Abstract
Determining response status in patients with myeloproliferative neoplasms is a complex problem requiring the integration of both structured and unstructured data elements from disparate information systems. By applying multiple techniques, a collaborative team of informatics professionals and research personnel were able to determine which elements were amenable to automated extraction and which required expert adjudication. With this knowledge in mind, we were able to build a system that joins together programmatically-derived and manually-abstracted data elements to facilitate response assessment – an important end point in clinical and translational research in this disease area.
Keywords: clinical research informatics, secondary use, cancer, natural language processing, data mining, computable phenotype
I. INTRODUCTION
Myeloproliferative neoplasms (MPNs) constitute a group of malignancies of the hematopoietic stem cells that share certain clinical features. MPNs include essential thrombocytosis (ET), polycythemia vera (PV), myelofibrosis (MF), chronic myeloid leukemia (CML), and others. In assessing the effectiveness of treatments for these conditions, the International Working Group – Myeloproliferative Neoplasms Research and Treatment (IWG-MRT) and the European LeukemiaNet (ELN) have issued a serious of consensus document reports detailing criteria for determining response in various MPNs, including MF [1]. Response is determined according to multiple factors, including blast count in bone marrow biopsy, cellularity, fibrosis, splenomegaly, transfusion dependence, platelet count, and others. Response may vary along the course of a patient’s treatment as the patient is induced on various lines of chemotherapy, responds, enters remission, relapses, and progresses. However, it remains the standard metric in assessing the efficacy of differing modes of treatment for myeloproliferative neoplasms, as well as other liquid tumors.
The current gold standard for determining response in clinical trials requires manual review of the patient’s electronic health record (EHR) by trained personnel. As seen in Table I, while some of the data required for the assessment of response exist in a structured fashion, some exist in free text (e.g. biopsy reports). As such, research personnel must review the patient’s chart manually, entering the required data elements into a research data capture tool to facilitate the assessment of response status – a time-consuming and difficult process.
TABLE I.
ELEMENTS REQUIRED TO ASSESS RESPONSE
| Data element | Structure |
|---|---|
| Blast count | Semi-structured – regularly expressed free text in bone marrow biopsy report |
| Laboratory values | Structured – tabular format in EHR |
| Cellularity | Unstructured – documented in free text in bone marrow biopsy report with high degree of lexical variation |
| Fibrosis | Unstructured – documented in free text in bone marrow biopsy report with high degree of lexical variation |
| Splenomegaly | Unstructured – while ICD-9/10 codes exist, most often documented in free text in progress note |
| Cytogenetics | Semi-structured using International System for Human Cytogenetic Nomenclature |
| Genomic data | Structured – HL7 feed from genomic information system (GIS) |
| Transfusion dependence | Structured – observations from inpatient electronic health record |
Computable phenotyping, a process by which informaticians use structured definitions to mine data from the EHR in order to determine which patients meet specific clinical criteria, is an established approach towards translating difficult-to-define clinical concepts into concrete, computable categories.
We hypothesized that by using existing data extraction techniques piloted as part of efforts to create a research data repository [2] for the Richard T. Silver Myeloproliferative Neoplasms Center at Weill Cornell Medicine (WCM), including development of an instance of the Observational Medical Outcomes Partnership Common Data Model (OMOP CDM) [3] and custom natural language processing (NLP) pipelines utilizing the Leo platform [4], we could alleviate the burden of manual chart review and data extraction by extracting as many of the elements as possible, allowing research personnel to focus their efforts on expert adjudication rather than manual chart review and data entry. Theoretically, by extracting all of the required elements for the IWG-MRT definition of response, the Center could develop a computable phenotype for response, operationalizing the group’s response definition as an algorithm that could be applied to structured data extracted from the EHR, along with NLP-derived elements extracted from unstructured data.
II. METHODS
A. Setting
Weill Cornell Medicine (WCM) is an academic medical center on Manhattan’s Upper East Side. An academic staff of approximately 1000 physicians treat patients at more than 20 sites in New York City, utilizing the EpicCare Ambulatory EHR. The Richard T. Silver, M.D. Myeloproliferative Neoplasms Center (hereafter the Center) conducts patient care as well as cutting-edge clinical research designed to understand the cause, progression and treatment of MPNs.
In conjunction with the Research Informatics (RI) division of WCM’s Information Technologies and Services department, the Center has embarked on the creation of a research data repository (RDR) designed to facilitate the integration of EHR data from multiple systems to facilitate cohort discovery, data collection, and analysis. [2] The RDR contains patient data from both research and clinical systems, including data captured in REDCap and an instance of the OMOP CDM. To support research personnel’s efforts to assess patient response, RI and Center researchers have implemented strategies designed to support extraction of the elements required to assess response, dependent on the state in which the data exists (as detailed in Table I).
B. Structured Data Extraction
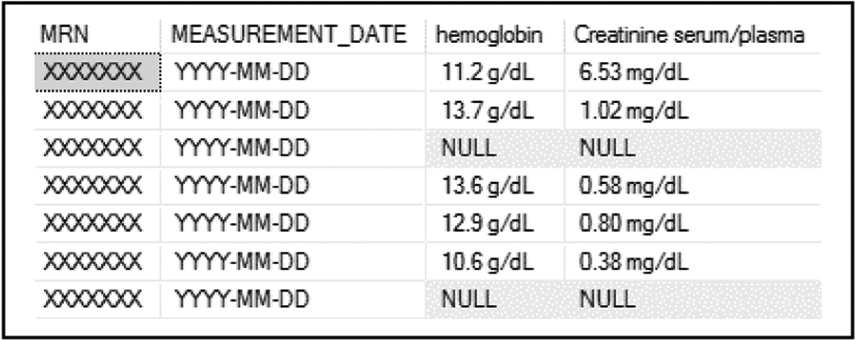
Extraction of the structured data elements used to assess response is the most straightforward component of our approach. Using structured query language (SQL) queries, we were able to pivot structured laboratory results data from the Center’s instance of the OMOP CDM in such a fashion as to allow research personnel to view the data on a longitudinal basis, determining when individual patients had certain hematological values, including absolute neutrophil count, platelet count, and others (see Fig. 1).
Fig. 1.
Extraction of structured laboratory values from OMOP CDM.
Other structured data elements, including next-generation sequencing data and transfusions, were similarly amenable to structured extraction and transformation from their native source systems. However, extraction of many of the other elements required the application of techniques with a significantly higher degree of methodological and technological sophistication. To extract blast counts from bone marrow biopsies, we initially employed a series of SQL queries dependent on regular expressions to extract blast counts, which, despite the appearance of regular structure, enter our EHR as freetext from an ancillary pathology application. After initial review, we determined that this approach did not have the requisite sensitivity or specificity to extract blast counts with sufficient rigor to support systematic assessment of response in MPN patients, in part due to lexical variation inherent in bone marrow biopsy reports.
C. Natural Language Processing
To further support the extraction of elements of interest from bone marrow biopsies, the RDR team and the Center research personnel engaged in an iterative definitional process to identify the elements from the bone marrow biopsy required to determine response, as detailed in Table II. These included blast count, cellularity, and fibrosis – however, each bone marrow biopsy included multiple observations, both from the aspirate and the clot section, necessitating section detection and tagging to extrapolate the source of the observation and label it accordingly. Furthermore, cellularity was recorded both on a quantitative (20%, 30%, etc) and a qualitative (hypo-, normo-, hyper-) basis, whereas fibrosis was recorded both on a quantitative and qualitative basis from both reticulin and Masson trichrome staining – all distinctions with clinical significance. Ultimately, we identified ten distinct target concepts requiring structured extraction.
TABLE II.
BONE MARROW BIOPSY NLP TARGETS
| Concept | Data type |
|---|---|
| Biopsy blast count | Numeric |
| Biopsy cellularity – quantitative | Numeric |
| Biopsy cellularity- qualitative | Categorical |
| Biopsy fibrosis – grade | Categorical |
| Biopsy fibrosis – qualitative - reticulin | String |
| Biopsy fibrosis – qualitative - trichrome | String |
| Aspirate blast count - differential | Numeric |
| Aspirate cellularity – quantitative | Numeric |
| Aspirate cellularity – qualitative | Categorical |
| Aspirate blast count – flow cytometry | Numeric |
After identifying the target concepts, the RDR team developed a natural language pipeline using Leo to extract the target concepts on a per-report basis and extracted the results into a SQL Server environment for validation by Center personnel.
D. Manual Data Capture
Other elements required for the assessment of response do not lend themselves as neatly to natural language processing techniques. Splenomegaly, for example, can be identified either from mentions within a progress note – complicated by the need for sophisticated negation detection approaches – or by specific dimensions denoted in imaging report. Likewise, karyotypes are theoretically determined by a standardized grammar – the International System for Human Cytogenetic Nomenclature. However, human error and imperfect adherence to the theoretical structure of the notation makes structured decomposition according to regular expression or computed techniques a difficult technique. While we hope to implement NLP techniques that can extract these concepts, the Center research personnel are expert in determining these values from a patient’s chart. Using REDCap, an established electronic data capture system, they record these values on a structured basis for individual patients.
III. RESULTS
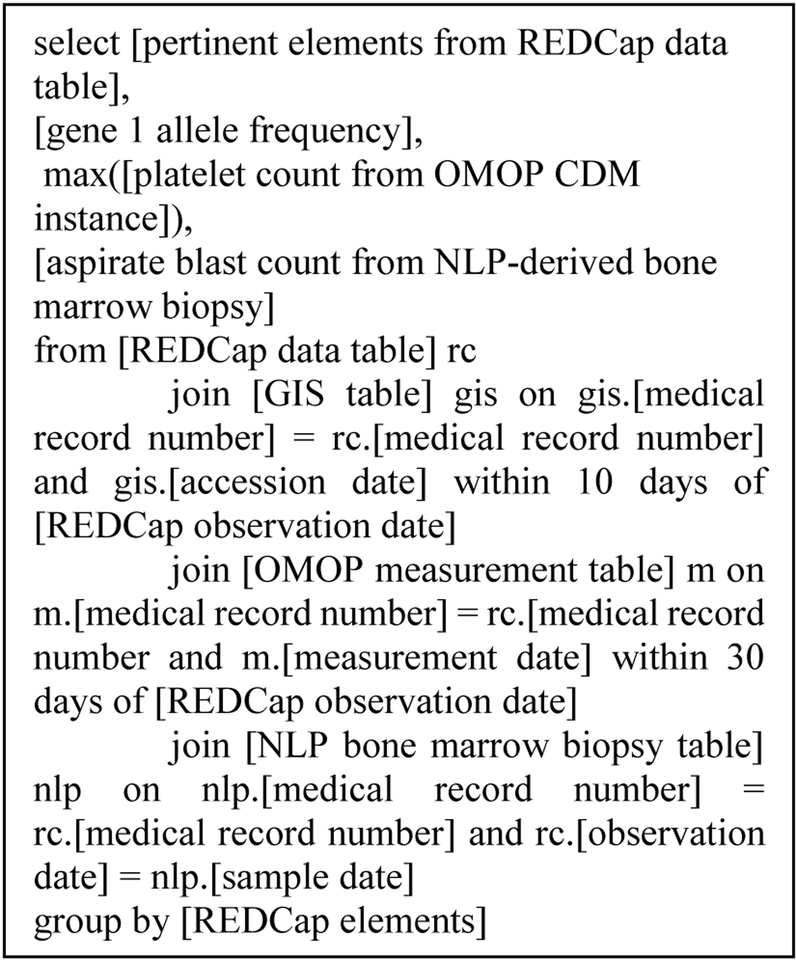
Utilizing a Microsoft SQL Server-based approach, we are able to integrate the various data elements in order to facilitate the structured assessment of response. Research personnel actively enter cytogenetics data and splenomegaly, as well as other pertinent data elements, into a REDCap project. The RDR team regularly extracts data from this REDCap project into a tabular format in the SQL Server environment as part of the SUPER data ingestion process [5]. Upon accession to the SQL server environment, the REDCap data is pivoted and loaded into a data mart using a stored procedure. It is then subject to ad hoc SQL queries from both the RDR team and the Center personnel that join manually abstracted data to both structured data from the OMOP CDM/genomic information systems and to structured natural language processing data extracted from freetext, as detailed in Fig. 2.
Fig. 2.
Simple pseudocode demonstrating join from manually abstracted REDCap data to automatically extracted data from EHR and GIS
Utilizing this technique, Center personnel can easily join data elements that require manual abstraction with elements that are amenable to structured extraction in an agile, modular fashion. By configuring the SQL query, parameters can easily be adjusted to widen or narrow temporal windows and integrate additional components as needed. Once the requisite data elements have been aggregated, they can then conduct expert adjudication to determine response status at a given time and enter it into REDCap to facilitate clinical and translational research.
IV. CONCLUSIONS
Despite the efforts we detail here, it is important to emphasize that we are not certain of the feasibility, or even the desirability, of a purely computable approach to a response phenotype for response in MPNs. We recognize the intrinsic complexity of the data elements required to compute response, and the institutional barriers towards implementing the level of structured data capture that would be required to fully enable the algorithmic determination of response – for the foreseeable future, expert determination will still be key in determining response. However, expert adjudication does not necessarily extend to data entry – elements that are amenable to structured extraction should be subject to this process in order to ensure that research personnel are focusing their efforts on processes that leverage the unique human ability to resolve ambiguity and parse ambiguous clinical narratives. Supplementing manually gathered REDCap data with data extracted from the EHR offers the potential of allowing research personnel to focus their efforts these tasks, rather than copy-pasting data from the EHR into an electronic data capture form.
We recognize multiple limitations to this work, not least including the inherent difficulty of natural language processing in this domain and the continuing requirement for extensive human effort, as well as the lack of formal validation of our NLP pipeline for extracting structured data from bone marrow biopsies. Future efforts will focus on the formal validation of existing natural language processing techniques, as well as the extension of the Leo pipeline to capture the data elements that still require expert adjudication – particularly with an eye towards a structured decomposition of ISCN karyotype annotation.
Working along these lines, we aim to develop a human-supervised system rather than a human-dependent system. We also hope to expand this technique beyond the domain of myeloproliferative neoplasms, as facilitating the detection of response status holds the potential to benefit the research enterprise in multiple domains. While this particular use case is tailored to MPNs, multiple liquid tumor disease areas share similar response features. The application of a computable phenotype supported with both structured data and the output of NLP processes could have significant benefit in reducing the effort required to determine a significant clinical endpoint in this domain.
ACKNOWLEDGMENTS
We thank the Research Informatics team (Prakash Addekkanatthu, Marcos Davila, Steven Flores, Xiaobo Fuld, Joseph Kabariti, David Kraemer, Ryan McGregor, Sean Pompea, Julian Schwartz, and Jacob Weiser) for their contributions to the efforts detailed herein.
This study received support from NewYork-Presbyterian Hospital (NYPH) and Weill Cornell Medical College (WCMC), including the Clinical and Translational Science Center (CTSC) (UL1 TR000457) and Joint Clinical Trials Office (JCTO).
Contributor Information
Evan Sholle, Information Technologies & Services, Weill Cornell Medicine, New York, NY.
Spencer Krichevsky, Department of Medicine, Weill Cornell Medicine, New York, NY.
Joseph Scandura, Department of Medicine, Weill Cornell Medicine, New York, NY.
Claudia Sosner, Department of Medicine, Weill Cornell Medicine, New York, USA.
Thomas R. Campion, Jr., Department of Healthcare Policy and Research, Weill Cornell Medicine, New York, USA.
REFERENCES
- [1].Tefferi A, Cervantes F, Mesa R, Passamonti F, Verstovsek S, Vannucchi AM, et al. Revised response criteria for myelofibrosis: International Working Group-Myeloproliferative Neoplasms Research and Treatment (IWG-MRT) and European LeukemiaNet (ELN) consensus report. Blood. 2013;122:1395–8.J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Hripcsak G, Albers DJ. Next-generation phenotyping of electronic health records. J Am Med Inform Assoc. 2013;20(1):117–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Sholle ET, Bollapragada R, Campion TR. Research data repositories: a tailored approach to secondary use of electronic health record data. AMIA Jt Summits Transl Sci Proc; 2016; San Francisco, CA [Google Scholar]
- [4].Observational Health Data Sciences and Informatics. Data Standardization [Internet]. Washington, DC: Observational Heatlh Data Sciences and Informatics; [cited 2017. Sep 25]. Available from: https://www.ohdsi.org/data-standardization/. [Google Scholar]
- [5].Sholle ET, Kabariti J, Johnson SB, Leonard JP, Pathak J, Varughese VI, Cole CC, Campion TR. Secondary use of patients’ electronic records (SUPER): an approach for meeting specific data needs of clinical and translational researchers. AMIA Annu Symp Proc; 2017; Washington, DC. [PMC free article] [PubMed] [Google Scholar]