS)G4>dcr2;pdf-RNAi | 23.4 (0.09) (19)* | 16.4 (0.44) | 59%** |
| tim(UAS)G4>dcr2;pdf-RNAi/itp-RNAi | – (–) (7) | – (–) | 33%** |
Percentage of rhythmicity and period lengths of ITP-KD flies and PDF-KD flies were statistically compared with the data for the two respective genetic controls.
*Significant differences in period length (for statistical values, see main text).
**Highly significant differences in the percentage of rhythmic flies.
Overexpression of ITP with timG4 impairs rhythmic behavior
Although the presence of ITP does not seem to be necessary for robust free-running rhythms, this does not exclude the possibility that ITP influences rhythmicity. High ectopic levels of PDF in the dorsal brain (close to its usual terminals) have been shown to disrupt the internal communication among the clock neurons, causing complex rhythms up to arrhythmic behavior (Helfrich-Förster et al., 2000 and Wülbeck et al., 2008). Therefore, we took a comparable approach to what was done for PDF and generated a UAS-ITP construct, which allowed the overexpression of ITP with different GAL4 (G4) lines. We chose several well characterized driver lines that are specific to the neuronal clock system, but also broadly expressing drivers. The overexpression success was verified by antibody staining with anti-ITP (Fig. 7).
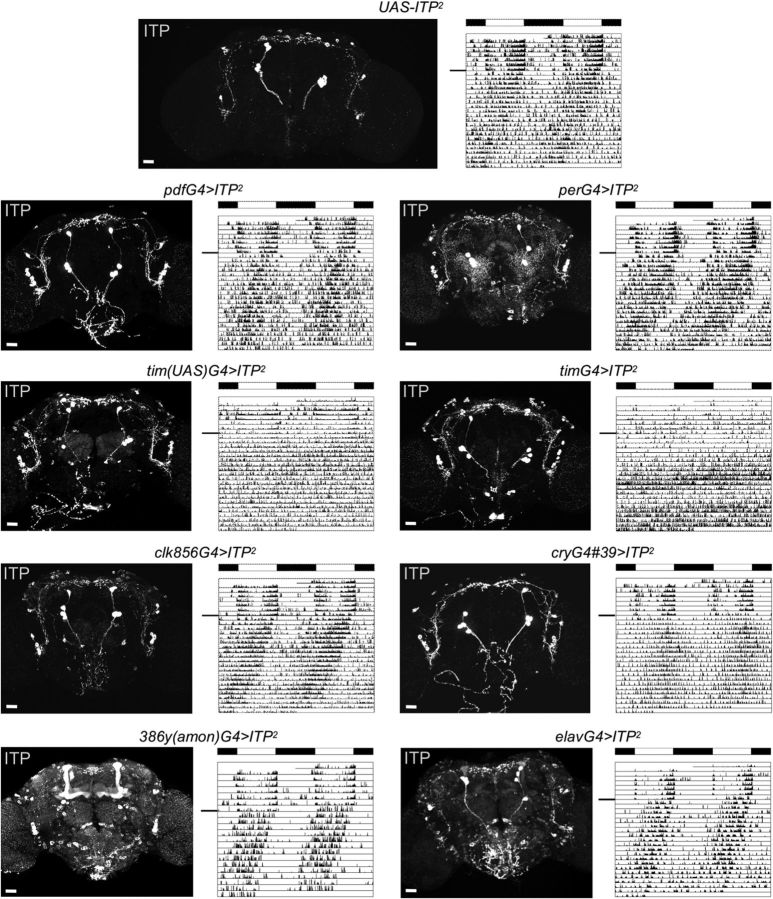
Figure 7.
Overexpression of ITP with different driver lines. Confocal pictures depict anti-ITP staining in heterozygous UAS-ITP2 controls (top) and ITP-overexpressing adult male brains. One individual representative double-plotted actogram is depicted for each genotype (black line indicates the transition from LD 12:12 to DD). Overexpression of ITP with tim(UAS)G4 and timG4 impaired rhythmicity, whereas overexpression with all other driver lines did not affect rhythmicity. T = 20°C; black and white bars indicate the light regime in LD 12:12 (100 lux). Scale bars, 10 μm.
In general, we were able to overexpress ITP ectopically with all driver lines that we used (Fig. 7, confocal images). Focusing on the clock neurons, we counterstained ITP-overexpressing brains with anti-VRI and anti-PDF (data not shown) and found that all clock neuron clusters were able to synthesize ITP. Overexpression with pdfG4, cryG4#39, and clk856G4 was rather specific to the neuronal clock network or a part of it (Fig. 7). Overexpression of ITP using tim(UAS)G4, timG4, or perG4 included not only clock neurons, but also structures like the antennal lobes, fan-shaped body, or ellipsoid body (Fig. 7). The very broad driver lines elav-GAL4 and 386y(amon)G4 showed even more, nearly pan-neuronal overexpression of ITP (Fig. 7).
Interestingly, when we compared the locomotor rhythms in LD 12:12 and DD in the different ITP-overexpressing genotypes, we only found differences in control flies using tim(UAS)G4 and timG4. These phenotypes were severe and identical in both driver lines in that flies barely showed any M and E activity bouts in LD and were almost completely arrhythmic in DD (tim(UAS)G4>ITP2 χ2 = 73,5097, p < 0.0001; timG4>ITP2 χ2 = 56,7964, p < 0.0001; Fig. 7, Table 2). Overexpression with none of the other drivers had any effect on rhythmicity or period length.
Table 2.
Rhythmicity data of ITP-overexpressing flies and controls in DD
| Genotype | Period in hours (SEM) (n rhythmic flies) | Power (SEM) | Rhythmic flies |
|---|---|---|---|
| UAS-ITP2/+ (ITP2) | 23.4 (0.07) (30) | 29.6 (1.2) | 100% |
| tim(UAS)G4/+ | 24.1 (0.04) (30) | 39.9 (2.30) | 97% |
| tim(UAS)G4>ITP2 | 25.0 (0.25) (2) | 19.8 (2.16) | 7%** |
| timG4/+ | 24.6 (0.11) (23) | 22.2 (1.18) | 74% |
| timG4>ITP2 | – (–) (0) | – (–) | 0%** |
| pdfG4/+ | 24.4 (0.06) (30) | 31.7 (1.59) | 97% |
| pdfG4>ITP2 | 23.7 (0.10) (31) | 21.4 (0.70) | 97% |
| perG4 | 25.3 (0.16) (31) | 32.1 (2.03) | 97% |
| perG4>ITP2 | 23.7 (0.04) (29) | 29.1 (1.66) | 100% |
| clk856G4/+ | 23.8 (0.04) (32) | 41.0 (1.89) | 100% |
| clk856G4>ITP2 | 23.3 (0.04) (32) | 35.2 (1.82) | 100% |
| cryG439/+ | 25.2 (0.16) (24) | 22.9 (1.60) | 75% |
| cryG439>ITP2 | 23.9 (0.06) (26) | 21.5 (1.21) | 81% |
| elavG4/+ | 23.7 (0.06) (25) | 23.8 (1.73) | 83% |
| elavG4>ITP2 | 23.6 (0.06) (20) | 28.7 (2.53) | 100% |
| 386y(amon)G4/+ | 23.1 (0.77) (31) | 29.1 (2.04) | 97% |
| 386y(amon)G4>ITP2 | 23.5 (0.05) (27) | 26.7 (1.19) | 100% |
Percentage of rhythmicity and period lengths of ITP-overexpressing strains were always compared with the respective GAL4-control and UAS-ITP2-control.
**Highly significant differences (for statistical values, see main text).
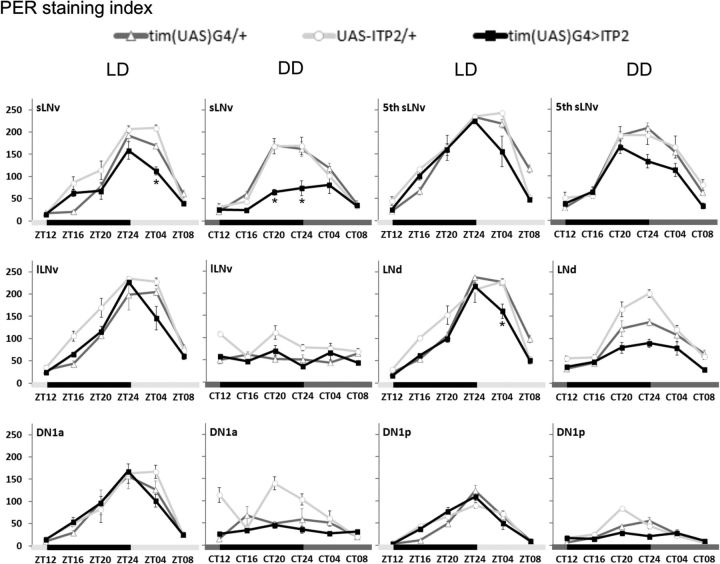
We first tested whether these arrhythmic phenotypes derive from a disruption of the molecular clock mechanism, possibly mediated by the direct action of ITP on the clock network. To do so, we immunostained brains of tim(UAS)G4>ITP2 flies and of the respective control genotypes with anti-PER every 4 h in LD and the third day in DD. In LD, the oscillation in PER staining intensity in most clock neuron clusters of tim(UAS)G4>ITP2 flies was not different from controls, neither were the maximum PER levels at ZT24 or ZT4 (ANOVA p > 0.05 from either one of the two controls), indicating that the PER protein cycling is normal in LD in ITP-overexpressing flies (Fig. 8). Only in the sLNv and LNd was PER amplitude slightly but significantly reduced (ANOVA p < 0.041), pointing to a weakened entrainment of these neurons. In DD, the amplitude of PER cycling was already reduced in some clock neurons of the control flies, but remained clearly cyclic in the sLNv, the fifth sLNv, and the LNd (sLNv: tim(UAS)G4 ANOVA F(5,26) = 31.079 p < 0.001, UAS-ITP2 ANOVA F(5,26) = 18.678 p < 0.001; fifth sLNv: tim(UAS)G4 ANOVA F(5,26) = 67.528 p < 0.001, UAS-ITP2 ANOVA F(5,26) = 14.002 p < 0.001; LNd: tim(UAS)G4 ANOVA F(5,26) = 17.647 p < 0.001, UAS-ITP2 ANOVA F(5,26) = 46.147 p < 0.001; Fig. 8). In tim(UAS)G4>ITP2 flies, we also found significant PER cycling in these three groups of clock neurons (sLNv: ANOVA F(5,25) = 4.438, p = 0.005; fifth sLNv: ANOVA F(5,25) = 19.582, p < 0.001; LNd: ANOVA F(5,25) = 9.040, p < 0.001), but the cycling amplitude (PER level at CT20 or CT24 or both) was largely decreased in the sLNv (ANOVA CT20 p < 0.005; CT24 p < 0.005) and decreased to the border of significance in the LNd (ANOVA CT24 p = 0.052) compared with controls (Fig. 8). Therefore, PER cycling in DD was not completely abolished in ITP-overexpressing flies, just dampened in its amplitude.
Figure 8.
PER staining intensity in clock neurons in LD 12:12 and DD in ITP-overexpressing flies and controls. Adult male brains were stained with anti-PER after entrainment to LD 12:12 (100 lux, 20°C). Flies were collected at different ZTs in LD 12:12 and at different CTs on the third day in DD. CTs indicate the time points when the light would have been on or off with respect to the previous LD cycle. Staining intensity in different clock neuron clusters was quantified in at least 5 brains per time point (n ≥ 5). PER cycling in behaviorally arrhythmic ITP-overexpressing flies (tim(UAS)G4>ITP2; black) was reduced in amplitude (PER levels at ZT04) in the sLNv and the LNd compared with control strains (light and dark gray) in LD. In DD, PER protein was still clearly cycling in sLNv, fifth sLNv, and LNd in both control flies and ITP-overexpressing flies. In the latter genotype, the amplitude of the oscillations (PER level at CT20 or CT24) was, however, again significantly reduced in the sLNv and the LNd, although the difference in the LNd was just above the significance level. Black and light gray bars indicate the LD light regime; black and dark gray bars indicate subjective night and day in DD. Error bars indicate SEM. *Significant differences (p < 0.05) in PER levels between the experimental strain and both controls at the respective time points.
Rhythm of ITP and PDF release seems affected in behaviorally arrhythmic ITP-overexpressing flies
Because clock protein cycling within the clock neurons was not completely impaired in behaviorally arrhythmic ITP-overexpressing flies, we assume that ITP may mainly act downstream of the clock on behavior-controlling target structures inside the brain.
To localize putative ITP targets that could possibly be responsible for the severe phenotype in ITP-overexpressing flies using timG4 and tim(UAS)G4, we compared the anti-ITP-staining pattern of these behaviorally arrhythmic flies with behaviorally rhythmic ITP-overexpressing flies. 386(amon)G4>ITP2 and elavG4>ITP2 flies showed high ITP expression virtually everywhere in the brain, but staining was especially high in the mushroom bodies and in the subesophageal ganglion (Fig. 7). perG4>ITP2 flies showed quite high ITP staining in the central complex and the antennal lobes. Nevertheless, all of these lines remained rhythmic, indicating that ITP does not evoke behavioral arrhythmicity by affecting these parts of the brain.
We then focused on the comparison of the arrhythmic tim(UAS)G4>ITP2 flies with the rhythmic perG4>ITP2 flies because these had a similar strong ITP expression in the clock neurons and especially in the PI projections, where we had discovered a daily rhythm in ITP staining. To investigate whether this rhythm is disturbed in the behaviorally arrhythmic flies but still present in the behaviorally rhythmic flies, we immunostained the two genotypes plus their relevant controls with anti-ITP and anti-PDF at ZT20 (when ITP levels had been high and PDF levels had been low in wild-type flies) and ZT02 (when ITP levels had been low, but PDF levels high in wild-type flies). We found that all control flies showed the expected significant differences in ITP and PDF staining intensity (Fig. 9A,B). The same was true for the perG4>ITP2 flies; as expected, these flies had very high ITP levels in the PI, but ITP-staining intensity was still cyclic (Fig. 9A). This was very different in tim(UAS)G4>ITP2 flies, in which we could not detect any significant difference in ITP staining at the two time points. ITP remained always similarly high, pointing to a constant release of ITP into the PI (Fig. 9A). We conclude that a constant release of ITP into the PI may disturb circadian rhythmicity, but as soon as the release cycles, behavioral rhythmicity is maintained even if ITP expression in the terminals is higher than normal, as we found in perG4>ITP2 flies. Interestingly, PDF cycling seemed also to be affected in tim(UAS)G4>ITP2 flies. The staining difference between ZT2 and ZT20 was smaller than in the other strains. PDF remained rather high in the middle of the night, when it was low in the controls (Fig. 9B). This may be partly caused by a changed projection pattern of the lLNv (Fig. 9C,D). In ∼60% of tim(UAS)G4>ITP2 flies, some fibers from the lLNv followed the projections of the s-LNv into the dorsal brain and terminated in the PI (χ2 = 25.55; p < 0.001; Fig. 9C). PDF in these lLNv terminals remained constantly high and may have led to a more or less constant PDF release that diminished the PDF rhythm from the sLNv. Furthermore, previous studies have shown that ectopic PDF fibers in this region provoked complex or arrhythmic activity rhythms in DD (Helfrich-Förster et al., 2000; Wülbeck et al., 2008). Putatively, the arrhythmicity of tim(UAS)G4>ITP2 flies is caused by a combination of constant ITP and PDF release into the dorsal brain.
Figure 9.

ITP and PDF cycling in the dorsal brain terminals in ITP-overexpression flies. A, tim(UAS)G4>ITP2 flies lack a significant difference in ITP staining at ZT2 and ZT20 in the PI. B, The difference in PDF-staining intensity between the two time points is reduced in tim(UAS)G4>ITP2 flies. C, tim(UAS)G4>ITP2 flies show a higher percentage of aberrant PDF-fibers in the PI than the other fly strains. D, Typical brain of a tim(UAS)G4>ITP2 fly stained with anti-PDF at ZT2 showing aberrant fibers stemming from the lLNv in the PI (cell bodies not in the picture). *Significant differences between the two time points in each genotype. Scale bar, 10 μm. n ≥ 10 for each time point and genotype.
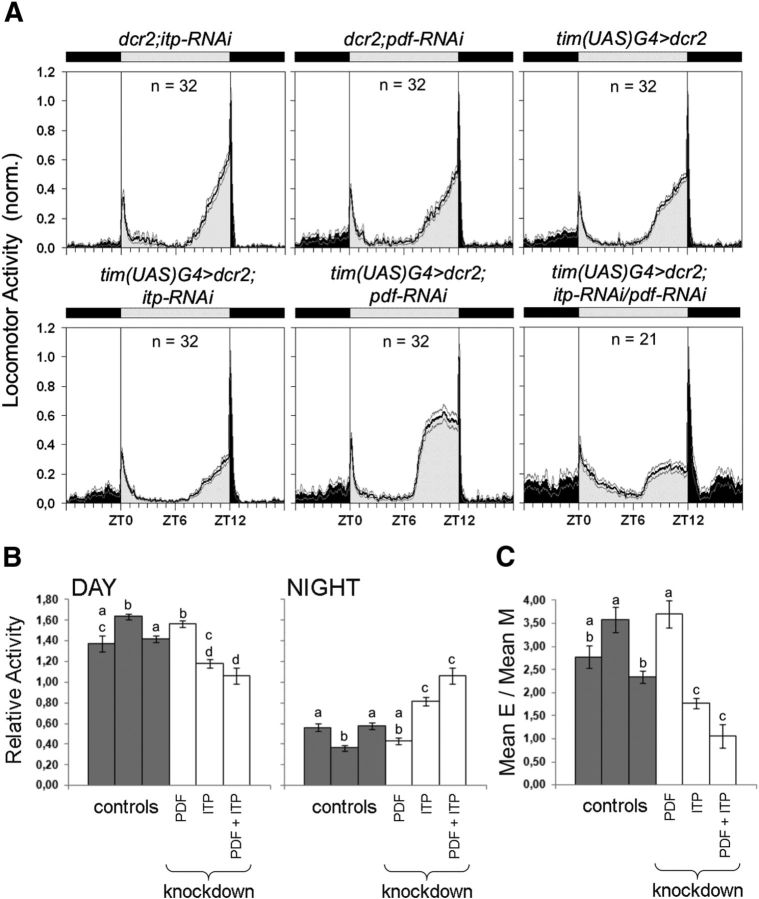
ITP/PDF-double-KD makes flies arrhythmic and hyperactive in DD
Because the results with tim(UAS)G4>ITP2 flies already point to an interaction of ITP and PDF in the control of rhythmic behavior, we generated ITP/PDF-double-KD flies (tim(UAS)G4>dcr2;itp-RNAi/pdf-RNAi) and compared their rhythmic behavior with the single-KD flies. The single ITP-KD had only mild effects on the free-running rhythms of the flies (Fig. 6): The percentage of rhythmic flies was the same as in the controls, only period was slightly but significantly longer (see also Table 1). In agreement with previous studies (Shafer and Taghert, 2009), the single PDF-KD had much more severe effects on rhythmicity than the ITP-KD: tim(UAS)G4>dcr2;pdf-RNAi flies were to a significantly lower amount rhythmic compared with tim(UAS)G4>dcr2 and dcr2;pdf-RNAi flies (χ2 = 30.072; p < 0.0001) and the remaining rhythmic flies showed weak short free-running periods (Kruskal–Wallis H(2) = 16.506; p < 0.001; Wilcoxon pairwise comparisons: PDF-KD to tim(UAS)G4>dcr2 p = 0.021; PDF-KD to dcr2;pdf-RNAi p = 0.003; Fig. 6, Table 1). This behavior largely mimicked that of Pdf0 mutants (Renn et al., 1999). The simultaneous KD of PDF and ITP further reduced rhythmicity, although the number of arrhythmic flies turned out not to be significantly higher than in the PDF-KD flies (χ2 = 3.441; p < 0.0636; Fig. 6). Nevertheless, the residual rhythms in the ∼30% ITP/PDF-double-KD flies were clearly different from the PDF-KD flies. Activity was clustered in irregular activity bouts (Fig. 6C) and periodogram analysis revealed multiple weak periods ranging from 10 to 35 h (Fig. 6D). There was no dominant period in any of the seven flies with such residual rhythmicity, making it impossible to calculate an average period. As can be seen in the actograms and periodograms, the rhythms of the ITP/PDF-double-KD flies looked much more disturbed than those of the single PDF-KD flies (Fig. 6C–E), allowing us to conclude that the double-KD strongly affects rhythmicity, rendering the flies virtually arrhythmic. In addition, the ITP/PDF-double-KD flies showed a significantly higher activity level than all other lines (Kruskal–Wallis H(5) = 54.746; p < 0.001; Fig. 6E).
In LD, ITP/PDF-double-KD flies combine the behavioral characteristics of the single-KD flies but also show effects on sleep
In LD conditions, the behavior of ITP/PDF-double-KD flies was less disturbed than under DD conditions. Most flies did still show a kind of M and E activity, but with clearly altered characteristics (Figs. 6, 10). Their E peak was as advanced as that of single PDF-KD flies (Fig. 10A, bottom row, right). Further, they revealed a reduced E peak (Kruskal–Wallis H(5) = 73,298, p < 0.001; Fig. 10C) and higher nocturnal activity, as did single ITP-KD flies (daytime activity: ANOVA F(5,171) = 16.787, p < 0.001; nighttime activity: ANOVA F(5,171) = 27.802, p < 0.001; Fig. 10B). Therefore, the effects of the single-KDs seem to add up in the double-KD flies. Nevertheless, we did also observe effects that were not present in the single-KD flies: ITP/PDF-double-KD flies had a less pronounced siesta, which is the typical midday break in activity observed in wild-type flies. In the double-KD flies, the activity after lights-on decreases only slowly, whereas in all other genotypes (including ITP- and PDF-single-KD flies), the activity quickly decreases after the lights-on reaction and stays at a relatively low level until the beginning of the E activity.
Figure 10.
Locomotor activity of ITP/PDF-double-KD flies and controls in LD 12:12. A, Average activity profiles were calculated for each genotype and were normalized to the highest activity value. PDF-KD flies (tim(UAS)G4>dcr2;pdf-RNAi) show the typical advanced E activity and reduced M activity. The same phenotypes can be seen in ITP/PDF-double-KD flies (tim(UAS)G4>dcr2;itp-RNAi/pdf-RNAi). In addition, these flies show a less pronounced siesta compared with the other genotypes. n = number of investigated flies; black areas indicates darkness, gray areas indicates light of 100 lux; black line = mean, gray lines = SEM; T = 20°C. B, Relative activity levels for day (left) and night (right) were calculated as mean beam crosses per minute during the light phase or the dark phase relative to the average of beam crosses during the whole day. Relative daytime and nighttime activities were significantly dependent on the genotype. In particular, ITP/PDF-double-KD flies showed a slight reduction in daytime activity. The tendency toward reduced daytime activity in ITP-KD flies (tim(UAS)G4>dcr2;itp-RNAi) was similar to the results of Figure 4B. Both ITP-KD flies and ITP/PDF-double-KD flies showed a significant increase in nighttime activity. C, Mean E activity (ZT06 to ZT18) was calculated in relation to mean M activity (ZT18 to ZT06) as in Figure 4C and was significantly dependent on the genotype. ITP-KD flies showed significantly less E activity than the controls and PDF-KD flies (cf. also Fig. 4C). E activity in ITP/PDF-double-KD flies was similarly reduced. The control strains in B and C (dark gray bars) are in the following order from left to right: tim(UAS)G4>dcr2, dc2;itp-RNAi, and dcr2;pdf-RNAi. Error bars indicate SEM. n ≥ 21 in all panels. Lowercase letters indicate significant differences (different letters indicate p < 0.05).
The lacking siesta and the higher nocturnal activity suggested that ITP/PDF-double-KD flies almost do not sleep. To investigate this, we analyzed sleep in LD (Fig. 11) in the same dataset that was used to calculate the LD activity profiles (Fig. 10). Neither ITP-KD nor PDF-KD alone affected the sleep profile, but the simultaneous KD of ITP and PDF clearly reduced sleep during the siesta and during the night (Fig. 11A). Therefore, the total amount of sleep during the light (ANOVA F(5,173) = 15.227, p < 0.001) and the dark phase (ANOVA F(5,173) = 30.670, p < 0.001) was significantly reduced in ITP/PDF-double-KD flies, but in none of the other strains (Fig. 11B), although the ITP-KD flies also showed higher nocturnal activity (Figs. 5, 10). Surprisingly, ITP/PDF-double-KD flies showed, on average, significantly more sleep bouts per day than all of the other genotypes (Kruskal–Wallis H(5) = 31.472, p < 0.001, p < 0.05 in pairwise comparisons; Fig. 11C). The average duration of these sleep bouts was, however, significantly shorter than in the controls or the single-KD flies (Kruskal–Wallis H(5) = 37.333, p < 0.001; p < 0.05 in pairwise comparisons; Fig. 11D).
Figure 11.
Sleep parameters of ITP/PDF-double-KD flies, ITP-KD, and PDF-single-KD flies and controls in LD 12:12. Sleep was defined as the average amount of time in which the flies did not cross the infrared light beam for at least 5 consecutive minutes. A, Daily average sleep profiles of ITP-KD flies (red), PDF-KD flies (blue), ITP/PDF-double-KD flies (magenta), and controls (different grays). ITP-KD flies do not show any differences in the sleep profile compared with controls. ITP/PDF-double-KD flies clearly sleep less during the night and during the first half of the day. B, Total amount of sleep during nighttime (full bars) and daytime (empty bars). ITP-KD flies do not differ from controls in total sleep. ITP/PDF-double-KD flies show significantly decreased nighttime and daytime sleep compared with all other genotypes. C, Average number of sleep bouts per day was increased in ITP/PDF-double-KD flies. D, ITP/PDF-double-KD flies show, on average, significantly shorter sleeping bouts than all other genotypes. *p < 0.05; **p < 0.001. Error bars indicate SEM. n ≥ 21 in all panels.
Discussion
In the present study, we show that the activity rhythms of Drosophila are not only dependent on the neuropeptide PDF (Renn et al., 1999), but are also clearly affected by the neuropeptide ITP. ITP promotes E activity and may therefore act as an output signal of the E oscillator cells. Under DD conditions, ITP has a mild period-shortening effect. Therefore, ITP somehow opposes the effects of PDF, which promotes M activity and has a predominantly period-lengthening effect under DD (Renn et al., 1999; Shafer and Taghert, 2009). Nevertheless, the effects of ITP under DD are relatively mild compared with PDF, which is necessary for robust rhythmicity. Notably, the double-KD of ITP and PDF almost completely disrupts circadian rhythmicity under DD, suggesting that the two neuropeptides are the clock's main output factors essential for rhythmicity under constant conditions. The two neuropeptides are also important for normal LD rhythms, whereby they seem to control different behavioral aspects: Whereas PDF strongly influences the activity phase of the flies, promoting their adaptation to long photoperiods (Yoshii et al., 2009), ITP has no such effects. ITP mainly influences the activity level of the flies, reducing nocturnal activity and enhancing diurnal E activity. Most interestingly, both peptides cooperate in controlling the flies' sleep. Whereas the single-KD of either PDF or ITP did not affect sleep at all, the double-PDF/ITP-KD strongly reduced sleep during the flies' siesta and night. In the following sections, we will discuss specific points in more detail.
ITP's rhythmic way of action
To function in a circadian fashion, the synthesis of a neuropeptide, its stability, or its receptor sensitivity can be under clock control. We have shown that ITP immunostaining is dramatically decreased inside of the clock neurons in ClkAR mutants, suggesting that the transcription of the itp gene might be regulated by CLK in the ITP+ clock neurons. Park et al. (2000) found a similar reduction in PDF immunostaining in ClkJrk mutants and identified an E-box (CACGTG) within the upstream regulatory region of the pdf gene. Nevertheless, pdf expression was independent of this E-box and pdf-mRNA levels were not cycling. We did not find any indications for the presence of E-boxes in the upstream region of the itp gene, indicating that ITP abundance is probably indirectly regulated by CLK, as is PDF. Similar to what was found for PDF (Park et al., 2000), we also did not find any significant cycling in ITP staining intensity in clock neuron cell bodies, but significant oscillations in staining intensity in the projection terminals. This suggests that PDF and ITP are continuously produced but rhythmically released from the axon terminals. ITP peaks in the middle of the night and the middle of the day. Assuming that peptide release occurs, when staining intensity decreases, we propose that ITP is most probably released from the clock neurons in the second half of the night and the second half of the day. Simultaneous analysis of PDF staining intensity in dorsal projection terminals of the same brains showed that PDF appears to be released in the middle of the day, in agreement with previous studies (Park et al., 2000).
Notably, PDF and ITP appear not only to have different release times, but also different release sites. Whereas the PDF fibers terminate in the PL close to the calyces of the mushroom bodies (Helfrich-Förster and Homberg, 1993), most ITP fibers terminate medially to the PDF fibers in the PI (see also Johard et al., 2009). Both mushroom bodies and the PI have been shown previously to control sleep (Yuan et al., 2006; Joiner et al., 2006; Pitman et al., 2006; Foltenyi et al., 2007; Crocker et al., 2010). Therefore, PDF and ITP may well interfere in the rhythmic control of sleep.
Clock-derived ITP promotes E activity and reduces nocturnal activity
RNA interference in combination with the GAL4/UAS-system is a powerful tool with which to disrupt gene expression in a spatially specified way. Both the KD of ITP and the KD of PDF were very efficient in our experiments, leaving both peptides undetectable by the antibodies in the relevant clock neurons.
We did not find any effects of ITP-KD on the timing of M and E activity bouts in LD, not even under long and short photoperiods (data not shown). Therefore, ITP does not seem to be involved in general entrainment mechanisms and the adaptation to changing photoperiods. We found, however, effects of ITP-KD on activity levels, especially during the evening and the night. The E activity of a wild-type fly occurs mainly during the light phase before lights-off, whereas the M anticipation before lights-on constitutes a large portion of the fly's M activity. Activity of ITP-KD flies was reduced during daytime and increased during nighttime relative to their overall average activity. Consistent with this, ITP-KD flies showed significantly less E activity in relation to their M activity compared with controls. Therefore, we conclude that ITP, deriving from the E oscillator cells, normally promotes E activity and reduces nighttime activity.
Knocking down ITP and PDF together phenocopied both characteristics of PDF-KD flies and ITP-KD flies. ITP/PDF-double-KD flies showed the same advance in E peak phase as was typical for PDF-KD flies. Conversely, the E peak amplitude was decreased and nocturnal activity increased compared with PDF-KD flies, as was the case when ITP was knocked down alone. Therefore, we conclude that PDF and ITP—independently of each other—control activity phase and levels, respectively.
ITP might slightly shorten the circadian free-running period in DD
In a previous study, we showed that the ablation of the NPF+ clock neurons lengthens the circadian free-running period in DD by 0.7 h and advances the E activity in LD (Hermann et al., 2012). Knocking down NPF via RNAi was not completely efficient and thus did not show any effect on LD or DD behavior (Hermann et al., 2012). The npfG4 line that we used for the cell ablation experiments in this former study had included the two ITP+ clock neurons. Interestingly, we demonstrate now that the KD of ITP within these cells also slightly (by 0.2 h) but significantly prolongs the circadian free-running period in DD. Although this effect on period was marginal, it suggests that the lack of ITP has been partly responsible for the period lengthening when the NPF+ cells were ablated. Therefore, ITP may normally act as a weak period-shortening factor leading to a slightly longer period when ITP signaling is disrupted. Conversely, Pdf01 or PDF-KD flies show shortened free-running rhythms in DD (Renn et al., 1999; Shafer and Taghert, 2009). Therefore, both peptides may have opposing effects on the period length, which could be a mechanism of fine-tuning clock neuron synchronization or rhythm output.
ITP might target mainly clock output sites to control rhythmic behavior
The ITP receptor and its expression pattern are so far unknown. Therefore, it is unclear whether ITP works within the clock network, as was revealed for PDF (Im and Taghert, 2010), or on clock output sites. The results of ITP overexpression with tim(UAS)G4 speak more for a role of ITP downstream of the clock, because the flies still retained almost normal PER cycling even though they were behaviorally arrhythmically. Interestingly, ITP overexpression with perG4 did not provoke arrhythmicity, although per and tim are expressed in the same clock neurons and perG4 even expresses in more ectopic places than tim(UAS)G4. The only difference that we found between the two lines was that rhythmic ITP accumulation in the PI terminals was abolished after ITP overexpression with tim(UAS)G4, but not with perG4. This suggests a crucial role of rhythmic ITP release into the PI, suggesting that neurons downstream of ITP locate there.
We can currently not exclude a feedback of ITP on certain clock neurons. First of all, ITP overexpression reduced the amplitude of PER cycling in the sLNv and the LNd, as well as that of PDF cycling in the sLNv terminals. Second, the ITP-KD slightly lengthened the free-running period. Third, the occurrence of multiple weak periods in the ITP/PDF-double-KD flies may point to a strong reduction of synchrony among the clock neurons' oscillations. A role in maintaining internal synchrony of the clock neurons is known for PDF (Peng et al., 2003; Lin et al., 2004; Shafer et al., 2008; Yoshii et al., 2009), and ITP may cooperate with PDF in keeping the clock neurons synchronous. All of these findings suggest a role of ITP in the clock network, in addition to its most likely effects downstream of the clock. Future studies will have to reveal which neurons in the brain express the ITP receptor and whether the sLNv and the LNd, the molecular PER cycling of which is reduced in tim(UAS)G4>ITP2 flies, are among them.
This is the first study demonstrating a role of ITP in the control of behavioral rhythms in D. melanogaster. We propose a role for ITP in the clock that is partly complementary and partly cooperative with PDF.
Footnotes
This work was funded by the European Commission (FP6 IP EUCLOCK), the German Research Foundation (Grant Fo-207/12-1; Collaborative Research Center Grant SFB 1047 “Insect timing” INST 93/784-1), and Japan Society for the Promotion of Science (KAKENHI Grant-in-Aid for Scientific Research 25840121). We thank the Bloomington stock center; the Vienna Drosophila RNAi Center; J.C. Hall, M.W. Young, M. Kaneko, O.T. Shafer, F. Rouyer, and C. Wegener for providing fly lines; D.R. Nässel, P.E. Hardin, R. Stanewsky, and the Developmental Studies Hybridoma Bank for providing antibodies; A. Fiala for the pUAST vector; and M. Schlichting and P. Menegazzi for practical assistance.
The authors declare no competing financial interests.
References
- Allada R, White NE, So WV, Hall JC, Rosbash M. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93:791–804. doi: 10.1016/S0092-8674(00)81440-3. [DOI] [PubMed] [Google Scholar]
- Blau J, Young MW. Cyclic vrille expression is required for a functional Drosophila clock. Cell. 1999;99:661–671. doi: 10.1016/S0092-8674(00)81554-8. [DOI] [PubMed] [Google Scholar]
- Bywalez W, Menegazzi P, Rieger D, Schmid B, Helfrich-Förster C, Yoshii T. The dual-oscillator system of Drosophila melanogaster under natural-like temperature cycles. Chronobiol Int. 2012;29:395–407. doi: 10.3109/07420528.2012.668505. [DOI] [PubMed] [Google Scholar]
- Crocker A, Shahidullah M, Levitan IB, Sehgal A. Identification of a neural circuit that underlies the effects of octopamine on sleep:wake behavior. Neuron. 2010;65:670–681. doi: 10.1016/j.neuron.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damulewicz M, Rosato E, Pyza E. Circadian regulation of the Na+/K+-ATPase alpha subunit in the visual system is mediated by the pacemaker and by retina photoreceptors in Drosophila melanogaster. PLoS One. 2013;8:e73690. doi: 10.1371/journal.pone.0073690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dircksen H. Insect ion transport peptides are derived from alternatively spliced genes and differentially expressed in the central and peripheral nervous system. J Exp Biol. 2009;212:401–412. doi: 10.1242/jeb.026112. [DOI] [PubMed] [Google Scholar]
- Dircksen H, Tesfai LK, Albus C, Nässel DR. Ion transport peptide splice forms in central and peripheral neurons throughout postembryogenesis of Drosophila melanogaster. J Comp Neurol. 2008;509:23–41. doi: 10.1002/cne.21715. [DOI] [PubMed] [Google Scholar]
- Foltenyi K, Greenspan RJ, Newport JW. Activation of EGFR and ERK by rhomboid signaling regulates the consolidation and maintenance of sleep in Drosophila. Nat Neurosci. 2007;10:1160–1167. doi: 10.1038/nn1957. [DOI] [PubMed] [Google Scholar]
- Glossop NR, Houl JH, Zheng H, Ng FS, Dudek SM, Hardin PE. VRILLE feeds back to control circadian transcription of Clock in the Drosophila circadian oscillator. Neuron. 2003;37:249–261. doi: 10.1016/S0896-6273(03)00002-3. [DOI] [PubMed] [Google Scholar]
- Goda T, Mirowska K, Currie J, Kim MH, Rao NV, Bonilla G, Wijnen H. Adult circadian behavior in Drosophila requires developmental expression of cycle, but not period. PLoS Genet. 2011;7:e1002167. doi: 10.1371/journal.pgen.1002167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grima B, Chélot E, Xia R, Rouyer F. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature. 2004;431:869–873. doi: 10.1038/nature02935. [DOI] [PubMed] [Google Scholar]
- Gummadova JO, Coutts GA, Glossop NR. Analysis of the Drosophila Clock promoter reveals heterogeneity in expression between subgroups of central oscillator cells and identifies a novel enhancer region. J Biol Rhythms. 2009;24:353–367. doi: 10.1177/0748730409343890. [DOI] [PubMed] [Google Scholar]
- Helfrich C, Engelmann W. Evidences for circadian rhythmicty in the per0 mutant of Drosophila melanogaster. Z Naturforsch. 1987;42c:1335–1338. doi: 10.1515/znc-1987-11-1231. [DOI] [PubMed] [Google Scholar]
- Helfrich-Förster C. The locomotor activity rhythm of Drosophila melanogaster is controlled by a dual oscillator system. Journal of Insect Physiology. 2001;47:877–887. doi: 10.1016/S0022-1910(01)00060-9. [DOI] [Google Scholar]
- Helfrich-Förster C, Homberg U. Pigment-dispersing hormone-immunoreactive neurons in the nervous system of wild-type Drosophila melanogaster and of several mutants with altered circadian rhythmicity. J Comp Neurol. 1993;337:177–190. doi: 10.1002/cne.903370202. [DOI] [PubMed] [Google Scholar]
- Helfrich-Förster C, Täuber M, Park JH, Mühlig-Versen M, Schneuwly S, Hofbauer A. Ectopic expression of the neuropeptide pigment-dispersing factor alters behavioral rhythms in Drosophila melanogaster. J Neurosci. 2000;20:3339–3353. doi: 10.1523/JNEUROSCI.20-09-03339.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich-Förster C, Shafer OT, Wülbeck C, Grieshaber E, Rieger D, Taghert P. Development and morphology of the clock-gene-expressing lateral neurons of Drosophila melanogaster. J Comp Neurol. 2007;500:47–70. doi: 10.1002/cne.21146. [DOI] [PubMed] [Google Scholar]
- Hermann C, Yoshii T, Dusik V, Helfrich-Förster C. Neuropeptide F immunoreactive clock neurons modify evening locomotor activity and free-running period in Drosophila melanogaster. J Comp Neurol. 2012;520:970–987. doi: 10.1002/cne.22742. [DOI] [PubMed] [Google Scholar]
- Hermann C, Saccon R, Senthilan PR, Domnik L, Dircksen H, Yoshii T, Helfrich-Förster C. The circadian clock network in the brain of different Drosophila species. J Comp Neurol. 2013;521:367–388. doi: 10.1002/cne.23178. [DOI] [PubMed] [Google Scholar]
- Im SH, Taghert PH. PDF receptor expression reveals direct interactions between circadian oscillators in Drosophila. J Comp Neurol. 2010;518:1925–1945. doi: 10.1002/cne.22311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johard HA, Yoishii T, Dircksen H, Cusumano P, Rouyer F, Helfrich-Förster C, Nässel DR. Peptidergic clock neurons in Drosophila: ion transport peptide and short neuropeptide F in subsets of dorsal and ventral lateral neurons. J Comp Neurol. 2009;516:59–73. doi: 10.1002/cne.22099. [DOI] [PubMed] [Google Scholar]
- Joiner WJ, Crocker A, White BH, Sehgal A. Sleep in Drosophila is regulated by adult mushroom bodies. Nature. 2006;441:757–760. doi: 10.1038/nature04811. [DOI] [PubMed] [Google Scholar]
- Kempinger L, Dittmann R, Rieger D, Helfrich-Förster C. The nocturnal activity of fruit flies exposed to artificial moonlight is partly caused by direct light effects on the activity level that bypass the endogenous clock. Chronobio Int. 2009;26:151–166. doi: 10.1080/07420520902747124. [DOI] [PubMed] [Google Scholar]
- Lin Y, Stormo GD, Taghert PH. The neuropeptide pigment-dispersing factor coordinates pacemaker interactions in the Drosophila circadian system. J Neurosci. 2004;24:7951–7957. doi: 10.1523/JNEUROSCI.2370-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majercak J, Sidote D, Hardin PE, Edery I. How a circadian clock adapts to seasonal decreases in temperature and day length. Neuron. 1999;24:219–230. doi: 10.1016/S0896-6273(00)80834-X. [DOI] [PubMed] [Google Scholar]
- Menegazzi P, Yoshii T, Helfrich-Förster C. Laboratory versus nature: the two sides of the Drosophila circadian clock. J Biol Rhythms. 2012;27:433–442. doi: 10.1177/0748730412463181. [DOI] [PubMed] [Google Scholar]
- Park JH, Helfrich-Förster C, Lee G, Liu L, Rosbash M, Hall JC. Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proc Natl Acad Sci U S A. 2000;97:3608–3613. doi: 10.1073/pnas.97.7.3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Stoleru D, Levine JD, Hall JC, Rosbash M. Drosophila freerunning rhythms require intercellular communication. PLoS Biol. 2003;1:E13. doi: 10.1371/journal.pbio.0000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschel N, Helfrich-Förster C. Setting the clock–by nature: circadian rhythm in the fruitfly Drosophila melanogaster. FEBS Lett. 2011;585:1435–1442. doi: 10.1016/j.febslet.2011.02.028. [DOI] [PubMed] [Google Scholar]
- Picot M, Cusumano P, Klarsfeld A, Ueda R, Rouyer F. Light activates output from evening neurons and inhibits output from morning neurons in the Drosophila circadian clock. PLoS Biol. 2007;5:e315. doi: 10.1371/journal.pbio.0050315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitman JL, McGill JJ, Keegan KP, Allada R. A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature. 2006;441:753–756. doi: 10.1038/nature04739. [DOI] [PubMed] [Google Scholar]
- Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell. 1999;99:791–802. doi: 10.1016/S0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- Rieger D, Stanewsky R, Helfrich-Förster C. Cryptochrome, compound eyes, Hofbauer-Buchner eyelets, and ocelli play different roles in the entrainment and masking pathway of the locomotor activity rhythm in the fruit fly Drosophila melanogaster. J Biol Rhythms. 2003;18:377–391. doi: 10.1177/0748730403256997. [DOI] [PubMed] [Google Scholar]
- Rieger D, Shafer OT, Tomioka K, Helfrich-Förster C. Functional analysis of circadian pacemaker neurons in Drosophila melanogaster. J Neurosci. 2006;26:2531–2543. doi: 10.1523/JNEUROSCI.1234-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid B, Helfrich-Förster C, Yoshii T. A new ImageJ plug-in “ActogramJ” for chronobiological analyses. J Biol Rhythms. 2011;26:464–467. doi: 10.1177/0748730411414264. [DOI] [PubMed] [Google Scholar]
- Shafer OT, Taghert PH. RNA-interference knockdown of Drosophila pigment dispersing factor in neuronal subsets: the anatomical basis of a neuropeptide's circadian functions. PLoS One. 2009;4:e8298. doi: 10.1371/journal.pone.0008298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer OT, Kim DJ, Dunbar-Yaffe R, Nikolaev VO, Lohse MJ, Taghert PH. Widespread receptivity to neuropeptide PDF throughout the neuronal circadian clock network of Drosophila revealed by real-time cyclic AMP imaging. Neuron. 2008;58:223–237. doi: 10.1016/j.neuron.2008.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanewsky R, Frisch B, Brandes C, Hamblen-Coyle MJ, Rosbash M, Hall JC. Temporal and spatial expression patterns of transgenes containing increasing amounts of the Drosophila clock gene period and a lacZ reporter: mapping elements of the PER protein involved in circadian cycling. J Neurosci. 1997;17:676–696. doi: 10.1523/JNEUROSCI.17-02-00676.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoleru D, Peng Y, Agosto J, Rosbash M. Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature. 2004;431:862–868. doi: 10.1038/nature02926. [DOI] [PubMed] [Google Scholar]
- Vanin S, Bhutani S, Montelli S, Menegazzi P, Green EW, Pegoraro M, Sandrelli F, Costa R, Kyriacou CP. Unexpected features of Drosophila circadian behavioral rhythms under natural conditions. Nature. 2012;484:371–375. doi: 10.1038/nature10991. [DOI] [PubMed] [Google Scholar]
- Wülbeck C, Grieshaber E, Helfrich-Förster C. Pigment-dispersing factor (PDF) has different effects on Drosophila's circadian clocks in the accessory medulla and the dorsal brain. J Biol Rhythms. 2008;23:409–424. doi: 10.1177/0748730408322699. [DOI] [PubMed] [Google Scholar]
- Yoshii T, Wülbeck C, Sehadova H, Veleri S, Bichler D, Stanewsky R, Helfrich-Förster C. The neuropeptide pigment-dispersing factor adjusts period and phase of Drosophila's clock. J Neurosci. 2009;29:2597–2610. doi: 10.1523/JNEUROSCI.5439-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii T, Rieger D, Helfrich-Förster C. Two clocks in the brain: an update of the morning and evening oscillator model in Drosophila. Prog Brain Res. 2012;199:59–82. doi: 10.1016/B978-0-444-59427-3.00027-7. [DOI] [PubMed] [Google Scholar]
- Yuan Q, Joiner WJ, Sehgal A. A sleep-promoting role for the Drosophila serotonin receptor 1A. Curr Biol. 2006;16:1051–1062. doi: 10.1016/j.cub.2006.04.032. [DOI] [PubMed] [Google Scholar]