Abstract
Single molecule fluorescence energy transfer methods allow us to determine the complete structural landscape between the donor and acceptor fluorophores introduced on the protein of interest. This method is particularly attractive to study ion channel proteins as single molecule current recordings have been used to study the function of these proteins for several decades. Here we describe the smFRET method used to study glutamate receptors.
Keywords: Single molecule FRET, fluorescence, NMDA receptor, glutamate receptor
1. Introduction
The first biological x-ray crystallography experiments characterizing myoglobin and hemoglobin not only opened our eyes to the relation of function and form in biology, but also revealed the utility of structural biology for studying the evolutionary origins of proteins and for designing useful drugs. X-ray crystallography and cryo-electron microscopy have since emerged as the front runners in the field of structural biology and have provided new insight into all fields of biological science. These techniques will without question continue to lead the way in structural biology. They both, however, suffer from the limitation that they lack the ability to provide the complete structural landscape, dynamics in terms of transitions between states and energetics of transitions between states. This limitation in current methods has created a niche for studying the structure of unresolvable molecules in addition to validating and studying the true dynamics of previously resolved x-ray and cryo-EM models (1–2). Fluorescent spectroscopy, in particular single-molecule fluorescent resonance energy transfer (smFRET), has emerged as the leading method for this role (3–5).
smFRET has become of particular interest to groups studying highly dynamic proteins and proteins composed of multiple domains acting in concert to perform mechanically complex motions. Initial efforts have focused on characterizing the structure of DNA/RNA molecules, DNA/RNA protein complexes, enzymes, signaling proteins, and ion channels (3–21). Ionotropic glutamate receptors (iGluRs) are a family of ion channels that serve as a perfect subject for smFRET studies (1–5,8,21). iGluRs are multimeric, have four domains with little functional overlap, operate with millisecond kinetics, and have previously-resolved x-ray and cyro-EM models.
iGluRs are tetrameric proteins with two extracellular domains, a transmembrane domain (TMD), and an intercellular C-terminal domain. iGluRs are divided by their selective agonists into AMPA, kainite, and NMDA subgroups, and some subgroups can express as homomers or heteromers depending on the subunits used. The extracellular amino terminal domain (NTD) and ligand binding domain (LBD) are of particular interest for smFRET studies due to the complex rearrangements proposed during the operation of the protein. In this chapter we will discuss the approach to and execution of smFRET to study the conformational dynamics of iGluRs.
1.1. Förster resonance energy transfer
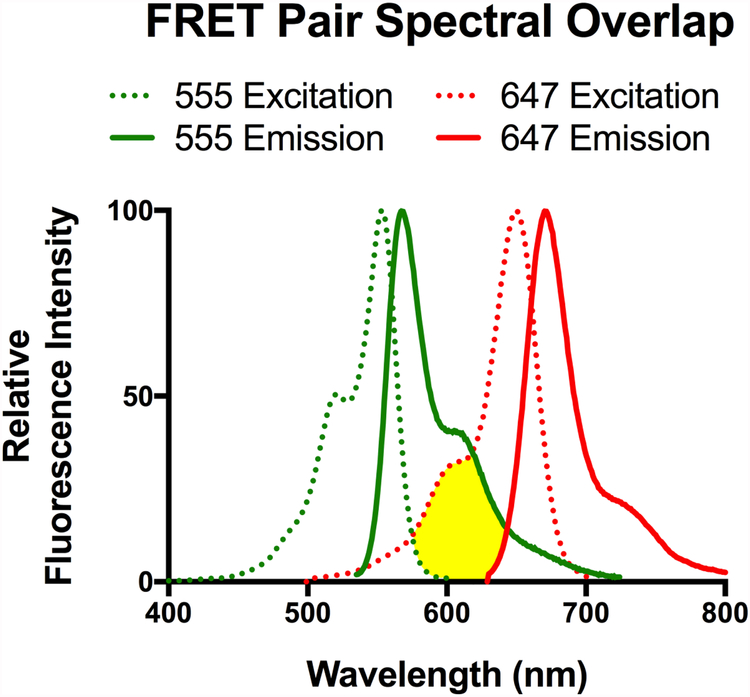
Förster (or fluorescent) resonance energy transfer (FRET) is a term describing the non-radiative transfer of energy from a donor fluorophore to an acceptor fluorophore. This relay of energy relies on the overlap of the donor and acceptor spectra, specifically that the emission of the donor overlaps with the excitation of the acceptor as seen in Figure 1. This phenomenon is distance-dependent where the closer the fluorophore pairs are in space the more likely they are to FRET. The efficiency of energy transfer is related to the inter-dye distance (RDA) and the distance of half-maximal energy transfer for a given fluorophore set (R0), as seenin equation 1. The effective distance of FRET for fluorophores ranges between 10 and 100 Angstroms, making FRET ideal for measuring distances at the protein level.
Figure 1:
Spectral data for the excitation (Dotted) and emission (Solid) wavelengths of Alexa 555 (Green) and Alexa 647 (Red).
| Eq. 1 |
When choosing a fluorophore pair for a FRET experiment the most important factors to consider are the distance intended to be measured and the R0 of the fluorophore pair (see Note 1). The factors contributing to the R0 of a fluorophore pair are the orientation factor, k; the quantum yield of the donor, ΦD; the spectral overlap integral, J; and the refractive index of the media, n. The relationship between these values and the R0 is shown in equation 2. There are many detailed reviews on the physics of FRET if additional information is needed (22).
| Eq. 2 |
1.2. Single-molecule Förster resonance energy transfer
In traditional FRET experiments, a measurement is made on a large population of molecules, and the signal obtained is an ensemble their collective behavior. This is useful when looking at large protein movements and proximity; however, these measurements provide an ensemble average and the complete structural landscape along with transitions between the states cannot be resolved. To circumvent this, total internal reflection microscopy, 2-photon excitation microscopy, and confocal microscopy have been adopted to bring measurements to the single molecule level of both immobilized and freely diffusing molecules as well as in living cells. These advantages of smFRET have vastly improved our ability to characterize sub-millisecond dynamics in protein, DNA, and RNA in addition to resolving protein subunit stoichiometry. However, collecting data with such detail does not come without its caveats. Data analysis has become the most difficult part of smFRET and methods for collecting and analyzing data have had to evolve. The advent of multiparameter fluorescence detection (MFD), time correlated single photon counting (TCSPC), pulsed interleaved excitation (PIE), and fluorescence correlation spectroscopy (FCS) have vastly improved the potential quality of smFRET data, discussed further in the data analysis section. Instruments capable of collecting data using MFD, FCS, PIE and TCSPC simultaneously are now available. The collective use of these techniques has greatly improved our ability to resolve the inherent complications of fluorophore photophysics and movements from true FRET signal (23). In this chapter we will focus on the smFRET measurements using confocal imaging on slide attached iGluR.
1.3. Experimental design
When designing a smFRET experiment studying iGluRs, the labeling strategy used will depend on the subunit being studied and the intended measurement. The main design step is site selection and considerations here will be the subunit arrangement, distance to be measured, and fluorophore pairs.
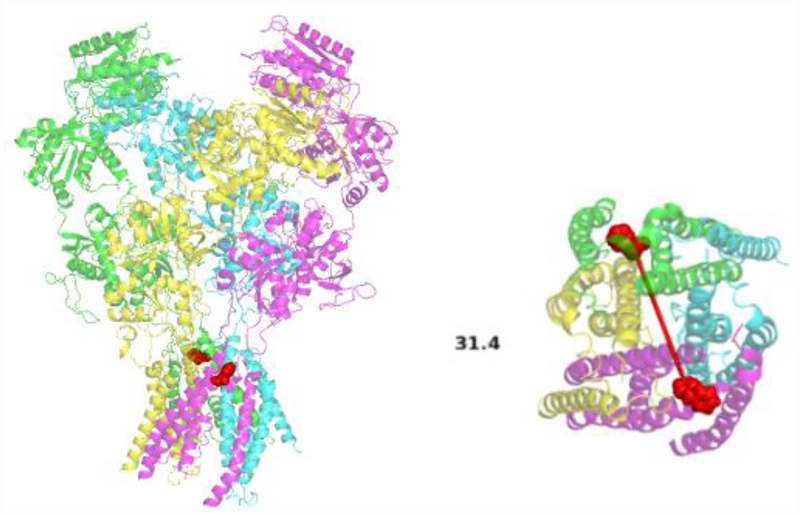
In a heteromeric iGluR such as the NMDA receptor there will be two sets of the subunits arranged in a defined orientation. By taking this into account it is possible to pick a surface-exposed amino acid position on either of the subunits used that is 0.5-1X the R0 of the fluorophore pair. In this case labeling with a site specific cysteine is recommended. For example, in Figure 2 the heteromeric NMDA receptor is shown with transmembrane residues selected to measure the change in distance across the axis of the pore. Only the highlighted residues will be labelled and measured.
Figure 2:
A structural model of the NMDA receptor showing phenylalanine 554 (Red) as the fluorescent labeling site for measurements across the ion pore (PDB:5UP2) (27), side view (left) and top view (right).
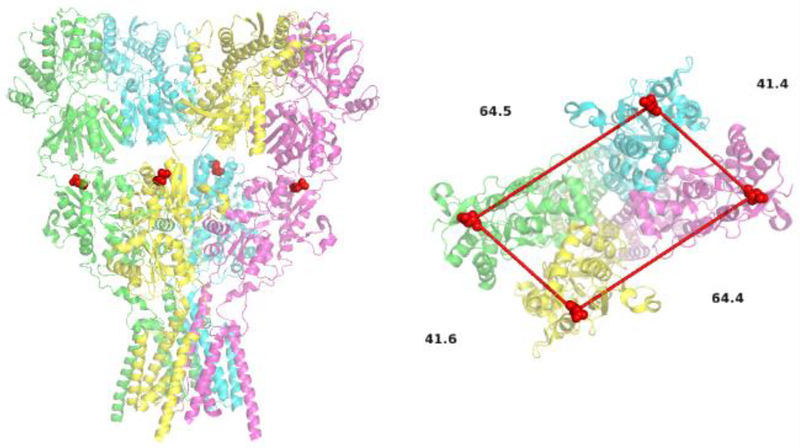
If using a homomeric iGluR such as in homomeric AMPA receptors, choosing sites for study could be more complicated. This is the result of having the site of interest on all four subunits. In this case, it is required that the site of interest has one high FRET distance 0.5–1X the R0 and one distance ideally well above the R0 of the fluorophore pair for a given state. As seen in Figure 3, the site chosen will have a primary FRET distance of 41 Å with an additional distance of 64 Å across dimer pairs. This ensures that the main FRET component, 41 Å, is 0.5-1X the R0 and will be easily resolved from the longer component. In this chapter we will focus on the use of heteromeric iGluRs.
Figure 3:
A structural model of the AMPA receptor showing aspartic acid 473 (Red) as the fluorescent labeling site for measurements between the LBDs (PDB:5VHZ) (28), side view (left) and top view (right).
When studying full length iGluRs, immobilizing the protein on slide via antibody pull-down is preferred (see Note 2). Any antibody with high specificity will work; it is recommended, however, to introduce an affinity tag at the C-terminus to minimize the effect of antibody binding on protein function. When using a new antibody it is critical to run western blots to confirm specificity of the antibody (see Note 3). This chapter will focus on the use of immobilized molecules.
1.4. Cysteine labeling
The best option for fluorophore labeling for smFRET is by introducing a cysteine at the site of interest. This allows use of many commonly available thiol-reactive fluorophores and retains high expression levels. This does however require that all the native non-disulfide bonded cysteines be mutated to provide a clean background. Once this is achieved, the expression and function of the cysteine-less construct must be verified (see Note 4). When approaching the design of an experiment it is ideal that one choose sites that will show a change in distance and will also have one effective FRET distance. It is not always possible to achieve the ideal labeling strategy in every region of the protein, or the measurement may need to be made within the subunit itself. If either of these is the case, utilizing unnatural amino acids allows for additional site specific labeling.
1.5. Unnatural amino acid labeling
Unnatural amino acids are advantageous in that one retains the native coding sequence of the protein; however additional plasmids must be maintained for use and expression of the protein needs to be verified by western blotting and function of the protein verified. The principle behind utilizing unnatural amino acids is using a tRNA and aminoacyl-tRNA synthetase that have been artificially evolved to suppress the amber stop codon (TAG). The TAG can be inserted by mutagenesis into any position along the protein and, when co-transfected with plasmids containing the tRNA and aminoacyl-tRNA synthetase, will be suppressed by the incorporation of the unnatural amino acid into the peptide chain (see Note 5) (24).
2. Materials
2.1. Protein Expression
HEK 293 cells maintained at 37°C and 5% CO2
DMEM media with high glucose, L-glutamine, 1 unit/mL Penicillin, 1 ng/mL Streptomycin, and 10% FBS, without sodium pyruvate
Phosphate buffered saline
Trypsin-EDTA: 0.25% Trypsin, 1 mM EDTA
Plasmid DNA at a concentration preferably > 1 μg/μl
Polyplus Jetprime transfection buffer
Polyplus Jetprime transfection reagent
If expressing NMDAR’s, APV and DCKA are needed in the media to inhibit receptor function
2.2. Protein Sample Preparation
Extracellular Buffer (ECB): 135 mM NaCl, 3 mM KCl, 2 mM CaCl2, 20 mM Glucose, and 20 mM HEPES
Alexa Fluor 555 C2 Maleimide (Thermo Fisher Scientific)
Alexa Fluor 647 C2 Maleimide (Thermo Fisher Scientific)
Solubilization Buffer: 2 mM Cholesteryl Hydrogen Succinate, 1% Lauryl Maltose Neopentyl Glycol, and protease inhibitor cocktail from Pierce protease inhibitor tablets (Thermo Fisher Scientific) in 1X PBS
2.3. Slide Preparation
Glass coverslips, 20mm × 20mm
Silicone Spacer Templates (Grace Biolabs)
Methanol
Liqui-Nox phosphate-free detergent (Alconox, Inc.)
Acetone
Tl-1 Solution: 4.3% NH4OH and 4.3% H2O2
Compressed Nitrogen
Compressed Oxygen
Vectabond Reagent (Vector Laboratories)
Overnight PEG Solution: 10 mM NaHCO3, 0.25% w/w NHS-PEG4-Biotin (Biotinylated PEG), 50 mM mPEG-Succinimidyl Carbonate (mPEG-SC) (Laysan Bio)
Short Chain PEG Solution: 25 mM MS(PEG)4 Methyl-PEG-NHS-Ester Reagent (Short-chain PEG) (Thermo Fisher Scientific), 0.1M NaHCO3
Hybriwell Hybridization System adhesive chambers (Grace Biolabs)
Press Fit Tubing Connectors (Grace Biolabs)
10X smFRET Imaging Buffer: 2 mM Cholesteryl Hydrogen Succinate and 10 mM N-Dodecyl-beta-Maltoside Detergent in 10X PBS
Streptavidin Solution: 0.2mg/mL Streptavidin in 1X smFRET Imaging Buffer
Primary Antibody Solution: 10nM Anti-iGluR Primary Antibody in 1X PBS
Secondary Antibody Solution: 10 nM Anti-Primary Biotinylated Secondary Antibody in 1X PBS
Bovine Serum Albumin (BSA) Solution: 0.1 mg/mL BSA in 1X PBS
ROXS Solution: 3.3% Glucose, 0.1 mg/mL glucose oxidase, 0.01 mg/mL catalase, 1mM ascorbic acid, and 1mM methyl viologen in 1X smFRET Imaging Buffer. Ligands can be added to the ROXS solution, if desired.
3. Methods
3.1. Preparation of Slides for smFRET
Submerge the silicone templates in methanol in a beaker. Place the beaker in an ultrasonic bath and sonicate the templates for 30 minutes. Transfer the templates into a 50 mL tube containing clean methanol and vortex the templates. Finally, store the templates in clean methanol.
Place 20×20mm glass slides in a slide holder and submerge the holder in a beaker containing a solution of soapy water (use phosphate-free soap). Place the beaker in an ultrasonic bath and sonicate the slides for ten minutes. Wash the slides with purified water three times. Repeat the 10 minute sonication step with the slides in a beaker of acetone, and wash the slides three times with purified water.
Prepare the Tl-1 solution in a beaker and place the beaker in a 60–80° C water bath. Place the slide holder in the beaker and allow the Tl-1 solution to boil for five minutes. Wash the slides with purified water, dry them using a jet of nitrogen, and place them in a metal slide holder.
Place the slide holder with the slides in a Plasma Cleaner such as the Harrick Plasma PDC-32G Plasma Cleaner. Turn on the vacuum pump and allow the chamber pressure to drop below 200 mTorr. Flush the chamber with oxygen 3 times, allowing the pressure to stabilize below 200 mTorr after each flush. Conduct plasma cleaning for two min, remove the vacuum, and extract the slides.
Mix 1 mL vectabond and 40 mL acetone, and submerge the slide holder with the slides in the vectabond/acetone solution for 5 min. Wash the slides with purified water for 1 min, dry the slides with nitrogen, and place the slides on a clean delicate task wipe. These slides can be stored under vacuum for up to five days. The vectabond only lasts two weeks after it has been opened and must be stored under nitrogen. The vectabond/acetone solution should only be used during the day on which it was made.
Remove the silicone templates from the methanol in which they are stored and dry them with nitrogen. Place one template on each slide, making an effort to center the template on the slide. Ensure that there are no bubbles trapped under the template. On the back of the slide, mark the location of the oblong area that the template encircles.
Make the Overnight PEG Solution as described above. Add 50 μl of the Overnight PEG Solution to the well in the middle of each slide. Place the slides on a damp delicate task wipe inside a petri dish. Allow the slides to incubate overnight in the dark at room temperature.
Next day wash slides with purified water, dry with gentle nitrogen flow, apply 50 μl Short Chain PEG Solution to each slide and incubate at room temperature for 2–3 hours. Remove the template from the slide and store the template in methanol. Wash the slide with purified water and dry with nitrogen.
Apply the elliptical chambers to the slides such that the chambers align with the treated areas of the slides. Ensure that there are no bubbles trapped underneath the chamber. Apply the tubing connectors to the ports on the chambers. Mark one port as the inlet port (see Note 6).
Make the streptavidin solution as described above. Add 36 μL to the inlet port of each slide (see Note 7). Ensure that the solution fills the chamber and there are no bubbles trapped in the chamber. Also ensure the solution exits the outlet port. Allow the slides to incubate at room temperature for ten minutes. Wash the chambers by adding 60 μL of PBS three times to each chamber.
Apply 60 μL of secondary antibody solution to each chamber twice. Incubate 20 minutes at 4°C. Wash the chambers by adding 60 μL of PBS two times to each chamber. Apply 60 μL of primary antibody solution to each chamber twice. Incubate 20 minutes at 4°C. Wash the chambers by adding 60 μL of PBS two times to each chamber. Apply 60 μL of BSA Solution to each chamber once. Incubate 20 minutes at 4°C. Wash the chambers by adding 60 μL of PBS two times to each chamber. Prepare the ROXS solution during these previous incubation steps.
Dilute the protein by a factor of 4–5 by adding 1X buffer (made from 10X buffer described above). Apply 90 μL of diluted protein sample (preparation described below) to each chamber twice. Incubate 20 minutes at 4°C. Wash the chambers by adding 100 μL of ROXS solution one time to each chamber and image (see Note 8).
3.2. Preparation of Protein Sample
Use Jetprime transfection reagent to transfect two 10 cm plates of approximately 50% confluent HEK293 cells with DNA construct of interest. Allow cells to express protein of interest for 24–48 hours.
Visually inspect the cells to ensure that they look healthy. Scrape the cells off of the plates and transfer them into one 50 mL conical vial. Pellet the cells in a centrifuge at 1100 × g for three minutes at 23°C. Decant the media away from the pelleted cells. Wash the cells three times with ECB. During the first wash, transfer the cells to a 15 mL conical vial. After three washes, resuspend the cells in 3 mL of ECB. Add fluorophores such that the final concentration in 3 mL will be 600 nM donor and 2.4 μM acceptor. Rotate the cells for 30–60 minutes at room temperature (see Note 9).
After the one hour incubation, wash the cells three times with ECB. Resuspend the cells in 2 mL of solubilization buffer (see Note 10). Nutate the cell suspension for one hour at 4°C.
Transfer the crude lysate to an ultracentrifuge tube and make a balance tube. Spin the tubes at 100000 × g for one hour at 4°C.
Collect the supernatant containing the labeled protein of interest. Use this supernatant to make the diluted protein solution that will be applied to the slides (see Note 11).
3.3. Data Collection
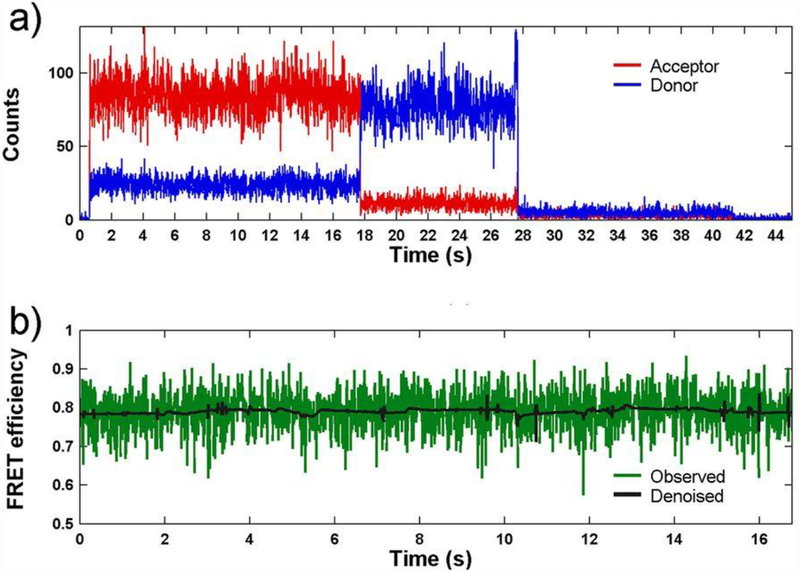
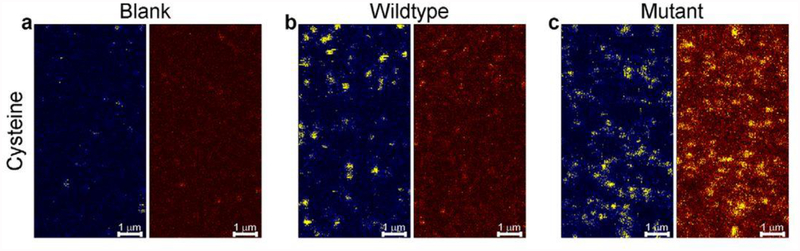
A confocal microscope is used for data collection. An example confocal system that is used for such smFRET experiments is the PicoQuant MicroTime 200 Fluorescence Lifetime Microscope. Scan a 20 μm × 20 μm area of the slide to identify molecules for imaging, then record the fluorescence intensity of each molecule until the donor and acceptor fluorophores undergo photobleaching (see Note 12). Example traces showing anticorrelation between the donor and acceptor are shown in Figure 4. To ensure that the fluorescence is from the receptor tagged at the site of interest, one will need to perform control experiments with no protein, protein with a single labeled site, and protein with two labeled sites as shown in Figure 5.
Figure 4:
(a) A representative smFRET trace measured from an agonist binding domain of AMPA receptor showing raw trajectories of the donor (blue) and acceptor (red) photons as a function of time. (b) The resulting calculated FRET trajectory (green) and its denoised counterpart (black). This research was originally published in the Journal of Biological Chemistry (3) © the American Society for Biochemistry and Molecular Biology.
Figure 5:
smFRET slide imaging showing the specificity of the labeling strategy. Blue pane corresponds to donor channel, and brown pane is FRET channel (acceptor frequency emission with donor frequency excitation). As seen in (a) there is no labeling when protein is not applied to the slide. In (b), the appearance of donor signal is seen when protein with single cysteine labeled with fluorophores is applied to the slide. In (c), the appearance of FRET is seen when protein with two cysteine labeling sites labeled with fluorophores is applied to the slide. This research was originally published in the Journal of Biological Chemistry (29) © the American Society for Biochemistry and Molecular Biology.
3.4. Data Analysis
For data analysis, export the donor and acceptor traces as ASCII files. Then, calculate the corrected FRET efficiencies (EA) according to equations 3 and 4:
| Eq. 3 |
| Eq. 4 |
where ID is the intensity of donor fluorescence, IA is the intensity of acceptor fluorescence, γ is a correction factor accounting for the efficiencies of the detectors used and for the quantum yield of the fluorophores, η is the efficiency of a given detector, and Φ is the quantum yield of a given fluorophore. Once corrected FRET efficiencies have been calculated, plot the histograms depicting the distribution of various FRET efficiencies and analyze those histograms using any graph software.
To identify states in the single molecule trajectory, the Hidden Markov Modeling (HaMMy) analysis (25) or the State Transition and State Identification (STaSI) analysis can be used (26).
4. Notes
Note 1: The fluorophore pair that will be used should be selected to have an R0 that is approximately double the distance that will be measured. Care must be taken to ensure that the fluorophores selected have spectral overlap so that FRET can occur. Furthermore, keep in mind that the distance between two given sites can change as a result of conformational rearrangements within the protein; this makes it necessary to ensure that the R0 of the fluorophore pair will allow measurements across the full range of possible distances.
Note 2: Instead of using antibodies to pulldown the protein of interest, the protein can be engineered to include a twin-Strep tag. This allows the protein to be pulled down directly by the streptavidin on the slide and allows one to forego the secondary antibody treatment, the primary antibody treatment, and the BSA treatment.
Note 3: When using antibodies to pull down the protein of interest onto the slide, one must ensure that the only species of protein bound by that antibody in the cell lysate is the protein of interest. Nonspecific binding of other proteins to the slide will increase the background fluorescence on the slide and lower the signal to noise ratio.
Note 4: For any new mutant proteins that one uses in an smFRET experiment, the expression and function of that protein must be verified. Western blotting with a previously validated antibody can confirm the expression of the mutant protein. It is also necessary to compare the function of the mutant protein to that of the wild type protein using electrophysiological measurements. The function of the mutant protein should be at least partially preserved; if the mutant has only a partial loss of function, that is acceptable.
Note 5: It is highly recommended to perform a screen of various transfection conditions when using the unnatural amino acid experimental design. Because there are several constructs that will need to be transfected into the cells, it is necessary to empirically determine the ratios of DNA that will result in maximal expression.
Note 6: When applying solution to the slide, designate one port as the inlet and use that port for all solution applications. Also, the outlet port should be the port that is more aligned to the chamber ports underneath so that the solutions will be able to exit the chamber easily.
Note 7: When applying solution to the slide, ensure that no bubbles enter the chamber. Bubbles in the chamber can cause patchy labeling of the slide. Beware when applying solutions to the slide that bubbles are often found at the tip of the pipet that are barely discernible by eye, and these bubbles must not be injected into the slide chamber.
Note 8: Once the first protein has been applied to the slide, the slide will only remain usable for a few hours. The proteins on the slide begin to degrade immediately and no useful data will be obtainable from the slides once that degradation is complete. Therefore, be ready to image the slides as soon as they are complete.
Note 9: The donor and acceptor fluorophores should be mixed with each other in a separate tube before being mixed with the sample. This will prevent one fluorophore from labeling the majority of the sites before the other fluorophore is added.
The amount of time that the sample is incubated with the fluorophores can affect the degree of labeling that occurs. When examining homomeric proteins, if one sees that the majority of molecules are exhibiting more than two photobleaching steps, one might consider labeling for a shorter amount of time in future experiments.
The donor to acceptor ratio of 1:4 that is listed in the protocol is one that has been used for heteromeric NMDA receptors. It is possible with other receptors or other experimental designs that the optimal donor to acceptor ratio will be different and will need to be determined. For example, in experiments using homomeric receptors, a one to one donor to acceptor ratio was seen to produce a greater proportion of doubly-labeled FRETing molecules. Each investigator will need to optimize the donor to acceptor ratio that is best for his/her experimental setup. Avoid exposing the fluorophores to excess light. Ambient light in the lab will not bleach the fluorophores from a short exposure, but prolonged exposure to light should be avoided by covering the sample tube with foil.
Note 10: Once the solubilization buffer has been added to the cells, the sample must be kept either on ice or at 4°C to prevent protein degradation.
It is crucial to use fresh protease inhibitors in the solubilization buffer, because all of the endogenous proteases from the HEK cells are still present in the sample. Without potent protease inhibitors, the sample will be rapidly degraded.
Note 11: Once the ultracentrifugation step is complete, the sample should be applied to the slide as soon as possible. Therefore one should time the slide preparation to line up with the protein preparation so that the slide and protein will be ready simultaneously.
Note 12: When first imaging a new slide, the immersion oil between the objective and the slide will be equilibrating to a stable volume. Additionally, the temperature of the slide will be equilibrating with the surrounding air. These two processes will affect the focus of the scope on the slide and cause that focus to drift. One will need to constantly refocus the scope for approximately 10–15 min or until the drifting of the focus has ceased.
Acknowledgements
This project was supported by NIH grants R35GM122528 (VJ) and F31GM130035 (RJD) and by the Houston Area Molecular Biophysics Training Program NIH-2T32 GM008280-26 (DBL).
5. References
- 1.Sirrieh RE, MacLean DM, Jayaraman V. (2015) A conserved structural mechanism of NMDA receptor inhibition: A comparison of ifenprodil and zinc. The Journal of General Physiology 146(2):173–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MacLean DM, Ramaswamy SS, Du M, Howe JR, Jayaraman V. (2014) Stargazin promotes closure of the AMPA receptor ligand-binding domain. The Journal of General Physiology 144(6):503–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramaswamy S, Cooper D, Poddar N, et al. (2012) Role of conformational dynamics in α-amino-3-hydroxy-5-methylisoxazole-4-propionic acid (AMPA) receptor partial agonism. Journal of Biological Chemistry 287(52):43557–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shaikh SA, Dolino DM, Lee G, et al. (2016) Stargazin Modulation of AMPA Receptors. CellReports 17(2):328–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dolino DM, Chatterjee S, MacLean DM, et al. (2017) The structure-energy landscape of NMDA receptor gating. Nature Chemical Biology [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker KA, Lamichhane R, Lamichhane T, Rueda D, Cunningham PR. (2016) Protein- RNA Dynamics in the Central Junction Control 30S Ribosome Assembly. Journal of Molecular Biology 428(18):3615–31. [DOI] [PubMed] [Google Scholar]
- 7.Bal M, Zaika O, Martin P, Shapiro MS. (2008) Calmodulin binding to M-type K+ channels assayed by TIRF/FRET in living cells. The Journal of Physiology 586(9):2307–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dolino DM, Rezaei Adariani S, Shaikh SA, Jayaraman V, Sanabria H. (2016) Conformational Selection and Submillisecond Dynamics of the Ligand-binding Domain of the N-Methyl-d-aspartate Receptor. Journal of Biological Chemistry 291(31):16175–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gomes G-N, Gradinaru CC. (2017) Insights into the conformations and dynamics of intrinsically disordered proteins using single-molecule fluorescence. Biochimica et Biophysica Acta [DOI] [PubMed] [Google Scholar]
- 10.Gouridis G, Schuurman-Wolters GK, Ploetz E, et al. (2015) Conformational dynamics in substrate-binding domains influences transport in the ABC importer GlnPQ. Nature Publishing Group 22(1):57–64. [DOI] [PubMed] [Google Scholar]
- 11.Kempe D, Cerminara M, Poblete S, Schöne A, Gabba M, Fitter J. (2017) Single-Molecule FRET Measurements in Additive-Enriched Aqueous Solutions. Analytical Chemistry 89(1):694–702. [DOI] [PubMed] [Google Scholar]
- 12.Kim J-Y, Kim C, Lee NK. (2015) Real-time submillisecond single-molecule FRET dynamics of freely diffusing molecules with liposome tethering. Nature Communications 6:6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinac B (2017) Single-molecule FRET studies of ion channels. Progress in Biophysics and Molecular Biology [DOI] [PubMed] [Google Scholar]
- 14.McLoughlin SY, Kastantin M. (2013) Single-molecule resolution of protein structure and interfacial dynamics on biomaterial surfaces. PNAS [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song C-X, Diao J, Brunger AT, Quake SR. (2016) Simultaneous single-molecule epigenetic imaging of DNA methylation and hydroxymethylation. PNAS 113(16):4338–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stockmar F, Kobitski AY, Nienhaus GU. (2016) Fast Folding Dynamics of an Intermediate State in RNase H Measured by Single-Molecule FRET. The Journal of Physical Chemistry B 120(4):641–9. [DOI] [PubMed] [Google Scholar]
- 17.Wang S, Vafabakhsh R, Borschel WF, Ha T, Nichols CG. (2016) Structural dynamics of potassium-channel gating revealed by single-molecule FRET. Nature Publishing Group 23(1):31–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, Liu Y, DeBerg HA, et al. (2014) Single molecule FRET reveals pore size and opening mechanism of a mechano-sensitive ion channel. eLife 3:e01834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warhaut S, Mertinkus KR, Höllthaler P, et al. (2017) Ligand-modulated folding of the full-length adenine riboswitch probed by NMR and single-molecule FRET spectroscopy. Nucleic Acids Research 45(9):5512–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landes CF, Rambhadran A, Taylor JN, Salatan F, Jayaraman V. (2011) Structural landscape of isolated agonist-binding domains from single AMPA receptors. Nature Chemical Biology 7(3):168–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cooper DR, Dolino DM, Jaurich H, et al. (2015) Conformational transitions in the glycine-bound GluN1 NMDA receptor LBD via single-molecule FRET. Biophysical Journal 109(1):66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roy R, Hohng S, Ha T. (2008) A practical guide to single-molecule FRET. Nature Methods 5(6):507–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sisamakis E, Valeri A, Kalinin S, Rothwell PJ, Seidel CAM (2010) Accurate Single-Molecule FRET Studies Using Multiparameter Fluorescence Detection. Methods in Enzymology 475:455–513 [DOI] [PubMed] [Google Scholar]
- 24.Ye S, Köhrer C, Huber T, et al. (2008) Site-specific incorporation of keto amino acids into functional G protein-coupled receptors using unnatural amino acid mutagenesis. The Journal of Biological Chemistry 283(3):1525–33. [DOI] [PubMed] [Google Scholar]
- 25.McKinney SA, Joo C, Ha T. (2006) Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophysical Journal 2006;91(5):1941–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shuang B, Cooper D, Taylor JN, et al. (2014) Fast Step Transition and State Identification (STaSI) for Discrete Single-Molecule Data Analysis. The Journal of Physical Chemistry Letters 5(18):3157–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lü W, Du J, Goehring A, Gouaux E. (2017) Cryo-EM structures of the triheteromeric NMDA receptor and its allosteric modulation. Science 355(6331). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Twomey EC, Yelshanskaya MV, Grassucci RA, Frank J, Sobolevsky AI (2017) Structural Bases of Desensitization in AMPA Receptor-Auxiliary Subunit Complexes. Neuron 94(3):569–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dolino DM, Cooper D, Ramaswamy S, Jaurich H, Landes CF, Jayaraman V. (2015) Structural dynamics of the glycine-binding domain of the N-methyl-D-aspartate receptor. Journal of Biological Chemistry 290(2):797–804. [DOI] [PMC free article] [PubMed] [Google Scholar]