Damage-free structures of the resting state of one of the most studied copper nitrite reductases were obtained independently by X-ray free-electron laser (XFEL) and neutron crystallography, representing the first direct comparison of neutron and XFEL structural data for any protein. In addition, damage-free structures of the reduced and nitrite-bound forms have been obtained to high resolution from cryogenically maintained crystals by XFEL crystallography.
Keywords: copper-containing nitrite reductases, neutron crystallography, X-ray free-electron lasers
Abstract
Copper-containing nitrite reductases (CuNiRs) that convert NO2 − to NO via a CuCAT–His–Cys–CuET proton-coupled redox system are of central importance in nitrogen-based energy metabolism. These metalloenzymes, like all redox enzymes, are very susceptible to radiation damage from the intense synchrotron-radiation X-rays that are used to obtain structures at high resolution. Understanding the chemistry that underpins the enzyme mechanisms in these systems requires resolutions of better than 2 Å. Here, for the first time, the damage-free structure of the resting state of one of the most studied CuNiRs was obtained by combining X-ray free-electron laser (XFEL) and neutron crystallography. This represents the first direct comparison of neutron and XFEL structural data for any protein. In addition, damage-free structures of the reduced and nitrite-bound forms have been obtained to high resolution from cryogenically maintained crystals by XFEL crystallography. It is demonstrated that AspCAT and HisCAT are deprotonated in the resting state of CuNiRs at pH values close to the optimum for activity. A bridging neutral water (D2O) is positioned with one deuteron directed towards AspCAT Oδ1 and one towards HisCAT N∊2. The catalytic T2Cu-ligated water (W1) can clearly be modelled as a neutral D2O molecule as opposed to D3O+ or OD−, which have previously been suggested as possible alternatives. The bridging water restricts the movement of the unprotonated AspCAT and is too distant to form a hydrogen bond to the O atom of the bound nitrite that interacts with AspCAT. Upon the binding of NO2 − a proton is transferred from the bridging water to the Oδ2 atom of AspCAT, prompting electron transfer from T1Cu to T2Cu and reducing the catalytic redox centre. This triggers the transfer of a proton from AspCAT to the bound nitrite, enabling the reaction to proceed.
1. Introduction
The highly brilliant undulator beamlines at modern synchrotron facilities have facilitated the structure determination of biological molecules and their complexes at high resolution using conventional synchrotron-radiation crystallography (SRX). The brilliance of the X-rays at some of the state-of-the-art crystallographic beamlines has enabled this to be achieved using much smaller (10–30 µm) crystals than was anticipated at the turn of the century. These gains have come at the expense of an increased absorbed X-ray dose per unit volume and the potential for concomitant radiolysis and radiation damage (Garman, 2010 ▸; Yano et al., 2005 ▸; Horrell et al., 2016 ▸). Biological molecules and their complexes that use the redox properties of transition metals are particularly sensitive to radiation damage and X-ray-induced chemical changes (Yano et al., 2005 ▸; Horrell et al., 2016 ▸). This poses a serious limitation to obtaining damage-free structures of redox enzymes at sufficiently high resolutions that the enzyme mechanism can be defined at the chemical level, such as changes resulting from one-electron reduction or the geometry and position of the chemical substrate. The recent advent of XFEL crystallography using femtosecond X-ray pulses provides a new opportunity to obtain damage-free structures (Suga et al., 2014 ▸, 2017 ▸), adding to neutron crystallography and NMR, which have remained the only radiation-damage-free structural probes for decades, albeit with their own particular limitations (Blakeley et al., 2015 ▸; Blakeley, 2009 ▸; Luchinat & Banci, 2017 ▸).
Copper nitrite reductases (CuNiRs) are a highly conserved family of enzymes involved in the first committed step in the denitrification pathway: the reduction of nitrite (NO2 −) in a one-electron two-proton reaction to form the gaseous product nitric oxide (NO; Zumft, 1997 ▸). The enzymes are homotrimeric, with two cupredoxin-like domains in each monomer that harbour one type 1 copper (T1Cu) and one type 2 copper (T2Cu) site. The catalytic T2Cu site is located at the interface between two monomers, with a (His)3–H2O ligand coordination, while the T1Cu site is located near the surface of the protein. The two copper ions are hard-wired via a 12.1 Å Cys–His bridge through which an electron required for catalysis is transferred when the substrate NO2 − binds to the oxidized T2Cu, displacing the bound water ligand of the resting enzyme. NO2 − accesses the active site via a hydrophobic channel of ∼6 Å in width that is formed at the monomer–monomer boundary. Two residues, Asp98 (AspCAT) and His255 (HisCAT) in Achromobacter cycloclastes CuNiR (AcNiR) numbering, that are conserved in all CuNiRs and are located either side of the T2Cu, are required for enzymatic activity. AspCAT is hydrogen-bonded to the copper H2O ligand and also to HisCAT via an H2O bridge. This hydrogen-bonding network is preserved when nitrite binds, and mutagenic, mechanistic and computational studies are consistent with its involvement in the provision of the two protons for the reaction. However, an unresolved issue in mechanistic studies is where the two protons that are required for catalysis originate from. Protonated nitrite is an unlikely source since when bound to Cu2+ it is energetically unstable, decaying to the damaging NO+. In addition to its role in proton delivery, AspCAT is involved in modulating nitrite binding. In AcNiR the AspCAT residue has been observed in two different conformations: one termed ‘proximal’, in which it is oriented towards the T2Cu and linked to His255 via a water molecule, and a second position termed ‘gatekeeper’, in which it is oriented away from the T2Cu and hydrogen-bonded to the H2O ligand of the T2Cu (Antonyuk et al., 2005 ▸). Synchrotron-based X-ray structural studies have shown that nitrite binds to the oxidized T2Cu site of CuNiR to displace the H2O ligand. In most structures, the binding mode is a bidentate η2-O,O binding (Solomon et al., 2014 ▸), with a single example of η1-O binding observed in an enzyme from a thermophile (Fukuda et al., 2014 ▸). A number of different orientations of η2-O,O-bound nitrite have been observed, both in the resting enzyme, which shift with pH, and also in X-ray-induced turnover. In the case of AcNiR the serial structures used to construct a structural movie, which were obtained by low-dose MSOX (multiple serial structures from one crystal) at 190 K, revealed a ‘top-hat’ (vertical O-bidentate) conformation of NO2 − in the first frame of structures, which then flipped to a ‘side-on’ conformation with near-equivalent Cu—O1, Cu—O2 and Cu—N distances in subsequent frames, prior to bond breakage and product formation (Horrell et al., 2018 ▸). Although this may indicate that the first stage of substrate utilization may involve a conformational change, pH-dependence of the NO2 − binding geometry has been observed for some CuNiRs from other species (Zhao et al., 2002 ▸; Abraham et al., 1997 ▸; Jacobson et al., 2007 ▸).
Significant efforts have been made using XFEL crystallography to obtain damage-free structures of the resting state as well as a number of catalytically important forms for a number of CuNiRs [NiRs from Alcaligenes faecalis (AfNiR; Fukuda, Tse, Nakane et al., 2016 ▸), Geobacillus thermodenitrificans (GtNiR; Fukuda, Tse, Suzuki et al., 2016 ▸) and Alcaligenes xylosoxidans (AxNiR; Halsted et al., 2018 ▸)]. In the resting-state serial femtosecond crystallography (SFX) structures of AfNiR and GtNiR, a chloride and a sodium ion originating from the crystallization or purification protocols were found in the T2Cu pocket, respectively. For AxNiR, serial femtosecond rotational crystallography (SF-ROX) revealed an unprecedented dioxo ligand bound to the T2Cu site, which was interpreted to rationalize the oxidase activity of some CuNiRs and was considered to represent a trapped intermediate of the oxidase reaction awaiting the delivery of a second electron to enable turnover to produce the product H2O2 (Halsted et al., 2018 ▸).
Here, using SF-ROX, damage-free structures of AcNiR have been obtained in the as-isolated oxidized, chemically reduced and NO2 −-bound forms. These are complemented by a damage-free room-temperature (RT) neutron crystallographic structure of the as-isolated perdeuterated enzyme. As such, unprecedented information is obtained on the nature of the ligands of T2Cu in the resting state and the protonation states of the catalytic residues. Neutron diffraction studies provide the positions of D atoms, allowing the direct determination of the protonation states of the protein residues and water molecules in the catalytic pocket for the first time, both of which are necessary in order to understand the enzyme mechanism. We unequivocally establish that in the resting state the T2Cu of AcNiR is liganded by a single, neutral water molecule. Moreover, the neutron structure shows AspCAT to be unprotonated, which is consistent with the consensus view (Solomon et al., 2014 ▸), but, contrary to expectation, shows that HisCAT is also deprotonated at the N∊2 position. These observations, together with damage-free SF-ROX structures of the reduced and substrate-bound forms, provide new insights into the catalytic steps of these important enzymes. These structures are also likely to provide a firm basis for detailed computational chemistry efforts.
2. Methods
2.1. Production of recombinant AcNiR crystals
The nirK gene from A. cycloclastes with codon optimization for expression in Escherichia coli was acquired from GenScript and cloned into a pET-26b(+) plasmid. The plasmid was transformed into E. coli BL21 (DE3) cells via heat shock and the transformant was cultured on KanR lysogeny broth (LB) agar to isolate individual colonies. 500 ml LB supplemented with 30 µg ml−1 kanamycin was inoculated with a single colony and was incubated with shaking at 37°C. Protein overexpression was induced with 2 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) and 1 mM CuSO4. Incubation continued for 24 h, after which the cells were harvested by centrifugation and resuspended in 20 mM Tris–HCl pH 7.5, 0.1 mg ml−1 lysozyme before being disrupted by sonication. The lysate was collected by centrifugation and dialyzed against 20 mM Tris–HCl pH 7.5, 2 mM CuSO4, followed by dialysis against water. The lysate was loaded onto a DEAE-Cellulose column equilibrated with 20 mM Tris–HCl pH 7.5, which was subsequently washed with 20 mM Tris–HCl pH 7.5 followed by 100 mM Tris–HCl pH 7.5. AcNiR was eluted from the column using an NaCl gradient from 100 to 250 mM in 20 mM Tris–HCl pH 7.5. 4 M ammonium sulfate was used to completely precipitate the AcNiR, which was dissolved in 10 mM HEPES–NaOH pH 6.5. The AcNiR was concentrated to 50 mg ml−1 and was crystallized by hanging-drop vapour diffusion against a 1:1 ratio of 1.2 M ammonium sulfate and 100 mM citrate buffer pH 5.0. Crystals with a pyramidal shape grew to ∼0.7 × 0.7 × 0.7 mm in size.
2.2. SF-ROX crystal treatment, data collection and processing
The harvested crystals of AcNiR were soaked in cryoprotectant, cryocooled by plunging into liquid nitrogen and maintained at 77 K after cooling. SF-ROXOX crystals were soaked in 3.4 M ammonium sulfate, 100 mM citrate buffer pH 5.0 for 10 s. SF-ROXNIT crystals were soaked in 3.4 M sodium malonate pH 5.0, 100 mM sodium nitrite for 10 s. SF-ROXRED crystals were soaked in 3.4 M sodium malonate pH 5.0, 100 mM sodium ascorbate for 30 min, after which their colour changed from green to colourless. SF-ROX data collection was carried out on BL2 EH3/4b at SACLA at 100 K as described previously (Hirata et al., 2014 ▸; Halsted et al., 2018 ▸). The X-ray energy was set to 10 keV and the pulses were of <10 fs in duration. The sample was positioned 10 mm downstream of the XFEL focal point, which gave a beam size at the sample position of 2.2 × 4.5 µm. The XFEL beam was attenuated to 5.7 × 1010 photons per pulse using a 100 µm thick aluminium X-ray attenuator. The crystals were rotated 0.1° and translated 50 µm between each snapshot. X-ray diffraction images were collected on an MX225-HS CCD detector (Rayonix) with a camera length of 110 mm. The same data-collection and processing procedure was used for all three data sets. Hit finding, indexing and integration were performed using CrystFEL (v.0.6.3; White et al., 2016 ▸), with the inner, middle and outer integration radii set to four, five and seven pixels, respectively. After the resolution of the indexing ambiguity, Bragg intensities were merged using the Monte Carlo method with frame scaling. Refinement was carried out in REFMAC5 (Murshudov et al., 2011 ▸) using the resting-state AcNiR structure (PDB entry 2bw4; Antonyuk et al., 2005 ▸) as the starting model with riding H atoms and isotropic B factors. Coot (Emsley et al., 2010 ▸) was used for manual model building between rounds of refinement. Double conformations of the side chains were assigned where 2F o − F c electron-density maps showed them clearly. The occupancies of the different conformers were chosen by examining the levels of the OMIT (F o − F c) electron-density maps and refined B factors. The final quality of the models was assessed using MolProbity (Chen et al., 2010 ▸). Data-processing and refinement statistics are given in Table 1 ▸.
Table 1. SF-ROX data-processing and refinement statistics.
Values in parentheses are for the highest resolution shell.
| SF-ROXOX | SF-ROXNIT | SF-ROXRED | |
|---|---|---|---|
| No. of crystals | 75 | 62 | 33 |
| Images collected | 1867 | 1257 | 581 |
| Images merged | 1377 | 1039 | 410 |
| Data collection | |||
| Space group | P213 | P213 | P213 |
| a = b = c (Å) | 94.95 | 94.92 | 94.61 |
| α = β = γ (°) | 90 | 90 | 90 |
| Resolution (Å) | 54.82–1.50 (1.54–1.50) | 54.80–1.50 (1.54–1.50) | 54.62–1.60 (1.64–1.60) |
| R split † (%) | 11.5 (90.4) | 10.6 (85.3) | 15.8 (70.8) |
| 〈I/σ(I)〉 | 6.3 (2.0) | 6.6 (2.3) | 5.4 (2.7) |
| CC1/2 ‡ | 0.980 (0.157) | 0.984 (0.288) | 0.957 (0.384) |
| Completeness (%) | 100.0 (100.0) | 100.0 (100.0) | 100.0 (100.0) |
| Multiplicity | 220.9 (77.2) | 154.7 (50.8) | 66.1 (44.3) |
| Wilson B factor (Å2) | 14.9 | 14.5 | 17.5 |
| Refinement | |||
| No. of unique reflections | 45883 (2276) | 45846 (2275) | 37489 (1858) |
| R work/R free (%) | 14.4/17.7 | 14.3/17.1 | 16.7/19.9 |
| No. of atoms | |||
| Protein | 2608 | 2595 | 2580 |
| Ligand/ion | 37 | 34 | 59 |
| Water | 425 | 418 | 279 |
| B factors (Å2) | |||
| Protein | 18.9 | 18.7 | 21.9 |
| Cu | 16.5 | 16.2 | 19.5 |
| SO4 2− | 33.2 | 41.5 | |
| NO2 − | 18.5 | ||
| Malonate | 37.2 | 30.1 | |
| Water | 30.0 | 30.0 | 32.7 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.013 | 0.013 | 0.015 |
| Bond angles (°) | 1.612 | 1.594 | 1.534 |
| PDB code | 6gsq | 6gt0 | 6gt2 |
2.3. In-house laboratory-source data collection and processing
Data sets for the resting state at a variety of pH values were obtained at the Barkla X-ray Laboratory of Biophysics using a Rigaku FR-E+ SuperBright rotating-anode generator with an EIGER R 4M detector. The same experimental setup was used to obtain a room-temperature structure from a perdeuterated crystal. For room-temperature data collection, the crystal was mounted in the cryoloop and protected from drying using MicroRT Tubing (MiTeGen) containing reservoir solution. Crystals of AcNiR were soaked for 15 min in 3.1 M sodium malonate, 100 mM NaNO2 at pH 5.0, 5.5, 6.0 and 6.5. For each pH value, data sets were collected with 15 s exposure time and a 1.0° oscillation step per image, with a total of 60 images per data set. The images were processed using XDS (Kabsch, 2010 ▸) and AIMLESS (Evans & Murshudov, 2013 ▸), and refinement, model building and validation were carried out in the same manner as for the SF-ROX structures. Data-processing and refinement statistics can be found in Supplementary Table S1.
2.4. Production of perdeuterated AcNiR crystals
An E. coli cell pellet containing AcNiR expressed under perdeuterated conditions was produced by the Deuteration Laboratory at the Institut Laue–Langevin (Haertlein et al., 2016 ▸). 5 g of the pellet was resuspended in lysis buffer consisting of 20 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.1 mg ml−1 lysozyme, 1 µg ml−1 protease inhibitors, 0.1 µg ml−1 DNAse and stirred on ice for 30 min. The cells were subsequently disrupted by sonication and the lysate was collected by centrifugation. The cell lysate was dialyzed against 20 mM Tris–HCl pH 7.5, 1 mM CuSO4 followed by the addition of 2 µl H2O2 and further dialysis against the same buffer without any additives. The lysate was loaded onto a hydroxyapatite column equilibrated with 100 mM Tris–HCl pH 7.5 and washed with a mixture of 6 mM Tris–HCl pH 7.5 and 2 mM potassium phosphate buffer pH 7.5 before being eluted using a potassium phosphate buffer gradient from 10 to 150 mM. The eluted fraction was applied onto a DEAE-Sepharose column pre-equilibrated with 20 mM Tris–HCl pH 7.5 and was eluted using an NaCl gradient from 50 to 250 mM. The eluted fraction was subsequently applied onto a HiLoad 16/600 Superdex 200 size-exclusion chromatography column (GE Life Sciences) pre-equilibrated with 150 mM NaCl in 20 mM Tris–HCl pH 7.5. In the final stage, AcNiR was precipitated using 4 M ammonium sulfate and the pellet was resuspended in 50 mM MES–NaOH pD 6.9. The pH values were determined using a conventional pH meter and the pHobs reading was corrected as described in Schowen & Schowen (1982 ▸). AcNiR was crystallized by hanging-drop vapour diffusion against a 1:1 ratio of 1.1 M ammonium sulfate and 100 mM sodium acetate pD 5.4. For crystallization, 5 µl protein solution at a concentration of 20 mg ml−1 was mixed with 5 µl reservoir solution; crystallization was initiated by adding microcrystals 2 h after the crystallization was set up. Two additional drops of equivalent size were added in one week and were merged with the nucleated drop after equilibration. It took three weeks for the crystal to reach its final size. A large single pyramid-shaped crystal of ∼0.9 × 0.4 × 1.0 mm was mounted in a 2 mm diameter capillary and stored for neutron data collection.
2.5. Neutron data collection and structural refinement
Neutron diffraction data were collected at RT to 1.8 Å resolution from a perdeuterated crystal of AcNiR (∼0.36 mm3 in volume) using the quasi-Laue neutron diffractometer LADI-III (Blakeley et al., 2010 ▸) at the Institut Laue–Langevin. A total of 20 images of 18 h exposure time each were collected from four different crystal orientations. These data were indexed and integrated using LAUEGEN (Campbell et al., 1998 ▸), wavelength-normalized using LSCALE (Arzt et al., 1999 ▸) and scaled and merged using the CCP4 program SCALA (Winn et al., 2011 ▸). Previously, D-exchanged crystals of similar volume had been used to collect diffraction data that extended to only 2.3 Å resolution (Blakeley et al., 2015 ▸), illustrating the benefits of using perdeuterated samples.
A 1.9 Å resolution X-ray data set collected at RT from a perdeuterated crystal was used as the starting model for neutron structural refinement. Using Ready_Set! from the PHENIX software suite (Adams et al., 2010 ▸), D atoms were added to the residues at calculated positions in preparation for structural refinement using phenix.refine. After initial rigid-body refinement, several rounds of maximum-likelihood-based refinement of individual coordinates and individual B factors against the neutron data were performed while applying restraints from the X-ray structure of the perdeuterated crystal, the data for which were collected at room temperature using an in-house X-ray generator, to maintain the geometry of the copper sites. After every round the model was visually inspected and manipulated in Coot (Emsley & Cowtan, 2010 ▸) using both positive and negative F o − F c and 2F o − F c nuclear scattering-length density maps to guide the modelling of solvent and protein D atoms. The final model contained 179 water molecules that were observed as full D2O molecules, along with ten water molecules that were rotationally disordered and thus were included as O atoms only. Data-processing and refinement statistics can be found in Table 2 ▸.
Table 2. Neutron data-processing and refinement statistics for neutronOX .
Values in parentheses are for the highest resolution shell.
| Data collection | |
| Wavelength range (Å) | 3.05–4.00 |
| No. of images | 20 |
| Setting spacing (°) | 7 |
| Average exposure time (h) | 18 |
| Space group | P213 |
| a = b = c (Å) | 97.98 |
| α = β = γ (°) | 90 |
| Resolution (Å) | 40–1.80 (1.90–1.80) |
| R p.i.m. (%) | 6.3 (12.7) |
| 〈I/σ(I)〉 | 7.9 (3.7) |
| Completeness (%) | 85.5 (69.8) |
| Multiplicity | 6.5 (2.9) |
| Refinement | |
| No. of unique reflections | 24728 |
| R work/R free (%) | 23.17/27.64 |
| No. of atoms | |
| Total | 5659 |
| Protein | 5109 |
| Cu | 2 |
| D2O | 182 D2O [546 atoms] |
| O | 2 |
| B factors (Å2) | |
| Protein | 15.2 |
| Cu | 8.6 |
| Water | 20.2 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.004 |
| Bond angles (°) | 0.884 |
| PDB code | 6gtj |
3. Results
3.1. Resting-state structures of AcNiR determined by SF-ROX and neutron crystallography
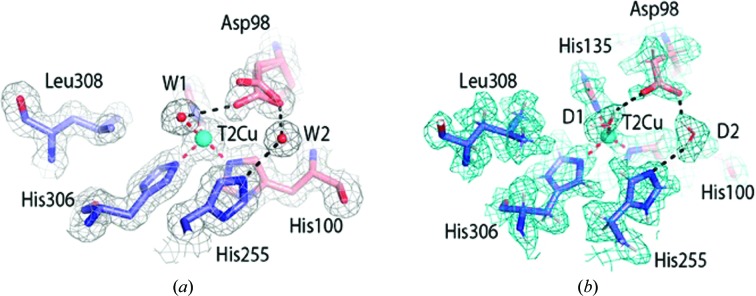
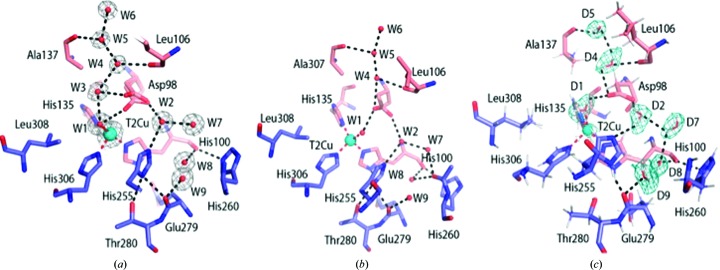
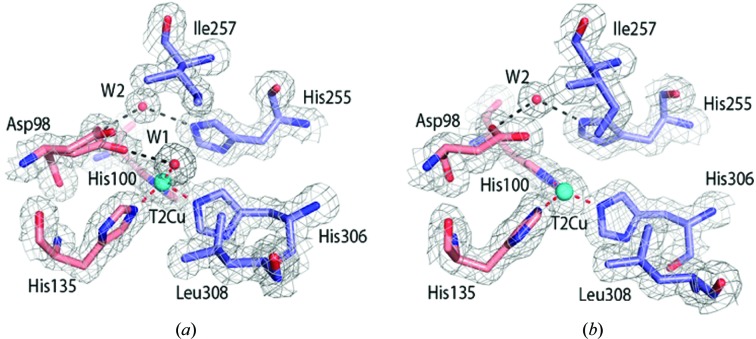
Resting-state structures of AcNiR were obtained using both SF-ROX at 100 K and neutron crystallography at RT and were refined to 1.5 and 1.8 Å resolution, respectively (Tables 1 ▸ and 2 ▸). The SF-ROXOX structure was compared with the 0.90 Å resolution synchrotron-radiation (SR) structure of AcNiR (Antonyuk et al., 2005 ▸), revealing conservation of the overall structure with an all-protein-atom r.m.s.d. of 0.29 Å. The T2Cu is ligated by a single, highly ordered water molecule (W1) bound in a distorted tetrahedral geometry relative to the histidine plane. The active-pocket residues HisCAT and Ile257 (IleCAT) were in similar positions; however, there were marked differences in the positioning of the AspCAT residue and its hydrogen-bonding network. In the SF-ROXOX structure the proximal conformation of the AspCAT side chain has two variants of the proximal conformation compared with a single proximal conformation in the synchrotron structure (Figs. 1 ▸ and 2 ▸). The additional conformation is formed by a 34° rotation around the Oδ1 atom, with the carboxyl Oδ2 atom forming a hydrogen bond to W1 at 3 Å, while the Oδ2 atom of the original conformation makes two hydrogen bonds to two water molecules, W3 and W4, one of which, W3, subsequently hydrogen-bonds to the T2Cu-ligated W1. These water molecules are part of the ordered water network in the substrate-entry channel. Both conformations are hydrogen-bonded via the Oδ1 atom to water W2, linking His255 to Asp98 (Fig. 2 ▸).
Figure 1.
The T2Cu site of AcNiR determined by SF-ROX and neutron crystallography. (a) The T2Cu in SF-ROXOX is ligated by a single water molecule (W1) hydrogen-bonded to the Asp98 residue. Asp98 (AspCAT) is visible in two conformations in the proximal position, with the residue rotating around the fixed Oδ1 atom. AspCAT is subsequently hydrogen-bonded to the linking water (W2), which is hydrogen-bonded to His255. Water molecules are shown as red spheres. (b) The T2Cu catalytic site determined by neutron crystallography. The protonation states of the T2Cu site residues are clearly seen, along with the orientations of the catalytic D2O molecule (D1) and the proton-sharing D2O molecule D2. There is no expected protonation of His255 and Asp98, while a D2 molecule connects His255 and Asp98. The 2F o − F c electron-density map is contoured at the 1σ level and is shown as a grey mesh. The 2F o − F c nuclear scattering-length density map is contoured at the 1σ level and is shown as a cyan mesh. Atoms are coloured by element, with different colour schemes used for the different chains. The T2Cu is shown as a cyan sphere and D2O water molecules are shown as red and white sticks. Metal-coordinating bonds are shown as red dotted lines. Selected hydrogen bonds are shown as black dotted lines.
Figure 2.
Water structure in the catalytic pocket and substrate-entry channel. (a) In the structure of oxidized AcNiR determined by SF-ROX, Asp98 (AspCAT) has a dual conformation; the usual proximal conformation hydrogen-bonds to two waters (W3 and W4), while the distorted proximal position, which is observed for the first time, hydrogen-bonds directly to the T2Cu water ligand. The waters (W3 and W4) are part of the ordered water network in the substrate-entry channel. Both proximal conformations of AspCAT are hydrogen-bonded via Oδ1, with the water W2 linking His255 (HisCAT) to AspCAT. 2F o − F c electron density is contoured at the 1σ level and is shown as a grey mesh. (b) In the atomic resolution crystal structure (PDB entry 2bw4) the proximal conformation is hydrogen-bonded to the ligated water W1A with an occupancy of 0.8. Water W1B with an occupancy of 0.2 is not shown for simplicity. (c) In the neutronOX structure, AspCAT is in a single proximal conformation. The 2F o − F c nuclear scattering-length density map is contoured around selected heavy waters at the 1σ level and is shown as a teal mesh. Atoms are coloured by element, with different colour schemes used for the different chains. The T2Cu is shown as a cyan sphere, D2O water molecules are shown as red and white sticks and water molecules are shown as small red spheres. Metal-coordinating bonds are shown as red dotted lines. Selected hydrogen bonds are shown as black dotted lines.
The neutron structure was modelled using the 1.9 Å resolution room-temperature X-ray structure of the perdeuterated protein obtained in this study. Subsequently, the structure was refined against the neutron data only, with restraints from the starting model to compensate for the weaker nuclear scattering length from Cu and S atoms. In contrast, the deuterons of the deuterated enzyme and the water molecules have neutron scattering lengths that are similar to those of C atoms. This makes the deuterons of histidine and water, for example, very visible in the neutron map. A water molecule appears as three atoms with similar densities. In contrast, H/D atoms are essentially invisible at the typical resolutions of X-ray structures. Even the subatomic resolution structure of AcNiR at better than 0.9 Å resolution was unable to provide the positions of many of the key H atoms in the catalytic pocket (Blakeley et al., 2015 ▸). Furthermore, the information in these very high-resolution SR structures is compromised as a significantly high X-ray dose is required that results in changes from the dose-dependent solvated electrons. The neutron structure determined here to 1.8 Å resolution provides the location of deuterons in the catalytic core and its associated water network for the first time (Figs. 1 ▸ and 2 ▸). The T2Cu is coordinated by a neutral D2O molecule similar to W1 in the distorted tetrahedral position observed in the SF-ROXOX structure.
In the atomic resolution SR structure the T2Cu has a tetrahedral coordination, with W1 hydrogen-bonded to the Oδ2 atom of the proximal AspCAT at a distance of 2.8 Å. The position of W1 in the SR structure differs from that in damage-free structures (Fig. 2 ▸). The neutron structure clearly shows W1 to be a D2O molecule rather than a D3O+ or OD− ion, which have previously been suggested as possible alternatives. The AspCAT residue in the neutronOX structure adopts a single proximal conformation and is bonded to a neutral heavy water D2O, which is also hydrogen-bonded to HisCAT. Compared with the SF-ROXOX structure, the His255 plane undergoes a 20° rotation. The neutron data revealed an ordered network of heavy water molecules around HisCAT, and also revealed hydrogen-bonding of the deuterated His255 N∊1 atom to the carbonyl O atom of Glu279 only [Fig. 2 ▸(b)]. An unresolved question in mechanistic studies of CuNiRs is the origin of the protons that are required for the reduction of NO2 −. Several studies involving intramolecular electron-transfer rates and pH-dependent activity, together with computational studies, have suggested the involvement of protonated AspCAT and HisCAT in providing the two protons during catalysis (Ghosh et al., 2009 ▸). Our neutron structure provides unequivocal data on the protonation states of active-site residues in the resting state of the CuNiR enzymes for the first time. The nuclear density maps clearly reveal that neither of these residues are protonated at pD 5.4, where the activity of the enzyme is at a maximum, while the bridging D2O has its two O—D bonds directed towards AspCAT and HisCAT. There is a chain of fully deuterated waters within hydrogen-bonding distance of each other, close to the liganded water at the T2Cu (Fig. 2 ▸).
The T1Cu site in the neutron structure shows no change in its copper geometry compared with the SF-ROX structure, but the second-sphere amino acid Met141 adopts a single conformation in the neutronOX structure as opposed to a dual conformation in the SF-ROXOX structure. Most of the differences in backbone structural alignment are found in an area of surface loop adjacent to Met141 consisting of residues 187–206, with an all-protein-atom r.m.s.d. of 1.02 Å (Supplementary Fig. S1). The loop is fully occupied and ordered in the neutron structure compared with the partially disordered loop in the SF-ROXOX structure. This loop is associated with the binding of the cognate partner protein cytochrome c 551 (Nojiri et al., 2009 ▸).
3.2. SF-ROX structures of the NO2 −-bound form of AcNiR
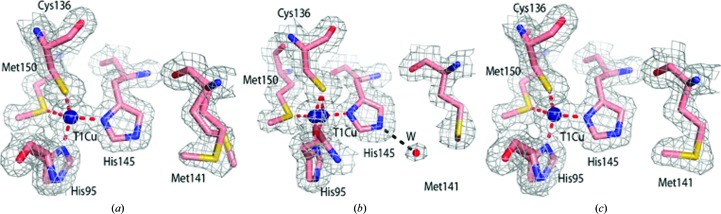
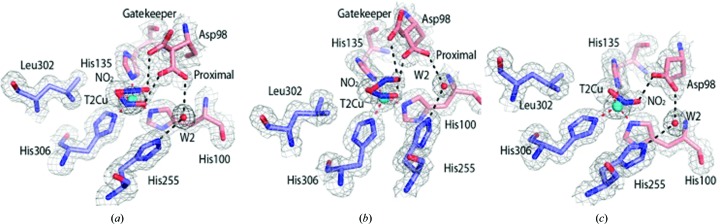
Upon NO2 − soaking of crystals of the oxidized enzyme, no changes in the geometry of the T1Cu site were observed in the SF-ROXNIT structure determined at 1.5 Å resolution (Table 1 ▸). Met141 is stabilized in a single conformation, covering His145 [Figs. 3 ▸(a) and 3 ▸(c)]. A large patch of positive electron density was observed at the T2Cu site, and NO2 − was initially assigned with full occupancy with a ‘side-on’ binding mode in view of the recent MSOX results (Horrell et al., 2018 ▸). This, however, did not fully satisfy the electron density, and the density was finally assigned as NO2 − bound in both ‘side-on’ and ‘top-hat’ conformations in almost equal proportions (Supplementary Fig. S2). The O1 atoms of ‘top-hat’ and ‘side-on’ NO2 − are separated by 1.3 Å. A partial-occupancy water (W4) is present at the position of the proximal Asp98 Oδ1 when in the gatekeeper conformation and is hydrogen-bonded to the bridging water W2. The observation of both conformations of nitrite in the damage-free SF-ROX structure raises an important question regarding the origin of the conformational changes observed during enzyme turnover in the initial frames of MSOX structures. Consistent with the occupancy of the two conformations observed in SF-ROXNIT, AspCAT adopts the proximal and gatekeeper conformations with equal occupancy [Fig. 4 ▸(a)]. Based on the possibility of steric interaction, the proximal AspCAT conformation coincides with ‘side-on’ NO2 −, while the gatekeeper conformation matches the ‘top-hat’ mode. The distorted proximal conformation seen in the SF-ROXOX structure is not visible here. In the atomic resolution SR structure of nitrite-bound AcNiR (PDB entry 2bwi; Antonyuk et al., 2005 ▸), where significant radiolysis would be expected to have occurred, the NO2 − ion takes up an intermediate position between the dual conformations observed here in the SF-ROX structure.
Figure 3.
The damage-free T1Cu site in the SF-ROX structures of AcNiR in (a) oxidized, (b) reduced and (c) nitrite-bound forms. 2F o − F c electron density is contoured at the 1σ level and is shown as a grey mesh. Atoms are coloured by element. The T1Cu is shown as a dark blue sphere and water molecules are shown as small red spheres. Metal-coordinating bonds are shown as red dotted lines. Selected hydrogen bonds are shown as black dotted lines.
Figure 4.
NO2 −-bound T2Cu site of AcNiR. (a) NO2 − is bound to the T2Cu in two conformations in the SF-ROXNIT structure: top-hat and side-on conformations with equal occupancy (50% each). Asp98 is visible in both proximal and gatekeeper conformations, with the gatekeeper conformation corresponding to the top-hat NO2 − in the SF-ROXNIT structure. (b) Conformation of nitrite at pH 5.0 obtained using a low-dose home source. (c) At pH 6.5 only a single side-on conformation is visible corresponding to a single Asp98 (AspCAT) proximal position. The half-occupancy water molecule is also bound to T2Cu in the same conformation as in the SF-ROXOX structure. 2F o − F c electron density is contoured at the 1σ level and is shown as a grey mesh. Atoms are coloured by element, with different colour schemes used for the different chains. The T2Cu is shown as a cyan sphere and water molecules are shown as small red spheres. Metal-coordinating bonds are shown as red dotted lines. Selected hydrogen bonds are shown as black dotted lines.
3.3. SF-ROX structures of chemically reduced AcNiR
Despite the wealth of structures of CuNiRs, there are very few structures of the reduced form of the enzyme. The best resolution structure available for a reduced copper nitrite reductase is that from A. faecalis, which was determined to 1.85 Å resolution some ten years ago (Wijma et al., 2007 ▸). There is no XFEL structure of the reduced form of the enzyme from any species.
The structure of AcNiR in the chemically reduced state (SF-ROXRED) obtained using 33 large colourless crystals was refined to a resolution of 1.6 Å (Table 1 ▸). The SF-ROXRED T1Cu site showed a marked difference from the SF-ROXOX structure, with two positions of the copper refined with occupancies of 0.7 and 0.3, respectively [Fig. 3 ▸(b)]. As the T2Cu site is fully reduced (as indicated by the absence of liganded water), both positions of T1Cu represent the reduced status of copper. Met141 is positioned in a single conformation away from His145, allowing a water molecule to fill the free space, making strong hydrogen bonds to both Met141 and His145. The loop (residues 187–206) undergoes a significant movement compared with that in the SF-ROXOX structure (Supplementary Fig. S1).
W1 is lost from the T2Cu site on chemical reduction, producing a tricoordinate T2Cu site with three histidine residues ligating the copper. The T2Cu also drops 0.5 Å into the histidine plane upon reduction. The electron density of the side chain of Ile257 revealed that the CD1 side chain flips down to partially occupy the active-site cavity space vacated by the water ligand [Fig. 5 ▸(b)]. The Ile257 CD1–T2Cu distance decreases from 5.2 to 3.6 Å, reducing the volume of and increasing the steric restraints on the active-site cavity. The distorted proximal Asp98 conformation seen in the SF-ROXOX structure is not visible here, with the residue adopting the original proximal conformation. The bridging water connecting AspCAT to HisCAT is positioned as in the SF-ROXOX structure [Fig. 5 ▸(a)]. The loss of water at the T2Cu in the SF-ROXRED structure and the colourless nature of the crystals confirm that this structure represents the damage-free structure of the chemically reduced enzyme. In X-ray radiolysis experiments T1Cu is reduced but T2Cu remains four-coordinate with the ligated water ligand intact (Hough et al., 2008 ▸); however, movement of the the T1Cu loop (residues 187–206) is again observed (PDB entry 2vm4).
Figure 5.
The T2Cu sites of oxidized and reduced AcNiR determined by SF-ROX. (a) Oxidized T2Cu site with two conformations of Asp98, water W1 bound to T2Cu, and Ile257 allowing space for this water. (b) Reduced T2Cu site (SF-ROXRED). The T2Cu water ligand is lost upon reduction. Only a single Asp98 (AspCAT) conformation is present. The Ile257 side chain flips down to partially fill the space vacated by the water ligand. 2F o − F c electron density is contoured at the 1σ level and is shown as a grey mesh. Atoms are coloured by element, with different colour schemes used for the different chains. The T2Cu is shown as a cyan sphere and water molecules are shown as small red spheres. Metal-coordinating bonds are shown as red dotted lines. Selected hydrogen bonds are shown as black dotted lines.
3.4. pH-dependence of nitrite conformation
Given the different binding modes of nitrite in the MSOX and SF-ROXNIT structures, we investigated the pH dependence of the nitrite conformation using the in-house copper-anode X-ray generator at the Barkla Laboratory equipped with an EIGER R 4M detector. The highly efficient photon-counting detector together with low-dose data collection allows complete data collection without the conversion of nitrite to NO, thus allowing the determination of NO2 −-bound AcNiR structures at a variety of pH values. The resolution limit of these data sets was restricted to 1.5 Å owing to the geometrical constraints of the in-house experimental setup (Supplementary Table S1). The structure at pH 5.0 was comparable to the SF-ROXNIT structure, with both ‘top-hat’ and ‘side-on’ conformations of NO2 − with 0.5 occupancy each. The AspCAT residue has two conformations, with the gatekeeper conformation corresponding to the ‘top-hat’ binding mode of NO2 − [Figs. 4 ▸(a) and 4 ▸(b)]. At pH 5.5 both the NO2 − and AspCAT conformations are present in equal proportions, but several changes are noticeable in the structure. The Met141 residue protecting His145 from water binding at the short distance has a single conformation [Supplementary Fig. S3(a)]. At pH 5.5 Met141 adopts two conformations with half occupancy each. This allows a partial water molecule to hydrogen-bond directly to His145 and create a water network to the protein surface which ends close to the low-density loop region. At pH 6.0 the original conformation of Met141 is lost, the water hydrogen-bonded to His145 is fully occupied and the side chain of Trp144 flips 180°. A major movement occurs in the external loop [residues 192–207; Supplementary Figs. S4(a) and 4(c)], with residues 195–201 regaining almost full occupancy. The crystal structure of AxNiR complexed with cytochrome c 551 (PDB entry 2zon) shows the AxNiR–Cyt c 551 interface aligned directly on top of the equivalent loop [Supplementary Fig. S4(d); Nojiri et al., 2009 ▸]. No changes are visible in the T2Cu geometry. Finally, at pH 6.5 few differences are observed around the T1Cu apart from both conformations of Trp144 being present. The outer loop is fully stabilized in its new conformation. The T2Cu site is changed significantly, with a single conformation of AspCAT, and NO2 − is in a side-on conformation [Fig. 4 ▸(c)].
3.5. Protonation of the active-site residues in CuNiR
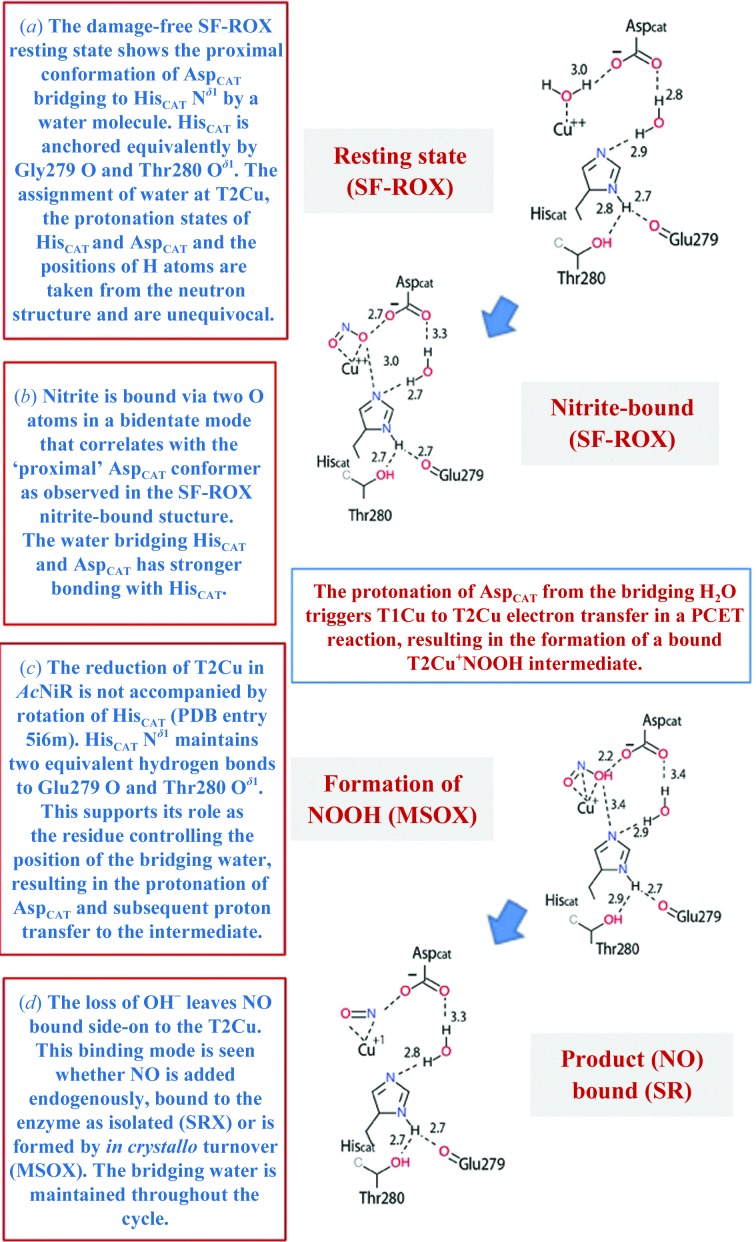
The consensus view of the resting state of CuNiRs at pH values close to the optimum for activity is that AspCAT is not protonated and HisCAT is fully protonated, with the two residues bridged by a hydrogen-bonded water molecule (Ghosh et al., 2009 ▸). In our AcNiR neutronOX structure the Oδ1 and Oδ2 atoms of AspCAT were not deuterated, as expected (Figs. 1 ▸ and 2 ▸), but, contrary to expectation, HisCAT lacked a deuteron at the N∊2 position as well. The linking water (D2) is positioned with one deuteron directed towards Asp98 Oδ1 and one directed towards HisCAT N∊2. Moreover, the T2Cu-ligated water (D1) can clearly be modelled as a D2O molecule (as opposed to a D3O+ or an OD− ion, which have been suggested previously as possible alternatives). The water (D2) linking HisCAT to Oδ1 of AspCAT restricts the movement of the unprotonated AspCAT. The bridging water is too distant to form a hydrogen bond to the O atom of the bound nitrite that interacts with AspCAT. We suggest that when NO2 − binds, a proton is transferred from this water to the Oδ2 atom of AspCAT, resulting in an increase in the reduction potential to facilitate electron transfer from T1Cu to T2Cu (Fig. 6 ▸). In the complex with the reduced T2Cu, the proton is transferred from AspCAT to bound nitrite and the second proton is donated from the bridging water of HisCAT. This residue has been shown to rotate on reduction of the T2Cu site and has a proposed role as a redox-coupled switch for proton transfer (Brenner et al., 2009 ▸; Leferink et al., 2011 ▸, 2012 ▸; Fukuda, Tse, Nakane et al., 2016 ▸). The structure also shows that the fourth ligand of the T2Cu is D2O, which is consistent with proton-uptake studies, which established that two protons coupled to electron transfer are required for turnover (Brenner et al., 2009 ▸). Synthetic copper complexes are able to carry out efficient NO2 − reduction with the addition of a proximal carboxylate group, analogous to AspCAT, to form part of the copper(II) coordination sphere (Cioncoloni et al., 2018 ▸). From a mechanistic viewpoint, our data are consistent with the binding of NO2 − to the oxidized T2Cu, resulting in displacement of the coordinated water ligand and triggering the protonation of AspCAT via the bridging water to initiate a proton-coupled electron-transfer (PCET) reaction and subsequent catalysis (Brenner et al., 2009 ▸; Ghosh et al., 2009 ▸).
Figure 6.
Structure-based mechanism.
4. Discussion
A surprising feature of the previously reported damage-free XFEL structures of several CuNiRs was the absence of a water ligand to the T2Cu site. For AfNiR, the resting-state SFX structure had a chloride ion originating from the purification protocol ligated to T2Cu (Fukuda, Tse, Nakane et al., 2016 ▸). In the case of GtNiR, a sodium ion was present in the T2Cu pocket along with a low-occupancy copper (Fukuda, Tse, Suzuki et al., 2016 ▸). For AxNiR, the SF-ROX structure revealed an unprecedented dioxo species bound to the T2Cu site in the resting state (Halsted et al., 2018 ▸), as anticipated for some time in view of a number of CuNiRs having a significant oxidase/superoxide dismutase (SOD) activity.
In contrast, our SF-ROX structure of as-isolated AcNiR reported here shows that the T2Cu is ligated to a water molecule. It is the first time that this catalytically important water has been observed in a crystallographic structure obtained using femtosecond pulses from an X-ray laser. We have validated the presence of water by obtaining a 1.8 Å resolution neutron structure of a perdeuterated protein in which the water (as D2O) exhibits clear density for three atoms for both the catalytic and the bridging water molecules. Both damage-free structures show the AcNiR T2Cu to be coordinated by three histidine residues and a single water molecule ligated in a distorted tetrahedral geometry. The distorted proximal position of AspCAT seen only in our SF-ROX structure shortens the hydrogen bond between the Oδ2 atom of Asp98 and the T2Cu water ligand W1 from 3.5 to 3 Å.
A comparison between the SR and SF-ROX structures of NO2 −-bound AcNiR reveals differences at the T2Cu site. NO2 − binding to the oxidized T2Cu site has been observed in both ‘side-on’ and ‘top-hat’ modes when X-ray radiolysis is used to the drive NO2 − reduction (Horrell et al., 2016 ▸). SFX structure determination of AfNiR revealed a single full-occupancy NO2 − molecule bound in the ‘top-hat’ position that flips to the ‘side-on’ position in SR structures (Fukuda, Tse, Nakane et al., 2016 ▸). It was suggested that ‘top-hat’ to ‘side-on’ conversion occurs following the photoreduction of the T1Cu and the transfer of an electron across the Cys–His bridge, and that the ‘side-on’ conformation may represent the initial intermediate species in the catalytic reaction. This explanation is not consistent with our observations for the SF-ROX structure, in which NO2 − is present in both ‘side-on’ and ‘top-hat’ binding modes. This structure, which was obtained using single-shot XFEL pulses of pulse length <10 fs, represents a time-frozen structure in which no radiolysis can take place owing to the speed of data collection, as the X-ray pulses are shorter than even the vibrational or rotational frequencies. Both of these binding modes are also visible in low-dose data sets collected using our in-house X-ray source at a range of pH values up to pH 6.5. We therefore suggest that the generation of solvated electrons in crystallo by X-ray radiolysis produces a change of the pH in the active-site micro-environment of CuNiRs, shifting the geometry of AspCAT and therefore affecting the NO2 −-binding mode. It has been suggested that HisCAT has a role as a redox-coupled switch for proton transfer (Fukuda, Tse, Nakane et al., 2016 ▸), which is consistent with computational and biophysical studies showing that protonation is required for the rate-limiting intramolecular electron-transfer reaction (Ghosh et al., 2009 ▸; Leferink et al., 2011 ▸; Lintuluoto & Lintuluoto, 2018 ▸). Here, we observed no protonation of N∊2 of HisCAT at pD 5.4, while the linking water is neutral, suggesting that an internal change in pH is required to transfer the proton from the water (W2) to HisCAT. The increase in pH causes a conformational change to the ‘side-on’ mode, enabling PCET-based reduction of nitrite (Fig. 6 ▸).
Even though the neutron structure was very helpful in defining the protonation states of key residues in the resting state, we note that the method has significant limitations owing to (i) lower resolution, (ii) lower completeness of data owing to Laue geometry, (iii) significantly weaker scattering lengths and cross-sections for heavier protein atoms (sulfur) and metals such as copper compared with 2H (Supplementary Fig. S5) and (iv) its applicability to smaller unit cells (<130 Å). XFEL-based crystallographic methods (SFX, SF-ROX, mix-and-inject SFX etc.) are thus currently the only methods for obtaining ‘damage-free’ structures at resolutions at which atomic details are visible with the accuracy that is necessary to define the chemistry surrounding redox centres and associated chemical reactions. Like any X-ray method, the sensitivity decreases in direct proportion to Z (atomic number) and hence has limitations in detecting biologically important H atoms. Combining the two approaches for the resting state has enabled us to define the protonation states of key residues experimentally for the first time.
5. Concluding remarks
Structural biology continues to benefit from an expanding toolkit, the principles of which are underpinned by rigorous physics, as is demonstrated here, where unprecedented insight into the enzyme species involved in proton delivery/substrate binding in CuNiR turnover has been gained by combining results from neutron, X-ray laser, modern synchrotron and in-house laboratory X-ray sources. Neutron crystallography has remained the only radiation-damage-free macromolecular structural probe, but the advent of femtosecond crystallography with X-ray free-electron lasers provides a new opportunity in which damage-free structures can be probed using much smaller crystals and for more complex macromolecules, including membrane proteins and multi-protein complexes (Suga et al., 2014 ▸, 2017 ▸; Hirata et al., 2014 ▸; Nango et al., 2016 ▸; Nogly et al., 2018 ▸). For redox enzymes, X-ray crystallography using femtosecond X-ray lasers provides a unique opportunity to obtain damage-free structures both at cryogenic and ambient temperatures at the resolution that is needed to understand the chemistry of catalysis. The damage-free structure of the resting state of a copper nitrite reductase (CuNiR) was defined using neutron and XFEL structural data and represents the first direct comparison of neutron and XFEL structural data for any protein. The structural insights gained here will have a direct impact on computational chemistry and synthetic biology efforts for understanding proton-coupled electron-transfer events (Ghosh et al., 2009 ▸) and for the design of synthetic compounds and peptides with such catalytic properties for environmental and biomedical applications (Cioncoloni et al., 2018 ▸; Koebke et al., 2018 ▸; Hedison et al., 2019 ▸).
Supplementary Material
PDB reference: Achromobacter cycloclastes CuNiR, resting state, SF-ROX structure, 6gsq
PDB reference: resting state, neutron structure, 6gtj
PDB reference: pH 5, resting state, low dose, 6gti
PDB reference: pH 5.5, resting state, low dose, 6gtk
PDB reference: pH 6, resting state, low dose, 6gtl
PDB reference: pH 6.5, resting state, low dose, 6gtn
PDB reference: nitrite-bound, SF-ROX structure, 6gt0
PDB reference: chemically reduced, SF-ROX structure, 6gt2
Supplementary Figures and Table. DOI: 10.1107/S2052252519008285/ec5013sup1.pdf
Data for neutron structure of AcNiR.: https://doi.org/10.5921/ILL-DATA.8-01-418
Acknowledgments
Thomas P. Halsted was supported by the RIKEN–Liverpool Partnership awarded to Masaki Yamamoto and S. Samar Hasnain. Data collection at SACLA took place with support from proposal No. 2017B8028 led by Hideo Ago. Neutron data collection (https://doi.org/10.5921/ILL-DATA.8-01-418) using LADI-III at ILL took place with support from proposal 8-01-418 awarded to S. Samar Hasnain and Svetlana V. Antonyuk. We thank all of the staff of SACLA BL2 at the RIKEN SPring-8 Centre, and of the D-LAB and the LADI-III beamline at the Institut Laue–Langevin. Author contributions were as follows. SVA, RRE, MY and SSH conceived and designed the project. TPH expressed, purified and crystallized AcNiR. TPH, KH, CG, RS, HA, GU and SSH collected the SF-ROX data. TPH and KY performed the SF-ROX data processing. TPH performed the SF-ROX structure determination and refinement. TPH and SVA purified and crystallized perdeuterated AcNiR. MPB collected and processed the neutron data. MPB and SVA performed the neutron structure refinement. TPH collected the pH-dependent data using the in-house facility at the Barkla X-ray Laboratory of Biophysics and performed the structure determination and refinement. TPH, SVA and SSH wrote the manuscript with contributions from all authors. All authors reviewed the manuscript.
Funding Statement
This work was funded by Biotechnology and Biological Sciences Research Council grants BB/R000220/1, BB/L006960/1, and BB/N013972/1. Science and Technology Facilities Council grant .
References
- Abraham, Z. H. L., Smith, B. E., Howes, B. D., Lowe, D. J. & Eady, R. R. (1997). Biochem. J. 324, 511–516. [DOI] [PMC free article] [PubMed]
- Adams, P. D., Afonine, P. V., Bunkóczi, G., Chen, V. B., Davis, I. W., Echols, N., Headd, J. J., Hung, L.-W., Kapral, G. J., Grosse-Kunstleve, R. W., McCoy, A. J., Moriarty, N. W., Oeffner, R., Read, R. J., Richardson, D. C., Richardson, J. S., Terwilliger, T. C. & Zwart, P. H. (2010). Acta Cryst. D66, 213–221. [DOI] [PMC free article] [PubMed]
- Antonyuk, S. V., Strange, R. W., Sawers, G., Eady, R. R. & Hasnain, S. S. (2005). Proc. Natl Acad. Sci. USA, 102, 12041–12046. [DOI] [PMC free article] [PubMed]
- Arzt, S., Campbell, J. W., Harding, M. M., Hao, Q. & Helliwell, J. R. (1999). J. Appl. Cryst. 32, 554–562.
- Blakeley, M. P. (2009). Crystallogr. Rev. 15, 157–218.
- Blakeley, M. P., Hasnain, S. S. & Antonyuk, S. V. (2015). IUCrJ, 2, 464–474. [DOI] [PMC free article] [PubMed]
- Blakeley, M. P., Teixeira, S. C. M., Petit-Haertlein, I., Hazemann, I., Mitschler, A., Haertlein, M., Howard, E. & Podjarny, A. D. (2010). Acta Cryst. D66, 1198–1205. [DOI] [PubMed]
- Brenner, S., Heyes, D. J., Hay, S., Hough, M. A., Eady, R. R., Hasnain, S. S. & Scrutton, N. S. (2009). J. Biol. Chem. 284, 25973–25983. [DOI] [PMC free article] [PubMed]
- Campbell, J. W., Hao, Q., Harding, M. M., Nguti, N. D. & Wilkinson, C. (1998). J. Appl. Cryst. 31, 496–502.
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Cioncoloni, G., Roger, I., Wheatley, P. S., Wilson, C., Morris, R. E., Sproules, S. & Symes, M. D. (2018). ACS Catal. 8, 5070–5084.
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Evans, P. R. & Murshudov, G. N. (2013). Acta Cryst. D69, 1204–1214. [DOI] [PMC free article] [PubMed]
- Fukuda, Y., Tse, K. M., Lintuluoto, M., Fukunishi, Y., Mizohata, E., Matsumura, H., Takami, H., Nojiri, M. & Inoue, T. (2014). J. Biochem. 155, 123–135. [DOI] [PubMed]
- Fukuda, Y., Tse, K. M., Nakane, T., Nakatsu, T., Suzuki, M., Sugahara, M., Inoue, S., Masuda, T., Yumoto, F., Matsugaki, N., Nango, E., Tono, K., Joti, Y., Kameshima, T., Song, C., Hatsui, T., Yabashi, M., Nureki, O., Murphy, M. E. P., Inoue, T., Iwata, S. & Mizohata, E. (2016). Proc. Natl Acad. Sci. USA, 113, 2928–2933. [DOI] [PMC free article] [PubMed]
- Fukuda, Y., Tse, K. M., Suzuki, M., Diederichs, K., Hirata, K., Nakane, T., Sugahara, M., Nango, E., Tono, K., Joti, Y., Kameshima, T., Song, C., Hatsui, T., Yabashi, M., Nureki, O., Matsumura, H., Inoue, T., Iwata, S. & Mizohata, E. (2016). J. Biochem. 159, 527–538. [DOI] [PMC free article] [PubMed]
- Garman, E. F. (2010). Acta Cryst. D66, 339–351. [DOI] [PMC free article] [PubMed]
- Ghosh, S., Dey, A., Sun, Y., Scholes, C. P. & Solomon, E. I. (2009). J. Am. Chem. Soc. 131, 277–288. [DOI] [PMC free article] [PubMed]
- Haertlein, M., Moulin, M., Devos, J. M., Laux, V., Dunne, O. & Forsyth, V. T. (2016). Methods Enzymol. 566, 113–157. [DOI] [PubMed]
- Halsted, T. P., Yamashita, K., Hirata, K., Ago, H., Ueno, G., Tosha, T., Eady, R. R., Antonyuk, S. V., Yamamoto, M. & Hasnain, S. S. (2018). IUCrJ, 5, 22–31. [DOI] [PMC free article] [PubMed]
- Hedison, T. M., Shenoy, R. T., Iorgu, A. I., Heyes, D. J., Fisher, K., Wright, G. R., Hay, S., Eady, R. R., Antonyuk, S. V., Hasnain, S. S. & Scrutton, N. S. (2019). ACS Catal. 9, 6087–6099. [DOI] [PMC free article] [PubMed]
- Hirata, K., Shinzawa-Itoh, K., Yano, N., Takemura, S., Kato, K., Hatanaka, M., Muramoto, K., Kawahara, T., Tsukihara, T., Yamashita, E., Tono, K., Ueno, G., Hikima, T., Murakami, H., Inubushi, Y., Yabashi, M., Ishikawa, T., Yamamoto, M., Ogura, T., Sugimoto, H., Shen, J.-R., Yoshikawa, S. & Ago, H. (2014). Nat. Methods, 11, 734–736. [DOI] [PubMed]
- Horrell, S., Antonyuk, S. V., Eady, R. R., Hasnain, S. S., Hough, M. A. & Strange, R. W. (2016). IUCrJ, 3, 271–281. [DOI] [PMC free article] [PubMed]
- Horrell, S., Kekilli, D., Sen, K., Owen, R. L., Dworkowski, F. S. N., Antonyuk, S. V., Keal, T. W., Yong, C. W., Eady, R. R., Hasnain, S. S., Strange, R. W. & Hough, M. A. (2018). IUCrJ, 5, 283–292. [DOI] [PMC free article] [PubMed]
- Hough, M. A., Antonyuk, S. V., Strange, R. W., Eady, R. R. & Hasnain, S. S. (2008). J. Mol. Biol. 378, 353–361. [DOI] [PubMed]
- Jacobson, F., Pistorius, A., Farkas, D., De Grip, W., Hansson, Ö., Sjölin, L. & Neutze, R. (2007). J. Biol. Chem. 282, 6347–6355. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Karplus, P. A. & Diederichs, K. (2015). Curr. Opin. Struct. Biol. 34, 60–68. [DOI] [PMC free article] [PubMed]
- Koebke, K. J., Yu, F., Salerno, E., Van Stappen, C., Tebo, A. G., Penner-Hahn, J. E. & Pecoraro, V. L. (2018). Angew. Chem. Int. Ed. 57, 3954–3957. [DOI] [PMC free article] [PubMed]
- Leferink, N. G. H., Eady, R. R., Hasnain, S. S. & Scrutton, N. S. (2012). FEBS J. 279, 2174–2181. [DOI] [PubMed]
- Leferink, N. G. H., Han, C., Antonyuk, S. V., Heyes, D. J., Rigby, S. E. J., Hough, M. A., Eady, R. R., Scrutton, N. S. & Hasnain, S. S. (2011). Biochemistry, 50, 4121–4131. [DOI] [PubMed]
- Lintuluoto, M. & Lintuluoto, J. M. (2018). Metallomics, 10, 565–578. [DOI] [PubMed]
- Luchinat, E. & Banci, L. (2017). IUCrJ, 4, 108–118. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Nango, E., Royant, A., Kubo, M., Nakane, T., Wickstrand, C., Kimura, T., Tanaka, T., Tono, K., Song, C., Tanaka, R., Arima, T., Yamashita, A., Kobayashi, J., Hosaka, T., Mizohata, E., Nogly, P., Sugahara, M., Nam, D., Nomura, T., Shimamura, T., Im, D., Fujiwara, T., Yamanaka, Y., Jeon, B., Nishizawa, T., Oda, K., Fukuda, M., Andersson, R., Båth, P., Dods, R., Davidsson, J., Matsuoka, S., Kawatake, S., Murata, M., Nureki, O., Owada, S., Kameshima, T., Hatsui, T., Joti, Y., Schertler, G., Yabashi, M., Bondar, A.-N., Standfuss, J., Neutze, R. & Iwata, S. (2016). Science, 354, 1552–1557. [DOI] [PubMed]
- Nogly, P., Weinert, T., James, D., Carbajo, S., Ozerov, D., Furrer, A., Gashi, D., Borin, V., Skopintsev, P., Jaeger, K., Nass, K., Båth, P., Bosman, R., Koglin, J., Seaberg, M., Lane, T., Kekilli, D., Brünle, S., Tanaka, T., Wu, W., Milne, C., White, T., Barty, A., Weierstall, U., Panneels, V., Nango, E., Iwata, S., Hunter, M., Schapiro, I., Schertler, G., Neutze, R. & Standfuss, J. (2018). Science, 361, eaat0094. [DOI] [PubMed]
- Nojiri, M., Koteishi, H., Nakagami, T., Kobayashi, K., Inoue, T., Yamaguchi, K. & Suzuki, S. (2009). Nature (London), 462, 117–120. [DOI] [PubMed]
- Schowen, K. B. & Schowen, R. L. (1982). Methods Enzymol. 87, 551–606. [PubMed]
- Solomon, E. I., Heppner, D. E., Johnston, E. M., Ginsbach, J. W., Cirera, J., Qayyum, M., Kieber-Emmons, M. T., Kjaergaard, C. H., Hadt, R. G. & Tian, L. (2014). Chem. Rev. 114, 3659–3853. [DOI] [PMC free article] [PubMed]
- Suga, M., Akita, F., Hirata, K., Ueno, G., Murakami, H., Nakajima, Y., Shimizu, T., Yamashita, K., Yamamoto, M., Ago, H. & Shen, J.-R. (2014). Nature (London), 517, 99–103. [DOI] [PubMed]
- Suga, M., Akita, F., Sugahara, M., Kubo, M., Nakajima, Y., Nakane, T., Yamashita, K., Umena, Y., Nakabayashi, M., Yamane, T., Nakano, T., Suzuki, M., Masuda, T., Inoue, S., Kimura, T., Nomura, T., Yonekura, S., Yu, L.-J., Sakamoto, T., Motomura, T., Chen, J.-H., Kato, Y., Noguchi, T., Tono, K., Joti, Y., Kameshima, T., Hatsui, T., Nango, E., Tanaka, R., Naitow, H., Matsuura, Y., Yamashita, A., Yamamoto, M., Nureki, O., Yabashi, M., Ishikawa, T., Iwata, S. & Shen, J.-R. (2017). Nature (London), 543, 131–135.
- White, T. A., Barty, A., Stellato, F., Holton, J. M., Kirian, R. A., Zatsepin, N. A. & Chapman, H. N. (2013). Acta Cryst. D69, 1231–1240. [DOI] [PMC free article] [PubMed]
- White, T. A., Mariani, V., Brehm, W., Yefanov, O., Barty, A., Beyerlein, K. R., Chervinskii, F., Galli, L., Gati, C., Nakane, T., Tolstikova, A., Yamashita, K., Yoon, C. H., Diederichs, K. & Chapman, H. N. (2016). J. Appl. Cryst. 49, 680–689. [DOI] [PMC free article] [PubMed]
- Wijma, H. J., MacPherson, I., Farver, O., Tocheva, E. I., Pecht, I., Verbeet, M. P., Murphy, M. E. P. & Canters, G. W. (2007). J. Am. Chem. Soc. 129, 519–525. [DOI] [PubMed]
- Winn, M. D., Ballard, C. C., Cowtan, K. D., Dodson, E. J., Emsley, P., Evans, P. R., Keegan, R. M., Krissinel, E. B., Leslie, A. G. W., McCoy, A., McNicholas, S. J., Murshudov, G. N., Pannu, N. S., Potterton, E. A., Powell, H. R., Read, R. J., Vagin, A. & Wilson, K. S. (2011). Acta Cryst. D67, 235–242. [DOI] [PMC free article] [PubMed]
- Yano, J., Kern, J., Irrgang, K.-D., Latimer, M. J., Bergmann, U., Glatzel, P., Pushkar, Y., Biesiadka, J., Loll, B., Sauer, K., Messinger, J., Zouni, A. & Yachandra, V. K. (2005). Proc. Natl Acad. Sci. USA, 102, 12047–12052. [DOI] [PMC free article] [PubMed]
- Zhao, Y., Lukoyanov, D. A., Toropov, Y. V., Wu, K., Shapleigh, J. P. & Scholes, C. P. (2002). Biochemistry, 41, 7464–7474. [DOI] [PubMed]
- Zumft, W. G. (1997). Microbiol. Mol. Biol. Rev. 61, 533–616. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: Achromobacter cycloclastes CuNiR, resting state, SF-ROX structure, 6gsq
PDB reference: resting state, neutron structure, 6gtj
PDB reference: pH 5, resting state, low dose, 6gti
PDB reference: pH 5.5, resting state, low dose, 6gtk
PDB reference: pH 6, resting state, low dose, 6gtl
PDB reference: pH 6.5, resting state, low dose, 6gtn
PDB reference: nitrite-bound, SF-ROX structure, 6gt0
PDB reference: chemically reduced, SF-ROX structure, 6gt2
Supplementary Figures and Table. DOI: 10.1107/S2052252519008285/ec5013sup1.pdf
Data for neutron structure of AcNiR.: https://doi.org/10.5921/ILL-DATA.8-01-418