Fibroblast growth factor receptor 3 (FGFR3) is a major potential actionable target in urothelial bladder cancer (BC). We found that FGFR3 mutations appeared conserved in primary BC and corresponding lymph-node metastases. We also showed that the deep part of the tumor needs to be assessed if neoadjuvant anti-FGFR3 treatment is considered. This suggests that personalized anti-FGFR3 therapy may improve BC treatment in the perioperative setting.
Keywords: FGFR3, mutations, heterogeneity, bladder, cancer, targeted therapy
Abstract
Background
Fibroblast growth factor receptor 3 (FGFR3) is an actionable target in bladder cancer. Preclinical studies show that anti-FGFR3 treatment slows down tumor growth, suggesting that this tyrosine kinase receptor is a candidate for personalized bladder cancer treatment, particularly in patients with mutated FGFR3. We addressed tumor heterogeneity in a large multicenter, multi-laboratory study, as this may have significant impact on therapeutic response.
Patients and methods
We evaluated possible FGFR3 heterogeneity by the PCR-SNaPshot method in the superficial and deep compartments of tumors obtained by transurethral resection (TUR, n = 61) and in radical cystectomy (RC, n = 614) specimens and corresponding cancer-positive lymph nodes (LN+, n = 201).
Results
We found FGFR3 mutations in 13/34 (38%) T1 and 8/27 (30%) ≥T2-TUR samples, with 100% concordance between superficial and deeper parts in T1-TUR samples. Of eight FGFR3 mutant ≥T2-TUR samples, only 4 (50%) displayed the mutation in the deeper part. We found 67/614 (11%) FGFR3 mutations in RC specimens. FGFR3 mutation was associated with pN0 (P < 0.001) at RC. In 10/201 (5%) LN+, an FGFR3 mutation was found, all concordant with the corresponding RC specimen. In the remaining 191 cases, RC and LN+ were both wild type.
Conclusions
FGFR3 mutation status seems promising to guide decision-making on adjuvant anti-FGFR3 therapy as it appeared homogeneous in RC and LN+. Based on the results of TUR, the deep part of the tumor needs to be assessed if neoadjuvant anti-FGFR3 treatment is considered. We conclude that studies on the heterogeneity of actionable molecular targets should precede clinical trials with these drugs in the perioperative setting.
introduction
Radical cystectomy (RC) has been the gold standard for the treatment of invasive, non-metastatic, urothelial carcinoma of the bladder (UCB) for >50 years. Despite major surgery, 5-year survival rate still only ranges from ±75% in pT2N0 to ±25% in pN+ UCB [1, 2]. Perioperative (neoadjuvant and adjuvant) platinum-based combination chemotherapy has only marginally (5%–7% overall survival benefit for neoadjuvant chemotherapy) improved patient's prognosis [3–5]. Consequently, better systemic treatment is urgently needed to improve clinical outcomes for invasive UCB.
Activating oncogenic mutations of FGFR3 were identified >10 years ago in UCB [6]. Interestingly, FGFR3 mutations were predominantly found in genetically stable UCB with a favorable prognosis [7]. Moreover, FGFR3 and TP53 mutations rarely coincide and FGFR3 mutations are, even in advanced UCB, most of the time accompanied by fewer molecular alterations than FGFR3 wild-type tumors [7–10]. This indicates that FGFR3 is also a major potential actionable target in a subgroup of advanced UCB [9–11]. Furthermore, preclinical in vitro and in vivo data show that anti-FGFR3 therapy slows down tumor growth, especially in FGFR3-mutated tumors [12]. However, the heterogeneity of FGFR3 status within a tumor or a patient has not been adequately addressed and may negatively impact therapeutic response [11].
We report a large multicenter, multi-laboratory study investigating the heterogeneity of the FGFR3 mutations in invasive UCB. We analyzed paired samples (superficial and deep compartments of the same lesion) of primary transurethral resection (TUR) of 61 patients. We also analyzed paired samples from RC and positive lymph nodes (LN+) of 614 patients who were treated for cN0M0-UCB without prior systemic chemotherapy and/or radiotherapy. FGFR3 expression was also analyzed by immunohistochemistry (IHC) in a subgroup of patients.
materials and methods
study populations
Three cohorts of patients with UCB were established to study the heterogeneity of FGFR3 mutation status in UCB. In total, 10 different hospitals were involved in the treatment of the patients and molecular analyses were done in four different laboratories.
cohort of TUR
To evaluate intratumor FGFR3 mutation heterogeneity, we studied a cohort of 61 patients who underwent a primary TUR for UCB. All tumors were primary UCB. The procedures were carried out in two hospitals (Toronto; n = 26 and Leeds; n = 35) between 1993 and 2006. Mean age at diagnosis was 70.3 years (SD 8.3 years); 15/61 patients were female. All TUR specimens contained muscle as assessed by a pathology review (THvdK and PH). For each case, a superficial and deep part of the same tumor specimen was separately dissected from the tissue-block or blank slides for DNA isolation and subsequent FGFR3 mutation analysis. All DNA samples of the 61 TURs were analyzed in both laboratories (Toronto and Leeds) and the results were the same. An additional four TUR cases, in which multiple parts of the same superficial (n = 3) or invasive (n = 1) areas were available, were analyzed in Toronto.
cohorts of RC
The second (International) cohort included 494 patients treated with RC including a pelvic lymph-node dissection for cN0M0 (staged with at least abdominal CT and chest X-ray) UCB in four hospitals in Amsterdam, the Netherlands (n = 204); Toronto, Canada (n = 104); Dallas, TX, USA (n = 132) and Turku, Finland (n = 54). A previous diagnosis of noninvasive UCB was allowed. Mean age at RC was 65.1 years (SD 10.8 years); 121/494 patients were female. Patients were treated between 1986 and 2012 by RC without prior neoadjuvant chemotherapy or pelvic radiation. Of these patients, 83/494 (17%) received adjuvant chemotherapy. A pathology review was done by JdJ, JS (Amsterdam) and THvdK (Toronto, Dallas and Turku). Node samples were available for reliable FGFR3 analysis in 117/155 pN+ cases. The laboratory in Amsterdam analyzed the 204 RC cases from Amsterdam, and the 290 RC cases from Toronto, Dallas and Turku were all analyzed in Toronto.
In the third (French) cohort, 120 cN0M0-UCB patients treated in five French hospitals for locally advanced pT3/pT4 (n = 100) and/or pN+ (n = 99) UCB were identified. All these patients were treated by RC including a pelvic lymph-node dissection and adjuvant platinum-based chemotherapy between 2000 and 2009 at the Henri Mondor Hospital, Créteil (n = 36); the Gustave Roussy Institute, Villejuif (n = 28); the Curie Institute, Paris (n = 7); the Claudius Regaud Institute, Toulouse (n = 28) and Bergonié Institute, Bordeaux (n = 21). Mean age at RC was 62.1 years (SD 9.1 years); 16/120 patients were female. A previous diagnosis of noninvasive UCB was allowed. None of the patients had prior neoadjuvant chemotherapy or pelvic radiation. A central pathology review was done by YA. Node samples were available for reliable FGFR3 analysis in 84/99 pN+ cases. The laboratory in Créteil analyzed all the RC cases of the French cohort.
clinicopathological data collection
The clinicopathological characteristics, treatment and follow-up data were retrospectively collected. Tumors were staged according to the 2009 TNM classification [13] and graded according to WHO criteria. Local ethics committees and/or translational research boards approved the three experimental protocols and, if applicable, patients provided written informed consent for central collection of their tissue specimens and clinical data for research purposes.
tissue (TUR and RC) specimens and DNA extraction
Hematoxylin and eosin slides served as templates for the manual macrodissection procedure on the formalin-fixed, paraffin-embedded tissue-block or blank slides. The dissected samples contained a minimum of 70% tumor cells, as assessed by histological examination. DNA was extracted from the tissues according to the manufacturer's protocols using the DNeasy® Tissue Kit in the TUR and international RC cohorts. In the French RC cohort, the Maxwell® 16 FFPE Plus LEV DNA Purification Kit and an automated Maxwell® platform (Promega®) were used for DNA isolation.
FGFR3 mutation analysis
FGFR3 mutation analysis was done using the PCR-SNaPshot method in all laboratories. Details of this method were reported previously [14, 15]. Briefly, three regions (exons 7, 10 and 15), frequently mutated and representing at least 99% of activating oncogenic FGFR3 mutations in UCB, were simultaneously amplified by PCR. After removing excess primers and deoxynucleotides, specific SNaPshot primers were annealed to the PCR products, separated by capillary electrophoresis and analyzed in an automatic sequencer (Prism® 3100 genetic analyzer). With this PCR-SNaPshot method, a total of 11 known oncogenic FGFR3 mutations can be detected. The codon numbering refers to the cDNA open reading frame of the FGFR3b isoform expressed in epithelia [6].
FGFR3 expression analysis
FGFR3 expression could be studied with IHC in 357/494 cystectomy specimens and in 72/117 paired RC/LN+ from the International cohort (a subset from Amsterdam, Toronto and Turku). Standard tissue micro-array technology was used in both laboratories [16]. The available cases were routinely processed with a monoclonal antibody against FGFR3 (FGFR3 B9, Santa Cruz, CA). Positive and negative controls were included in each run. Slides were assessed by BWGvR and THvdK (Toronto) and by BWGvR and JS (Amsterdam). A semi-quantitative scoring system was used: 0, negative; 1, faint/normal; 2, moderate positivity; 3, strong positivity. FGFR3 overexpression was defined by a score of 2 or 3 as previously described [15, 17, 18].
statistics
SPSS®, version 20, was used for data documentation and analysis. χ2 statistics were used to analyze possible associations between FGFR3 status and pathological variables. Statistical significance was set at P < 0.05.
results
Within the TUR cohort, FGFR3 mutations were detected in 13/34 T1 and 8/27 ≥T2 UCB, respectively. Comparing paired superficial and deep parts, no discordance was found within the T1-TUR samples (Figure 1A), whereas discordance was observed in half of the cases within the ≥T2-TUR samples with only 4/8 FGFR3 mutations in the invasive area (Figure 1B). In another four TUR cases (one with mutation), multiple samples from same area (three multiple superficial and one multiple invasive areas) were analyzed as a control experiment. We found no difference among these samples.
Figure 1.
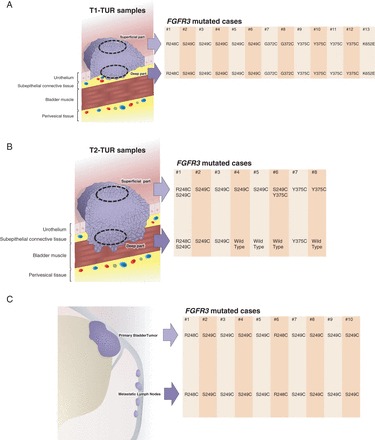
The distributions of fibroblast growth factor receptor 3 (FGFR3) mutations in the superficial and deep compartments of the 61 (34 T1 and 27 ≥T2) patients included in the transurethral resection (TUR) cohort and in the 614 radical cystectomy patients with 201 paired cystectomies and metastatic nodes available. (A) The 13 mutated cases in 34 paired T1-TUR samples are displayed. Both parts (superficial and deep) were wild type in 21 cases. (B) The eight mutated cases in paired ≥T2-TUR samples are displayed. Both parts (superficial and deep) were wild type in 19 cases. (C) The 10 mutated cases in paired cystectomies and metastatic nodes are displayed. The cystectomy and metastatic node were both wild type in 191 cases.
Within the RC cohort, FGFR3 status was known for 614 RC, of which 254 (41%) were pN+. Of the 254 LN+ cases, FGFR3 status was available for 201 (79%) paired RC/LN+ samples. In the 614 cystectomies, 67 (11%) FGFR3 mutations were detected, of which 54 were pN0 (Table 1). supplementary Table S1, available at Annals of Oncology online summarizes the distribution of mutations for the International and French RC cohorts, respectively. In supplementary Table S2, available at Annals of Oncology online, the types of FGFR3 mutations, with S249C (67%) as the most frequent one, are listed. Table 2 presents the clinicopathological characteristics of the 13 patients with an FGFR3 mutation and pN+ UCB. In the 201 paired RC/LN+ samples, the same FGFR3 mutation was detected in the cystectomy and LN+ specimen (Figure 1C). Discordance between the 201 paired samples was not observed (specificity: 100%). The presence of an FGFR3 mutation was associated with lower pT-stage (P < 0.001) and pN0 (P < 0.001) at RC (Table 1).
Table 1.
The distribution of samples according to the primary tumors pathologic pT-stage and FGFR3 mutation status among either N0 or N+ cases in the radical cystectomy cohort
| pTa, pT1, pTis | pT2 | pT3 | pT4 | Total | |
|---|---|---|---|---|---|
| N0 | |||||
| Wild type | 47 | 93 | 118 | 48 | 306 |
| Mutated | 23 | 11 | 14 | 6 | 54 |
| N+ | |||||
| Wild type | 5 | 46 | 127 | 63 | 241 |
| Mutated | 0 | 0 | 6 | 7 | 13 |
| Total | 75 | 150 | 265 | 124 | 614 |
FGFR3 mutations were associated with lower pT-stage (P < 0.001) and pN0 (P < 0.001) at radical cystectomy.
Table 2.
Clinical and pathological characteristics of patients with pN+ UC and a FGFR3 mutation detected in cystectomy and/or positive lymph node.
| Patient | Age | Gender | Histology | Pathological stage | WHO1973 grade | AC | Relapse | Relapse type | Vital status | Follow-up (years) | Disease status | FGFR3 mutation | Mutated samples |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Int711 | 73 | M | UC | pT4aN2 | 3 | No | Yes | DM | Dead | 1.7 | DOD | S249C | T, N |
| Int1008 | 44 | F | UC + SCC | pT4aN2 | 3 | No | No | – | Alive | 0.7 | FOD | S249C | T |
| Int1015 | 39 | M | UC | pT4aN2 | 3 | No | Yes | DM | Alive | 1.1 | FOD | S249C | T |
| Int1028 | 78 | F | UC | pT4aN2 | 2 | Yes | No | – | Alive | 1.5 | FOD | S249C | T, N |
| Int3097 | 56 | F | UC | pT3bN2 | 3 | No | Yes | DM | Dead | 1.2 | DOD | R248C | T, N |
| Int3113 | 78 | M | UC + SCC | pT3aN2 | 3 | No | No | – | Alive | 9.4 | FOD | S249C | T, N |
| Int3125 | 75 | M | UC | pT3aN1 | 3 | No | Yes | DM | Dead | 1.1 | DOD | S249C | T |
| Int3180 | 62 | F | UC + SCC | pT4bN2 | 3 | No | Yes | DM | Dead | 0.4 | DOD | S249C | T, N |
| Int3280 | 81 | F | UC | pT3bN2 | 3 | No | Yes | DM | Dead | 1.9 | DOD | R248C | T, N |
| VCA023 | 77 | M | UC | pT3aN1 | 3 | Yes | No | – | Alive | 2 | FOD | S249C | T, N |
| VCA045 | 46 | M | UC | pT3bN2 | 3 | Yes | Yes | DM | Dead | 3.6 | DOD | S249C | T, N |
| VCA047 | 56 | M | UC | pT4aN1 | 3 | Yes | No | – | Alive | 11 | FOD | S249C | T, N |
| VCA090 | 59 | M | UC | pT4aN2 | 3 | Yes | Yes | DM | Dead | 2.6 | DOD | S249C | T, N |
In three cases, the node sample was not available.
AC, adjuvant chemotherapy; M, male; F, female; UC, urothelial carcinoma; UC + SCC, urothelial carcinoma with squamous differentiation; T, tumor; N, node; DM, distant metastasis; FOD, free of disease; DOD, dead of disease.
Finally, FGFR3 expression was studied with IHC in 357/614 cystectomy specimens (Table 3). In 280 RC, FGFR3 expression was normal and no mutation was found. We found 70 RC with overexpression, of whom 37 had a mutation. In seven cases, we found an FGFR3 mutation with normal expression at IHC (Table 3). IHC samples were available for 72/201 paired RC/LN+ cases (Table 4). FGFR3 expression was concordant in 64/72 (89%) cystectomy and LN+ specimens.
Table 3.
FGFR3 expression and FGFR3 mutation (cystectomy specimens) in a subset of 357/494 cases from the international radical cystectomy cohort
|
FGFR3 expression in cystectomy |
Total | ||
|---|---|---|---|
| Normal | Overexpression | ||
| FGFR3 mutation in cystectomy | |||
| Wild type | 280 | 33 | 313 |
| Mutated | 7 | 37 | 44 |
| Total | 287 | 70 | 357 |
Table 4.
FGFR3 expression in cystectomy and corresponding metastatic lymph nodes in a subset of 72/117 pN+ cases from the international radical cystectomy cohort
| FGFR3 expression in cystectomy |
Total | ||
|---|---|---|---|
| Normal | Overexpression | ||
| FGFR3 expression in positive node | |||
| Normal | 57 | 4 | 61 |
| Overexpression | 4 | 7 | 11 |
| Total | 61 | 11 | 72 |
discussion
In metastatic UCB, several targeted therapies have been evaluated as second-line treatment [19], but none of them has made it into the clinical practice so far. Although the development of effective inhibitors (including anti-FGFR3 treatment) still is at an early stage, FGFR3 is a very promising actionable target in UCB [9–12, 19]. Comparable with other malignancies, targeted therapy has shown significant activity in only a minority of UCB patients [10–12, 19]. Reasons for this limited activity may include the diverse genomic landscape of UCB [10], the absence of molecular tumor analysis before test drug administration [12] and lack of adequate studies addressing intratumor/patient heterogeneity of potential actionable targets [11]. Considering cN0M0 patients in the perioperative setting, molecular tumor analysis and heterogeneity assessment are pivotal before administering a drug against an actionable target. To our knowledge, the present study is the first to address tumor heterogeneity for the perioperative setting in UCB with TUR and RC/LN+ specimens.
FGFR3 activation mostly occurs via oncogenic mutations [6–12], occasionally by rearrangements [10, 20] and also via overexpression by other mechanisms such as copy number gain [10, 15, 17]. Less is known about FGFR3 intratumor/patient heterogeneity in UCB [21]. The main purpose of our multicenter, multi-laboratory study was to address this heterogeneity for the perioperative setting of invasive UCB. Previous small, single-center, single laboratory studies have shown an ∼80% concordance in multiple synchronous and metachronous noninvasive UCB [17, 21]. Furthermore, recent important preclinical work provided a cellular and genetic basis for this diversity in UCB [22]. In our study on TUR samples, we showed that FGFR3 mutation status may differ between the superficial and invasive part of one tumor. So far, only one previous study reported on FGFR3 heterogeneity in superficial and deep invasive parts at TUR [17]. Within 18 mutated UCB, 9 had the same mutation in the two compartments, 8 had mutation only in the most superficial area and 1 had different mutations in the two parts. However, the authors were not sure that samples were from the same lesion in the bladder. In the present TUR series, the same tumor was analyzed. It was notable that we found four cases with an FGFR3 mutation in the superficial part but not in the deep part of the same ≥T2 tumor. Conversely, we did not observe a difference in FGFR3 status in 201 RC and LN+ samples of our RC cohort. Therefore, it is likely that, at RC, the deep part of the tumor has been analyzed and that the superficial part was already removed by the preceding TUR. The mutation frequency at RC (11%) also corresponded to that of the deep part of the ≥T2-TUR cohort (15%). The frequency of FGFR3 mutations (12%) in the TCGA cohort of 131 high-grade muscle-invasive UCB (mostly cystectomy specimens) was also comparable with our cohort. This implies that the deep part of the tumor at TUR needs to be assessed if neoadjuvant anti-FGFR3 treatment is considered.
Our study showed that, if a mutated clone progresses in MI-UCB, the FGFR3 mutation is conserved in the invasive compartment and also in the metastatic node, despite the notion that not all the lesions in the RC cohorts were primary (first diagnosis) UCB. We also reported that the FGFR3 mutation was associated with lower T-stage and pN0 at RC. Others have already reported that FGFR3 mutations are also in muscle invasive urothelial carcinoma of the bladder (MI-UCB) most of the time not accompanied by many other molecular alterations [8, 10]. Taken together, all these findings suggest that anti-FGFR3 treatment may have significant clinical impact in the perioperative setting for a relative small subgroup of MI-UCB patients.
FGFR3 expression is another way to explore FGFR3 activity. Turo et al. [18] recently reported a heterogeneity study using FGFR3 expression by IHC without FGFR3 mutation evaluation. In their cohort, paired RC/LN+ samples were available for IHC analysis in 106/150 pN+-UCB and concordance was found in 79/106 (75%) cases. We here reported IHC concordance in 64/72 (89%). Previous IHC studies showed that ∼40% of invasive FGFR3 wild-type tumors overexpress FGFR3, suggesting an alternative mechanism to activate FGFR3 [10, 15, 17]. In our RC series, only 10% of wild-type cases showed overexpression (Tables 3 and 4). One of the reasons for this lower percentage might be that we analyzed RC specimens and consequently deeper parts of the tumor than in the previous studies. Nevertheless, we cannot exclude that a small subset of patients with wild-type tumors may still benefit from anti-FGFR3 treatment. On the other hand, we showed that FGFR3 mutation analysis was extremely robust across four laboratories. IHC is likely more prone to observer variability than FGFR3 mutation analysis, making it less appropriate to assess FGFR3 heterogeneity within a tumor or metastases of a patient. Future study should focus on how to combine FGFR3 mutation, translocation and copy number status with FGFR3-IHC to guide optimal personalized anticancer treatment.
In conclusion, we found that FGFR3 mutations appeared conserved in primary bladder cancer and corresponding lymph-node metastases. Hence, anti-FGFR3 treatment may have significant clinical impact in the adjuvant setting. We also showed that the deep part of the tumor needs to be assessed if neoadjuvant anti-FGFR3 treatment is considered. Our data on tumor heterogeneity suggest that personalized anti-FGFR3 therapy may improve bladder cancer treatment of a relatively small, well-selected subgroup of invasive UCB patients. Studies on the heterogeneity of actionable molecular targets should precede clinical trials with these drugs in the perioperative setting.
funding
Financial support was given by the University of Toronto, a grant from the Dutch Cancer Society –KWF Kankerbestrijding, the European Urological Scholarship Programme, a program grant from Cancer Research UK and the French Urological Association. Specific grant numbers are not applicable.
disclosure
The authors have declared no conflicts of interest.
Supplementary Material
acknowledgements
The authors acknowledge all patients who contributed tissue for research. The authors thank Simone Russell, Rati Vajpeyi, Sally Hanna, Roni Sambas and Cynthia Kuk (Toronto) and Floris Groenendijk, Renate de Groot and Chantall Curial (Amsterdam) for help and advice. We thank the Core Facility for Molecular Pathology and Bio-banking of the Netherlands Cancer Institute—Antoni van Leeuwenhoek and the Tissue Bank Unit—PRB Mondor for their assistance. We also thank the pathologists who provided tissue blocks for the multicenter French cohort. The authors would like to acknowledge the International Bladder Cancer Network (IBCN) and the Bladder Cancer Advocacy Network (BCAN) for providing the platform to collaborate on this project. The Regional Ethics Board of the University Health Network, Toronto (02-0515-C and 08-0263-T) gave approval. The Translational Research Board of the Netherlands Cancer Institute—Antoni van Leeuwenhoek Hospital (CFMPB 160) approved the study. Approval was obtained from the Leeds East Research Ethics Committee. The Regional Ethics Board of Ile-de-France IX (Comité de protection des Personnes—Ile-de-France IX, Créteil) approved the study (11-052).
references
- 1. Stein JP, Lieskovsky G, Cote R et al. Radical cystectomy in the treatment of invasive bladder cancer: long-term results in 1,054 patients. J Clin Oncol 2001; 19: 666–675. [DOI] [PubMed] [Google Scholar]
- 2. Ghoneim MA, Abdel-Latif M, el Mekresh M et al. Radical cystectomy for carcinoma of the bladder: 2720 consecutive cases 5 years later. J Urol 2008; 180: 121–127. [DOI] [PubMed] [Google Scholar]
- 3. Griffiths G, Hall R, Sylvester R et al. International phase III trial assessing neoadjuvant cisplatin, methotrexate, and vinblastine chemotherapy for muscle-invasive bladder cancer: long-term results of the BA06 30894 trial. J Clin Oncol 2011; 29: 2171–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zargar H, Espiritu PN, Fairey AS et al. Multicenter assessment of neoadjuvant chemotherapy for muscle-invasive bladder cancer. Eur Urol 2015; 67: 241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sternberg CN, Skoneczna I, Kerst JM et al. Immediate versus deferred chemotherapy after radical cystectomy in patients with pT3-pT4 or N+ M0 urothelial carcinoma of the bladder (EORTC 30994): an intergroup, open-label, randomised phase 3 trial. Lancet Oncol 2015; 16: 76–86. [DOI] [PubMed] [Google Scholar]
- 6. Cappellen D, De Oliveira C, Ricol D et al. Frequent activating mutations of FGFR3 in human bladder and cervix carcinomas. Nat Genet 1999; 23: 18–20. [DOI] [PubMed] [Google Scholar]
- 7. van Rhijn BW, Vis AN, van der Kwast TH et al. Molecular grading of urothelial cell carcinoma with fibroblast growth factor receptor 3 and MIB-1 is superior to pathologic grade for the prediction of clinical outcome. J Clin Oncol 2003; 21: 1912–1921. [DOI] [PubMed] [Google Scholar]
- 8. Neuzillet Y, Paoletti X, Ouerhani S et al. A meta-analysis of the relationship between FGFR3 and TP53 mutations in bladder cancer. PLoS ONE 2012; 7: e48993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Iyer G, Al-Ahmadie H, Schultz N et al. Prevalence and co-occurrence of actionable genomic alterations in high-grade bladder cancer. J Clin Oncol 2013; 31: 3133–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014; 507: 315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lerner SP. Targeted therapies for metastatic bladder cancer. J Urol 2015; 193: 8–9. [DOI] [PubMed] [Google Scholar]
- 12. Mazzola CR, Siddiqui KM, Billia M, Chin J. Dovitinib: rationale, preclinical and early clinical data in urothelial carcinoma of the bladder. Expert Opin Investig Drugs 2014; 23: 1553–1562. [DOI] [PubMed] [Google Scholar]
- 13. Edge S, Byrd D, Compton C. AJCC Cancer Staging Manual, 7th edition New York, NY: Springer; 2010. [Google Scholar]
- 14. van Oers JM, Lurkin I, van Exsel AJ et al. A simple and fast method for the simultaneous detection of nine fibroblast growth factor receptor 3 mutations in bladder cancer and voided urine. Clin Cancer Res 2005; 11: 7743–7748. [DOI] [PubMed] [Google Scholar]
- 15. Neuzillet Y, van Rhijn BW, Prigoda NL et al. FGFR3 mutations, but not FGFR3 expression and FGFR3 copy-number variations, are associated with favourable non-muscle invasive bladder cancer. Virchows Archiv 2014; 465: 207–213. [DOI] [PubMed] [Google Scholar]
- 16. van Rhijn BW, Catto J, Goebel PJ et al. Molecular markers for urothelial bladder cancer prognosis: towards implementation in clinical practice. Urol Oncol 2014; 32: 1078–1087. [DOI] [PubMed] [Google Scholar]
- 17. Tomlinson DC, Baldo O, Harnden P, Knowles MA. FGFR3 protein expression and its relationship to mutation status and prognostic variables in bladder cancer. J Pathol 2007; 213: 91–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Turo R, Harnden P, Thygesen H et al. FGFR3 expression in primary invasive bladder cancers and matched lymph node metastases. J Urol 2015; 193: 325–330. [DOI] [PubMed] [Google Scholar]
- 19. Rouanne M, Loriot Y, Lebret T, Soria JC. Novel therapeutic targets in advanced urothelial carcinoma. Crit Rev Oncol Hematol 2015; 10.1016/j.critrevonc.2015.10.021. [DOI] [PubMed] [Google Scholar]
- 20. Williams SV, Hurst CD, Knowles MA. Oncogenic FGFR3 gene fusions in bladder. Hum Mol Genet 2013; 22: 795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gerlinger M, Catto JW, Orntoft TF et al. Intratumour heterogeneity in urologic cancers: from molecular evidence to clinical implications. Eur Urol 2015; 67: 729–737. [DOI] [PubMed] [Google Scholar]
- 22. Van Batavia J, Yamany T, Molotkov A et al. Bladder cancers arise from distinct urothelial sub-populations. Nat Cell Biol 2014; 16: 982–991. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


