Abstract
Neural progenitor cells (NPCs) have regenerative capabilities that are activated during inflammation. We aimed at elucidating how NPCs, with special focus on the spinal cord-derived NPCs (SC-NPCs), are affected by chronic inflammation modeled by experimental autoimmune encephalomyelitis (EAE). NPCs derived from the subventricular zone (SVZ-NPCs) were also included in the study as a reference from a distant inflammatory site. We also investigated the transcriptional and functional difference between the SC-NPCs and SVZ-NPCs during homeostatic conditions. NPCs were isolated and propagated from the SVZ and cervical, thoracic, and caudal regions of the SC from naive rats and rats subjected to EAE. Using Affymetrix microarray analyses, the global transcriptome was measured in the different NPC populations. These analyses were paralleled by NPC differentiation studies. Assessment of basal transcriptional and functional differences between NPC populations in naive rat revealed a higher neurogenic potential in SVZ-NPCs compared with SC-NPCs. Conversely, during EAE, the neurogenicity of the SC-NPCs was increased while their gliogenicity was decreased. We detected an overall increase of inflammation and neurodegeneration-related genes while the developmentally related profile was decreased. Among the decreased functions, we isolated a gliogenic signature that was confirmed by differentiation assays where the SC-NPCs from EAE generated fewer oligodendrocytes and astrocytes but more neurons than control cultures. In summary, NPCs displayed differences in fate-regulating genes and differentiation potential depending on their rostrocaudal origin. Inflammatory conditions downregulated gliogenicity in SC-NPCs, promoting neurogenicity. These findings give important insight into neuroinflammatory diseases and the mechanisms influencing NPC plasticity during these conditions.
Keywords: differentiation, experimental autoimmune encephalomyelitis, gliogenesis, microarray, neural stem cells, neurogenesis
Introduction
Neural stem/progenitor cells (NPCs) are present in the mammalian CNS throughout the neuroaxis (Weiss et al., 1996; Kehl et al., 1997; McKay, 1997; Doetsch et al., 1999; Johansson et al., 1999), and they respond to various pathological stimuli (Brundin et al., 2003; Packer et al., 2003; Wong et al., 2004; Covacu et al., 2006; Danilov et al., 2006; Thored et al., 2006; Ziv et al., 2006; Johansson et al., 2008; Whitney et al., 2009). However, it is evident that the NPC response varies with the type and duration of the CNS insult. Chronic inflammation is a hallmark of many autoimmune diseases, such as multiple sclerosis (MS). Studies on NPC in the context of chronic inflammation have so far focused on NPC niches in the brain (Picard-Riera et al., 2002; Monje et al., 2003; Pluchino et al., 2008; Tepavčević et al., 2011), whereas the spinal cord NPCs (SC-NPCs) have been mostly observed during traumatic injury models (Frisén et al., 1995; Mothe and Tator, 2005). In previous studies, we demonstrated that SC-NPCs are mobilized during chronic inflammation, modeled by experimental autoimmune encephalomyelitis (EAE) induced in the Dark Agouti (DA) rat (Brundin et al., 2003; Danilov et al., 2006). This EAE model is characterized by demyelinated, immune-infiltrated lesions, and the disease development resembles the human MS disease (Storch et al., 1998; Pierson et al., 2012). In this study, we aimed at elucidating how SC-NPCs are affected on a transcriptional and functional level by chronic inflammation.
To answer these questions, we isolated and in vitro propagated NPCs from the different rostrocaudal levels of the DA rat spinal cord (SC) and also from the subventricular zone (SVZ). We measured the NPCs' global transcriptome and, in parallel, assessed their differentiation ability. The study was initiated with a comparison between SC-NPCs and SVZ-NPCs from naive (unimmunized) rats, where the SVZ-NPCs served as a neurogenic reference. Second, we performed the same transcriptional and functional analyses on NPC populations generated from rats with EAE and compared them with NPCs from naive rats. SVZ-NPCs were used in this case as a reference from a distant inflammatory site.
Our results reveal basal longitudinal neuroxis-dependent differences in the NPCs from naive animals where the neurogenic capacity of the NPCs decreased in a rostrocaudal manner and was replaced by an increasing gliogenicity. This was supported by both transcriptome analyses and functional differentiation assays. In inflammation, the most adversely induced gene expression changes were found in the NPCs isolated from the caudal part of the SC. We detected an increase in neurodegeneration and inflammation-related genes and a general downregulation of developmentally related genes. Moreover, in the SC-NPCs, the gliogenic gene signature was downregulated after neuroinflammation. The differentiation assays revealed decreased gliogenic differentiation and increased neurogenic differentiation in SC-NPCs from all three rostrocaudal levels. In all, our results have important implications for the understanding the diversity of NPC populations in the CNS and their behavior during chronic inflammation.
Materials and Methods
Experimental animals and EAE induction.
DA female rats 7–8 weeks old (Scanbur B&K; http://www.scanbur.eu/) were kept at the animal facility at Karolinska University Hospital. All animal experiments were performed in accordance with the Swedish ethical regulations (Stockholms Norra Djurförsöksetiska Nämnd).
Recombinant myelin oligodendrocyte glycoprotein (rMOG; amino acids 1–125 from the N terminus) was prepared as previously described (Amor et al., 1994). Female rats between 10 and 11 weeks of age were anesthetized with isoflurane (Forane; Abbott Laboratories) and immunized subcutaneously at the dorsal tail base with 200 μl inoculum containing 20 μg rMOG in saline emulsified 1:1 with IFA (Sigma-Aldrich). The rats were clinically assessed daily for signs of EAE from day 9 until day 30–40 postimmunization. The clinical symptoms were scored as follows: 0, no clinical signs of EAE; 1, tail weakness or tail paralysis; 2, hindlimb paraparesis; 3, hindlimb paralysis; 4, tetraplegia; and 5, death.
Cell culture.
Brains and SCs were harvested from naive animals and from EAE-diseased animals with clinical score 2–3 at 30–40 d postimmunization For the microarray expression analysis, only animals with disease reaching score 3 were used and killed at day 40 postimmunization NPC cultures were isolated and propagated from the SVZ and different levels of the SC, which were defined as follows: cervical (CER = above T2), thoracic (THOR = T2-T12), and caudal (CAUD = below T12); for indication of segments, see Figure 1. The SVZ-derived NPCs were isolated according to a modified protocol by Johansson et al. (1999). Briefly, SVZ biopsies were isolated and the cells dissociated as for the SC tissue. For isolation of the SC-NPCs, the SC was divided rostrocaudally and the meninges were peeled off before mechanical and enzymatic dissociation using 200 U/ml DNase (Sigma-Aldrich) and 10 U/ml papain (Worthington). To remove the myelin debris the cells were resuspended in 0.9 m sucrose in Hanks balanced salt solution (Invitrogen) and washed. The cells were cultured in propagation medium, composed of DMEM/F-12 containing B27 supplement (Invitrogen), penicillin (100 U/ml), and streptomycin (100 μg/ml) (Invitrogen; http://www.invitrogen.com), 20 ng/ml epidermal growth factor (EGF, Sigma-Aldrich; http://www.sigmaaldrich.com), and 20 ng/ml basic fibroblast growth factor (bFGF, R&D systems). The NPC cultures were propagated and passaged twice and used in experiments as single cells after the second passage. For differentiation, single-cell suspensions were seeded onto poly-d-lysine-coated plates (Sigma-Aldrich) and cultured for 5–7 d in medium lacking EGF and bFGF but supplemented with 1% FCS (Invitrogen).
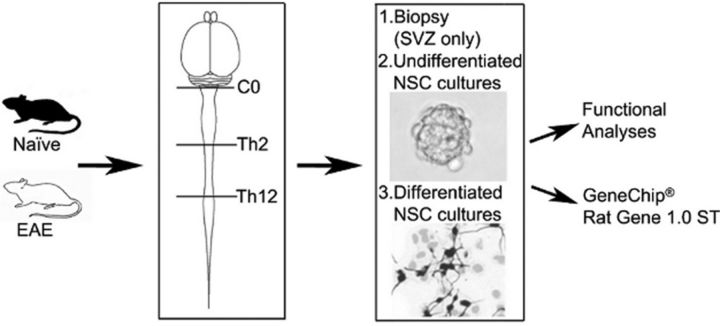
Figure 1.
Experimental setup. NPCs were isolated from the SVZ and from the cervical, thoracic, and caudal SC segments of naive DA rats or rats with EAE. C0, Th2, and Th12 denote the vertebrae levels between which the mentioned SC segments were cut: cervical, between C0 and Th2, thoracic, between Th2 and Th12 and caudal, below Th12. Total RNA was purified from these NPC cultures, both undifferentiated and differentiated, and also from freshly isolated SVZ biopsies. The global transcriptome was quantified using Affymetrix GeneChipRAT Gene 1.0 ST arrays (n = 3, 1 array/rat/culture). Undifferentiated and differentiated NPCs were also used in differentiation analyses.
Microarray analysis, sample preparation, and data analysis.
Gene expression was measured in the following experimental groups: NPC cultures (undifferentiated or differentiated) isolated from the SVZ and CER, THOR, and CAUD parts of the SC and from the SVZ biopsy before NPC culturing (Fig. 1). Within each of these groups, three naive controls and three EAE individuals were compared. The array platform used was Affymetrix GeneChip RAT Gene ST 1.0; one microarray was used for each individual and each NPC culture. Array hybridization and basic data processing were performed at the Bioinformatics and Expression Analysis Core facility at Karolinska Institutet, Stockholm, Sweden. The basic data processing involved background signal correction using the GC composition-based background correction algorithm, array normalization with global median and signal summarization using the probe logarithmic intensity error estimation (plier); all steps were performed in the GeneChip Expression console from Affymetrix. Statistical significance between naive and EAE groups was calculated using two-sided unpaired Student's t test. The false discovery rate (FDR) was calculated using the q-value plugin for R, and an FDR level of 5% was set. The functional analysis and canonical pathway analysis of the entire dataset was generated with Ingenuity Pathway Analysis (IPA, Qiagen, http://www.qiagen.com/Ingenuity). Molecules from the dataset that met the signal intensity cutoff of ≥50 and passed a 5% FDR level were considered for the analysis. To determine the p value of the association between the dataset and a function, disease, or canonical pathway, Fisher's exact test and Benjamini and Hochberg correction for multiple testing were used. To identify differentially expressed genes between NPC groups from naive animals, one-way ANOVA with adjusted Bonferroni correction was performed in Multiple Experimental Viewer (MeV) (Saeed et al., 2003). Functional clustering was performed in WEB-based Gene Set Analysis Toolkit (WEBGESTALT) (Duncan et al., 2010) and/or DAVID Bioinformatics Resources 6.7 (Huang da et al., 2009). The data discussed in this manuscript have been deposited in NCBI's Gene Expression Omnibus and has the GEO Series accession number GSE46373.
Quantitative real-time PCR.
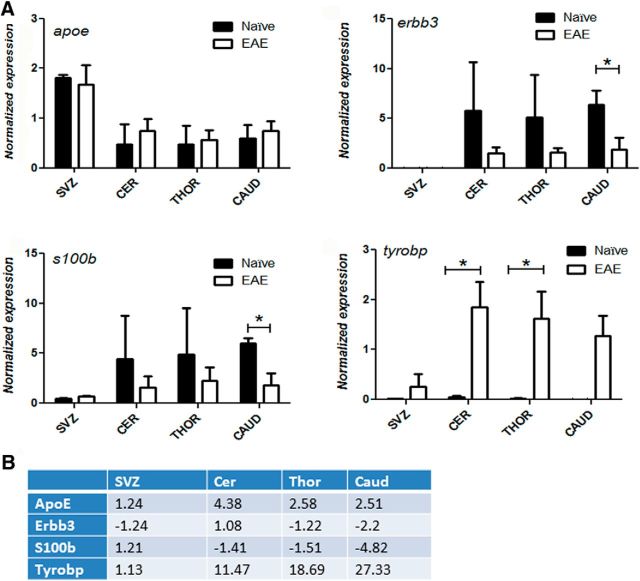
RNA was purified using an RNeasy mini kit (QIAGEN), and cDNA was prepared using the iScript kit (Bio-Rad). Quantitative real-time PCR (qPCR) was performed using a Bio-Rad iQ5 iCycler Detection System with a two-step PCR using SYBR Green (Bio-Rad). Expression levels corrected for amplification efficiency and normalized to housekeeping gene expression (Gapdh or β-actin) were analyzed using iQ5 v2.0 software (Bio-Rad). The primers used for SYBR Green reactions are listed in Table 1. The expression pattern detected with the gene array was verified with qPCR for the following genes: ApoE, Erbb3, S100β, and Tyrobp (Figure 2).
Table 1.
Sequences of primers for quantitative real-time PCRa
| Target | Forward primer | Reverse primer |
|---|---|---|
| Gapdh | 5′-TCAACTACATGGTCTACATGTTCCAG-3′ | 5′-TCCCATTCTCAGCCTTGACTG-3′ |
| β-actin | 5′-CGTGAAAAGATGACCCAGATCA-3′ | 5′-AGAGGCATACAGGGACAACACA-3′ |
| ApoE | 5′-ACACAGGAACTGACGGTACTGAT-3′ | 5′-GTGTTTACCTCGTTGCGGTACT-3′ |
| Erbb3 | 5′-CGTCATGCCAGATACACACC-3′ | 5′-CCCAGAGAAGCACTGAGGTC-3′ |
| S100B | 5′-TTCCATCAGTATTCAGGGAGAGA-3′ | 5′-CCATAAACTCCTGGAAGTCACAC-3′ |
| Tyrobp | 5′-TACAGGCCCAGAGTGACAATTAC-3′ | 5′-CTGTATACTTCTGGCCTCTGACC-3′ |
aThe primers were designed to span exon–exon borders by using the Primer3Plus software (http://primer3plus.com/cgi-bin/dev/primer3plus.cgi).
Figure 2.
A, Quantitative PCR validation of expression of ApoE, Erbb3, S100β, and Tyrobp in NPCs isolated from the different CNS regions of naive rats and rats with EAE (n = 2–4). Error bars indicate mean ± SEM. *p < 0.05 (unpaired t test). **p < 0.01 (unpaired t test). B, The corresponding fold change of expression values EAE/naive detected in the gene array. The expression pattern detected with the gene arrays could be verified with quantitative PCR for most of the genes in most of the experimental groups, with the exception of Apoe.
Colorimetric measurement of nitrite.
After the first passage, NPC cultures supernatants (naïve, n = 23; EAE, n = 27) were collected and nitrite levels were measured using Griess Reagent (Sigma-Aldrich) according to the manufacturer's instructions.
Immunohistochemistry.
Cells were differentiated on poly-d-lysine hydrobromide (Sigma-Aldrich) coated glasses, fixed with 4% PFA in PBS (Bie&Berntsen A-S), blocked in PBS/0.1% saponin/10% goat serum, and incubated with the primary antibody overnight. After washing, the secondary antibody was applied for 1 h at room temperature. Antibodies and their corresponding dilution used were as follows: rabbit anti-Sox2 (Millipore) 1:500, mouse anti-nestin (Millipore Bioscience Research Reagents) 1:100, mouse anti-Ascl1 (BD Biosciences PharMingen), rabbit anti-Olig2 (Abcam) 1:250, rabbit anti-glial fibrillary acidic protein (GFAP) 1:1000 (Dako), mouse anti-galactocerebroside (Gal C) 1:100 (Millipore), mouse anti-β-III tubulin (Tuj) 1:100 (Millipore), mouse-anti rat CD11b 1:200 (Millipore), Cy3 donkey anti-mouse 1:1000 (Jackson ImmunoResearch Laboratories), Alexa-488 donkey anti-rabbit 1:500 (Invitrogen), Alexa-594 goat anti-mouse IgG 1:100 (Invitrogen), Alexa-488 goat-anti mouse IgG (Invitrogen) 1:200, and Alexa-594 goat anti-mouse IgG (Invitrogen) 1:200. For visualizing all cells, the nuclei were counterstained with DAPI (Invitrogen). Labeled cells were visualized and photographed using a fluorescence (Leica DFC 320 Microsystems) and confocal microscope (Leica TCS SP5 Confocal Microscopy System).
After immunohistochemistry, GFAP+, β-III tubulin+, Gal C+, and CD11b+ cells were counted and presented as percentage of the total number of DAPI+ cells. The number of animals used for each assessment was 4–7 in each group (naive and EAE) for all immunostainings.
Western blot.
NPCs were differentiated in medium without mitogens but supplemented with 1% FCS for 5–7 d, and total cell homogenates were made using RIPA buffer (Invitrogen). Cell homogenates were separated by SDS-PAGE at 180 V and transferred to a nitrocellulose membrane (1 h at 100 V). After blocking in PBS/Tween (0.01%) with 5% nonfat milk, the membrane was incubated with primary antibody at 4°C overnight. After washing, the secondary antibodies were added to the membrane for 1 h at room temperature. Antibodies used were as follows: rabbit anti-GFAP 1:1000, mouse anti-Gal C 1:300, β-actin 1:2000, mouse anti-Tuj 1:50, swine anti-rabbit HRP 1:500 (Dako), and goat anti-mouse 1:1000 (Dako). Bands were detected using enhanced chemiluminescence Western Blotting Detection kit (GE Healthcare Bio-Sciences). The bands were scanned (Umax PowerLook 1120), and the net intensity was measured using the ImageJ software (http://rsbweb.nih.gov/ij). Each protein was examined in NPCs isolated from 3 controls and 3 diseased animals.
Results
NPCs from healthy animals show regional differences: higher neurogenic capacity in SVZ-NPCs compared with SC-NPCs
During the development of the CNS, the NPCs are anatomically and temporally exposed to different cues along the neuroaxis and have been reported to develop regional characteristics (Shihabuddin et al., 1997; Kulbatski et al., 2007; Kelly et al., 2009; Kulbatski and Tator, 2009). We wanted to use a global approach to identify the gene expression differences between adult NPCs from the SVZ and the SC. The SVZ is a germinal niche and the residing SVZ-NPCs have high neurogenic competence (Lois and Alvarez-Buylla, 1994), thus providing a “neurogenic reference” for the gene profiling analysis. We isolated NPCs from the SVZ and different rostrocaudal levels of the SC from healthy, nonimmunized (here referred to as “naive”) DA rats and measured the global transcriptome using Affymetrix GeneChip Rat Gene 1.0 ST arrays (Fig. 1). The measurements were performed both in undifferentiated and differentiated cultures.
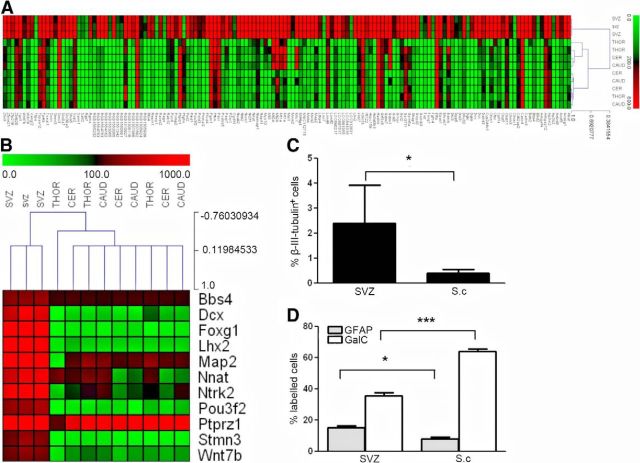
In undifferentiated NPCs from naive animals, we identified 187 differentially expressed genes between the SVZ-NPC and SC-NPCs (one-way ANOVA and Dunn's post hoc test, p < 0.001; Fig. 3A). Interestingly, 183 of the differentially expressed genes had a higher expression level in the SVZ compared with the SC-NPCs, whereas only four genes had higher expression levels in the SC (Fig. 3B). Further gene enrichment analysis on the gene set with higher expression in the SVZ identified a cluster of 14 genes associated with CNS development (GO:0007417), generation of neurons (GO:0048699), neurogenesis (GO:0022008), and neuronal differentiation (GO:0030182) (Figure 3C). These genes included the following: Lhx2, Notch3, Ptk2, Cxcr4, Rorb, and Emx2 (Fig. 3C).
Figure 3.
Genes with differential expression in undifferentiated SVZ-NPCs versus SC-NPCs. A, Heat map showing expression levels of differentially expressed genes in undifferentiated SVZ versus SC-NPCs from naive animals. B, Differentially expressed genes with higher expression in SC-NPCs. C, Neurogenesis-associated genes differentially expressed between SVZ and SC-NPCs. To generate the heat maps, the MeV software was used. Color coding bars represent level of expression: red represents high expression; green represents low expression levels.
In the gene set from the differentiated NPCs from naive animals, we identified 146 differentially expressed genes that had a higher expression level in the NPCs isolated from SVZ compared with SC-NPCs (Fig. 4A). Also, here, we identified a set of neuronal cell-related genes associated with cell functions found in differentiated, mature neurons, mirroring the differentiated state of the NPC cultures (Fig. 4B). Among these genes were as follows: Dcx and Map2, expressed in neuroblasts; and FoxG1, Lhx2, Nnat, and Pou3f2, which promote neurogenesis (Pang et al., 2011).
Figure 4.
Genes with differential expression in differentiated SVZ versus SC-NPCs. A, Heat map representing expression levels of differentially expressed genes in differentiated SVZ versus SC-NPCs from naive animals. B, Neurogenesis-associated genes differentially expressed between SVZ and SC-NPCs. C, Percentage of β-III tubulin+ neurons, (D) GFAP+ astrocytes, and GalC+ oligodendrocytes in differentiated NPCs from SVZ and SCs of naive animals. To generate the heat maps, the MeV software was used. Color coding bars represent level of expression.
In parallel to the array assay, NPCs from SVZ and SC were differentiated, labeled for neuronal and glial markers, and quantified. As shown in Figure 4, SVZ-derived NPCs generated significantly more neurons (p < 0.05) and more GFAP+ astrocytes (p < 0.05) than SC-NPCs (Fig. 4, C and D, respectively). Conversely, the SC-NPCs generated significantly more oligodendrocytes (p < 0.001; Figure 4D).
Together, our transcriptional and functional analyses from NPCs cultured from naive rats clearly demonstrated that there were distinct differences between SVZ and SC-NPC populations (i.e., SVZ-derived NPCs displayed a stronger neurogenic fate than their SC counterparts, which were more gliogenic).
Neurogenic Ascl1+ SC-NPC population is threefold higher in EAE than in control
To investigate how chronic inflammation influenced the transcriptional profile of NPCs, we compared gene expression levels in undifferentiated or differentiated NPCs from EAE with NPCs from naive rats (Figure 1). As a general read for the inflammatory status of the harvested tissue used for cell culture isolation, the levels of nitric oxide derivatives were measured in the culture supernatants before the first culture passage (Aktan, 2004). The nitrite levels were significantly higher in NPC cultures derived from EAE versus naive animals (p < 0.05; Fig. 5A) but did not differ significantly between different EAE-derived cultures, indicating that the in vitro inflammatory status of the NPC cultures derived from different SC levels of EAE rats did not differ significantly. We also quantified the numbers of CD11b+ microglia/macrophages in the NPC cultures. Compared with other immune cells, the microglia/macrophage population has the highest probability of survival in the NPC culture conditions and could therefore influence the results. Thus, NPC cultures from both EAE and control animals were immune-labeled for CD11b, and the percentage of the CD11b+ cells was quantified. CD11b+ cells were present in the NPC cultures, but their numbers were not significantly elevated in EAE compared with naive controls (Fig. 5B).
Figure 5.
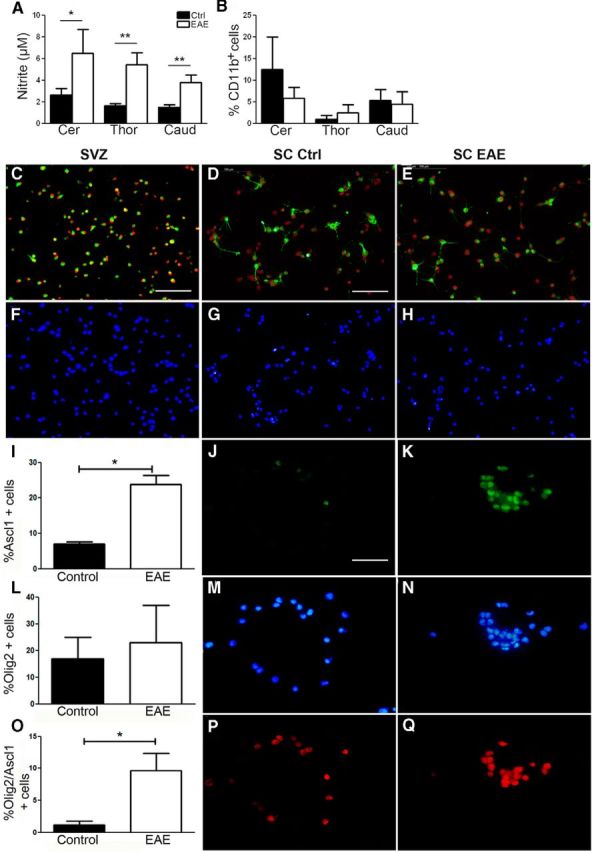
Inflammatory status of the SC-NPC cultures. A, Nitrite levels measured in supernatants from cultures of NPC isolated from naive (black bars) and EAE (white bars) animals (naive controls, n = 23; EAE, n = 27). B, The percentage of CD11b+ cells in undifferentiated NPC cultures (n = 3). C–H, Sox2 and Nestin immunolabeling of SVZ-NPCs (C), SC-NPCs from control animals (D), SC-NPCs from EAE (E), and corresponding DAPI labeling of nuclei (F–H). I–K, Quantification of Ascl1+ cells in SC-NPCs from control and EAE (n = 3) (I). Immunolabeling for Ascl1 (green) in control SC-NPC (J) and EAE SC-NPCs (K) and their corresponding DAPI-stained nuclei (M, N). L–Q, Quantification of Olig2+ cells in SC-NPCs from control and EAE (n = 2) (L). Immunolabeling for Olig2 (red) in control SC-NPCs (P) and EAE SC-NPCs (Q) and their corresponding DAPI-stained nuclei (M, N). O, Quantification of Olig2+/Ascl1+ double-positive SC-NPCs from controls and EAE. Data are mean ± SEM. *p < 0.05 (Student's t test). **p < 0.01 (Student's t test). Scale bars: C, F, 125 μm; D–H, 100 μm; J–Q, 25 μm.
We proceeded with the characterization of the NPC cultures (Figure 5C–Q). Up to 90% of the SVZ-NPCs and SC-NPCs were double-positive for Sox2 and Nestin, indicating that, at least in regards to these markers, the cultures were homogeneous and were kept in an immature state. The level of Sox2/Nestin immunolabeling was similar in SC-NPC cultures from control and EAE animals and also in SVZ-NPCs (Figs. 5C–H). We also assessed the expression of Olig2 (Takebayashi et al., 2000) and Ascl1 (Sommer et al., 1996), transcription factors involved in oligodendrogenesis and neurogenesis, respectively. The percentage of Ascl1+ cells was threefold higher in EAE-derived SC-NPCs compared with control, indicating that EAE has a beneficial effect on this population (Figure 5I–K). The percentage of Olig2+ cells was not significantly different in control and EAE-derived SC-NPC cultures (Figure 5L,P,Q). On the other hand, the percentage of Olig2+Ascl1+ double-positive cells was significantly increased (p < 0.05) in EAE-derived SC-NPCs than in controls (Figure 5O).
Neuroinflammation decreases the gliogenic gene profile of SC-NPCs
Even though the different SC regions were similar in regards to inflammatory status in vitro, significant gene expression changes (FDR ≤ 0.05 and fold change ≥ 1.2 or fold change ≤ −1.2) were detected only in one experimental group, namely, the caudal undifferentiated NPC (Fig. 6; pdf format expandable).
Figure 6.
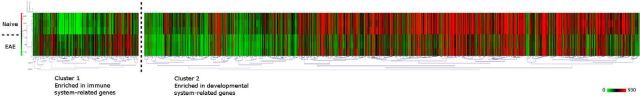
Heat map showing the levels of expression of genes with significant (FDR < 0.05 FC ≥ 1.2 or FC ≤ −1.2) expression changes in the caudal NPCs from EAE versus naive animals (n = 3). Two major clusters are shown: cluster 1 with enriched immune-related genes and cluster 2 enriched in developmental process-related genes. MeV platform was used to perform unsupervised hierarchal clustering (Pearson's correlation).
To identify coregulated genes among the significantly regulated genes from caudal NPCs, we used the MeV platform and could detect two major gene clusters: one with higher expression in NPCs from EAE and the second with higher expression in NPCs from naive controls. We used the WEBGESTALT platform to functionally classify these coregulated genes and could identify that the genes with higher expression in EAE were enriched in immune-related genes, whereas the genes with higher expression in controls were enriched in developmental process-related genes (Fig. 6).
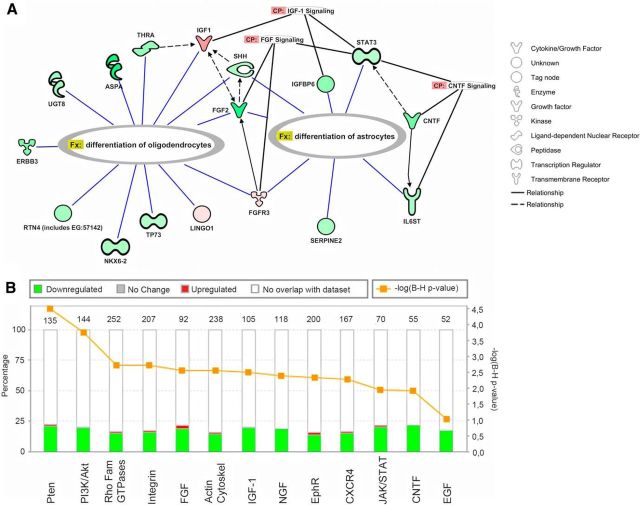
To obtain a better understanding of the underlying biological processes, we performed IPA analysis on the entire caudal NPC dataset. IPA confirmed the results from the WEBGESTALT analysis; the inflammatory gene signature was increased, whereas the developmentally related gene signature was decreased in EAE (Table 2). A subset of the most decreased functions were related to cell viability, branching/neuritogenesis, and lipid metabolism, whereas the most increased functions involved neurodegeneration and inflammation (Table 2). Interestingly, when focusing our analysis on nervous system-related genes, a subset of the most significant functions that emerged involved glial-related functions, such as myelination, quantity of Schwann cells, survival and morphology of oligodendrocytes, and quantity and proliferation of neuroglia (Table 3). Within these functions, we could detect genes with pivotal function in astrogliogenesis (Cntf, Stat3, Fgf3, and Shh), oligodendrogenesis (Shh, Nkx6-2, Erbb3, and Fgf2), and oligodendrocyte differentiation (Thra, Lingo1, Rtn4, p73, and Aspa) (Fig. 7; Table 4). The collective expression changes of these genes predicted a downregulation of the function they regulated (i.e., a decrease in astrogliogenesis, oligodendrogenesis, and oligodendrocyte maturation). Further strengthening this prediction, we detected that several canonical pathways involved in gliogenesis were downregulated: CNTF, IGF1, FGF, and JAK/STAT (Fig. 7; Table 5). In all, the transcriptome analysis pointed to an increase in neurodegeneration-related genes and a decrease in the gliogenic potential of SC-NPCs.
Table 2.
Functional groups predicted to be affected during EAE in caudal NPCsa
| Functions annotation | p value | Prediction | z-score | No. of molecules |
|---|---|---|---|---|
| Formation of cellular protrusions | 8.38E-10 | Decreased | −3.34 | 124 |
| Growth of neurites | 3.87E-08 | Decreased | −2.88 | 77 |
| Morphogenesis of neurites | 4.23E-05 | Decreased | −2.33 | 51 |
| Branching of neurites | 2.18E-04 | Decreased | −2.22 | 34 |
| Dendritic growth/branching | 7.32E-04 | Decreased | −2.19 | 25 |
| Neuritogenesis | 3.27E-06 | Decreased | −2.09 | 75 |
| Differentiation of cells | 1.70E-10 | Decreased | −3.22 | 287 |
| Inflammation of organ | 6.10E-04 | Increased | 4.20 | 108 |
| Synthesis of lipid | 2.04E-05 | Decreased | −2.49 | 104 |
| Synthesis of phospholipid | 2.54E-04 | Decreased | −2.27 | 30 |
| Metabolism of membrane lipid derivative | 9.81E-06 | Decreased | −2.24 | 63 |
| Neurodegeneration | 2.35E-07 | Increased | 2.28 | 41 |
| Degeneration of axons | 4.74E-05 | Increased | 2.55 | 15 |
| Degeneration of neurites | 2.99E-05 | Increased | 2.72 | 16 |
| Apoptosis of neurons | 4.23E-05 | Increased | 4.02 | 59 |
| Cell survival | 1.00E-07 | Decreased | −5.43 | 185 |
| Quantity of cells | 9.97E-11 | Decreased | −5.09 | 259 |
| Cell viability | 1.10E-06 | Decreased | −5.05 | 168 |
aEAE-regulated genes in the caudal NPCs were annotated to functional clusters using IPA. Here we show the functional groups (column 1) that have the strongest prediction rates to be either decreased or increased during EAE. The prediction rate is defined by the z-score, and the predictions with a z-score >2 or <−2 were deemed significant. A negative z-score shows a down-regulated function, whereas a positive z-score shows an up-regulated function. The last column shows the number of molecules annotated to a certain function.
Table 3.
Functional groups belonging to the Nervous System Development and Function category that were affected in caudal NPCs from EAEa
| Functions annotation | p value | Activation | Activation z-score | No. of molecules |
|---|---|---|---|---|
| Myelination | 6.59E-10 | 0.003 | 47 | |
| Growth of neurites | 1.28E-05 | Decreased | −2.875 | 77 |
| Morphology of nervous system | 1.28E-05 | 134 | ||
| Outgrowth of neurites | 3.78E-05 | Decreased | −2.558 | 68 |
| Regeneration of neurites | 1.48E-04 | −0.047 | 15 | |
| Quantity of neuroglia | 1.80E-04 | −0.213 | 22 | |
| Neuritogenesis | 4.13E-04 | Decreased | −2.088 | 75 |
| Quantity of Schwann cells | 5.48E-04 | −1.53 | 7 | |
| Morphology of nervous tissue | 6.18E-04 | 92 | ||
| Development of CNS | 6.39E-04 | −1.452 | 96 | |
| Regeneration of axons | 1.37E-03 | 0.095 | 12 | |
| Myelination of cells | 1.46E-03 | 17 | ||
| Accumulation of neuroglia | 2.09E-03 | 0 | 5 | |
| Quantity of CNS cells | 2.27E-03 | −0.894 | 27 | |
| Morphogenesis of neurites | 2.48E-03 | Decreased | −2.326 | 51 |
| Proliferation of neuroglia | 3.63E-03 | −0.176 | 23 | |
| Morphology of CNS | 4.39E-03 | 80 | ||
| Length of neurites | 4.63E-03 | Decreased | −2.243 | 15 |
| Proliferation of neuroblasts | 5.72E-03 | −0.915 | 17 | |
| Abnormal morphology of PNS glial cells | 5.95E-03 | 8 | ||
| Abnormal morphology of neuroglia | 6.27E-03 | 19 | ||
| Abnormal morphology of oligodendrocytes | 7.09E-03 | 6 | ||
| Branching of neurites | 8.59E-03 | Decreased | −2.223 | 34 |
| Axonogenesis | 9.14E-03 | −0.036 | 32 | |
| Quantity of cholinergic neurons | 9.90E-03 | −0.896 | 5 | |
| Regeneration of nervous tissue | 9.90E-03 | −0.728 | 7 | |
| Morphology of axons | 9.90E-03 | 18 | ||
| Differentiation of neurosphere cells | 1.09E-02 | −0.438 | 6 | |
| Growth of axons | 1.31E-02 | −0.172 | 24 | |
| Auditory evoked potential | 1.35E-02 | Decreased | −2.485 | 16 |
| Morphology of neuroglia | 1.43E-02 | 22 | ||
| Motor function | 1.54E-02 | −1.332 | 19 | |
| Morphology of brain | 1.64E-02 | 70 | ||
| Differentiation of neuroblasts | 1.79E-02 | −0.936 | 7 | |
| Survival of oligodendrocytes | 1.79E-02 | −0.068 | 7 | |
| Abnormal morphology of Schwann cells | 1.79E-02 | 7 | ||
| Abnormal morphology of sciatic nerve | 1.79E-02 | 7 | ||
| Development of brain | 1.89E-02 | −0.998 | 68 | |
| Abnormal morphology of axons | 1.89E-02 | 17 | ||
| Dendritic growth/branching | 1.95E-02 | Decreased | −2.188 | 25 |
| Function of excitatory synapses | 1.95E-02 | 3 |
aGenes in caudal NPCs that were EAE-regulated and belonged to the Nervous System Development and Function category were annotated to functional groups using IPA. Here we show the functional groups (column 1) that have the strongest prediction rates to be either decreased or increased during EAE. The prediction rate is defined by the z-score, and the predictions with a z-score >2 or <−2 were deemed significant. A negative z-score shows a down-regulated function, whereas a positive z-score shows an up-regulated function. The last column shows the number of molecules annotated to a certain function.
Figure 7.
A, Genes related to astroglial and oligodendroglial differentiation in caudal NPCs from EAE versus naive animals. Analysis and figure made in IPA. B, Significantly regulated canonical signaling pathways in NPCs from caudal SC-NPCs of EAE versus naive animals. Green represents downregulation of genes in EAE versus naive values; red represents upregulation of genes in EAE versus naive values. Transparent bars represent no overlap with the dataset. Values on top of each column indicate the total number of genes annotated to a certain pathway.
Table 4.
Genes involved in glial differentiation and supporting publicationsa
| Symbol | Affymetrix ID | FC | FDR | Family | Reference oligodendrocyte differentiation | Reference astrocyte differentiation |
|---|---|---|---|---|---|---|
| ASPA | 10744766 | −3.4 | 0.029 | Enzyme | Francis et al. (2006) | |
| CNTF | 10729024 | −2.5 | 0.042 | Cytokine | Tatebayashi et al. (1999); Setoguchi et al. (2004) | |
| ERBB3 | 10899839 | −2.2 | 0.027 | Kinase | Calaora et al. (2001) | |
| FGF2 | 10815026 | −3.2 | 0.047 | Growth factor | Viti et al. (2003) | |
| FGFR3 | 10777748 | 2.3 | 0.028 | Kinase | Oh et al. (2003) | Oh et al. (2003) |
| IGF1 | 10894695 | 10.6 | 0.045 | Growth factor | Galvin et al. (2010) | |
| IGFBP6 | 10899465 | −2.1 | 0.019 | Other | Russo et al. (2005) | |
| IL6ST | 10813007 | −1.6 | 0.025 | Transmembrane receptor | Nakashima et al. (1999) | |
| LINGO1 | 10917711 | 1.8 | 0.028 | Other | Mi et al. (2005); McDonald et al. (2011) | |
| NKX6-2 | 10726477 | −1.6 | 0.023 | Transcription regulator | Cai et al. (2005) | |
| RTN4 | 10774766 | −1.8 | 0.030 | Other | Pernet et al. (2008) | |
| SERPINE2 | 10929288 | −1.7 | 0.029 | Other | Cavanaugh et al. (1990) | |
| SHH | 10852676 | −1.6 | 0.020 | Peptidase | Nery et al. (2001); Tekki-Kessaris et al. (2001) | Okano-Uchida et al. (2004) |
| STAT3 | 10747506 | −1.5 | 0.034 | Transcription regulator | Rajan et al. (1998); Kamakura et al. (2004) | |
| THRA | 10738056 | −2.0 | 0.047 | Ligand-dependent nuclear receptor | Billon et al. (2002) | |
| TP73 | 10882050 | −1.5 | 0.037 | Transcription regulator | Billon et al. (2004) | |
| UGT8 | 10826607 | −1.8 | 0.033 | Enzyme | Bansal (1999) |
aData were retrieved from IPA. FC, Fold change EAE versus naive.
Table 5.
Significantly changed signaling pathways in caudal NPCs from EAE animalsa
| Ingenuity canonical pathways | −log (B-H p value) | Downregulated | No change | Upregulated | No overlap with dataset |
|---|---|---|---|---|---|
| PTEN signaling | 4.5E00 | 28/135 (21%) | 0/135 (0%) | 2/135 (1%) | 105/135 (78%) |
| PI3K/AKT signaling | 3.74E00 | 28/144 (19%) | 0/144 (0%) | 1/144 (1%) | 115/144 (80%) |
| Axonal guidance signaling | 2.82E00 | 54/440 (12%) | 0/440 (0%) | 8/440 (2%) | 378/440 (86%) |
| Reelin signaling in neurons | 2.81E00 | 16/82 (20%) | 0/82 (0%) | 4/82 (5%) | 62/82 (76%) |
| Signaling by Rho family GTPases | 2.72E00 | 37/252 (15%) | 0/252 (0%) | 4/252 (2%) | 211/252 (84%) |
| Integrin signaling | 2.72E00 | 32/207 (15%) | 0/207 (0%) | 4/207 (2%) | 171/207 (83%) |
| FGF signaling | 2.56E00 | 17/92 (18%) | 0/92 (0%) | 3/92 (3%) | 72/92 (78%) |
| Actin cytoskeleton signaling | 2.56E00 | 33/238 (14%) | 0/238 (0%) | 4/238 (2%) | 201/238 (84%) |
| IGF-1 signaling | 2.49E00 | 20/105 (19%) | 0/105 (0%) | 1/105 (1%) | 84/105 (80%) |
| NGF signaling | 2.38E00 | 22/118 (19%) | 0/118 (0%) | 0/118 (0%) | 96/118 (81%) |
| Ephrin receptor signaling | 2.32E00 | 27/200 (14%) | 0/200 (0%) | 4/200 (2%) | 169/200 (85%) |
| CXCR4 signaling | 2.27E00 | 25/167 (15%) | 0/167 (0%) | 2/167 (1%) | 140/167 (84%) |
| JAK/Stat signaling | 1.94E00 | 14/70 (20%) | 0/70 (0%) | 1/70 (1%) | 55/70 (79%) |
| CNTF signaling | 1.91E00 | 12/55 (22%) | 0/55 (0%) | 0/55 (0%) | 43/55 (78%) |
aTable shows the significantly EAE-regulated canonical pathway in caudal NPCs. The analysis was performed using IPA. The p value is corrected for multiple comparisons using the Benjamini–Hochberg correction. The remaining columns show the number of molecules belonging to a certain pathway that were downregulated (column 3), unchanged (column 4), or upregulated (column 5) in our dataset. The percentages that these molecules constitute from the total number of molecules known to be involved in a certain pathway are shown in parentheses. The last column shows the number and percentage of the remaining molecules not regulated in our dataset but involved in a certain pathway.
Functional validation: increased neurogenesis and decreased oligodendrogenesis in SC-NPC isolated from rats with EAE
To functionally verify the microarray analyses, we determined the effect of EAE on the differentiation potential of NPCs by performing immunocytochemistry analysis and Western blotting for neuronal (β-III tubulin), astroglial (Gfap), and oligodendroglial (GalC) markers on differentiated cultures, derived from naive or EAE animals. The ability to differentiate into the three major cell types of the CNS (neurons, oligodendrocytes, and astrocytes) is the key feature of NPCs and could be affected in neuroinflammation. Figure 8A, B shows the decrease of the oligodendrocyte percentage in EAE-derived SC-NPCs (p < 0.05; Fig. 8B), and this was confirmed using Western blotting for GalC (Fig. 8C). Astrogliogenesis, shown in Figure 8D–F, was also decreased in SC-NPCs, which was consistent with the array results. Although the immunocytochemistry quantification showed a significant decrease of GFAP+ cells only in the thoracic NPCs, Western blotting showed a consistent decrease of the GFAP protein in all three SC groups (Fig. 8F). In contrast to the SC-NPCs, the SVZ-NPCs generated a higher percentage of astrocytes (Fig. 8D,E), whereas oligodendrocyte differentiation was not affected in SVZ-NPCs after inflammation.
Figure 8.
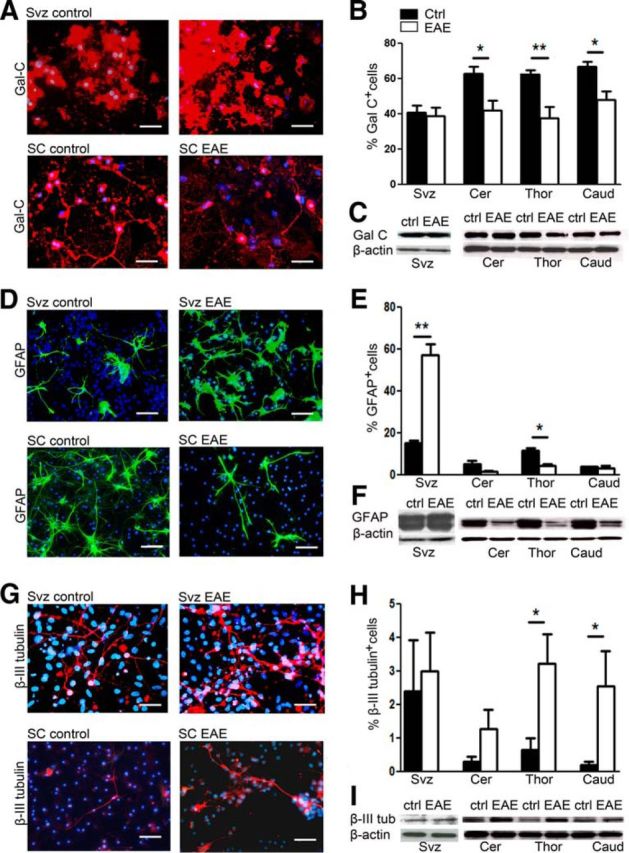
In vitro differentiation of NPCs from SVZ and SC of EAE and naive control animals. A–C, Oligodendrogenesis. A, Differentiated NPCs labeled for oligodendrocytic marker GalC+ (red). Nuclei stained with DAPI (blue). B, Percentage of oligodendrocytes (GalC+/DAPI+) in NPC cultures from SVZ (n = 4–6) and SC cultures (n = 5–7). C, Western blot analysis for GalC and β-actin used as loading control. D–F, Astrogliogenesis. D, Differentiated NPCs labeled for astrocytic marker GFAP (green). E, Percentage of astrocytes (GFAP+/DAPI+) in NPC cultures from SVZ (n = 4–6) and SC (n = 4). F, Western blot analysis for GFAP. G–I, Neurogenesis. G, Differentiated NPCs labeled for neuronal marker β-III tubulin. H, Percentage of neurons (β-III tubulin+/DAPI+) in NPC cultures from SVZ (n = 4–6) and SC (n = 4 or 5). I, Western blot analysis for β-III tubulin. Black bars represent naive controls; white bars represent EAE. Scale bar, 50 μm. Scale bar: G, 25 μm. Data are mean ± SEM. *p < 0.05 (nonparametric Mann–Whitney). **p < 0.01 (nonparametric Mann–Whitney).
In contrast to glial differentiation, the neuronal differentiation was increased in SC-NPC cultures from EAE animals (Figure 8G,I). The Western blot analysis for the neuronal marker, β-III tubulin, was also increased in SC cultures supporting the immunocytochemistry data (Fig. 8H,I). No statistical difference in neuronal differentiation was found between EAE and naive-derived progenitors obtained from the SVZ (Figure 8H).
In all, we demonstrate that EAE induces gene expression changes predominantly in the SC-NPCs, which functionally translates to a decrease of oligodendrocyte and astrocyte differentiation and an increase in neuronal differentiation.
Discussion
The aim of this study was to investigate how SC-NPCs were affected by chronic inflammation, modeled by EAE, an animal model for MS. To that end, we induced EAE in DA rats; and after 30–40 d of disease, we isolated and propagated NPCs from the SCs and SVZ, which was used as a reference from a distant inflammatory site. Using extensive gene expression analyses, we revealed that the most significant changes occurred in NPCs isolated from SC, the main site of inflammatory activity in this EAE model (for review, see Pierson et al., 2012). The transcriptional changes consisted of an upregulation of immune-related genes and an overall downregulation of developmentally/nervous system-related genes, especially that of gliogenesis-related genes. In the SC, functional analyses of the NPC differentiation potential supported this finding and showed a significant decrease in oligodendrocyte and astrocyte differentiation and an increase in neuronal differentiation. This suggests that, during EAE, the otherwise gliogenic SC-NPCs are pushed toward a more neurogenic fate.
SVZ versus SC, active versus nonactive sites of regeneration
In healthy CNS, the sites of regeneration are the SVZ and the hippocampus in the brain, which supply new neurons to the olfactory bulb and subgranular zone, respectively (Lois and Alvarez-Buylla, 1994; Gage, 2000). Importantly, under such healthy conditions, the SC is mostly quiescent. Our results obtained from healthy animals clearly demonstrated a neurogenic potential for SVZ-NPCs in which neurogenic gene expression signatures were significantly enriched compared with SC-NPCs. The neurogenic gene expression signature of the undifferentiated NPCs consisted of Emx2 and FoxG1, involved in telencephalic differentiation (Tao and Lai, 1992; Bishop et al., 2000) and, together with Lhx2, Cxcr4, and Egfr, have been identified by others in the neurogenic gene signature of embryonic cortical NPCs (Kelly et al., 2009). Lhx2, which inhibits astrogliogenesis and promotes neurogenesis in hippocampus (Subramanian et al., 2011), and FoxG1 were also found in the neurogenic gene signature of differentiated NPCs, together with Dcx and Map2, markers of neuroblasts and immature neurons, respectively (Kempermann et al., 2004; Ming and Song, 2005). The higher neurogenicity of the SVZ-NPCs compared with SC-NPCs was further corroborated by functional analyses of NPCs from naive controls revealing that, under comparable culture conditions, SVZ-NPCs generated a higher percentage of neurons (Fig. 4C). Importantly, and in agreement with earlier studies demonstrating the gliogenicity of the SC (Kulbatski et al., 2007; Kulbatski and Tator, 2009; Petit et al., 2011), we found that the SC-NPCs in turn generated a higher percentage of oligodendrocytes (Fig. 4D).
Inflammation alters the differentiation potential of SC-NPCs
Upon exposure to inflammation, the SC-NPC differentiation resulted in a decreased percentage of oligodendrocytes and increased percentage of neurons to the same levels as SVZ-NPC cultures (Fig. 8H). This differentiation effect could have occurred either through a selective effect upon a certain cell population or through an actual diversion of differentiation mechanisms. Speaking in favor of the cell population effect is the fact that we have a decrease in the glia population. Kulbatski et al. (2007), Kulbatski and Tator (2009), and Petit et al. (2011) have characterized the radial glia (RG) population, present at the pial surfaces of the SC. These RG cells have sphere-forming ability but are only producing glial-restricted progenitors and are also present in differentiated spheres. Moreover, these studies show that the RG are absent in the brain but in the SC their abundance increases gradually in a rostrocaudal direction. Thus, in our cultures, which were composed of both ependymal and pial cell subsets, a deleterious effect on radial glia would result in fewer glia cells in differentiated SC-NPCs. Immunolabeling of our differentiated SC-NPC cultures for BLBP, an RG marker, had indeed reveled that differentiated SC-NPC cultures from EAE animals were completely devoid of BLBP, whereas up to 30% of the corresponding controls were BLBP+ (data not shown). On the other hand, the increase of the Olig2+/Ascl1+ double-positive population (Fig. 5O) in the immature SC-NPCs cannot be explained by a deleterious effect on another cell population but rather by a possible instructive effect of EAE on neurogenesis.
It is well established that inflammation induces NPC activity, and various inflammatory factors have been shown to have priming effects on NPC differentiation (Butovsky et al., 2006; Covacu et al., 2006). In an earlier study using the same DA rat model of EAE, we observed BrdU+/NeuN+ cells that had originated from the ependymal regions of the SC and migrated into the inflammatory lesions (Danilov et al., 2006). Several cytokines and inflammatory factors have been shown to have neurogenic effects on NPCs, and one of them is IFNγ, a pivotal cytokine in the inflammatory reaction in EAE. Interestingly, IFNγ exerts opposite effects in the brain and SC inflammation, acting anti-inflammatory in the brain and proinflammatory in the SC (Wensky et al., 2005; Lees et al., 2008) and reviewed by Pierson et al. (2012). The discrepancy of the IFNγ effect in the different CNS compartments might explain results from Tepavčević et al. (2011). In a model of targeted EAE with periventricular brain lesions, this group showed that the inflammation rendered the SVZ-NPCs more gliogenic, producing higher numbers of Olig2+ cells and less neurogenic, generating fewer neuroblasts (Tepavčević et al., 2011). These findings contrast with our results on SC cells where the gliogenic preference of the SC-NPCs was decreased during inflammation, indicating that the NPC populations of the CNS are not only intrinsically different but respond differently to inflammation. In addition, the response of the NPCs to various types of inflammation might vary. For instance, we have compared our dataset with a study performed by Lee et al. (2013) who analyzed the regulation of stem cell-related genes in spinal cord injury. Fifteen genes overlapped between our datasets. All these genes were upregulated in the spinal cord injury, whereas in our dataset only three were upregulated while the rest of 12 were downregulated (data not shown), indicating that the outcome of the inflammatory exposure on the NPCs might have the opposite effect in spinal cord injury than in EAE.
The decreased gliogenic preference of NPCs in our analyses was supported by the decrease of several pivotal gliogenic factors and their signaling pathways. Among these signaling pathways, we counted CNTF, IGF1, and FGF. The expression of Igf1, which has been reported to support oligodendrocyte differentiation and maturation (Galvin et al., 2010), was increased 10-fold in EAE-exposed NPCs. However, all its downstream signaling pathways, among them JAK/STAT, also involved in CNTF signaling, were downregulated, possibly counteracting the benefit of the Igf1 upregulation. Importantly, loss of remyelination capacity, with underlying dysfunction in oligodendrocyte differentiation, is a hallmark of chronic MS (Patrikios et al., 2006). In addition to the functions associated with oligodendrocyte differentiation, the array data gave strong predictions associated with lipid metabolism and neurodegeneration. It is well established that, in MS, normal-appearing white matter can harbor degenerating axons, which can be one of the features involved in the pathology of chronic MS (Bjartmar et al., 2001). Recently, Laule et al. (2013) described the features of diffusely abnormal white matter in MS patients where loss of myelin lipids was accompanied by neurodegeneration. It is noteworthy that NPCs exposed a relatively short time to inflammation in vivo display such an extensive upregulation of gene signatures involved in neurodegeneration. This might reflect an in vivo situation where inflammation conditions the NPC's transcriptional program to support pathological mechanisms.
In the present study, we demonstrate that SC-NPCs differ from SVZ-NPCs transcriptionally and functionally. In the healthy CNS, the SC-NPCs were more gliogenic whereas the SVZ-NPCs were more neurogenic. The gliogenicity of the SC-NPCs was decreased by chronic inflammation that rendered the cells more neurogenic. This finding has important clinical implications for chronic inflammatory diseases, such as MS, where NPCs in the different CNS compartments might be affected differently by inflammation. Understanding the heterogeneity of NPCs and their response to inflammation is pivotal for understanding the disease etiology and serve as a starting point for future treatment designs.
Footnotes
This work was supported by the Swedish Research Council, Torsten and Ragnar Soderberg Foundation, Swedish Association of Persons with Neurological Disabilities, Lars Hierta Memorial Foundation, and Karolinska Institutet. We thank the staff, with special thanks to David Brodin, at the Bioinformatics and Expression Analysis Core facility at Karolinska Institutet, Stockholm, Sweden for valuable assistance with array hybridizations and basic data analysis; and Arvid Frostell for assistance with the artwork.
The authors declare no competing financial interests.
References
- Aktan F. iNOS-mediated nitric oxide production and its regulation. Life Sci. 2004;75:639–653. doi: 10.1016/j.lfs.2003.10.042. [DOI] [PubMed] [Google Scholar]
- Amor S, Groome N, Linington C, Morris MM, Dornmair K, Gardinier MV, Matthieu JM, Baker D. Identification of epitopes of myelin oligodendrocyte glycoprotein for the induction of experimental allergic encephalomyelitis in SJL and Biozzi AB/H mice. J Immunol. 1994;153:4349–4356. [PubMed] [Google Scholar]
- Bansal R, Winkler S, Bheddah S. Negative regulation of oligodendrocyte differentiation by galactosphingolipids. J Neurosci. 1999;19:7913–7924. doi: 10.1523/JNEUROSCI.19-18-07913.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billon N, Jolicoeur C, Tokumoto Y, Vennstrom B, Raff M. Normal timing of oligodendrocyte development depends on thyroid hormone receptor alpha 1 (TRalpha1) EMBO J. 2002;21:6452–6460. doi: 10.1093/emboj/cdf662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billon N, Terrinoni A, Jolicoeur C, McCarthy A, Richardson WD, Melino G, Raff M. Roles for p53 and p73 during oligodendrocyte development. Development. 2004;131:1211–1220. doi: 10.1242/dev.01035. [DOI] [PubMed] [Google Scholar]
- Bishop KM, Goudreau G, O'Leary DD. Regulation of area identity in the mammalian neocortex by Emx2 and Pax6. Science. 2000;288:344–349. doi: 10.1126/science.288.5464.344. [DOI] [PubMed] [Google Scholar]
- Bjartmar C, Kinkel RP, Kidd G, Rudick RA, Trapp BD. Axonal loss in normal-appearing white matter in a patient with acute MS. Neurology. 2001;57:1248–1252. doi: 10.1212/WNL.57.7.1248. [DOI] [PubMed] [Google Scholar]
- Brundin L, Brismar H, Danilov AI, Olsson T, Johansson CB. Neural stem cells: a potential source for remyelination in neuroinflammatory disease. Brain Pathol. 2003;13:322–328. doi: 10.1111/j.1750-3639.2003.tb00031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butovsky O, Ziv Y, Schwartz A, Landa G, Talpalar AE, Pluchino S, Martino G, Schwartz M. Microglia activated by IL-4 or IFN-gamma differentially induce neurogenesis and oligodendrogenesis from adult stem/progenitor cells. Mol Cell Neurosci. 2006;31:149–160. doi: 10.1016/j.mcn.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Cai J, Qi Y, Hu X, Tan M, Liu Z, Zhang J, Li Q, Sander M, Qiu M. Generation of oligodendrocyte precursor cells from mouse dorsal spinal cord independent of Nkx6 regulation and Shh signaling. Neuron. 2005;45:41–53. doi: 10.1016/j.neuron.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Calaora V, Rogister B, Bismuth K, Murray K, Brandt H, Leprince P, Marchionni M, Dubois-Dalcq M. Neuregulin signaling regulates neural precursor growth and the generation of oligodendrocytes in vitro. J Neurosci. 2001;21:4740–4751. doi: 10.1523/JNEUROSCI.21-13-04740.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanaugh KP, Gurwitz D, Cunningham DD, Bradshaw RA. Reciprocal modulation of astrocyte stellation by thrombin and protease nexin-1. J Neurochem. 1990;54:1735–1743. doi: 10.1111/j.1471-4159.1990.tb01228.x. [DOI] [PubMed] [Google Scholar]
- Covacu R, Danilov AI, Rasmussen BS, Hallén K, Moe MC, Lobell A, Johansson CB, Svensson MA, Olsson T, Brundin L. Nitric oxide exposure diverts neural stem cell fate from neurogenesis towards astrogliogenesis. Stem Cells. 2006;24:2792–2800. doi: 10.1634/stemcells.2005-0640. [DOI] [PubMed] [Google Scholar]
- Danilov AI, Covacu R, Moe MC, Langmoen IA, Johansson CB, Olsson T, Brundin L. Neurogenesis in the adult spinal cord in an experimental model of multiple sclerosis. Eur J Neurosci. 2006;23:394–400. doi: 10.1111/j.1460-9568.2005.04563.x. [DOI] [PubMed] [Google Scholar]
- Doetsch F, Caillé I, Lim DA, García-Verdugo JM, Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97:703–716. doi: 10.1016/S0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- Duncan DT, Prodduturi N, Zhang B. WebGestalt2: an updated and expanded version of the Web-based Gene Set Analysis Toolkit. BMC Conformations. 2010:1. [Google Scholar]
- Francis JS, Olariu A, McPhee SW, Leone P. Novel role for aspartoacylase in regulation of BDNF and timing of postnatal oligodendrogenesis. J Neurosci Res. 2006;84:151–169. doi: 10.1002/jnr.20866. [DOI] [PubMed] [Google Scholar]
- Frisén J, Johansson CB, Török C, Risling M, Lendahl U. Rapid, widespread, and longlasting induction of nestin contributes to the generation of glial scar tissue after CNS injury. J Cell Biol. 1995;131:453–464. doi: 10.1083/jcb.131.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage FH. Mammalian neural stem cells. Science. 2000;287:1433–1438. doi: 10.1126/science.287.5457.1433. [DOI] [PubMed] [Google Scholar]
- Galvin J, Eyermann C, Colognato H. Dystroglycan modulates the ability of insulin-like growth factor-1 to promote oligodendrocyte differentiation. J Neurosci Res. 2010;88:3295–3307. doi: 10.1002/jnr.22484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Johansson CB, Momma S, Clarke DL, Risling M, Lendahl U, Frisén J. Identification of a neural stem cell in the adult mammalian central nervous system. Cell. 1999;96:25–34. doi: 10.1016/S0092-8674(00)80956-3. [DOI] [PubMed] [Google Scholar]
- Johansson S, Price J, Modo M. Effect of inflammatory cytokines on major histocompatibility complex expression and differentiation of human neural stem/progenitor cells. Stem Cells. 2008;26:2444–2454. doi: 10.1634/stemcells.2008-0116. [DOI] [PubMed] [Google Scholar]
- Kamakura S, Oishi K, Yoshimatsu T, Nakafuku M, Masuyama N, Gotoh Y. Hes binding to STAT3 mediates crosstalk between Notch and JAK-STAT signalling. Nat Cell Biol. 2004;6:547–554. doi: 10.1038/ncb1138. [DOI] [PubMed] [Google Scholar]
- Kehl LJ, Fairbanks CA, Laughlin TM, Wilcox GL. Neurogenesis in postnatal rat spinal cord: a study in primary culture. Science. 1997;276:586–589. doi: 10.1126/science.276.5312.586. [DOI] [PubMed] [Google Scholar]
- Kelly TK, Karsten SL, Geschwind DH, Kornblum HI. Cell lineage and regional identity of cultured spinal cord neural stem cells and comparison to brain-derived neural stem cells. PLoS One. 2009;4:e4213. doi: 10.1371/journal.pone.0004213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempermann G, Jessberger S, Steiner B, Kronenberg G. Milestones of neuronal development in the adult hippocampus. Trends Neurosci. 2004;27:447–452. doi: 10.1016/j.tins.2004.05.013. [DOI] [PubMed] [Google Scholar]
- Kulbatski I, Tator CH. Region-specific differentiation potential of adult rat spinal cord neural stem/precursors and their plasticity in response to in vitro manipulation. J Histochem Cytochem. 2009;57:405–423. doi: 10.1369/jhc.2008.951814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulbatski I, Mothe AJ, Keating A, Hakamata Y, Kobayashi E, Tator CH. Oligodendrocytes and radial glia derived from adult rat spinal cord progenitors: morphological and immunocytochemical characterization. J Histochem Cytochem. 2007;55:209–222. doi: 10.1369/jhc.6A7020.2006. [DOI] [PubMed] [Google Scholar]
- Laule C, Pavlova V, Leung E, Zhao G, MacKay AL, Kozlowski P, Traboulsee AL, Li DK, Moore GR. Diffusely abnormal white matter in multiple sclerosis: further histologic studies provide evidence for a primary lipid abnormality with neurodegeneration. J Neuropathol Exp Neurol. 2013;72:42–52. doi: 10.1097/NEN.0b013e31827bced3. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Wu J, Chung J, Wrathall JR. SOX2 expression is upregulated in adult spinal cord after contusion injury in both oligodendrocyte lineage and ependymal cells. J Neurosci Res. 2013;91:196–210. doi: 10.1002/jnr.23151. [DOI] [PubMed] [Google Scholar]
- Lees JR, Golumbek PT, Sim J, Dorsey D, Russell JH. Regional CNS responses to IFN-gamma determine lesion localization patterns during EAE pathogenesis. J Exp Med. 2008;205:2633–2642. doi: 10.1084/jem.20080155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois C, Alvarez-Buylla A. Long-distance neuronal migration in the adult mammalian brain. Science. 1994;264:1145–1148. doi: 10.1126/science.8178174. [DOI] [PubMed] [Google Scholar]
- McDonald CL, Bandtlow C, Reindl M. Targeting the Nogo receptor complex in diseases of the central nervous system. Curr Med Chem. 2011;18:234–244. doi: 10.2174/092986711794088326. [DOI] [PubMed] [Google Scholar]
- McKay R. Stem cells in the central nervous system. Science. 1997;276:66–71. doi: 10.1126/science.276.5309.66. [DOI] [PubMed] [Google Scholar]
- Mi S, Miller RH, Lee X, Scott ML, Shulag-Morskaya S, Shao Z, Chang J, Thill G, Levesque M, Zhang M, Hession C, Sah D, Trapp B, He Z, Jung V, McCoy JM, Pepinsky RB. LINGO-1 negatively regulates myelination by oligodendrocytes. Nat Neurosci. 2005;8:745–751. doi: 10.1038/nn1460. [DOI] [PubMed] [Google Scholar]
- Ming GL, Song H. Adult neurogenesis in the mammalian central nervous system. Annu Rev Neurosci. 2005;28:223–250. doi: 10.1146/annurev.neuro.28.051804.101459. [DOI] [PubMed] [Google Scholar]
- Monje ML, Toda H, Palmer TD. Inflammatory blockade restores adult hippocampal neurogenesis. Science. 2003;302:1760–1765. doi: 10.1126/science.1088417. [DOI] [PubMed] [Google Scholar]
- Mothe AJ, Tator CH. Proliferation, migration, and differentiation of endogenous ependymal region stem/progenitor cells following minimal spinal cord injury in the adult rat. Neuroscience. 2005;131:177–187. doi: 10.1016/j.neuroscience.2004.10.011. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Yanagisawa M, Arakawa H, Taga T. Astrocyte differentiation mediated by LIF in cooperation with BMP2. FEBS Lett. 1999a;457:43–46. doi: 10.1016/S0014-5793(99)00997-7. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Wiese S, Yanagisawa M, Arakawa H, Kimura N, Hisatsune T, Yoshida K, Kishimoto T, Sendtner M, Taga T. Developmental requirement of gp130 signaling in neuronal survival and astrocyte differentiation. J Neurosci. 1999b;19:5429–5434. doi: 10.1523/JNEUROSCI.19-13-05429.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nery S, Wichterle H, Fishell G. Sonic hedgehog contributes to oligodendrocyte specification in the mammalian forebrain. Development. 2001;128:527–540. doi: 10.1242/dev.128.4.527. [DOI] [PubMed] [Google Scholar]
- Oh LY, Denninger A, Colvin JS, Vyas A, Tole S, Ornitz DM, Bansal R. Fibroblast growth factor receptor 3 signaling regulates the onset of oligodendrocyte terminal differentiation. J Neurosci. 2003;23:883–894. doi: 10.1523/JNEUROSCI.23-03-00883.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano-Uchida T, Himi T, Komiya Y, Ishizaki Y. Cerebellar granule cell precursors can differentiate into astroglial cells. Proc Natl Acad Sci U S A. 2004;101:1211–1216. doi: 10.1073/pnas.0307972100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer MA, Stasiv Y, Benraiss A, Chmielnicki E, Grinberg A, Westphal H, Goldman SA, Enikolopov G. Nitric oxide negatively regulates mammalian adult neurogenesis. Proc Natl Acad Sci U S A. 2003;100:9566–9571. doi: 10.1073/pnas.1633579100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ, Citri A, Sebastiano V, Marro S, Südhof TC, Wernig M. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476:220–223. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrikios P, Stadelmann C, Kutzelnigg A, Rauschka H, Schmidbauer M, Laursen H, Sorensen PS, Brück W, Lucchinetti C, Lassmann H. Remyelination is extensive in a subset of multiple sclerosis patients. Brain. 2006;129:3165–3172. doi: 10.1093/brain/awl217. [DOI] [PubMed] [Google Scholar]
- Pernet V, Joly S, Christ F, Dimou L, Schwab ME. Nogo-A and myelin-associated glycoprotein differently regulate oligodendrocyte maturation and myelin formation. J Neurosci. 2008;28:7435–7444. doi: 10.1523/JNEUROSCI.0727-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit A, Sanders AD, Kennedy TE, Tetzlaff W, Glattfelder KJ, Dalley RA, Puchalski RB, Jones AR, Roskams AJ. Adult spinal cord radial glia display a unique progenitor phenotype. PLoS One. 2011;6:e24538. doi: 10.1371/journal.pone.0024538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard-Riera N, Decker L, Delarasse C, Goude K, Nait-Oumesmar B, Liblau R, Pham-Dinh D, Baron-Van Evercooren A. Experimental autoimmune encephalomyelitis mobilizes neural progenitors from the subventricular zone to undergo oligodendrogenesis in adult mice. Proc Natl Acad Sci U S A. 2002;99:13211–13216. doi: 10.1073/pnas.192314199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson E, Simmons SB, Castelli L, Goverman JM. Mechanisms regulating regional localization of inflammation during CNS autoimmunity. Immunol Rev. 2012;248:205–215. doi: 10.1111/j.1600-065X.2012.01126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluchino S, Muzio L, Imitola J, Deleidi M, Alfaro-Cervello C, Salani G, Porcheri C, Brambilla E, Cavasinni F, Bergamaschi A, Garcia-Verdugo JM, Comi G, Khoury SJ, Martino G. Persistent inflammation alters the function of the endogenous brain stem cell compartment. Brain. 2008;131:2564–2578. doi: 10.1093/brain/awn198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan P, Gearan T, Fink JS. Leukemia inhibitory factor and NGF regulate signal transducers and activators of transcription activation in sympathetic ganglia: convergence of cytokine- and neurotrophin-signaling pathways. Brain Res. 1998;802:198–204. doi: 10.1016/S0006-8993(98)00611-8. [DOI] [PubMed] [Google Scholar]
- Russo VC, Schutt BS, Andaloro E, Ymer SI, Hoeflich A, Ranke MB, Bach LA, Werther GA. Insulin-like growth factor binding protein-2 binding to extracellular matrix plays a critical role in neuroblastoma cell proliferation, migration, and invasion. Endocrinology. 2005;146:4445–4455. doi: 10.1210/en.2005-0467. [DOI] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Setoguchi T, Kondo T. Nuclear export of OLIG2 in neural stem cells is essential for ciliary neurotrophic factor-induced astrocyte differentiation. J Cell Biol. 2004;166:963–968. doi: 10.1083/jcb.200404104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shihabuddin LS, Ray J, Gage FH. FGF-2 is sufficient to isolate progenitors found in the adult mammalian spinal cord. Exp Neurol. 1997;148:577–586. doi: 10.1006/exnr.1997.6697. [DOI] [PubMed] [Google Scholar]
- Sommer L, Ma Q, Anderson DJ. Neurogenins, a novel family of atonal-related bHLH transcription factors, are putative mammalian neuronal determination genes that reveal progenitor cell heterogeneity in the developing CNS and PNS. Mol Cell Neurosci. 1996;8:221–241. doi: 10.1006/mcne.1996.0060. [DOI] [PubMed] [Google Scholar]
- Storch MK, Stefferl A, Brehm U, Weissert R, Wallström E, Kerschensteiner M, Olsson T, Linington C, Lassmann H. Autoimmunity to myelin oligodendrocyte glycoprotein in rats mimics the spectrum of multiple sclerosis pathology. Brain Pathol. 1998;8:681–694. doi: 10.1111/j.1750-3639.1998.tb00194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian L, Sarkar A, Shetty AS, Muralidharan B, Padmanabhan H, Piper M, Monuki ES, Bach I, Gronostajski RM, Richards LJ, Tole S. Transcription factor Lhx2 is necessary and sufficient to suppress astrogliogenesis and promote neurogenesis in the developing hippocampus. Proc Natl Acad Sci U S A. 2011;108:E265–E274. doi: 10.1073/pnas.1101109108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebayashi H, Yoshida S, Sugimori M, Kosako H, Kominami R, Nakafuku M, Nabeshima Y. Dynamic expression of basic helix-loop-helix Olig family members: implication of Olig2 in neuron and oligodendrocyte differentiation and identification of a new member, Olig3. Mech Dev. 2000;99:143–148. doi: 10.1016/S0925-4773(00)00466-4. [DOI] [PubMed] [Google Scholar]
- Tao W, Lai E. Telencephalon-restricted expression of BF-1, a new member of the HNF-3/fork head gene family, in the developing rat brain. Neuron. 1992;8:957–966. doi: 10.1016/0896-6273(92)90210-5. [DOI] [PubMed] [Google Scholar]
- Tatebayashi Y, Iqbal K, Grundke-Iqbal I. Dynamic regulation of expression and phosphorylation of tau by fibroblast growth factor-2 in neural progenitor cells from adult rat hippocampus. J Neurosci. 1999;19:5245–5254. doi: 10.1523/JNEUROSCI.19-13-05245.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekki-Kessaris N, Woodruff R, Hall AC, Gaffield W, Kimura S, Stiles CD, Rowitch DH, Richardson WD. Hedgehog-dependent oligodendrocyte lineage specification in the telencephalon. Development. 2001;128:2545–2554. doi: 10.1242/dev.128.13.2545. [DOI] [PubMed] [Google Scholar]
- Tepavčević V, Lazarini F, Alfaro-Cervello C, Kerninon C, Yoshikawa K, Garcia-Verdugo JM, Lledo PM, Nait-Oumesmar B, Baron-Van Evercooren A. Inflammation-induced subventricular zone dysfunction leads to olfactory deficits in a targeted mouse model of multiple sclerosis. J Clin Invest. 2011;121:4722–4734. doi: 10.1172/JCI59145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thored P, Arvidsson A, Cacci E, Ahlenius H, Kallur T, Darsalia V, Ekdahl CT, Kokaia Z, Lindvall O. Persistent production of neurons from adult brain stem cells during recovery after stroke. Stem Cells. 2006;24:739–747. doi: 10.1634/stemcells.2005-0281. [DOI] [PubMed] [Google Scholar]
- Weiss S, Dunne C, Hewson J, Wohl C, Wheatley M, Peterson AC, Reynolds BA. Multipotent CNS stem cells are present in the adult mammalian spinal cord and ventricular neuroaxis. J Neurosci. 1996;16:7599–7609. doi: 10.1523/JNEUROSCI.16-23-07599.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wensky AK, Furtado GC, Marcondes MC, Chen S, Manfra D, Lira SA, Zagzag D, Lafaille JJ. IFN-gamma determines distinct clinical outcomes in autoimmune encephalomyelitis. J Immunol. 2005;174:1416–1423. doi: 10.4049/jimmunol.174.3.1416. [DOI] [PubMed] [Google Scholar]
- Whitney NP, Eidem TM, Peng H, Huang Y, Zheng JC. Inflammation mediates varying effects in neurogenesis: relevance to the pathogenesis of brain injury and neurodegenerative disorders. J Neurochem. 2009;108:1343–1359. doi: 10.1111/j.1471-4159.2009.05886.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viti J, Feathers A, Phillips J, Lillien L. Epidermal growth factor receptors control competence to interpret leukemia inhibitory factor as an astrocyte inducer in developing cortex. J Neurosci. 2003;23:3385–3393. doi: 10.1523/JNEUROSCI.23-08-03385.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong G, Goldshmit Y, Turnley AM. Interferon-gamma but not TNF alpha promotes neuronal differentiation and neurite outgrowth of murine adult neural stem cells. Exp Neurol. 2004;187:171–177. doi: 10.1016/j.expneurol.2004.01.009. [DOI] [PubMed] [Google Scholar]
- Ziv Y, Ron N, Butovsky O, Landa G, Sudai E, Greenberg N, Cohen H, Kipnis J, Schwartz M. Immune cells contribute to the maintenance of neurogenesis and spatial learning abilities in adulthood. Nat Neurosci. 2006;9:268–275. doi: 10.1038/nn1629. [DOI] [PubMed] [Google Scholar]