Abstract
We present a curriculum description, an initial student outcome investigation, and sample scientific results for a representative Course-Based Undergraduate Research Experience (CURE) that is part of the “Undergraduates Phenotyping Arabidopsis Knockouts” (unPAK) network. CUREs in the unPAK network characterize quantitative phenotypes of the model plant Arabidopsis from across environments to uncover connections between genotype and phenotype. Students in unPAK CUREs grow plants in a replicated block design and make quantitative measurements throughout the semester. This CURE enables students to answer plant science questions that draw from fields such as environmental science, genetics, ecology, and evolution. Findings indicate that this experience provides students with opportunities to make relevant scientific discoveries. Eighty percent of student datasets produced from the CURE met criteria for inclusion in the project database, indicative of student learning in data collection and analysis of quantitative plant traits. Student datasets uncovered novel effects of mutation on plant form. In addition, students’ science self-efficacy increased as a result of course participation, and faculty feedback on course implementation was positive. We present unPAK as a new network that supports CUREs and research experiences focused on collecting biological data made publicly available to the scientific community. The unPAK CUREs can be tailored to address instructor interests or pedagogical needs while involving students in research investigating quantitative plant phenotypes.
INTRODUCTION
National initiatives in biology education recognize the need for all undergraduate biology students to participate in research (1–3). Biology students who participate in research experience many fruitful outcomes, including increased persistence in science, increased science self-efficacy, lasting science learning, and increased understanding of research processes (4–11). Course-based undergraduate research experiences (CUREs) involve students in science research within the context of a course. By integrating research directly into the curriculum, CUREs enroll a greater number of students than can be accommodated by one-on-one mentorship alone (12–17). Here, we discuss a CURE module in which students explore links between genotype and phenotype through the measurement of quantitative plant traits (i.e., plant traits that fall on a continuous scale, e.g., growth); and traits that fall into categorical groups (e.g., alive or dead), which are measured qualitatively.
Facilitating students’ understanding of the link and distinction between genotype and phenotype and their interplay with the environment is an especially important goal in biology instruction, as is facilitating the understanding of structure, function, variation, and natural selection (2). Phenotypes are influenced by both genetics and the environment, and characterizing phenotypes often requires quantitative approaches. For example, the interaction between a human’s genotype and their environment throughout development, (e.g., childhood nutrition) can influence adult stature and vary among individuals, where stature is a quantitative trait (18). This is one example of a genotype-by-environment interaction that is central to agriculture, human health, and natural systems. Measuring quantitative traits and describing the influence of genetics and environment on phenotypes also exercises students’ use of quantitative reasoning, a key competency in Vision & Change (2, 18).
The Undergraduates Phenotyping Arabidopsis Knockouts (unPAK) network consists of more than 16 institutions in which students measure quantitative phenotypic traits on a variety of genotypes of the model plant Arabidopsis thaliana. Investigations of genotype and phenotype links are made possible in the model organism Arabidopsis due to the extensive library of available Arabidopsis T-DNA insertion mutants. In these plant lines, T-DNA (transferred DNA) is used to disrupt specific genes within the Arabidopsis genome. By growing non-mutant and mutant plants in two environmental treatments and measuring growth, size, and reproductive phenotypes (all quantitative traits), students and researchers can investigate how insertion mutations that disrupt gene function may alter plant responses to the environment. This makes it possible to determine whether that gene has a positive, negative, or neutral effect on Arabidopsis fitness under varying environmental conditions (19, 20). The Arabidopsis system makes it possible to obtain quantitative phenotypic data on a library of tens of thousands of T-DNA insertion mutants, with the goal of large-scale coverage of the nuclear genome (20, 21; http://arabidopsisunpak.org). Currently, more than 38,000 Salk T-DNA mutant lines (Salk Institute for Genomic Analysis [http://signal.salk.edu/tabout.html]) are available from the Arabidopsis Biological Resource Center (ABRC) (arabidopsis.org). Of these lines, ~9,000 are currently screened and curated in the unPAK stock center (for details see arabidopsisunpak.org, also see 20).
Instructors of CUREs can select mutant lines from those curated by unPAK, expose them to different environmental treatments, and measure quantitative traits associated with plant growth and fitness to test questions with a focus on ecology, evolution, plant science, or genomics. All unPAK students use the same standards for experimental design and measurement methodology and are subject to the same quality controls on data. unPAK CUREs are unique in that each course is supported by an education-research network consisting of instructors and students working toward the overarching goal of linking genotype to fitness phenotypes. Below, we describe the design and student outcomes of a representative unPAK CURE module from one of the campuses that is available for adoption.
Intended audience and prerequisite student knowledge
The CURE module presented here is currently offered at multiple institutions (see arabidopsisunpak.org for an updated list of participating institutions and classes). It has been successfully integrated into lower-division biology majors’ laboratory courses with a focus on ecology, evolution, plant science, or genetics. Here, we describe a version of the CURE module from the College of Charleston (CofC), where students have previously taken Introductory Biology. As this module is designed for integration into a lower-division class, students are not expected to have extensive biology knowledge and there are not multiple biology pre-requisites prior to the class. However, it is ideal if they have introductory-level knowledge of how to conduct literature searches, read primary literature, design simple experiments, manage data, conduct basic statistics, and graph. These skills are obtained by students in their first introductory biology course at the College of Charleston or in equivalent transfer courses. We also require a pre-or co-requisite introduction to statistics course. See the appendix for modifications of the CURE module which are also ready to be adopted that have been used at additional institutions in upper division biology courses.
Learning time
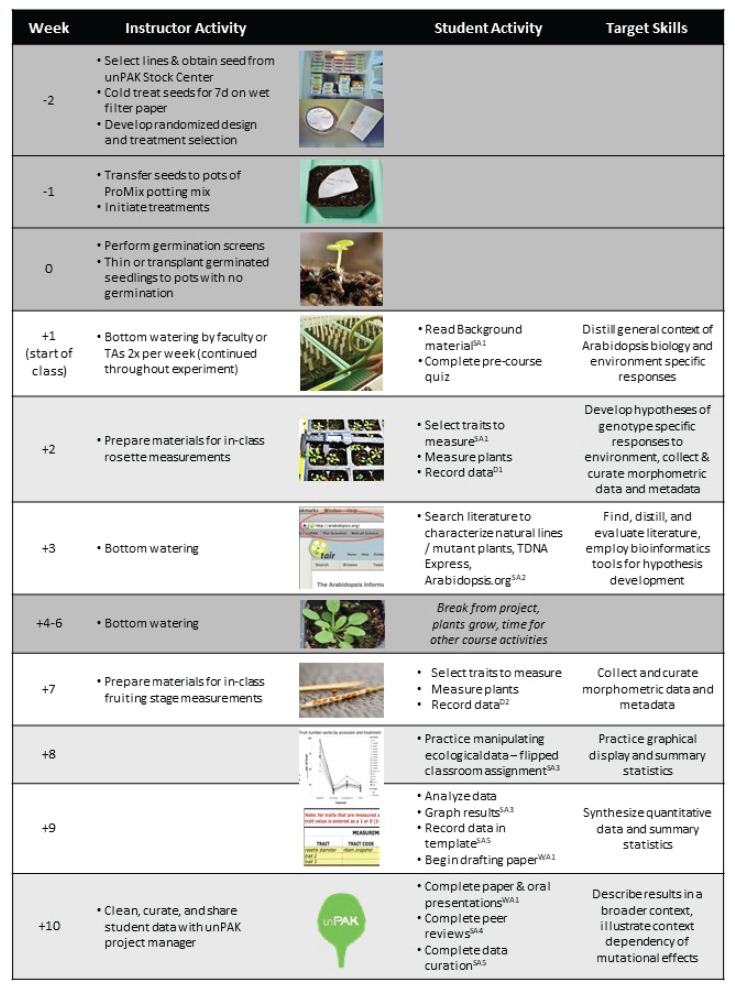
The CURE module is integrated into a course that meets weekly for one three-hour laboratory period and two 75-minute lectures for 14 weeks. Five laboratory periods over the course of the semester are dedicated entirely to the CURE module (weeks 1, 2, 3, 7, and 10, Fig. 1), and two other weeks are dedicated in part to facilitating CURE activities (weeks 8 and 9), making in-class time 20 to 25 hours. To match the life cycle of the plant, CURE activities begin in week 2 and end in week 11 of the 14-week semester. The plants require time to grow over the course of the activity; therefore, there is one three-week break (weeks 4 to 6) during the semester when plants are growing and other, non-CURE-related, learning activities are implemented. Likewise, weeks 11, 12, 13, and 14 are dedicated to activities that are not directly related to the CURE module. Work done out of class is scaffolded, such that early weeks have minor time commitments (<1 hour) and build toward the final writing assignment which requires ~5 hours for graphing, statistics, and writing.
FIGURE 1.
Student and instructor workflow for the unPAK CURE offering described in this work. Week numbering begins two weeks prior to the start of the semester (−2). Superscripts refer to the corresponding supplemental documents. Note that weeks 4 to 6 are a break from this module when other course goals can be completed. Photo credits: E.A. Cousins and A. Matthews.
Learning objectives
Upon completion of this CURE module, students will:
Demonstrate evidence of the ability to measure quantitative plant phenotypes, analyze quantitative data through graphing and statistical analyses, and communicate findings (Learning Objective [LO] 1)
Demonstrate an ability to explain the link between genotype and phenotype, potential influences of mutation on phenotype, and variation across genotypes in phenotypic responses across environments (LO2)
Engage in scientific research with the opportunity to discover something new to the scientific community (LO3)
Gain confidence in their ability to do scientific research (LO4)
PROCEDURE
Materials and unPAK Network
Pre-screened seeds of the Salk T-DNA mutant lines and appropriate control lines can be obtained by contacting unPAK (unpakstockcenter@gmail.com, or murrenc@cofc.edu). We present technical details about the lines and guidelines on experimental design in Appendix 1.
The selection of mutants from the unPAK stocks can be lines with mutations in genes of specific interest to the instructor or a random set of lines provided by un-PAK that have not been phenotyped (see Appendix 1 for further suggestions).
Student and faculty instructions
General description
The CURE module described here is framed in the context of genetic variation for phenotypic plasticity (variation among genetic lines in response to environmental variation, also known as genotype-by-environment effects), and human-influenced environmental change (e.g., increased temperature associated with global climate change). By examining how environmental treatments affect natural accessions and genetic mutants of the plant Arabidopsis thaliana, students have the opportunity to observe that different genotypes of the same species may respond differently to environmental conditions. Further, students observe that mutations can have environment-specific positive, negative, or null effects on complex plant quantitative traits and corresponding phenotypes, such as fitness. Students gain hands-on experience with phenotyping methods (i.e., measuring quantitative traits), experimental design, data management, statistical methods, graphical visualizations, literature searches, working with publicly available databases for model organisms (e.g., TAIR, http://arabidopsis.org, and T-DNA Express, http://signal.salk.edu/cgi-bin/tdnaexpress), and communicating research effectively to peers through writing and oral presentations (Fig. 1; Appendices 1 and 9).
Pre-CURE set-up and experimental design
Prior to the course, instructors prepare the experimental design and obtain seeds from the unPAK stock center for mutant, parental (Col-0), and natural lines. (See Appendix 1 for unPAK contact information and for additional instructor support information, including preCURE set up, control line use, suggestions on treatment implementation, tips on randomized complete block design, guidance on building experimental designs aligned with pedagogical and research goals, and managing space constraints; see also Fig. 1, weeks −2 and −1). Instructors vernalize (cold-treat) seeds on wet filter paper for a week (Fig. 1) and then transfer seeds to individual pots (see Appendix 1 for materials). Instructors grow plants in pots arranged in growth chambers in a randomized block design. After three to four weeks of growth and in the second week of the semester, each pair of students measures six replicates per treatment of two lines each, either 1) COL 70000 and a T-DNA mutant line or 2) a pair of natural accessions. Option 1 enables students to investigate reproductive effects of a disrupted gene (T-DNA mutant line) and environment-specific mutant effects. Option 2 enables students to compare two naturally-occurring Arabidopsis lines to view how natural genetic variation affects plant responses to environmental variation. All of the class’s plants constitute a single temporal block of a stratified random experimental design as a contribution to unPAK research efforts for specific traits, and students explore additional phenotypes based on their interests. Thus, multiple sections of a course across semesters contribute to replication of an experiment. Inclusion of the COL70000 wild-type and natural accessions allows for comparison of data collected across CUREs implemented within the unPAK network (Fig. 1, Appendices 2 and 9).
Student activities
During the first week of the module, students read background material to develop hypotheses of environment responses for their lines (Fig. 1; Appendix 2). During the second week, students make initial measurements of quantitative plant phenotypes, specifically rosette diameter (Appendices 1 and 3), which follow standard unPAK protocols and can later contribute to the curated central unPAK database (see http://arabidopsisunpak.org for details). Between measurements, while plants are growing, students expand literature searches and refine their hypotheses as they develop expertise (Fig. 1; Appendix 6). Instructors may engage students in activities unrelated to unPAK, with other learning goals, during this period. Also during this period, instructors water and tend to plants; however, this could easily be done by students at institutions where students have regular access to growth facilities. Students measure fruit production in week seven. Because the students observe plants both in early growth stages (class week 2; Appendices 2 and 3) and late growth stages (class week 7; Appendices 4 and 5), they have the opportunity to observe how plant traits at distinct life stages are affected by the environmental treatments and whether the two genotypes respond differently to the treatments. In week nine, students graph and analyze their data (Appendix 6), using these results to produce a primary literature style paper on their findings (Fig. 1; Appendices 7 and 9). Throughout the module, instructors emphasize the use of the data to advance scientific knowledge within the Arabidopsis community and describe how careful measurements and well-curated data can help to further that knowledge.
Data quality control
As mentioned above, students are required to measure two unPAK-defined phenotypes, rosette diameter and fruit production, following specific protocols for measurement (Appendix 1). Through this process, students gain experience with formal protocols useful for addressing questions of genotype by environment. Students’ datasets receive two peer reviews, an instructor review, and an unPAK database manager review prior to inclusion into the central unPAK database (arabidopsisunpak.org). To validate student data and check for consistency and quality, datasets are a) checked for the direction of the overall sample response to treatment (e.g., if plants were observed to be larger in one environment, we would expect the general student dataset to reflect that direction), b) screened for extreme outliers indicative of typing errors, c) examined for order of magnitude differences in trait values for a particular group indicative of errors in measurement units (e.g., cm versus mm), and d) checked for matches between the plant IDs students submit and the plant IDs assigned to them. When multiple sections of students measure the same plants, as often happens in unPAK courses with multiple lab sections, measurements are evaluated for consistency. If students fail to follow measurement protocols, do not properly submit data and metadata in the unPAK template (Appendix 9), have incomplete columns or metadata missing, or include units in the column with quantitative values instead of recording them in the meta-data, the data fail quality control steps and are not included in the database. This process contributes to quality control in data submission (Appendix 9) and confirms that appropriate control lines were included in each experiment. This is essential to ensure the quality and utility of the data once it is in the database where it will be used by other unPAK researchers for further analyses or to inform future research. Successful completion of the project and communication of findings indicates successful accomplishment of learning objectives 1 and 2. Individual student assessment is based on a final presentation of results in a literature-style paper (Appendices 6 and 8).
Suggestions for determining student learning
LO1: Demonstrate evidence of the ability to measure quantitative plant phenotypes, analyze quantitative data through graphing and statistical analyses, and communicate findings
Students’ successful collection of plant quantitative phenotypes, record-keeping, graphing, statistical analyses, and submission of data to the unPAK database constitute evidence of this objective. All students collect data on rosette diameter and fruit number after being trained in the use of calipers and in plant growth patterns particular to this species. As formative feedback on students’ measurement efficacy, peer partners check an early subset of each other’s measurements by observing their partner, commenting on technique, and suggesting any necessary improvements or seeking additional instructor guidance. Students’ data are then subject to the rigorous quality control procedures described above. Given the level of scrutiny each sample is subject to, successful completion of the above steps and submission of complete data lines to the database can be taken as evidence of success in learning how to measure quantitative plant phenotypes and communicate findings.
LO2: Demonstrate an ability to explain the link between genotype and phenotype, potential influences of mutation on phenotype, and variation across genotypes in phenotypic responses across environments
Students are evaluated as to whether they have accomplished this objective using their final writing assignment, a scientific-style paper detailing their rationale, methods, and results. In the paper introduction, students must discuss prior research linking their mutants or natural populations (genotypes) to traits characterized in the literature (phenotypes). Then, in the results section, students must report on their findings, describing how their chosen mutants or natural populations (genotypes) responded (phenotypic responses) to variation in environments in their text and use graphical and statistical procedures. Finally, students are required to interpret and explain their results, including discussing the links between genotype, phenotype, and environment in the discussion (see Appendix 7). Students are then graded on their ability to make these connections (see Appendix 7 rubric), indicating their successful ability to explain the link between genotype and phenotype and responses across environments.
LO3: Engage in scientific research with the opportunity to discover something new to the scientific community
The opportunity to make a scientific discovery is an important design feature of CUREs for three reasons: 1) students have the potential to actually make a discovery that might contribute to science, 2) pursuing a discovery that is new and relevant to the scientific community may motivate students to engage, since the prospect of discovery is exciting, and 3) pursuing an unknown answer to a scientific question, regardless of whether students support their hypothesis or make a new “discovery,” necessitates student engagement with the uncertainty inherent to science. Students must be able to logically explain their own results; they cannot look to past research or experiments to find the correct answer as they may be able to do in more traditional labs. This leads to learning regardless of the results of the experiment. The discovery scale in the Laboratory Course Assessment Survey (LCAS) measures students’ opportunities to make relevant discoveries (17). This scale consists of five items that ask students to report on whether they had opportunities to make new discoveries or develop new ideas of relevance to the scientific community. Response options for each question range from 1 (strongly disagree) to 6 (strongly agree). This scale was given to students at the end of the unPAK CURE, and the course average was compared with scores collected from a national sample of CUREs using t-tests in order to assess whether students reported opportunities to make scientifically relevant discoveries and whether these opportunities were comparable with those offered by other CUREs.
LO4: Gain confidence in their ability to do scientific research
Development of students’ science self-efficacy is an important outcome as it is correlated with students’ long-term persistence in STEM (22, 23). Chemers’s science self-efficacy scale consists of six items that ask students to rate their confidence in their ability to do scientific tasks on a scale of 1 (not at all confident) to 5 (absolutely confident) (24). We used a pre-/post-test approach to measure changes in science self-efficacy over the duration of the course. We assessed differences in students’ scores for self-efficacy using a matched-pairs t-test performed on pre- and post-test data.
To assess LO3 and LO4, we followed appropriate protocols for informed consent and confidentiality in data collection (IRB, College of Charleston # IRB-2014-08-29-154322).
Sample plant data
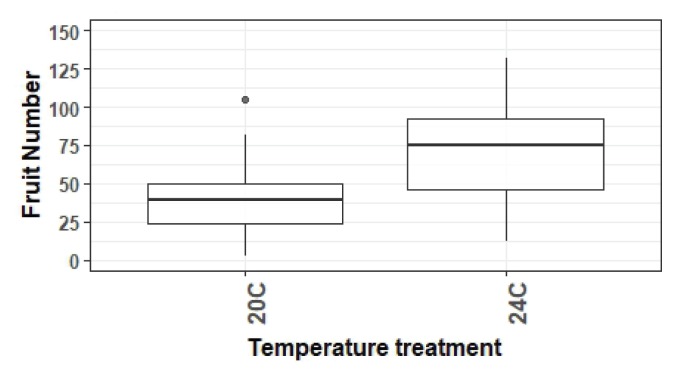
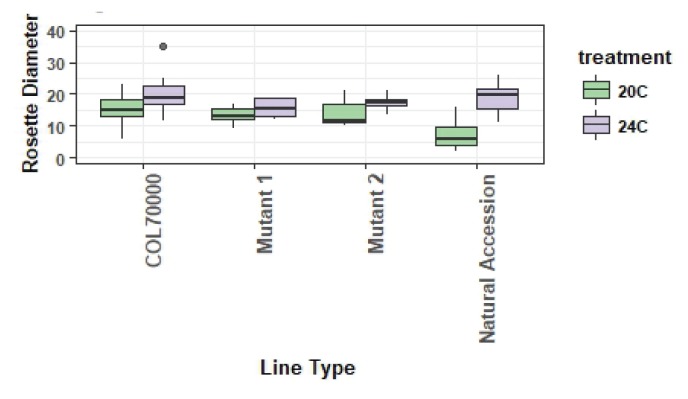
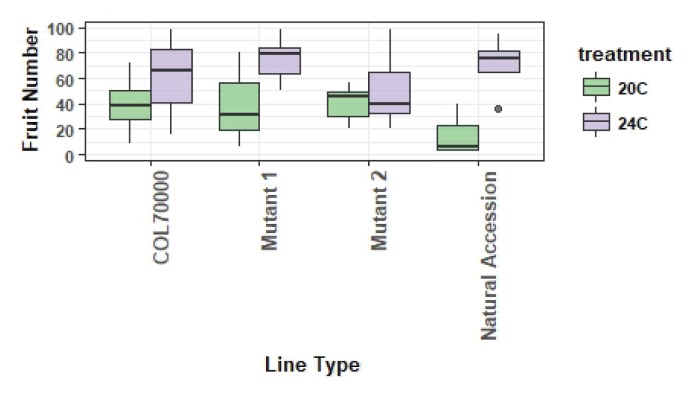
As an example of successful quantitative data collection, graphing, and analysis that occurs in unPAK CUREs, we provide a subsample of data submitted to the unPAK project manager that were reviewed and met data quality control standards. Analysis of variance was used to evaluate whether plant quantitative phenotypes differed between temperature treatments in a student-collected CURE dataset and whether genotypes differed in response to environment (Figs. 2 and 3). When necessary to meet assumptions of analysis of variance, data were log10 transformed. For this example, differences between treatments were detected for traits at both life stages measured by rosette diameter (F1,167 = 4.8, p = 0.03) and fruit number (F1,167 = 79.9, p < 0.0001; Fig. 3). Performance at elevated temperatures was higher for both traits. Additionally, mutant and control lines varied in their response to environmental treatments (Fig. 3a, F2,167 = 16.8, p < 0.0001; Fig. 3b, F1,158 = 53.6, p < 0.0001). One mutant line that was not responsive to elevated temperature had increased performance in comparison with the control line, whereas another mutant had significantly increased performance in comparison with the control only at the elevated temperature (Fig. 3b). These results exemplify the potential of unPAK to uncover phenotypic variation among genotypes and in response to different environments for various A. thaliana mutants. Data collected by CURE students (beyond the example here) are flagged in the arabidopsisunpak.org database as additional examples.
FIGURE 2.
Fruit number varies by temperature environment. Example student-collected data from plants grown at 20°C and 24°C. Boxplot features include the median (line) 25th and 75th percentiles (edges of the box); whiskers are 1.5× the interquartile range, and outliers are denoted by dots.
FIGURE 3a.
Rosette diameter for control (Col70000), 2 example Salk T-DNA mutant lines, and example natural accession lines for an example set of student-collected data that vary by temperature environment. Box plot features as in Figure 2.
FIGURE 3b.
Fruit number for control (Col70000), 2 example Salk T-DNA mutant lines, and natural accession lines for an example set of student-collected data that vary by temperature environment. Box plot features as in Figure 2.
Safety issues
This laboratory does not entail lab work with hazardous chemicals or materials (e.g., extraction and amplification of DNA), and direct hazards to human health are minimal. Handling of biological materials by students is limited to the physical measurement of plants and counting of plant materials (e.g., seeds). Handling of biological materials by instructors is limited to measurement and growth of plants in standard potting soil. Safety issues concern the movement and potential misuse of T-DNA seed-stocks. Environmental safety procedures to restrict seed movement out of the laboratory setting include sweeping/wiping the laboratory surfaces, use of floor sticky mats at doors (Appendix 1), completely covering trays as they are moved between growing space and classroom to restrict potential seed movement, and careful sterilization and cleaning between runs of trays and pots in growing spaces to omit seed movement. Following experiments, potting mix is autoclaved prior to disposal.
DISCUSSION
Field testing
The CURE was implemented in a foundation course taken by biology majors focused on ecology and evolution in which students perform science skills associated with unPAK and emphasize communication skills in science (e.g., reading, writing, data management, synthesis and presentation skills). The CURE was developed in 2012 and subsequently revised, then conducted in ~55 course sections across 11 institutions in class sizes of ~20 students. Evidence of student learning below describes a subset of sections that participated in the unPAK CURE.
Evidence of student learning
Evidence of LO1
Variation in plant phenotypes was uncovered in data collected by unPAK students in the classroom. Students contributed this data to a national database used by researchers across the unPAK network, demonstrating students’ ability to measure quantitative plant phenotypes and communicate their findings. Out of 123 pairs of student data reviewed from a random selection of sections between 2012 and 2016 at CofC, 80.5% of student pairs’ data files met data inclusion standards and were subsequently included in the unPAK database. This assessment underscores students’ success in achieving gains in their ability to execute quantitative phenotypic methods. Please see the sample plant data section for an example of student results that met data inclusion standards and indicate achieving LO1.
Evidence of LO2
Students successfully demonstrated an ability to explain the link between genotype and phenotype and variation across genotypes in phenotypic responses across environments. The final paper (see Appendix 7) is the most accurate and authentic measure of this learning outcome since students are required to construct arguments that link genotype, phenotype, and environment in the paper (see Suggestions for determining student learning section, above, for details). Any student failing to execute paper tasks linking genotype, phenotype, and environment is unlikely to receive above a maximum score of 66% on their final project (see the rubric and point structure in Appendix 7). Since 66% is considered a barely passing grade, passing the final project with a 70% or better indicates accomplishment of LO2. Of students who completed the assignment and the course (over 16 sections each with ~20 students per section), 97.7% received an acceptable passing grade on the final paper, indicating achievement of this outcome. Among the paper components most closely related to LO2, students most commonly struggled with the explanation of their results in the context of prior work, indicating that future work needs to focus on class examination of results and discussion sections of published literature. Future classes could incorporate more opportunities to practice this skill. For example, students might read additional relevant literature and engage in a class discussion about how the literature is important to their work, with specific coaching on how they might reference the literature in their final report. The module written assignment and activities represent 16% of the total points for the four-credit course at CofC.
Student survey data collection
Surveys with questions to assess discovery and science self-efficacy were distributed pre- and post-course to assess LO3 and LO4. Eighty-three percent of students given the opportunity to participate responded from one fall and one spring semester. We used t-tests to analyze the data and the Benjamini-Hochberg false discovery rate (FDR) correction to account for potential Type I error in conducting multiple t-tests (25). We report FDR-adjusted p values below.
Evidence of LO3
Students had opportunities to discover something new to the scientific community. The course provided students with opportunities to make relevant scientific discoveries comparable with students in a national sample of CUREs. The discovery and relevance scale used to measure students’ opportunities to make discoveries consisted of six questions with five possible responses each (summed score range: 5 to 30, Cronbach’s alpha = 0.87). Opportunities for relevant discovery reported by unPAK CURE students (mean = 25.14, sd = 5.16) did not differ significantly from students in the national sample of CUREs reported in previously published work by Corwin and colleagues (9, 17) (n = 72, mean = 24.35, sd = 4.04, p = 0.408). In addition, students showed significantly more opportunities for relevant discovery than students in a national sample of traditional labs (n = 60, mean = 20.77, sd = 5.82, p < 0.001). These results demonstrate that the unPAK CURE is similar to other CUREs nationally in offering opportunities for relevant discovery, which is predicted to increase students’ motivation and contribute to their persistence in science and, importantly, offers students the opportunity to work on a problem with an unknown answer. This constitutes a learning opportunity regardless of whether students’ results support a specific hypothesis (17). However, it is important to note that, overall since 2012, over 150 mutant lines have been screened in unPAK CUREs, generating new information that constitutes unPAK’s novel discoveries.
Evidence of LO4
Students gained confidence in their ability to do scientific research. The self-efficacy scale used to measure students’ changes in self-efficacy consisted of five questions with six possible responses each (summed score range: 6 to 30, Cronbach’s alpha = 0.86, 26) given to course sections of 20 across two semesters. Students made significant gains in science self-efficacy, with a pre-course mean score of 21.99 (sd = 4.71) and a post-course mean score of 24.20 (sd = 5.40; p < 0.001). Such an outcome is a common result of one-on-one mentored research participation and has been characterized as an indirect contributor to persistence in science, since it precedes outcomes that directly predict persistence (22, 23, 27).
Faculty feedback
After implementing the CURE, instructors (n = 6) were queried about their experience (28; further details in Appendix 11). We asked, “Why did you choose to implement a CURE? What goals did this module accomplish that a non–research based course might not have accomplished? What was the most important outcome of this module for your course?” All faculty agreed that the activities were valuable for their students and were well designed while offering flexibility. All stated they would use the CURE again in their course (Supplemental Table, Appendix 11).
Possible modifications
The multi-week module presented here can be modified to serve curricular needs for other foundation courses in biology, ecology, evolution, genetics, or plant science. Modifications can occur via selection of various mutant lines of interest for instruction or research (e.g., lines from a particular biochemical pathway), or by varying the growth environments (e.g., to focus on a topic of plant nutrition), or by emphasis on T-DNA insert position (intron vs. exon) or protein function to meet particular content or curricular goals. Inclusion of genetic assessments and advanced statistical approaches are additional examples of modifications, which may increase difficulty for more advanced students. For settings with limited growing space, students in different sections can measure the same plants. Findings from one semester could inform the instructor as to the next step in an inquiry, for example, changing environments with the same mutant lines.
CONCLUSIONS
This work reports on the curriculum of a new CURE through the unPAK network, with a focus on quantitative plant phenotypes. The CURE produces high-quality scientific results uncovering quantitative phenotypic responses associated with mutant lines in a common plant genetic background across environments. It increases students’ understanding of the genotype–phenotype link and interactions with the environment, provides students with opportunities to make discoveries that are relevant to the scientific community, and increases students’ science research self-efficacy. Thus, the unPAK CURE model can be added to the curricular repertoire of instructors aiming to help students achieve outcomes associated with participation in research while actively contributing to biology research projects.
unPAK CUREs achieve positive student, faculty, and science outcomes for relatively low cost and are accessible to many students. Modifications can be made to the mutants and environments examined in unPAK CUREs, making this model appropriate for other institutions wanting to maintain distinctive curricular goals while also offering more research opportunities to many students. Since previously existing broad-scale CURE curricula (e.g., 29, 30) have not yet focused on quantitative phenotypic traits, this CURE adds to the diversity of topics available for instruction in this pedagogical mode.
SUPPLEMENTAL MATERIALS
ACKNOWLEDGMENTS
We thank Erik Sotka, Yana Wieckowski, Claudia Alt, Abigail Zoger, and Cynthia Chang for assistance with the unPAK CURE. We are grateful for the support from NSF IOS-1052262 and IOS-1355106 to M.T.R., A.E.S., and C.J.M., IOS-1146977; IOS-1052323 to H.S.C., and IOS-1050153 and IOS-1354603 to M.J.W. The NSF did not participate in any components of data collection or manuscript preparation. Research reported was approved by IRB review boards or determined to be exempt. We thank Irfanul Alam, Erin Fried, and Gracen Mitrick for comments on an earlier version of the manuscript. We thank anonymous reviewers for comments on a previous version. The authors declare that there are no conflicts of interest.
Footnotes
Supplemental materials available at http://asmscience.org/jmbe
REFERENCES
- 1.National Research Council. BIO2010: Transforming Undergraduate Education for Future Research Biologists. The National Academies Press; Washington, DC: 2003. [PubMed] [Google Scholar]
- 2.American Association for the Advancement of Science. Vision and change in undergraduate biology education: a call to action. Washington, DC: 2011. [Accessed 28 December 2016]. Retrieved from: http://visionandchange.org/files/2011/03/Revised-Vision-and-Change-Final-Report.pdf. [Google Scholar]
- 3.President’s Council of Advisors on Science and Technology. Engage to excel: producing one million additional college graduates with degrees in science, technology, engineering, and mathematics. 2012. [Accessed 15 September 2014]. Retrieved from: www.whitehouse.gov/sites/default/files/microsites/ostp/pcast-engage-to-excel-final_feb.pdf.
- 4.Lopatto D. Survey of undergraduate research experiences (SURE): first findings. CBE Life Sci Educ. 2004;3:270–277. doi: 10.1187/cbe.04-07-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Russell SH, Hancock MP, McCullough J. Benefits of undergraduate research experiences. Science. 2007;316:548–549. doi: 10.1126/science.1140384. [DOI] [PubMed] [Google Scholar]
- 6.Lopatto D. Science in solution: the impact of undergraduate research on student learning. Council on Undergraduate Research and Research Corporation for Scientific Advancement; Washington, DC: 2010. [Google Scholar]
- 7.Seymour E, Hunter AB, Laursen SL, DeAntoni T. Establishing the benefits of research experiences for undergraduates in the sciences: first findings from a three-year study. Sci Educ. 2004;88:493–534. doi: 10.1002/sce.10131. [DOI] [Google Scholar]
- 8.Laursen SL, Hunter AB, Seymour E, Thiry H, Melton G. Undergraduate research in the sciences: engaging students in real science. John Wiley & Sons; San Francisco, CA: 2010. [Google Scholar]
- 9.Corwin LA, Graham MJ, Dolan EL. Modeling course-based undergraduate research experiences: an agenda for future research and evaluation. CBE Life Sci Educ. 2015;14:es1. doi: 10.1187/cbe.14-10-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodenbusch SE, Hernandez PR, Simmons SL, Dolan EL. Early engagement in course-based research increases graduation rates and completion of science, engineering, and mathematics degrees. CBE Life Sci Educ. 2016;15:ar20. doi: 10.1187/cbe.16-03-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corwin LA, Runyon CR, Ghanem E, Sandy M, Clark G, Palmer GC, Reichler S, Rodenbusch SE, Dolan EL. Effects of discovery, iteration, and collaboration in laboratory courses on undergraduates’ research career intentions fully mediated by student ownership. CBE Life Sci Educ. 2018;17:ar20. doi: 10.1187/cbe.17-07-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weaver GC, Russell CB, Wink DJ. Inquiry-based and research-based laboratory pedagogies in undergraduate science. Nat Chem Biol. 2008;4:577–580. doi: 10.1038/nchembio1008-577. [DOI] [PubMed] [Google Scholar]
- 13.Schultz PW, Hernandez PR, Woodcock A, Estrada M, Chance RC, Aguilar M, Serpe RT. Patching the pipeline reducing educational disparities in the sciences through minority training programs. Educ Eval Policy Anal. 2011;33:95–114. doi: 10.3102/0162373710392371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei CA, Woodin T. Undergraduate research experiences in biology: alternatives to the apprenticeship model. CBE Life Sci Educ. 2011;10:123–131. doi: 10.1187/cbe.11-03-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Auchincloss LC, Laursen SL, Branchaw JL, Eagan K, Graham M, Hanauer DI, Lawrie G, McLinn CM, Pelaez N, Rowland S, Towns M, Trautmann NM, Varma-Nelson P, Weston TJ, Dolan EL. Assessment of course-based undergraduate research experiences: a meeting report. CBE Life Sci Educ. 2014;13:29–40. doi: 10.1187/cbe.14-01-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bangera G, Brownell SE. Course-based undergraduate research experiences can make scientific research more inclusive. CBE Life Sci Educ. 2014;13:602–606. doi: 10.1187/cbe.14-06-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Corwin LA, Runyon C, Robinson A, Dolan EL. The laboratory course assessment survey: a tool to measure three dimensions of research-course design. CBE Life Sci Educ. 2015;14:ar37. doi: 10.1187/cbe.15-03-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frazer KA. Human genetic variation and its contribution to complex traits. Nat Rev Gen. 2009;10:241. doi: 10.1038/nrg2554. [DOI] [PubMed] [Google Scholar]
- 19.Rutter MT, Roles A, Conner JK, Shaw RG, Shaw FH, Schneeberger K, Ossowski S, Weigel D, Fenster CB. Fitness of Arabidopsis thaliana mutation accumulation lines whose spontaneous mutations are known. Evolution. 2012;66:2335–2339. doi: 10.1111/j.1558-5646.2012.01583.x. [DOI] [PubMed] [Google Scholar]
- 20.Rutter MT, Wieckowski YM, Murren CJ, Strand AE. Fitness effects of mutation: testing genetic redundancy in Arabidopsis thaliana. J Evol Biol. 2017;6:1124. doi: 10.1111/jeb.13081. [DOI] [PubMed] [Google Scholar]
- 21.O’Malley RC, Ecker JR. Linking genotype to phenotype using the Arabidopsis unimutant collection. Plant J. 2010;61:928–940. doi: 10.1111/j.1365-313X.2010.04119.x. [DOI] [PubMed] [Google Scholar]
- 22.Graham MJ, Frederick J, Byars-Winston A, Bunter AB. Increasing persistence of college students in STEM. Science. 2013;341:1455–1456. doi: 10.1126/science.1240487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Estrada M, Woodcock A, Hernandez PR, Schultz P. Toward a model of social influence that explains minority student integration into the scientific community. J Educ Psychol. 2011;103:206–222. doi: 10.1037/a0020743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chemers MM, Syed M, Goza BK, Zurbriggen EL, Bearman S, Crosby FJ, Morgan EM. The role of self-efficacy and identity in mediating the effects of science support programs (Technical Report No. 5) University of California; Santa Cruz, CA: 2010. [Google Scholar]
- 25.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc. 1995;57:289–300. [Google Scholar]
- 26.Cronbach LJ. Coefficient alpha and the internal structure of tests. Psychometrika. 1951;16:297–334. doi: 10.1007/BF02310555. [DOI] [Google Scholar]
- 27.Robnett RD, Chemers MM, Zurbriggen EL. Longitudinal associations among undergraduates’ research experience, self-efficacy, and identity. J Resin Sci Teach. 2015;52:847–867. doi: 10.1002/tea.21221. [DOI] [Google Scholar]
- 28.Maravasi M, Choudhury M, Binesh Vala N, Teplitski M. Fitness of antibiotic-resistant bacteria in the environment: a laboratory activity. J Microbiol Biol Educ. 2017;18(1):1–7. doi: 10.1128/jmbe.v18i1.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaffer CD, Alvarez C, Bailey C, Barnard D, Bhalla S, Chandrasekaran C, Chandrasekaran V, Chung HM, Dorer DR, Du C, Eckdahl TT, Poet JL, Frohlich D, Goodman AL, Gosser Y, Hauser C, Hoopes LL, Johnson D, Jones CJ, Kaehler M, Kokan N, Kopp OR, Kuleck GA, McNeil G, Moss R, Myka JL, Nagengast A, Morris R, Overvoorde PJ, Shoop E, Parrish S, Reed K, Regisford EG, Revie D, Rosenwald AG, Saville K, Schroeder S, Shaw M, Skuse G, Smith C, Smith M, Spana EP, Spratt M, Stamm J, Thompson JS, Wawersik M, Wilson BA, Youngblom J, Leung W, Buhler J, Mardis ER, Lopatto D, Elgin SC. The genomics education partnership: successful integration of research into laboratory classes at a diverse group of undergraduate institutions. CBE Life Sci Educ. 2010;9:55–69. doi: 10.1187/09-11-0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jordan TC, Burnett SH, Carson S, Caruso SM, Clase K, DeJong RJ, Dennehy JJ, Denver DR, Dunbar D, Elgin SC, Findley AM, Gissendanner CR, Golebiewska UP, Guild N, Hartzog GA, Grillo WH, Hollowell GP, Hughes LE, Johnson A, King RA, Lewis LO, Li W, Rosenzweig F, Rubin MR, Saha MS, Sandoz J, Shaffer CD, Taylor B, Temple L, Vazquez E, Ware VC, Barker LP, Bradley KW, Jacobs-Sera D, Pope WH, Russell DA, Cresawn SG, Lopatto D, Bailey CP, Hatful GH. A broadly implementable research course in phage discovery and genomics for first-year undergraduate students. mBio. 2014;5:e01051–13. doi: 10.1128/mBio.01051-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.