Abstract
Epigenome editing refers to the generation of precise chromatin alterations and their effects on gene expression and cell biology. Until recently, much of the efforts in epigenome editing were limited to tissue culture models of disease. However, the convergence of techniques from different fields including mammalian genetics, virology, and CRISPR engineering is advancing epigenome editing into a new era. Researchers are increasingly embracing the use of multicellular model organisms to test the role of specific chromatin alterations on development and disease. The challenge of successful live animal epigenomic editing will depend on a well-informed foundation of the current methodologies for cell-specific delivery and editing accuracy. Here we will review the opportunities for basic research and therapeutic applications.
Keywords: epigenomic editing, animal models, neuro-development, cancer, CRISPR, delivery
The Prospect of Live Animal Epigenomic Editing
Targeted alteration of chromatin and gene transcription in mammalian cells is a rapidly evolving field of research. Much effort in epigenome editing has focused on understanding the role of chromatin in cell function using tissue culture models. The contribution of this work, in conjunction with human epigenome sequencing studies, has reinforced the functional role of chromatin in normal development and disease. However, the frontier is shifting to live animal studies. The ability to precisely and accurately edit chromatin structure in multicellular organisms is becoming the system of choice to answer questions in biology and explore new avenues for therapeutic intervention. Such avenues of investigation will greatly advance our understanding of many areas in biology including aging, neurological function, cancer development, circadian biology, and multi-organ level functions.
This review of the seminal studies in live animal epigenetic editing will focus on CRISPR/dCas9 technology, but will also touch upon the other two well-known genome targeting platforms: zinc fingers (ZFs), and transcription activator-like effectors (TALEs). It will also discuss the effector domains that can be fused to these platforms to mediate transcriptional activation, repression, and long-range chromatin alterations. Considering that one of the biggest challenges to successful epigenome research in live animals is the delivery of the therapeutic agent, the choice of delivery form (viral, cell implant, transgene or macromolecular assemblies) and the delivery route (intracranial, intravenous, intrathecal, intraperitoneal) will be of particular focus. In addition, the prospect for basic biology research and therapeutic development will also be discussed.
CRISPR-based Epigenome Editing Technologies: Overview and Design Principles
Unlike gene editing, the goal of epigenome editing is to alter transcription without altering DNA sequences. Nuclease-based gene editing relies on the cellular repair of double-strand breaks, which can lead to unintended consequences such as chromosomal rearrangements [1,2], non-contiguous repair patches [3], integration of normally episomal viral vectors [3,4], apoptosis of damage-sensitive cells and selection of clones with impaired sensitivity to damage [3–6]. In contrast, altering epigenetic information is the primary method used by cells to cause long-term changes in the expression of their genes. To manually perform such alterations at specific genes ZFs, TALEs, and the rapidly advancing CRISPR/dCas system can be used. In epigenome editing, the catalytically dead nuclease (dCas9) has been the major player allowing precise binding without cleavage. Epigenome editors are developed by fusing genome targeting platforms to domains that function as writers or erasers of epigenetic modifications associated with activation or repression of gene transcription. Of the three available platforms, CRISPR has become the preferred form for genome [7] and epigenome editing [8,9].
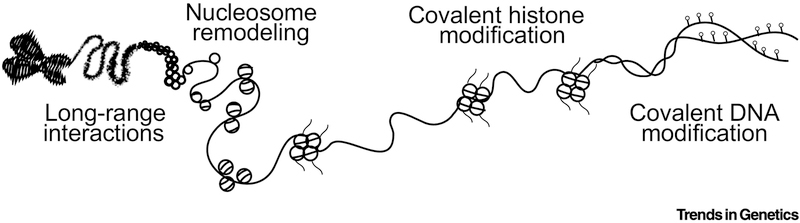
CRISPR is the platform of choice for technology development because of the ease of its two-component, RNA-programmable genome targeting feature [10]. CRISPR/dCas9 epigenome editing technologies fall into four non-mutually exclusive classes based on their primary biochemical action on chromatin (Figure 1). The first class of epigenome editors is those whose primary function is to alter chromatin topology and long-range chromosomal interactions. Members of this class include artificial proximity dimerizers fused to dCas9 [11] which can bring together distal sites in a chromosome, facilitate interchromosomal interaction, and localization of loci to specific nuclear sub-organelles. The second class of epigenome editors is nucleosome remodelers, which can result in nucleosome repositioning [12]. The third class of editors, and most documented result in covalent modifications on histones including changes in methylation and acetylation [13]. This class also includes dCas9 recombinant proteins fused to peptides that recruit cellular histone-targeting enzymes [14,15]. The fourth class of epigenome editors is those that covalently modify DNA through cytosine methylation [16]. Together these classes of epigenome editors can mediate a larger set of biochemical actions on chromatin that results in enhanced transcription or repression of target loci.
Figure 1. Classes of CRISPR/dCas9 Epigenome Modifications.
CRISPR-based epigenome editors mediate four distinct classes of modifications on chromatin based the domains utilized. Long-range interactions between different loci and nuclear compartments are facilitated by proximity interacting domains. Nucleosome remodeling is regulate enzymatic domains that mediate arrangement of nucleosomes on DNA. Covalent histone modifications is catalyzed by enzymatic domains that target amino acids on histones. Covalent DNA modifications is catalyzed by enzymatic domains that add or remove methyl groups from cytosine.
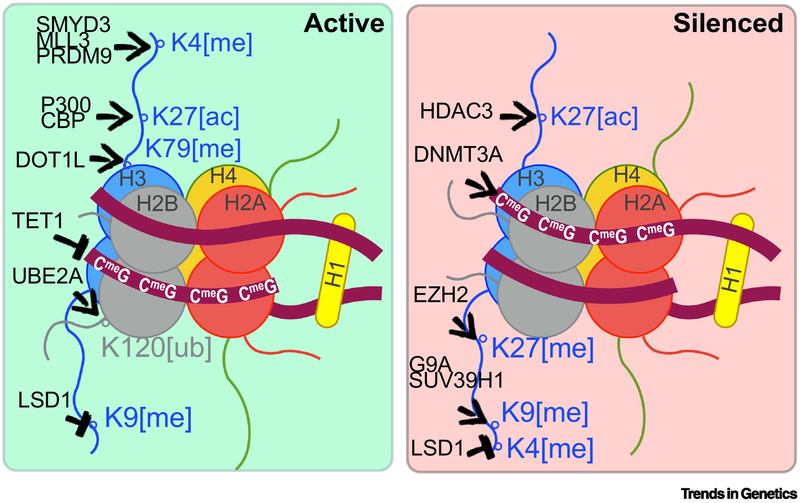
The effects of epigenome editors on transcription depend largely on the catalytic domains engineered into the CRISPR/dCas9 platform or the intermolecular interactions mediated by specific domains. For gene activation, dCas9 has been used to induce several modifications on histones and DNA associated with RNA Polymerase transcription (Figure 2, left panel). For gene repression, several modifications on histones and DNA have also been deposited by dCas9 epigenome editors (Figure 2, right panel). For live animal epigenome editing studies, several of these editors have been applied including the use of the KRAB domain for repression [14,15] and the VP64 [14] P65 [14], Rta [17], HSF1 [15] and VP64-P65-Rta (VPR) [14,17] domains for activation [18] These applications also include the use of the SunTag system, which is based on an epitope array linked dCas9 for recruitment of recombinant proteins fused with an intracellular single chain antibody (scFv). Important considerations for live animal studies include whether the target tissues possess the endogenous machinery for epigenetic editing or if dosage effects will confound experimental or therapeutic goals. Our discussion of the literature on live animal studies is guided by the mode of delivery: cell implantation, viral delivery, purified ribonucleic complexes and/or transgenesis.
Figure 2. Enzymatic domains for covalent histone and DNA modifications.
Several catalytic domains associated with gene activation (left) or silencing (right) have been paired with CRISPR-based epigenome editing. The enzymes and their DNA or amino acid targets on a nucleosome are shown in each panel. Open circles denote the targeted lysine and their position on the histone protein. The induced post-translational modification is denoted by brackets: me indicate methylation, ac indicates acetylation, and ub indicates ubiquitination. Arrows indicate deposition of the modification, and blunt-ended arrows indicate removal.
Live Animal Epigenome Editing by Cell Implantation
One of the most technically feasible approaches to investigate the effect of epigenetically edited genes in multicellular organisms is through cell implantation models. This approach provides a useful experimental link between tissue culture and live animal studies. It has the added advantage that human cells can be epigenetically edited and its consequence monitored in more physiologically relevant environments.
Three recent studies illustrate the application of this approach in understanding cancer biology and neuronal function. In one of the first studies involving live animal epigenetic reprogramming, TALEs were used to alter the expression of genes involved in hematopoiesis [19]. Specifically, in this study TALE binding arrays were fused to KRAB to silence c-kit and PU.1 in mouse bone marrow stem cells. After generating stable cells encoding the TALE-KRAB protein, the researchers introduced the cells into mice and monitored bone marrow repopulation. To monitor the fate of the edited cells once inside the mouse the team included fluorescent markers in the transgene design. This allowed the researchers to measure cell proliferation and monitor repopulation of the bone marrow. Although this study did not track the edited cells during differentiation of lymphoid and myeloid lineages, it was successful in provided a rational basis to study the role of precise epigenetic editing in hematopoiesis.
A similar study has used CRISPR-mediated epigenetic editing to targeting immune cells for the purpose of investigating the role of oncogenes and tumor suppressors in a mouse model of acute lymphoblastic leukemia [20]. The focus of this study was to epigenetically edit B lymphoma cell lines by inhibiting or activating genes involved in cancer growth, implant the cells into mice intravenously, and monitor B cell replication. Specifically, the team used lentivirus and retrovirus transduction to construct stable B lymphoma cell lines expressing dCas9-gRNA pairs for either suppression of transformation-related tumor protein 53 (Trp53) by KRAB or activation of Mgmt (O6 - methylguanine–DNA methyltransferase) by VP64. Edited cells were delivered by tail vein injection. Through this approach, the researchers were able to monitor several physiologically relevant phenotypes including accumulation of edited cells in lymph nodes, growth relative to non-targeting gRNA controls, resistance to cisplatin and temozolomide (TMZ), and survival of mice. The team also carried out the first application of epigenetic activation screens in live animals by co-infecting a population of fluorescently labeled lymphoma cells each carrying a gRNA targeting one of 25 putative genes involved in temozolomide (TMZ) resistance. That Chek2 was de-enriched in TMZ-treated mice relative to untreated controls suggested this gene increased sensitivity to drug treatment, which was validated in subsequent experiments. Together, this study is quiet successful in illustrating the versatility of CRISPR epigenetic editing in live animals, and the potential to conduct gain-of-function screens via CRISPR/dCas9.
A more recent study explored the role of epigenetic editing in live animals by using a cell implantation model in the mouse brain [21]. The goal was to reactivate the Fragile X Mental Retardation 1 (FMR1), the causative gene of Fragile X Syndrome (FXS). In most cases, the disease allele is silenced through epigenetic mechanisms that include DNA methylation of the promoter region [22]. To reactivate FMR1 in FXS neurons, a transgene encoding the components necessary for CRISPR-mediated recruitment of Tet methylcytosine deoxygenase 1 (Tet1) to the FMR1 gene was first introduced into iPSCs by lentivirus transduction. After verifying that Tet1 could lead to reactivation of FMR1 in iPSCs, the research team then implanted the cells into brains of recipient mice. Once in the brain, these cells were monitored for neuronal differentiation and continued FMR1 expression by immunohistochemistry assays. Even though several questions remain about the specificity of dCas9-Tet1, this study successfully demonstrates how it is possible to study the role of epigenetics in neuronal diseases through cell implantation assays.
There are several valuable advantages to live animal epigenomic studies by cell implantation. First, cell implantation studies enable testing the consequences of epigenetic editing in more relevant anatomical environments. Second, by encoding the implanted cells with fluorescent markers, the researchers were able to monitor the longevity of the epigenetic changes over the lifespan of the mouse. The potential for using the mouse as a “tissue culture vessel” by cell implantation of epigenetically edited cells offers a first step for researchers to translate experimental observations from the tissue culture hood to the mouse.
Live Animal Epigenome Editing by AAV Delivery
Of all the delivery methods explored to data, viral delivery has been the most extensively used to target a broad range of cells including blood cells, liver cells, and neurons. In this regard, even the most precise epigenome therapy is destined to fail if the delivery of the therapeutic is not achieved in the desired cell types. The brain remains the least therapeutically accessible organ in mammals because of the physiological barrier by specialized endothelial cells, also known as the blood-brain barrier. From all the available viral vectors for genetic therapy, recombinant adeno-associated viruses (rAAVs) are most commonly used for CNS disorders [23]. Transduction efficiency for different cell types varies between AAV serotypes and the route of administration [24]. AAV9 is particularly effective for transducing cortical neurons, whereas AAV8 is more efficient for transduction in astrocytes [25]. In many brain regions, AAVrh.10 is at least as efficient as rAAV9 [26].
One common application of viral vector delivery has been for TALE and ZF epigenome editing in the brain. This is partially due to fact that these proteins match the coding capacity of most viral vectors and the ability of these viral vectors to transduce many types in the brain [27–30]. One of the earliest applications of this method for live animal epigenome editing focused on activating expression of the glial cell line-derived neurotrophic factor (GDNF) gene in a rat model of Parkinson’s disease [31]. Noting that loss of GDNF expression is common feature in Parkinson’s disease, this study focused on characterizing a ZF-VP64 peptide that could specifically target the promoter of both the rat and human GDNF gene. The use of genome-wide expression analysis showed conclusively the specificity of the ZF-VP64 peptide. Furthermore, the use of AAV2 capsid variant and injection into the brain striatum of rats was sufficient enough to lead to rescue of motor function phenotypes. Although the study did not test the long-term of effect of this therapeutic approach, the potential of create long-last changes after repeated injection remains a possibility.
Outside of the brain, several proof-of-concept studies were recently published, which highlight how the combination of delivery route and choice of rAAV capsid mutant can be used to reach different target cells. One study CRISPR/dCas9 was used for activation of target genes by using a dual AAV system: co-injection of AAV9-dCas9 and AAV9-gRNA [32]. In the study, P65-HSF1 transcriptional activation peptides were used. The researchers tested several routes of delivery including intramuscular, intracranial, tail vein and facial vein. Optimizing their system for each delivery route allowed them to target genes whose function affect a broad range of physiological phenotypes including muscle mass and contraction by activation of Follistatin or Utrophin, kidney regeneration by activation of Il10 or Klotho, and liver cell reprogramming by activation of Pancreatic and duodenal homeobox 1. For transcriptional repression, another study used a dual AAV8 (a serotype which transduces preferably the liver) vector system to deliver dCas9-KRAB and a gRNA targeting Pcsk9. Pcsk9 is a regulator of LDL cholesterol levels in the liver and the epigenomic editing strategy showed 24-week long repression of the targeted gene in adult mice [33]. In order to target the human Frataxin (FXN) gene, a different study used TALE-VP64 [34]. Here, rAAV9 and intraperitoneal injection were used as the delivery route to reach several tissues including the liver, muscle, and heart of a transgenic mouse strain expressing human FXN. Presence of the TALE-VP64 in these tissues correlated with increased expression of FXN. More recently, a study combined the effectiveness of the TALE-mediated FXN targeting with the multimerization capability of the SunTag system to potently activate FXN in muscle and heart [35]. From these studies, two key advantages of live animal epigenomic editing by TALEs can be appreciated when compared with CRISPR/dCas9. First, TALE-mediated genome targeting is a one-component system, which facilitates delivery compared to the two-component CRISPR systems. Second, TALEs are much smaller than dCas9. This feature facilitates packaging and addition of effector domains, like the SunTag array.
To circumvent the packing limits of the rAAV when using CRISPR epigenome editing, a recent study used a split dCas9 system. In this study, split components were separately packaged into two AAV particles and a pair of intein peptides between the dCas9 fragments mediated reassembly in cells transduced with both viral particles [36]. The goal of this study was to correct retinal dystrophy disease in retinitis pigmentosa, a disease whose manifestation begins with degradation of rod photoreceptor cells in the eye. To circumvent disease development, the team focused on cellular reprogramming of disease-susceptible rod cells into disease-resistant cone photoreceptor cells in mice. This goal was achieved by repression of neural retina leucine zipper (Nrl), a master regulator of rod cell identity. Here, an AAV2-Y444F variant that displayed tropism for photoreceptor cells was delivered by subocular injections. Tissue histology and gene expression from injected mice displayed thicker retinal cell layer and a higher percentage of cone photoreceptor cells, as well as the rescue of photosensitivity and behavioral phenotypes associated with visual acuity. This study in particular highlights the elegance of epigenomic therapeutic approaches in which a disease associated with a specific cell type (rod photoreceptor cells) can be circumvented by reprogramming into resistant cells (cone photoreceptor cells) of similar function. In this case, the knowledge of Nrl as a master regular between rod and cone cells differentiation made this approach feasible.
Another approach to expanding AAV delivery cell specificity is by using cell-specific promoters to preferentially drive transgene expression. For neurons, the synapsin promoter has been used efficaciously [37,38]. An additional approach to deliver AAVs to the brain noninvasively is by generating BBB penetrating capsids. This advancement has been made possible by generating highly diverse capsid libraries through peptide insertion, homology-based recombination and error-prone PCR [39,40]. An unprecedented ability to transfer genes to the CNS in the adult mouse showed >40-fold enhancement with the AAV-PHP.eB version over the previous standard AAV9 [39,40]. Although it opened an exciting field for capsid improvement for BBB crossing, it has been shown that the AAV-PHP.eB capsids transduction potency is limited to C57Bl/6J mice [41]. These studies illustrate several important lessons for epigenetic editing in live animals. First, picking the AAV serotype with the desired tropism for the organ of interest and a cells-specific promoter for the target cells is an important first step in the success of the study. Second, the AAV capsid may require extra modifications for transduction efficiency and passing through anatomical barriers, which may lower AAV titer. Third, the coding capacity needs to be considered ahead in case the use of a dual AAV approach for delivery is required. In the case of large proteins like Cas9, smaller orthologs have now been reported [42]. Finally, for brain-wide distribution considering delivery methods like intrathecal slow infusion may provide a strong advantage [43].
Live Animal Epigenome Editing by Macromolecule Delivery
Injection of purified macromolecules including ZFs, TALEs, or dCas9-gRNA ribonucleic complexes is a promising alternative to AAV-based delivery. This approach offers more control over dosage and rapid pharmacodynamics. Several attempts to find the most efficient nanoparticle composition for nucleic acid and protein delivery to the specific target tissue has been explored through screens of complex libraries of lipid nanoparticles (see [44] for example), and the use of stable metal-protein complexes like CRISPR-Gold (see [45] for example). While work in this area has clearly gained interest in genome editing, the approach has not been widely adopted for epigenome editing. This could be explained by the fact that preparation of injection-quality macromolecules is more laborious and time-consuming. However, a recent study demonstrated the potential of this approach for a neurodevelopmental disease [46]. Using a purified ZF-KRAB peptide, this study silenced a repressor of the gene Ube3a whose absence causes Angelman Syndrome. The macromolecule was engineered with a red fluorescent protein and a cell-penetrating peptide, which allowed BBB passage and tracking of the ZF in live animals. Interestingly, the macromolecule displayed a large distribution over the brain and induced widespread Ube3a activation in the CNS. This study exemplifies the power of macromolecule-based epigenetic editing in live animals. It also offers a possible path for biological therapeutic interventions.
Transgenic Mouse Strains to Facilitate CRISPR Epigenome Editing
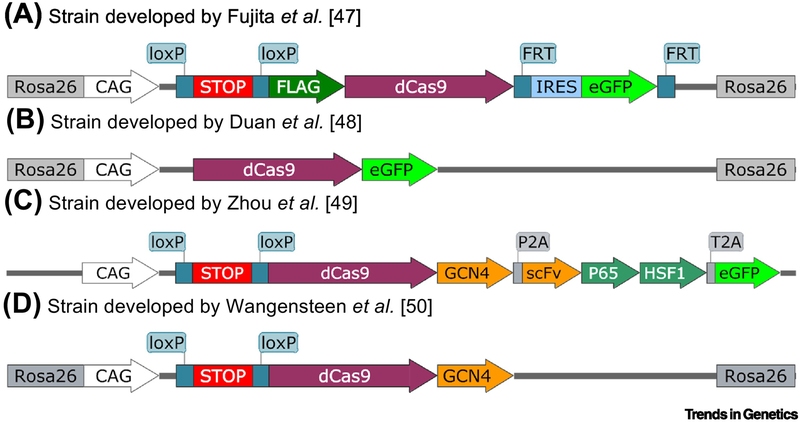
Investigators studying epigenome editing in live animals will benefit from the recently reported knock-in and transgenic mouse lines carrying dCas9 alone or paired with epigenomic editing domains. Since these strains lack gRNA sequences, researchers can deliver gRNA against their genes-of-interest for different targeting experiments. These strains will be of great use for basic biology and therapeutic development studies. The choice of strain will depend on the specific question and the characteristics sought by the researchers. Currently, there are four strains available for CRISPR epigenetic studies with different CRISPR components (Figure 3).
Figure 3. Sequences used to construct transgenic and knock-in mouse strains.
A. Strain with Cre-inducible expression of a FLAG-tagged dCas9 protein with FLP-controlled eGFP expression. B. Strain with constitutive expression of a GFP-tagged dCas9 protein. C. Strain with Cre-inducible expression of a dCas9-GCN4-epitope recombinant protein, followed by a detached single-chain antibody fused to P65 and HSF1 domains, and a detached eGFP protein. D. Strain with Cre-inducible expression of a dCas9-GCN4-epitope recombinant protein. Rosa26 denotes the locus on mouse chromosome 6. White arrow indicates a chimeric CAG promoter system. loxP-STOP-loxP denotes presence of stop codons and a poly-adenylation sequence flanked by loxP sites. P2A and T2A denote the location of exclusive intramolecular cleaving proteases for processing of the polypeptide. All mouse strains were constructed with strep pyogenes derived Cas9.
Two mouse strains are particularly useful to monitor chromatin in live mice when paired with AAV-delivered gRNAs (Figure 3A, 3B). These strains are Rosa26 knock-ins in which the transgene is driven by a CAG promoter system for high transcription in all tissues [47]. With the presence of a loxP-stop-loxP sequence between the promoter and coding sequence allows, Cre-mediated activation of the transgene in specific cell types or developmental time points can be obtained. This strain has been used for chromatin immunoprecipitation by isolating primary immune cells and transfecting them in tissue culture with plasmids expressing specific gRNAs. A similar strain was recently reported with the main difference being that dCas9 was fused to eGFP [48]. . This allowed the researchers to carry out cell imaging of specific loci in liver cells when AAV-packaged gRNAs were introduced by tail vein injection. Although imaging is limited to repetitive loci in which multiple dCas9-eGFP proteins can bind, these mouse strains offer the possibility to monitor specific loci by fluorescent-based assays in live animals in order to understand their changes in response to development, again, and cancer.
Two additional strains were recently reported that offer the potential to perturb transcriptional activity. Both of these strains are based on the CRISPR activation paradigm in which dCas9-gRNA complexes are used to tether specific transcriptional activation domains to sites of interest. One strain was constructed by introducing a transgene that included dCas9-SunTag fusion followed by an scFv antibody fragment fused to trans-activators P65 and HSF1, and a P2A-eGFP element (Figure 3C) [49]. The strain has been used for cell-specific reprogramming and multi-gene editing. Researchers interested in activating multiple genes simultaneously in the same cells will greatly benefit from this mouse strain since all the components except the gRNA are already encoded in the genome. A very similar strain for CRISPR activation was also recently reported [50]. This mouse strain was generated by homologous recombination into the Rosa26 locus and does not include the scFv portion. To validate this strain, the researchers examined whether they could carry out an epigenetic selection screen in live mice by selecting for genes whose activation would lead to the proliferation of liver cells. This mouse strain offers two key advantages. First, since this is a knock-in mouse strain, tracking alleles and crossing to other mouse strains is much easier. Thus, researchers focused on genetically defined diseases will benefit greatly from this mouse strain. Second, the scFv-activator coding sequence is not part of the transgene, allowing this mouse strain to be injected with AAVs encoding scFV-repressors allowing for CRISPR repression experiments.
Taken together, these mouse strains offer the potential to track and alter the transcriptional trajectory of specific genes across development and in physiological environments that are difficult to recreate in tissue culture. In these systems, viral transduction was the choice for gRNA delivery. The selection of injection route and the increasingly large number of capsid variants allows for cell type-specific delivery of gRNAs in mouse models of disease and development.
Concluding Remarks and Future Perspectives
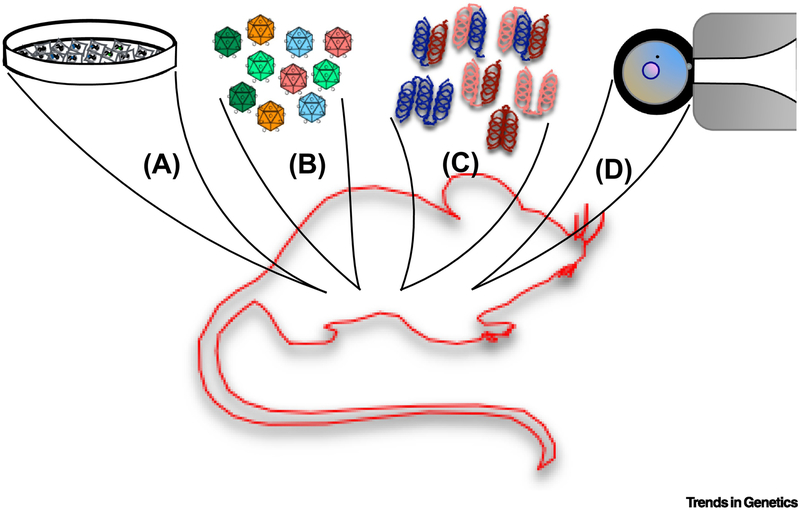
The study of epigenomic editing in live animals is about to enter a rapid growth phase thanks to the convergence of the technologies discussed. These technological advancements include the optimized AAV capsid variants for tissue and cell-specific tropism, the large catalog of CRISPR/dCas9 epigenome effector domains, and the growing number of transgenic and knock-in mice carrying epigenomic editing machinery. Along with cell implantation models and direct macromolecule injection, researchers have at least four modes to transition their studies from tissue culture to live rodent models (Figure 4).
Figure 4. Summary of Methods for Live Animal Epigenome Editing.
A. Epigenomic edited human or murnie cells can be implantation into model organisms and their differentiation, growth, localization monitored over time. B. To alter endogenous mouse cells, the epigenomic editing machinery can be packaged into AAV with mutant capsid variants for specific troprim. C. Alternatively, the epigenomic edited machinery can purified and ribonucleic-protein complexes can be delivered via different routes for targeted organ distribution. D. Genetically encoding dCas9 and epigenomic editing domains in the mouse genome by transgenesis can greatly facilitate the study of epigenomic editing in live animals.
Live animal epigenomic editing has several applications. One of those is modeling and treating brain disorders. Almost all diseases have a genetic component as well as an epigenetic contribution, in which DNA methylation, histone modifications, chromatin accessibility, miRNA expression, pre-mRNA splicing, or long noncoding RNAs are affected. Many of these epigenetic components play an important role in central nervous system disorders, either in neurodegenerative diseases like Alzheimer’s disease [51] and Parkinson’s disease [52], or neurodevelopmental diseases such as Fragile X syndrome [22], Rett Syndrome [53], Angelman Syndrome [54], and Prader-Willi Syndrome [55]. The latter group of listed neurodevelopmental disorders is also part of the autism spectrum disorders with symptoms varying in severity affecting cognitive plasticity, presenting broad impairments in communication and restricted, repetitive behaviors [56]. All of these disorders are monogenic, and disease manifestation and severity arise from dysregulation of downstream epigenetic pathways, making them excellent candidates for genetic and epigenetic therapies [57].
In cancer, the role of epigenetics continues to draw much attention. These efforts are informed, and motivated, by recent patient epigenome sequencing studies, which have uncovered profound changes in DNA methylation, genome-wide accessibility and localization of histone post-translational modifications in patient’s cancer tissues (recently reviewed in [58]). In a few well-studied cases, causal relations between oncogenesis and epigenomic deregulation have been established. These include the role of DNMT3A in acute myeloid leukemia [59] and PBMR1 chromatin remodeling in clear cell renal cell carcinoma [60], among others. Although tissue culture models have helped elucidate key events in cell division, many aspects of oncogenesis that cannot be modeled in a petri dish are still unknown. Live animal epigenome editing studies offer the potential to dissect the temporal dynamics from initially cell cycle deregulation to metastasis. Future live animal epigenome editing research will help elucidate how epigenomic dysregulation helps cancer cells overcome anatomical and immunological barriers at each stage of cancer growth.
The promise of epigenetic-based therapies is the ability to alter the expression of disease-associated gene networks without creating permanent mutations in the human genome. However, one of the clinical translation obstacles that CRISPR gene and epigenome editing approaches are facing is immunogenicity [61–63]. The most commonly used sources of Cas9 come from bacteria known to cause infections in humans [64]. Thus, work is needed to resolve this issue. This and many questions remain on the applications of epigenomic editing in live animals. The studies highlighted inspire the imagination on the potential of this technology but do not address the breath and optimization landscape still needs for reproducible and translatable applications (See Outstanding Questions Box). Live animal epigenetic editing studies will be a necessary catalyst to advance the development of this class of therapies into useful clinical treatments.
Table 1.
Ex vivo study summary on different Cas protein modular systems for epigenetic editing
| Cas protein | Modular systems | Delivery form and route | Gene targeted | Comments | Reference |
|---|---|---|---|---|---|
| dCas9Sp and dCas9Sa | CLOuD9 technology - dCas9 fused to induced proximity system that utilizes the plant phytohormone S-(+)-abscisic acid (ABA) and modified components of the plant ABA signaling pathway | Lentiviral transduction of K562 cells | β-globin gene locus control region which regulates the expression of the distant β-like globin genes through formation of a long-range chromatin loop | Authors developed a new method for chromatin loop reorganization. | [11] |
| dCas9Sa | FIRE-Cas9 system - enhanced dCas9–MS2 with Fkbp/Frb dimerizing fusion proteins | Lentiviral transduction of HEK 293 cells and mouse embryonic stem cells | Three loci upstream of the highly expressed CXCR4 (C-X-C motif chemokine receptor 4) gene and regulatory sequences upstream of the Oct4 | A reversible epigenome editing by endogenous chromatin regulators which is particularly suited to the analysis of endogenous multi-subunit chromatin regulator complexes. | [12] |
| dCas9Sp and dCpf1As | Epigenetic repressors fused (G9A, SUV39H1, Krüppel-associated box (KRAB), DNMT3A as well as the first targetable versions of Ezh2 and Friend of GATA-1 (FOG1) to dCas9 or Cpf1 | HCT116 cell line transfection | Promoters of HER2 and MYC and EPCAM genes | Screened different repressors and highlighted
the differences between them in achieving persistent
repression. Demonstrated that the dCpf1As fusions were not active when used to repress Her2 expression. |
[13] |
| cCas9Sp | dCas9-NLS-HA-BFP fused to either KRAB, CS or WRPW for repression or for activation: VP64 or p65 activation domain | Lentiviral transduction of GFP+ HEK293 cells | GFP, transferrin receptor (CD71) and C-X-C chemokine receptor type 4 (CXCR4) | Showed robust gene knockdown of both reporter and endogenous genes and improved their previously published work on CRISPRi as an alternate method to RNAi for repressing gene expression in mammalian cells. | [14] |
| dCas9Sp | structure-guided engineering of a CRISPR-Cas9 complex: combination of sgRNA 2.0, NLS-dCas9-VP64, and MS2-p65-HSF1: synergistic activation mediator (SAM) | Transient transfection experiments in Neuro2 cells and HEK293FT cells and lentiviral transduction of A375 cells | 12 genes which were previously found by several groups to be difficult to activate using dCas9-VP64 and individual sgRNA | Showed that SAM stimulated transcription at
least 2-fold for all genes and more than 15-fold for 8 out of 12
genes. Engineered sgRNA to incorporate protein-interacting aptamers for gene upregulation. |
[15] |
| dCas9Sp | dCas9-DNMT3A fusion | Transfection of HEK293T cells with lipofectamine and lentiviral transduction | CDKN2A, ARF and, Cdkn1a promoter | The developed system allows mechanistic studies of DNA methylation. | [16] |
| dCas9Sp | dCas9-VP64-p65-Rta (VPR) | HEK 293T cells and Neuro-2A cell transfection | NGN2 and NEUROD1 | Showed activation of endogenous coding and non-coding genes, targeted several genes simultaneously and stimulated neuronal differentiation of human induced pluripotent stem cells | [17] |
| dCas9 | dCas9-SunTag-DNMT3A vs dCas9-DNMT3A | Transfection of MCF-7 or HeLa cells | CTCF and NRF1 | Showed that the dCas9-SunTag-DNMT3A system can recruit multiple DNMT3A catalytic domains to a target site for editing DNA methylation at a much higher induction rate | [18] |
| dCas9 | dCas9-Dnmt or Tet | Lentiviral transduction of multiple FXS patient derived iPSCs and as in vitro derived FXS neurons | FMR1 reactivation: reverse the hypermethyl ation of CGG repeats at the FMR1 locus | Demonstrated evidence that demethylation of the CGG repeats is sufficient to reactivate FMR1 | [21] |
Table 2.
Summary of in vivo epigenetic editing studies. The epigenome editor used, genes modified, target tissues/cells, disease, and delivery mechanisms are listed.
| Epigenome Editor | Gene | Target | Disease or Condition | Delivery Form | Delivery Route | Comments | Reference |
|---|---|---|---|---|---|---|---|
| TALE-KRAB | c-Kit, PU.1 (Spi1) | Bone Marrow (BM) | BM transplantation | Cell implantation | Intravenous injection | Combined BM transplantation with a multicolor TALE-KRAB expression vector to knock down two targets genes in hematopoietic compartment in vivo. A technology for investigating functional roles of multiple gene targets. | [19] |
| dCas9-eGFP, dCas9-VP64 | Trp53, Mgmt | B-cell lymphoma, B-ALL leukemia | Lymphoma | Cell implantation | Intravenous injection | Showed dCas9-mediated gene level perturbation for modeling cancer progression and therapeutic relapse both in vitro and in mouse models. | [20] |
| dCas9-Tet1 | FMR1 | iPSC-derived neurons | Fragile X | Cell implantation | Intracranial injection/Neonatal engrafting | Showed sustained reactivation of FMR1 in the methylation edited FXS cells when injected into the P1 mouse brain for subsequent analysis one- or three-month post transplantation. | [21] |
| ZF-VP64 | GDNF | Glial cells | Parkinson’s disease | AAV2 | Striatum injection with convection-enhanced delivery | Demonstrated that the activation of endogenous GDNF is sufficient to protect against 6-OHDA lesion in rat, and, additionally, showed that the ZF-vp64 activates the endogenous human GDNF gene. | [31] |
| dCas9 expressing mice with MS2-P65-HSF1 | Fst, Il10, klotho, Pdx1, Utrn | Liver, Muscle and Kidney | Diabetes, muscular dystrophy, and acute kidney disease | AAV2/9 | Intravenous Intramuscular Intracranial | The authors showed that this system can activate genes by modulating histone marks rather than editing DNA sequences and claimed that it can be used to express genes to compensate for disease-associated genetic mutations, or to overexpress long non-coding RNAs, and GC-rich genes which has been difficult until now. | [32] |
| dSaCas9-KRAB | Pcsk9 | Liver | High cholesterol diseases | AAV8 | Intravenous | Authors discuss that one potential improvement of dSaCas9-KRAB system is that they need to deliver two components (the Cas9 and gRNA), which minimizes additional recruitment and potential immune response. | [33] |
| TALE-VP64 or TALE-SunTag VP64 | FXN | Liver, Heart, Muscle, Brain | Friedreich’s ataxia | AAV9 | Intraperitoneal injection | Authors discuss that the delivery of AAV9 could be improved with intracranial and intravenous injections in future studies. The delivery of AAV9 intraperitoneal ly is suboptimal if the target organ is also the brain. | [34], [35] |
| dCas9-KRAB, dCas9-VP64-Rta | Cd81, Afp, Nrl | Liver, Retina | Retinitis pigmentosa | AAV8, AAV2-Y444F | Intravenous, Subretinal | Utilized split-Cas9 system and showed up to 80% transcriptional repression and up to 6-fold transcriptional activation and showed efficacy of using AAV-KRAB-Cas9 in the context of gene therapy in a mouse model for retinitis pigmentosa. | [36] |
| ZF-KRAB | Ube3a | Brain | Angelman Syndrome | Protein | Intraperitoneal, Subcutaneous | A transient activation of Ube3a gene in the brain by using a ZF protein which when injected subcutaneous crosses the blood-brain barrier. | [46] |
| dCas9 | cmyc | T cells | N.A. | Transgenesis Retrovirus | N.A. | The generated transgenic animals represent a useful tool for enChIP analysis in primary mouse cells. | [47] |
| dCas9-eGFP KRAB | telomeres Trf1 | Liver | N.A. | Transgenesis, DNA | Microinject ion for the generation of the animals in mouse zygotes and hydrodynamic tail vein (intravenous) injection | The generated dCas9-EGFP knock-in mouse provides a useful tool to dissect genome functions, including chromatin dynamics in live animals. | [48] |
| dCas9-SunTag-P65-HSF1 | Ascl1, Neurog2, Neurod1, Dkk1, Hbb, many | Brain, Astrocytes, Fibroblasts | N.A. | Transgenesis, AAV8 and Lentivirus | Intracranial, Intravenous | The generated transgenic mouse model allows for a flexible screening for studying complex gene networks, including long noncoding RNAs and gain-of-function phenotypes in the nervous system. | [49] |
| dCas9-SunTag VP64 | Myc, Tnfrsf1a, Slc7a11, Tp53 | Liver | Liver injury and tumorigenesis | Transgenesis, and AAV8 | Intravenous | The authors envision that their system CRISPRa system will be useful for performing additional tissue‐ specific genetic screens as dCas9 can be activated in any tissue of interest. They showed hepatocyte specificity with an AAV- Cre virus, but viruses with other cell type–specific tropisms could be used to target other tissues. | [50] |
BOX 1: Design Parameters for Epigenome Editing.
Recruitment Designs and Current Tools
Two major design parameters affect the engineering of CRISPR/dCas9 epigenome platforms: Recruitment and Source. The recruitment parameter deals with the mode by which dCas9 enables localization of the epigenome editing machinery to the site of interest on DNA. Localization can be mediated by either direct covalent attachment to dCas9 or through non-covalent protein-protein or protein-RNA interactions. For live animal studies, recruitment by non-covalent attachment may allow for multimerization of epigenomic editing enzymes at specific loci, increase complexity of the epigenome machinery, and packaging into multiple viral vectors. The second design parameter deals with the source of the epigenome machinery: exogenous or endogenous. Exogenous design platforms are those in which a catalytic or functional domain is engineered into the transgene. Endogenous design platforms are those in which cellular encoded enzymes and chromatin organizers are re-localized to specific loci. The exogenous design platforms engineered into CRISPR include histone methyltransferases and demethylases: EZH2, G9A, SUV39H1 [13], DOT1L, PRDM9 [65], SMYD3 [66], MLL3SET [67] and LSD1 [68], histone acetyltransferases P300 [69], CBP [70]. Examples of endogenous design platforms by recruiting peptides are KRAB [14], HP1a [12], SID [71], FOG [13], C-terminal-MECP2 [72], VP64 [14], P65 [14], Rta [17], and HSF1[15]. Together these epigenetic editing tools offer many options to alter the epigenome in live animals for locus-specific biochemistry. The choice of recruitment modality and source of epigenetic machinery has a significant impact on the functionality of the epigenome editing system.
Future Applications of other Nucleases
Novel type of nucleases were introduced for RNA editing by separate teams led by Doudna [73] and Zhang. The latter team showed that a catalytically inactive Cas13b ortholog from Prevotella sp. is the most efficient and specific for mammalian cell applications [74]. The deactivated variant of Cas13b was fused to ADAR2 deaminase domain, which participates in the conversion of adenosine to inosine. This system was able to recognize a specific target sequence, whereas the ADAR2 element was performing the base conversion. These studies demonstrate an excellent new platform for precise RNA editing with broad applicability, and since then other smaller orthologues with robust activity in human cells have been characterized [75]. In the case of large proteins like Cas9, smaller orthologs have now been reported to show equal editing efficiency [42,76,77]. That is especially useful for AAV packaging.
Maximizing the packing size of AAVs
Given that several of the epigenetic tools, including some of the more commonly used nucleases, exceed the 3.5 kB AAV packaging limit, there have been several studies addressing the limiting packaging size. For the CRISPR/dCas system, smaller dCas9 orthologs [42,77,78], a split-dCas9 system [79], and dual AAV system [32,80] has been suggested. Future studies will show if dual AAV systems are as efficient in transducing target cells compared to the generation of cell specific AAV capsid libraries (for a recent review on AAV dual systems see [80]).
HIGHLIGHTS.
Epigenome editing and preclinical therapies can now be performed in live animals
Epigenome editing is mediated by effector domains fused to the editing tools to mediate transcriptional activation, repression, and long-range chromatin alterations
CRISPR technology, zinc fingers and transcription activator-like effectors (TALEs) each offers unique advantages in live animal epigenomic editing
Delivery form (viral, cell implant, transgene or macromolecular assemblies) and route (intracranial, intravenous, intrathecal, intraperitoneal) are two of the biggest challenges to successful epigenome research in live animals
ACKNOWLEDGMENTS
Supported was provided to JAG by NIH T32CA108459, JAG and DJS by DOD W81XWH-17-1-0200, and UB and DJS by the Foundation for Angelman Syndrome Therapeutics.
GLOSSARY
- Artificial proximity dimerizers
chemical inducers of dimerization which are designed to bind to two different proteins and bring them into close proximity in the presence of the dimerizer
- CRISPR/Cas
identified in bacteria as an antiviral defense strategy, now is utilized as a technology for gene editing which includes an enzyme called nuclease and a guide RNA which allows to target practically any sequence in the genome for genetic or epigenetic editing
- Erasers (epigenetic)
epigenetic marks can be removed by a group of enzymes called “erasers” that can reverse the influence of an epigenetic mark on gene expression
- HSF1
A potent trans-activator peptide known for recruiting endogenous transcription activation cellular machinery. Derived from a conserved fragment of mammalian heat shock protein 1
- Knock-in mouse strain
A mouse strains in which a transgene has been inserted at a specific site in the genome as single copy integration. Traditionally constructed by introducing foreign DNA (transgene) into embryonic stem cells by homologous recombination. Edited embryonic stem cells are then injected into a blastocyst and embryos are brought to term. Mosaic founders are tested for germline transmission of the transgene.
- KRAB
The Krüppel associated box domain is potent transcriptional repressor. The peptide is highly conserved across species and is known to interact with the KAP1 complex to mediate trimethylation of lysine 9 of histone 3.
- P65
A potent trans-activator peptide known for interacting with NF-kB and mediating transcription activation. Derived from a conserved fragment of mammalian RELA gene.
- SunTag system
a repeating peptide array that can recruit multiple copies of an antibody-fusion protein for signal amplification
- Rta
A potent trans-activator peptide known for interacting with OCT1 and SP1 in mediating transcription activation. Derived from a fragment of the R transactivator protein of Epstein-Barr Virus.
- Rosa26
is a specific site / locus in the mouse genome that is widely used for achieving generalized expression in the mouse and has been used in generating over a hundred knock-in cell lines. A human homolog of the mouse Rosa26 locus has been identified later and has been used to generate embryonic stem cell lines.
- TALEs
Transcription activator-like effectors are proteins which can be designed to modulate gene transcription. These proteins recognize and bind to DNA sequences based on a variable number of tandem repeats
- Transgenic mouse strain
A mouse strain in which a transgene has been inserted at random sites in the genome either as single copy or multi-copy integrations. Traditionally constructed by introducing foreign DNA (transgene) into single cell fertilized eggs and screening founders for transgene integration.
- VP64
A potent trans-activator peptide known for recruiting endogenous transcription activation cellular machinery. Composed of four copies of a fragment from viral protein 16 (VP16), which is derived from herpes simplex virus.
- Writers (epigenetic)
a group of enzymes which catalyzes the addition of chemical groups onto either histone tails or the DNA itself which affect gene expression and are also known as epigenetic marks
- Zinc Fingers
engineered proteins which behave like transcription factors with the ability for each finger module to recognize three to four bases of sequence, and by mixing and matching those modules almost any sequence can be targeted
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Korablev AN et al. (2017) Generation of megabase-scale deletions, inversions and duplications involving the Contactin-6 gene in mice by CRISPR/Cas9 technology. BMC Genet 18, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li J et al. (2015) Efficient inversions and duplications of mammalian regulatory DNA elements and gene clusters by CRISPR/Cas9. J. Mol. Cell Biol 7, 284–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kosicki M et al. (2018) Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol DOI: 10.1038/nbt.4192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang L et al. (2018) Meganuclease targeting of PCSK9 in macaque liver leads to stable reduction in serum cholesterol. Nat. Biotechnol 36, 717–725 [DOI] [PubMed] [Google Scholar]
- 5.Chiu A and Rao MS (2003) Human Embryonic Stem Cells, Springer Science & Business Media. [Google Scholar]
- 6.Haapaniemi E et al. (2018) CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat. Med 24, 927–930 [DOI] [PubMed] [Google Scholar]
- 7.Wright AV et al. (2016) Biology and Applications of CRISPR Systems: Harnessing Nature’s Toolbox for Genome Engineering. Cell 164, 29–44 [DOI] [PubMed] [Google Scholar]
- 8.Pulecio J et al. (2017) CRISPR/Cas9-Based Engineering of the Epigenome. Cell Stem Cell 21, 431–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thakore PI et al. (2016) Editing the epigenome: technologies for programmable transcription and epigenetic modulation. Nat. Methods 13, 127–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jinek M et al. (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morgan SL et al. (2017) Manipulation of nuclear architecture through CRISPR-mediated chromosomal looping. Nat. Commun 8, 15993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braun SMG et al. (2017) Rapid and reversible epigenome editing by endogenous chromatin regulators. Nat. Commun 8, 560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Geen H et al. (2017) dCas9-based epigenome editing suggests acquisition of histone methylation is not sufficient for target gene repression. Nucleic Acids Res 45, 9901–9916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbert LA et al. (2013) CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Konermann S et al. (2015) Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517, 583–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDonald JI et al. (2016) Reprogrammable CRISPR/Cas9-based system for inducing site-specific DNA methylation. Biol. Open 5, 866–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chavez A et al. (2015) Highly efficient Cas9-mediated transcriptional programming. Nat. Methods 12, 326–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pflueger C et al. (2018) A modular dCas9-SunTag DNMT3A epigenome editing system overcomes pervasive off-target activity of direct fusion dCas9-DNMT3A constructs. Genome Res 28, 1193–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Z et al. (2014) A multicolor panel of TALE-KRAB based transcriptional repressor vectors enabling knockdown of multiple gene targets. Sci. Rep 4, 7338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Braun CJ et al. (2016) Versatile in vivo regulation of tumor phenotypes by dCas9-mediated transcriptional perturbation. Proc. Natl. Acad. Sci. U. S. A 113, E3892–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu XS et al. (2018) Rescue of Fragile X Syndrome Neurons by DNA Methylation Editing of the FMR1 Gene. Cell 172, 979–992.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kraan CM et al. (2018) Epigenetics of fragile X syndrome and fragile X-related disorders. Dev. Med. Child Neurol DOI: 10.1111/dmcn.13985 [DOI] [PubMed] [Google Scholar]
- 23.Deverman BE et al. (2018) Gene therapy for neurological disorders: progress and prospects. Nat. Rev. Drug Discov 17, 767. [DOI] [PubMed] [Google Scholar]
- 24.Gray SJ et al. (2010) Viral vectors and delivery strategies for CNS gene therapy. Ther. Deliv 1, 517–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aschauer DF et al. (2013) Analysis of transduction efficiency, tropism and axonal transport of AAV serotypes 1, 2, 5, 6, 8 and 9 in the mouse brain. PLoS One 8, e76310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H et al. (2011) Several rAAV Vectors Efficiently Cross the Blood–brain Barrier and Transduce Neurons and Astrocytes in the Neonatal Mouse Central Nervous System. Mol. Ther 19, 1440–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garriga-Canut M et al. (2012) Synthetic zinc finger repressors reduce mutant huntingtin expression in the brain of R6/2 mice. Proc. Natl. Acad. Sci. U. S. A 109, E3136–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bustos FJ et al. Epigenetic editing of the Dlg4/PSD95 gene improves cognition in aged and Alzheimer’s disease mice., Brain, 140 (2017), 3252–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heller EA et al. (2016) Targeted Epigenetic Remodeling of the Cdk5 Gene in Nucleus Accumbens Regulates Cocaine- and Stress-Evoked Behavior. J. Neurosci 36, 4690–4697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heller EA et al. Locus-specific epigenetic remodeling controls addiction- and depression-related behaviors., Nature Neuroscience, 17 (2014), 1720–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laganiere J et al. An Engineered Zinc Finger Protein Activator of the Endogenous Glial Cell Line-Derived Neurotrophic Factor Gene Provides Functional Neuroprotection in a Rat Model of Parkinson’s Disease., Journal of Neuroscience, 30 (2010), 16469–16474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liao H-K et al. (2017) In Vivo Target Gene Activation via CRISPR/Cas9-Mediated Trans-epigenetic Modulation. Cell 171, 1495–1507.e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thakore PI et al. (2018) RNA-guided transcriptional silencing in vivo with S. aureus CRISPR-Cas9 repressors. Nat. Commun 9, 1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chapdelaine P et al. (2016) Development of an AAV9 coding for a 3XFLAG-TALEfrat#8-VP64 able to increase in vivo the human frataxin in YG8R mice. Gene Ther 23, 606–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cherif K et al. (2018) Increased Frataxin Expression Induced in Friedreich Ataxia Cells by Platinum TALE-VP64s or Platinum TALE-SunTag. Mol. Ther. Nucleic Acids 12, 19–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moreno AM et al. (2018) In Situ Gene Therapy via AAV-CRISPR-Cas9-Mediated Targeted Gene Regulation. Mol. Ther 26, 1818–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dashkoff J et al. (2016) Tailored transgene expression to specific cell types in the central nervous system after peripheral injection with AAV9. Mol Ther Methods Clin Dev 3, 16081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McLean JR et al. (2014) Widespread neuron-specific transgene expression in brain and spinal cord following synapsin promoter-driven AAV9 neonatal intracerebroventricular injection. Neurosci. Lett 576, 73–78 [DOI] [PubMed] [Google Scholar]
- 39.Chan KY et al. (2017) Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci 20, 1172–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Albright BH et al. (2018) Mapping the Structural Determinants Required for AAVrh.10 Transport across the Blood-Brain Barrier. Mol. Ther 26, 510–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hordeaux J et al. (2018) The Neurotropic Properties of AAV-PHP.B Are Limited to C57BL/6J Mice. Mol. Ther 26, 664–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim E et al. (2017) In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat. Commun 8, 14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bailey RM et al. (2018) Development of Intrathecal AAV9 Gene Therapy for Giant Axonal Neuropathy. Mol Ther Methods Clin Dev 9, 160–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paunovska K et al. (2018) A Direct Comparison of in Vitro and in Vivo Nucleic Acid Delivery Mediated by Hundreds of Nanoparticles Reveals a Weak Correlation. Nano Lett 18, 2148–2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee B et al. (2018) Nanoparticle delivery of CRISPR into the brain rescues a mouse model of fragile X syndrome from exaggerated repetitive behaviours. Nature Biomedical Engineering 2, 497–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bailus BJ et al. (2016) Protein Delivery of an Artificial Transcription Factor Restores Widespread Ube3a Expression in an Angelman Syndrome Mouse Brain. Mol. Ther 24, 548–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fujita T et al. (2018) Transgenic mouse lines expressing the 3xFLAG-dCas9 protein for enChIP analysis. Genes Cells 23, 318–325 [DOI] [PubMed] [Google Scholar]
- 48.Duan J et al. (2018) Live imaging and tracking of genome regions in CRISPR/dCas9 knock-in mice. Genome Biol 19, 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou H et al. (2018) In vivo simultaneous transcriptional activation of multiple genes in the brain using CRISPR-dCas9-activator transgenic mice. Nat. Neurosci 21, 440–446 [DOI] [PubMed] [Google Scholar]
- 50.Wangensteen KJ et al. (2018) Combinatorial genetics in liver repopulation and carcinogenesis with a in vivo CRISPR activation platform. Hepatology 68, 663–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Jager PL et al. (2014) Alzheimer’s disease: early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nat. Neurosci 17, 1156–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Navarro-Sánchez L et al. (2018) Epigenetic Study in Parkinson’s Disease: A Pilot Analysis of DNA Methylation in Candidate Genes in Brain. Cells 7, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.LaSalle JM Epigenetic Mechanisms in Rett Syndrome., Epigenetics: Current Research and Emerging Trends (2015), 199–216
- 54.Leung K (2009) Epigenetic Pathways in Angelman and Prader-Willi Syndromes. Targeted Protein Database DOI: 10.2970/tpdb.2009.235 [DOI] [Google Scholar]
- 55.Zink F et al. (2018) Insights into imprinting from parent-of-origin phased methylomes and transcriptomes. Nat. Genet 50, 1542–1552 [DOI] [PubMed] [Google Scholar]
- 56.Chahrour M et al. (2016) Current Perspectives in Autism Spectrum Disorder: From Genes to Therapy. J. Neurosci 36, 11402–11410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Benger M et al. (2018) Autism spectrum disorder: prospects for treatment using gene therapy. Mol. Autism 9, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bennett RL and Licht JD (2018) Targeting Epigenetics in Cancer. Annu. Rev. Pharmacol. Toxicol 58, 187–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ley TJ et al. (2010) DNMT3A mutations in acute myeloid leukemia. N. Engl. J. Med 363, 2424–2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.The Cancer Genome Atlas Research Network (2013) Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 499, 43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Colque-Navarro P et al. (2010) Levels of Antibody against 11 Staphylococcus aureus Antigens in a Healthy Population. Clin. Vaccine Immunol 17, 1117–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kolata JB et al. (2015) The Fall of a Dogma? Unexpected High T-Cell Memory Response toStaphylococcus aureusin Humans. J. Infect. Dis 212, 830–838 [DOI] [PubMed] [Google Scholar]
- 63.Mortensen R et al. (2015) Adaptive Immunity against Streptococcus pyogenes in Adults Involves Increased IFN-γ and IgG3 Responses Compared with Children. J. Immunol 195, 1657–1664 [DOI] [PubMed] [Google Scholar]
- 64.Chew WL (2018) Immunity to CRISPR Cas9 and Cas12a therapeutics. Wiley Interdiscip. Rev. Syst. Biol. Med 10, [DOI] [PubMed] [Google Scholar]
- 65.Cano-Rodriguez D et al. (2016) Writing of H3K4Me3 overcomes epigenetic silencing in a sustained but context-dependent manner. Nat. Commun 7, 12284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim J-M et al. (2015) Cooperation between SMYD3 and PC4 drives a distinct transcriptional program in cancer cells. Nucleic Acids Res 43, 8868–8883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yan J et al. (2018) Histone H3 lysine 4 monomethylation modulates long-range chromatin interactions at enhancers. Cell Res 28, 387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kearns NA et al. (2015) Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat. Methods 12, 401–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hilton IB et al. (2015) Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol 33, 510–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cheng AW et al. (2016) Casilio: a versatile CRISPR-Cas9-Pumilio hybrid for gene regulation and genomic labeling. Cell Res 26, 254–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Konermann S et al. (2013) Optical control of mammalian endogenous transcription and epigenetic states. Nature 500, 472–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yeo NC et al. (2018) An enhanced CRISPR repressor for targeted mammalian gene regulation. Nat. Methods 15, 611–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.East-Seletsky A et al. (2017) RNA Targeting by Functionally Orthogonal Type VI-A CRISPR-Cas Enzymes. Mol. Cell 66, 373–383.e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cox DBT et al. (2017) RNA editing with CRISPR-Cas13. Science 358, 1019–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Konermann S et al. (2018) Transcriptome Engineering with RNA-Targeting Type VI-D CRISPR Effectors. Cell 173, 665–676.e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hou Z et al. (2013) Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc. Natl. Acad. Sci. U. S. A 110, 15644–15649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ran FA et al. (2015) In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ibraheim R et al. (2018) All-in-one adeno-associated virus delivery and genome editing by Neisseria meningitidis Cas9 in vivo. Genome Biol 19, 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zetsche B et al. (2015) A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat. Biotechnol 33, 139–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McClements ME and MacLaren RE (2017) Adeno-associated Virus (AAV) Dual Vector Strategies for Gene Therapy Encoding Large Transgenes. Yale J. Biol. Med 90, 611–623 [PMC free article] [PubMed] [Google Scholar]