Abstract
The c-Myc oncogene (MYC) drives malignant progression, but also induces robust anabolic and proliferative programs leading to intrinsic stress. The mechanisms enabling adaptation to MYC-induced stress are not fully understood. We have uncovered an essential role for the transcription factor ATF4 in survival following MYC activation. MYC upregulates ATF4 by activating GCN2 kinase through uncharged tRNAs. Subsequently, ATF4 co-occupies promoter regions of over 30 MYC target genes, primarily those regulating amino acid and protein synthesis, including 4E-BP1, a negative regulator of translation. 4E-BP1 is essential to balance protein synthesis, relieving MYC-induced proteotoxic stress. 4E-BP1 activity is negatively regulated by mTORC1-dependent phosphorylation and inhibition of mTORC1 signaling rescues ATF4 deficient cells from MYC-induced ER stress. Acute deletion of ATF4 significantly delays MYC-driven tumor progression and increases survival in mouse models. Our results establish ATF4 as a cellular rheostat of MYC-activity, ensuring enhanced translation rates are compatible with survival and tumor progression.
Introduction
Cells utilize distinct stress response pathways to overcome environmental and physiological stresses. The Integrated Stress Response (ISR) pathway promotes cellular adaptation to various stresses such as viral infection, heme deprivation, hypoxia, nutrient deprivation and acidosis1. The ISR kinases, PKR-like ER kinase (PERK), general control non-derepressible 2 (GCN2), double-stranded RNA-dependent protein kinase (PKR) and heme-regulated eIF2α kinase (HRI), sense distinct stresses and catalyze phosphorylation of the α subunit of the eukaryotic initiation factor eIF2 (eIF2α). Phosphorylation of eIF2α at serine 51 attenuates general protein synthesis while enhancing the translation of select transcripts containing distinct regulatory sequences in their 5’ untranslated region2, 3, most notably that of the activating transcription factor 4 (ATF4). Once translated, ATF4 drives the transcription of genes involved in antioxidant response, autophagy, amino acid biosynthesis and transport4, 5.
The ability of cancer cells to adapt to cell-extrinsic and intrinsic stresses is critical for maintaining viability and growth. We and others previously showed that the ISR is essential in adaptation to extrinsic stresses present in the tumor microenvironment such as hypoxia and nutrient deprivation6–9. Cancer cells also experience intrinsic stress due to activation of oncogenes that increase bioenergetic processes10–12. In this context, amplification of the MYC oncogene, a frequent event in multiple human malignancies13 causes intrinsic stress due to enhanced protein synthesis and rewired metabolic pathways to meet the demands of rapid cell growth and proliferation14, 15. MYC upregulates protein synthesis by transactivating components of the translational machinery including initiation factors, ribosomes and tRNAs15 and targeting the translation machinery has proven to be effective in MYC-driven cancers16, 17. However, protein synthesis rates need to be controlled to a critical level to sustain survival of cancer cells during tumor development. For example, increased protein synthesis has to be accompanied by a concomitant increase in the folding capacity and size of the Endoplasmic Reticulum (ER) to avoid proteotoxicity18.
We previously demonstrated that MYC-induced increase in translation rates activates the PERK arm of ISR, which is required for supporting MYC induced transformation and survival11, primarily by activating cytoprotective autophagy and attenuating Ca2+ release from the ER11. Although ATF4 has been implicated in supporting survival of cancer cells experiencing a deficit of oxygen and nutrients, its role in oncogene-induced stress has not been well characterized.
Here, we show that optimal ATF4 expression upon MYC dysregulation requires both the PERK and GCN2 kinases, the latter being activated by excess uncharged amino acids produced by increased MYC activity. Induced ATF4, cooperatively with MYC, co-regulates a number of gene products including 4E-BP1 to fine-tune mRNA translation induced by MYC. Importantly, ablation of ATF4 results in increased ER stress and cytotoxicity which is attenuated by mTORC1 inhibition or expression of a dominant-negative 4E-BP1. Our results demonstrate the critical role of ISR signaling-induced ATF4 in supporting cell adaptation and survival during MYC-dependent tumor growth and progression.
Results
ATF4 is induced by MYC and promotes survival.
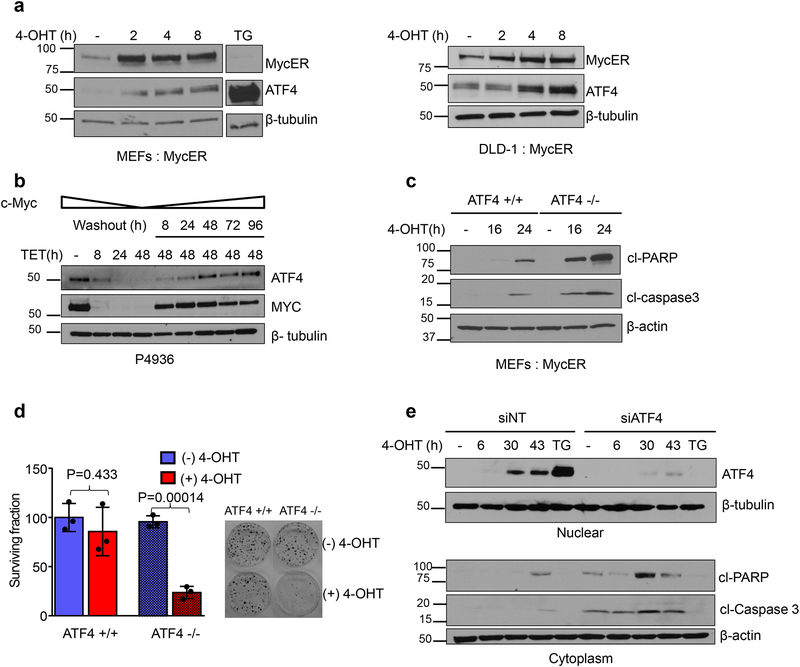
We previously reported that activation of MYC leads to phosphorylation of PERK and eIF2α11. To test whether MYC activation also induces ATF4, we analyzed ATF4 expression following MYC induction in DLD-1, human colon adenocarcinoma cells and mouse embryonic fibroblasts (MEFs) stably expressing a tamoxifen-inducible MYC chimera, MycER. Treatment with 4-hydroxy-tamoxifen (4-OHT) led to accumulation of nuclear MYC and expression of ATF4 in both cell lines (Fig. 1a, Supplementary Fig. 1a). Similarly, in the human Burkitt’s lymphoma cell line P493–6, in which expression of MYC is turned off by administration of tetracycline, suppression of MYC resulted in a concomitant decrease of ATF4. Upon restoration of MYC levels, ATF4 protein levels also recovered (Fig. 1b). These data indicate that elevated MYC induces ATF4 expression.
Figure 1. MYC induced ATF4 inhibits apoptosis and promotes survival.
a. Immunoblot of nuclear lysates from MEFs (left panel) and DLD-1 cells (right panel) expressing MycER were treated with 4-OHT to activate MYC. Thapsigargin (0.5μM for 4h) treated cells were used as a positive control for ATF4 induction. b. Immunoblot of nuclear lysates from P4936 (Tet-off MYC) cells treated with tetracycline (0.1ug/ml) or tetracycline was washed off for the indicated times. c. Immunoblot analysis of whole cell lysates from ATF4 +/+ and ATF4 −/− MEFs treated with 4-OHT for indicated times. d. Clonogenic survival was performed after activating MYC in MEFs, representative plates from three biological replicates are shown. Colonies were counted and surviving fraction is shown normalized to no treatment control. Error bars represent mean ± SD, two tailed student t-test. e. DLD-1: MycER cells were transfected with non-targeting siRNA or siRNA targeting ATF4. Cells were treated with 4-OHT and indicated proteins were assessed by immunoblotting. All immunoblots are representative of three biological replicates that showed similar results. Unprocessed scans of blots are shown in Supplementary Fig 7.
Notably, ablation of ATF4 in MEFs significantly enhanced MYC-induced cell death, highlighted by increased levels of the apoptosis markers including cleaved Poly-ADP ribose polymerase, cl-PARP, cleaved caspase 3 (Fig. 1c), and by reduced clonogenic survival (Fig. 1d). Similarly, knockdown of ATF4 in DLD-1 cells also markedly enhanced apoptosis following MYC activation (Fig. 1e). Collectively, these results highlight a significant role for ATF4 in promoting survival of transformed cells upon MYC activation.
PERK and GCN2 are required for optimal phosphorylation of eIF2α following MYC induction.
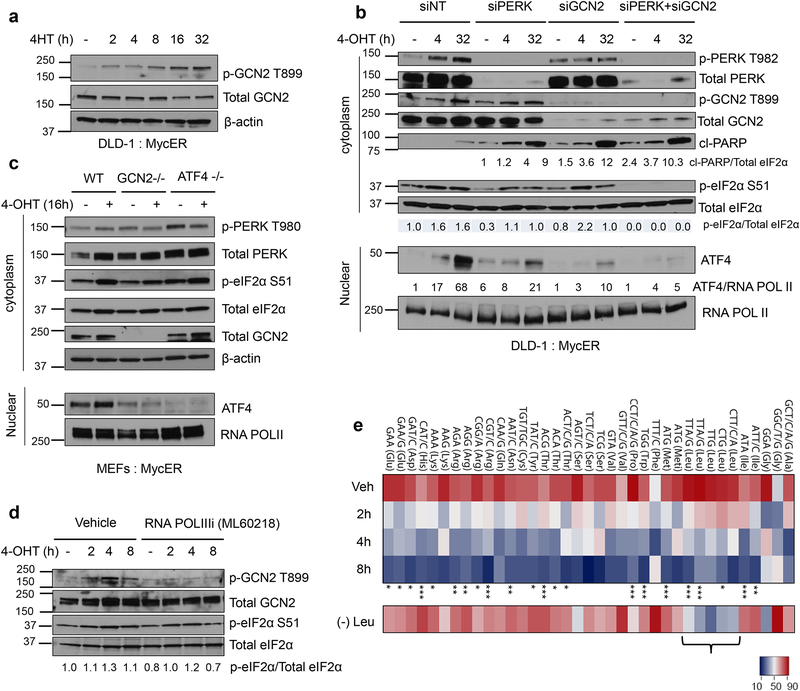
Activation of MYC enhances protein synthesis resulting in ER stress11, 19. Although PERK is primarily responsible for phosphorylating eIF2α during MYC activation, residual phosphorylation of eIF2α in PERK knockout cells prompted us to ask whether other ISR kinases phosphorylate eIF2α in the absence of PERK11. We focused on GCN2, a kinase we and others have shown to be activated in multiple solid tumors7, 20. Interestingly, we noted robust phosphorylation of GCN2 following MYC activation (Figs. 2a and 2b). Consistent with our previous report11, phosphorylation of eIF2α was reduced, but still present upon PERK knockdown (Fig. 2b). Notably, eIF2α phosphorylation was markedly reduced in the absence of both kinases, indicating functional compensation between these kinases upon MYC activation (Fig. 2b).
Figure 2. The amino acid sensor GCN2 is activated by uncharged tRNAs and is required for optimal activation of ATF4 upon MYC induction.
a. Representative immunoblot showing activation of GCN2 following MYC induction using cytoplasmic lysates. b. DLD-1: MycER cells were transfected with non-targeting siRNA or siRNA against PERK, GCN2 or both. ISR signaling and apoptosis were assessed by immunoblotting after MYC activation. c. Representative immunoblot analysis of cytoplasmic and nuclear lysates from GCN2 +/+, GCN2−/− and ATF4−/− MycER MEFs after 16hrs of MYC activation. d. DLD-1: MycER cells were pretreated with DMSO or 50μM RNA POLIII inhibitor (ML60218) for 2hrs prior to MYC activation. Indicated protein levels were measured by immunoblotting from cytoplasmic lysates. e. Microarray of aminoacyl–tRNAs of DLD-1: MycER cells after MYC induction at indicated times, four independent experiments, one-way ANOVA, *, p<0.05; **, p<0.01; ***, p< 0.001 (Refer to Statistics Source Data table for exact p values). (−) Leu denotes Leu-deprived cells and the marked tRNAs reading Leu codons are uncharged, n=1. tRNA probes are depicted with their cognate codon and the corresponding amino acid; Meti, initiator tRNAMet. Two different probes recognizing two different tRNALeu isodecoders that pair to the same codon TTA/G Leu codon but differ in their sequence outside the anticodon were used on the arrays. Brackets show decreased charging of tRNALeu isodecoders in Leu deprivation condition. All immunoblots are representative of three biological replicates showing similar results. Unprocessed scans of blots are shown in Supplementary Fig 7.
Consistent with ATF4 loss, knockdown of both PERK and GCN2 enhanced apoptosis following MYC activation (Fig. 2b). Decreased expression of either PERK or GCN2 also reduced ATF4 protein levels (Fig. 2b), and this effect was more prominent in GCN2 knockdown cells. Similarly, transformed GCN2 deficient MEFs failed to induce ATF4 following MYC activation (Fig. 2c). Interestingly, we observed higher activation of PERK in the absence of ATF4 suggesting increased ER stress in these cells (Fig. 2c). Because GCN2 deficient cells still displayed residual p-eIF2α, yet ATF4 protein was absent compared to GCN2 proficient cells, we hypothesized that GCN2 may be also regulating ATF4 at the transcriptional level. Indeed, induction of MYC resulted in a significant upregulation of ATF4 mRNA only in GCN2 proficient cells, though another well-characterized MYC target gene, ornithine decarboxylase (ODC1) was activated independently of GCN2 (Supplementary Fig. 1b). These results indicate that GCN2 is required not only for efficient ATF4 protein expression but also contributes to its mRNA transcription in response to MYC activation.
MYC increases the ratio of uncharged tRNAs leading to activation of GCN2.
Amino acid deficiency leads to accumulation of uncharged tRNAs which in turn bind to GCN2 resulting in its autophosphorylation and activation3. Activated GCN2 subsequently phosphorylates eIF2α, resulting in transient inhibition of general protein synthesis during amino acid deprivation conditions21. Therefore, we sought out to determine the mechanism of robust phosphorylation of GCN2 following MYC activation in cells grown in complete media (Fig. 2a). MYC has been shown to robustly enhance transcription of tRNAs in RNA POL III-dependent manner22 suggesting that newly synthesized, uncharged tRNAs might activate GCN2. To test this, we pretreated cells with a specific RNA polymerase III inhibitor ML6021823 to suppress tRNA synthesis prior to MYC activation. ML60218 markedly reduced GCN2 phosphorylation, indicating that RNA POL III mediated transcription of tRNAs is required for MYC activation of GCN2 (Fig. 2d).
RNA polymerase III regulates the transcription of other small structured RNAs including tRNAs22. To directly analyze the levels of aminoacyl-tRNAs following MYC activation, we used tRNA-tailored microarrays that distinguish charged vs uncharged tRNAs24. We found a striking and time-dependent increase in the levels of multiple uncharged tRNAs following MYC activation (Fig. 2e). There was also a pronounced increase in the overall abundance of tRNAs (Supplementary Fig. 1c); principal component analysis (PCA) revealed these changes were strongly associated with tRNAs charged with hydrophobic amino acids and in particular, with branched amino acids (Supplementary Fig. 1d). As expected, leucine deprivation led to increased uncharging of all leu tRNA isoacceptors, demonstrating the specificity of the assay (Fig. 2e). Furthermore, inhibition of RNA polymerase III markedly increased the fraction of charged tRNAs (Supplementary Fig. 1e) by reducing the total amount of all tRNAs (Supplementary Fig. 1f). These data indicate that MYC-induced RNA polymerase III dependent transcription of tRNAs leads to transient accumulation of uncharged tRNAs, thereby activating GCN2.
ATF4 and MYC have common (overlapping) DNA binding sites.
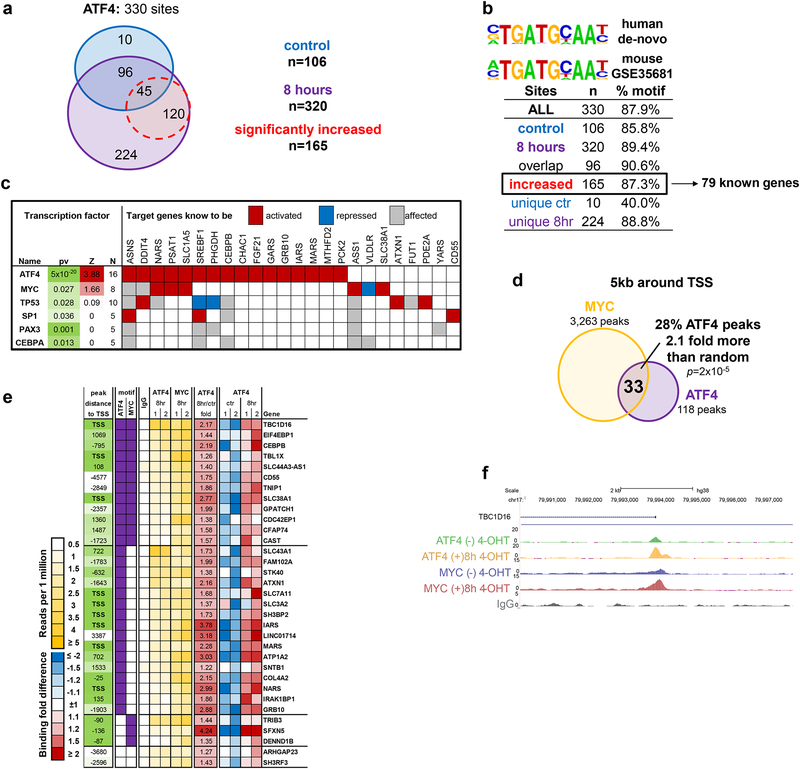
Since ATF4 is critical for survival following MYC activation (Fig. 1c–e), we performed chromatin immunoprecipitation (ChIP)-sequencing in DLD-1:MycER cells with or without MYC activation to map genes bound by ATF4 on a genome-wide scale. We identified 330 unique ATF4 binding sites that had ATF4 ChIP-seq signal significantly enriched (FDR<5%, at least 4-fold) over IgG control (Fig. 3a). Approximately 50% of the binding sites (165 out of 330) were significantly increased following MYC activation for 8 hrs (Fig. 3a). De-novo motif search revealed that ~90% of identified ATF4 binding sites contained a previously established mouse ATF4 binding motif25 (GSE35681) (Fig. 3b). From the 165 ATF4 binding sites, we identified 79 genes (with Entrez ID) where ATF4 occupied within 5kb from at least one gene transcription start site (TSS). Of this subset, 16 genes are previously well-characterized ATF4 targets4, 25 (Supplementary Fig. 2a). Analysis of the 79 gene list for functional and pathway enrichment showed significant overrepresentation of 11 functional categories and 10 pathways (Supplementary Fig. 2b). As expected, one of the pathways was the UPR confirming the well-characterized role of ATF4 in this pathway. Other key functional categories were amino acid transport and biosynthesis and tRNA synthetases underscoring the important role of ATF4 in protein synthesis (Supplementary Fig. 2b). We then analyzed the gene list for enrichment of known transcriptional regulators to identify potential ATF4 co-regulators (Fig. 3c). As expected, ATF4 itself was the top significant hit with 16 known targets and a Z-score indicating positive regulation (Z=3.88, based on the majority of targets being upregulated by ATF4). Intriguingly, the only other significant transcriptional co-regulator was MYC with a Z score of at least 1.5, higher than additional factors such as p53, SP1 and Pax3 that also had significant enrichment of targets.
Figure 3. ATF4 and MYC bind to common target genes.
a. ATF4 ChIP-seq identified 330 binding sites for ATF4. Binding sites occupied by ATF4 without 4-OHT and with 4-OHT are shown in blue and purple respectively. Binding sites that are significantly increased after 8 hours of MYC activation are shown in red. Significance of binding signal increase was estimated by HOMER algorithm (FDR<5%). b. Motif enriched within 330 ATF4 binding sites (human de-novo) and within ATF4 mouse experiment from GSE35681 dataset23. Table shows percent of binding sites containing the motif for different site groups. c. Transcription factors whose known targets were enriched among genes bound by ATF4, Z=z-score for predicted transcription activation state calculated by IPA based on number of targets shown to be activated by the TF. d. Overlap and its significance estimated by hypergeometric test of MYC and ATF4 bound genes in 8hr samples: sites co-bound by ATF4 and MYC within 5kb from genes’ TSS. e. List of all 33 genes from panel (d) that were bound by ATF4 and MYC at 8hrs of MYC activation. f. ChIP-seq signal track within TBC1D16 locus showing ATF4 and MYC binding peaks. ChIP-seq was performed twice in two independent experiments with similar results.
We then performed ChIP-seq for MYC to map its global binding following stimulation with 4-OHT in the same cells. This experiment identified 3263 peaks at 8hr of MYC induction that were within 5kb from a gene TSS, 33 of which overlapped with ATF4 binding sites (28% of all ATF4 peaks), a significant overlap (2.1 more than expected by chance, p=2×10−5 by hypergeometric test) suggesting similar mechanism regulating a significant subset of genes by both MYC and ATF4 (Fig. 3d). Moreover, the promoters of genes with functions in amino acid transport (SLC7A11, SLC38A1, SLC43A1) and tRNA charging (IARS, MARS, NARS) were occupied by both ATF4 and MYC (Fig. 3e). We validated the ChIP-seq results by ChIP-qPCR (Supplementary Fig. 2c). A ChIP-seq profile at a representative MYC and ATF4 target loci, TBC1D16, (Fig. 3e) shows overlapping binding of both MYC and ATF4 (Fig. 3f). The ChIP-seq result suggests that MYC and ATF4 share common target genes, most of which are involved in amino acid transport and tRNA charging process.
ATF4 suppresses mTORC1 dependent signaling to prevent proteotoxicity following MYC activation.
We then sought to determine the mechanism by which ATF4 exerts its pro-survival effects following MYC dysregulation. Given the known role of ATF4 in amino acid metabolism, antioxidant response, and fatty acid synthesis, we attempted to rescue ATF4 deficient cells by supplying metabolites in trans in culture media under conditions of MYC activation4, 5, 26. ATF4 deficient MEFs require the presence of non-essential amino acids and antioxidants to grow4. Supplementaiton with additional antioxidants or long chain fatty acids did not rescue ATF4 deficient cells (Supplementary Fig. 3a, 3b). Notably, supplementation with alpha-ketoglutarate (αKG), an intermediary of the TCA cycle, delayed apoptosis during earlier (16hr) but not later (24h) time points of MYC activation in ATF4 deficient MEFs (Supplementary Fig. 3a, 3c). These results suggest that MYC activation imposes metabolic stress which can be partially relieved by αKG. However, additional ATF4-dependent processes must be activated to maintain long-term cell viability.
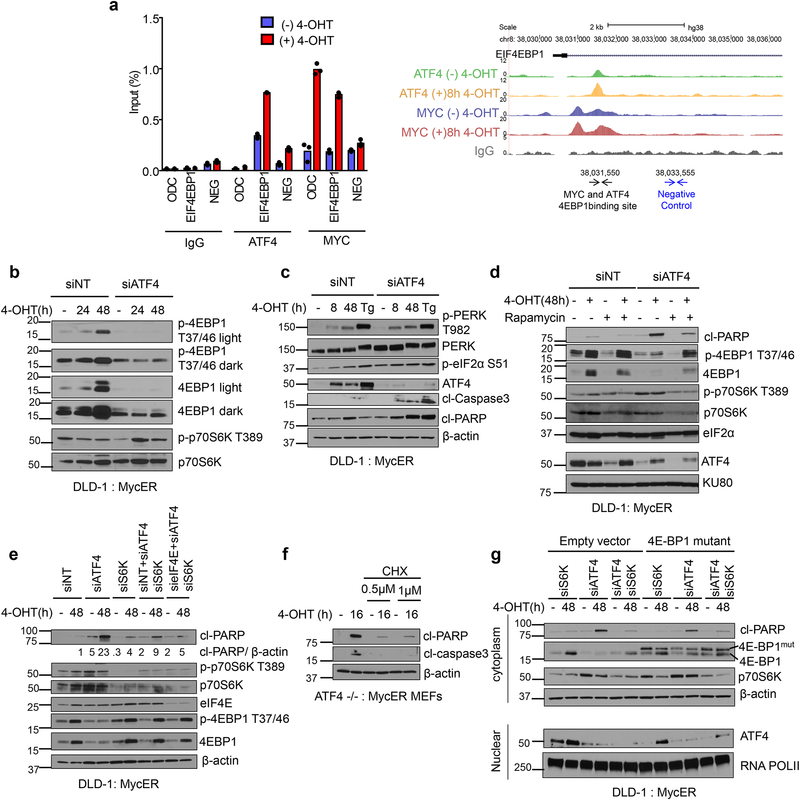
One of the targets identified by ChIP-seq to be bound by both MYC and ATF4 was the negative regulator of the major cap-binding protein eIF4E, EIF4EBP1 (4E-BP1), a downstream substrate of mammalian target of rapamycin complex 1 (mTORC1) (Fig 3e, Fig. 4a). mTORC1 integrates oncogenic stimuli into protein synthesis and cell growth signaling27 by promoting phosphorylation of p70S6K and 4E-BP128. Hyperphosphorylation of 4E-BP1 by mTORC1 leads to its dissociation from eIF4E, enhancing cap-dependent protein translation downstream of eIF4E29. We confirmed that both ATF4 and MYC bind to the intron of 4E-BP1 (Fig. 4a). Moreover, 4E-BP1 mRNA was reduced in the absence of ATF4 in both DLD-1 and MEFs (Supplementary Fig. 4a, 4b). We also observed that MYC activation enhanced levels of both phosphorylated and total 4E-BP1 (Fig. 4b, Fig. 4d, Fig. 4e, and Supplementary Fig. 4c), yet there was a marked reduction in 4E-BP1 accumulation in the absence of ATF4 (Fig. 4b, Fig. 4d, Fig. 4e, and Supplementary Fig. 4c) indicating that ATF4 is required for MYC-induced 4E-BP1. Intriguingly, loss of ATF4 resulted in upregulation of PERK phosphorylation (Fig. 4c), indicating ER stress activation. These results strongly suggest that 4E-BP1, downstream of the MYC-ATF4 axis, acts as a rheostat to set a protein synthesis level compatible with MYC oncogenic activity. ATF4 deficient cells also exhibited sustained phosphorylation of p70S6K (Fig. 4b and Supplementary Fig. 4c). We next examined whether mTORC1 suppression in ATF4 deficient cells impacted cell death during MYC activation. Indeed, inhibition of mTORC1 by rapamycin treatment led to a marked decrease in MYC-dependent apoptosis and enhanced clonogenic survival in ATF4 deficient cells (Fig. 4d, Supplementary Fig. 4c and Supplementary Fig. 4d).
Figure 4. ATF4 suppresses mTORC1 dependent signaling and inhibition of mTORC1 reduces cell death of ATF4 deficient cells following MYC activation.
a. ATF4 and MYC ChIP followed by qPCR at the EIF4EBP1 locus, n=3, technical replicates. Right schematic shows ATF4 and MYC peaks from ChIP-seq and locus for qPCR primers used. ChIP-seq was performed twice independently with similar results, neg=negative control. b. Representative Immunoblot of DLD-1, MycER cells assessing mTORC1 signaling in cells transfected with non- targeting siRNA or ATF4 siRNA. c. Representative immunoblot assessing ISR signaling in cells transfected with non-targeting siRNA or ATF4 siRNA. Tg, thapsigargin (0.5μM for 4hrs). d. Representative immunoblot of DLD-1 cells pretreated for 2 hours with 200nM Rapamycin followed by MYC activation. e. DLD-1: MycER cells were transfected with non-targeting siRNA or indicated siRNAs, and proteins were assessed by immunoblotting after MYC activation. f. ATF4−/−, MycER MEFs were pretreated with Cyclohexamide (CHX) for indicated times followed by MYC activation. g. DLD-1: MycER cells were transfected with non-targeting siRNA or indicated siRNAs, and proteins were assessed by immunoblotting after MYC activation. All immunoblots are representative of three biological replicates that showed similar results. Unprocessed scans of blots are shown in Supplementary Fig 7.
To further probe the link between ATF4 and mTORC1 effectors, we knocked down p70S6K and eIF4E, two major translational regulators downstream of mTORC1 signaling in the absence of ATF4. Consistent with the rapamycin treatment results, knockdown of p70S6K reduced cell death due to ATF4 deficiency when MYC was activated (Fig. 4e). Interestingly, combined knockdown of S6K and ATF4 resulted in feedback upregulation of 4E-BP1 and p-4EBP levels (Fig. 4e). Additionally, knockdown of both p70S6K and eIF4E further reduced cell death in the absence of ATF4 (Fig. 4e). This suggests that ATF4 is required for defenses against proteotoxicity in the context of hyperactive MYC-mTORC1 signaling funneled through p70S6K and eIF4E.
In support of this notion, inhibiting protein synthesis with low doses of cycloheximide led to a similar reduction in cell death in ATF4 deficient cells (Fig. 4f). However, we did not observe a general increase in rate of translation by 35S-Methionine and Cysteine labeling in the absence of ATF4, suggesting that synthesis of specific proteins rather than global translation is deregulated in ATF4 deficient cells and contributes to ER proteotoxicity (Supplementary Fig. 4e). Furthermore, treatment with a chemical chaperone 4-Phenylbutyric acid (4-PBA) which can protect against proteotoxicity under other stress contexts30, also promoted survival of ATF4 deficient cells following MYC activation (Supplementary Fig. 4f). Since 4E-BP1 levels were reduced in the absence of ATF4, we overexpressed a mutant version of 4E-BP1 that cannot be phosphorylated and inactivated by mTORC131. Expression of the 4E-BP1 mutant led to decreased apoptosis in ATF4 knockdown cells (Fig. 4g) which was further reduced when combined with knockdown of S6K (Supplementary Fig. 4g). Collectively, these data indicate that inhibiting both branches of mTORC1 signaling in ATF4-deficient cells reduces the demand of specific protein synthesis and promotes adaptation to hyperactive MYC-induced gene expression, mitigating proteotoxicity.
Inhibition of both PERK and GCN2 promotes survival of MYC driven lymphoma bearing mice.
To test the importance of the GCN2-eIF2α-ATF4 pathway in vivo, we used a well characterized mouse model of MYC-driven lymphoma, Eμ-myc32 which harbors MYC coupled to the IgH enhancer and succumb to aggressive B-cell lymphoma between 6–15 weeks of age32. Gcn2−/− mice are viable and fertile, unlike Atf4−/− mice which show several pathological abnormalities in utero or shortly after birth33–35. We generated Eμ-myc/+; GCN2+/+, Eμ-myc/+; GCN2+/− and Eμ-myc/+; GCN2−/− mice (Supplementary Fig. 5a). Surprisingly, loss of GCN2 did not affect tumor initiation or progression or impact overall mouse survival (Supplementary Fig. 5b). Assessment of ISR signaling in B cells from each genotype revealed robust PERK and eIF2α phosphorylation and ATF4 upregulation in B cell lymphoma compared to normal B cells from littermate controls, suggesting that PERK is likely compensating for loss of GCN2 (Supplementary Fig. 5c).
To test this notion, we transplanted either Eμ-myc/+; GCN2+/+ or Eμ-myc/+; GCN2−/− lymphomas (Supplementary Fig. 5d) into C57BL/6J mice and targeted PERK by using a potent and specific PERK inhibitor, LY-436. LY-4 treatment was well tolerated and did not affect body weight or pancreas weight37 but significantly reduced phosphorylation of PERK (Supplementary Figs. 5e, 5f and 5h). Inhibition of PERK in mice bearing Eμ-myc/+; GCN2−/− lymphomas, but not Eμ-myc/+; GCN2+/+ lymphomas, increased overall survival (Supplementary Fig. 5g). Consistent with our in vitro data, inhibition of both PERK and GCN2 significantly reduced phosphorylation of eIF2α in the lymphoma cells (Supplementary Figs. 5h and 5i). Unexpectedly, inhibition of PERK and GCN2 did not reduce ATF4 protein levels, suggesting an eIF2α-independent regulation of ATF4 in the Eμ-myc lymphoma model (Supplementary Fig. 5h). One possible explanation for the modest increase in survival of these mice is the decreased phosphorylation of eIF2α which can exacerbate ER stress in tumor cells due to unregulated protein translation38. These results underscore a strict selective pressure for maintenance of robust ATF4 levels in MYC-driven tumors.
Acute ablation of ATF4 in lymphoma cells significantly enhances tumor-free survival and overall survival.
The results with the Eμ-myc/+; GCN2−/− mice indicated that maintenance of ATF4 may be important for tumor progression. Therefore, we generated conditional knockout ATF4fl/fl mice and crossed the Eμ-myc/+ with Rosa26-CreERT2/+ mice39, which express a tamoxifen inducible Cre recombinase enzyme. Eμ-myc/+; Rosa26-CreERT2/+ were crossed with ATF4fl/fl mice to generate Eμ-myc/+; ATF4wt/wt; Rosa26-CreERT2/+ and Eμ-myc/+; ATF4fl/fl; Rosa26-CreERT2/+ genotypes (Supplementary Fig. 6a and Supplementary Fig. 6b). Following lymphoma onset, lymphoma cells were purified and allografted by intravenous injection into C57BL/6 syngeneic mice. The recipient mice were treated either with vehicle or tamoxifen to excise ATF4 specifically in the lymphoma cells (Fig 5b). Remarkably, excision of ATF4 significantly extended lymphoma-free and overall survival (Fig. 5b, 5c). This effect was not due to tamoxifen treatment because mice with Eμ-myc/+; ATF4wt/wt; Rosa26-CreERT2/+ lymphomas displayed no significant difference in lymphoma-free or overall survival (Supplementary Fig. 6c and Fig. 5c).
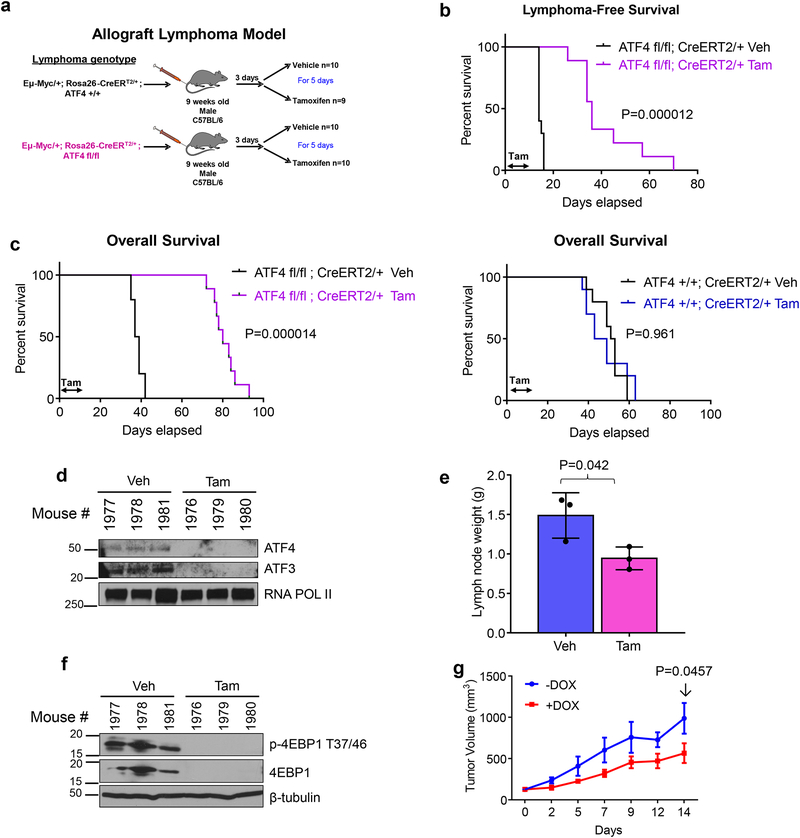
Figure 5. Acute ablation of ATF4 significantly delays MYC driven Lymphomagenesis and promotes survival of MYC driven lymphoma bearing mice.
a. Schematic showing allograft lymphoma model, where lymphoma cells are injected via tail vein into 9-weeks old C57BL/6J mice. Three days after lymphoma engraftment, mice are randomized to receive either vehicle or tamoxifen treatment by oral gavage for 5 days. b. Kaplan-Meier analysis for lymphoma-free survival of mice bearing Eμ-myc; ATF4fl/fl lymphoma treated with either vehicle (veh) or tamoxifen (tam) for 5 days. n=9 for Eμ-myc; ATF4fl/fl lymphoma tamoxifen group, all other groups n=10. Kaplan-Meier curves were analyzed by two-tailed log-rank test. c. Kaplan-Meier analysis for overall survival of mice bearing Eμ-myc; ATF4fl/fl lymphoma (left) or Eμ-myc; ATF4+/+ lymphoma (right). Kaplan-Meier curves were analyzed by two-tailed log-rank test. d. Immunoblot of B cells isolated from Eμ-myc; ATF4fl/fl lymphoma bearing mice a day after the last day of tamoxifen treatment. e. Lymph node weight of mice in panel d, n=3 per group, error bars represent mean ± SD, two tailed student t-test. f. Immunoblot of lymphoma lysates from mice in panel e. For d and f, lysates from three different mice per treatment were used. g. DLD-1, MycER, i-shATF4 cells were transplanted into 12-week-old nude mice and tumor size is shown. One (−) Dox mouse had to be sacrificed on day 12 because tumor reached maximum size limit. n=4 (−) Dox and n=4 (+) Dox. Two-way ANOVA, error bars represent mean± SEM. Unprocessed scans of blots are shown in Supplementary Fig 7.
To further assess the requirement for ATF4 in lymphoma progression, we treated a separate cohort of mice as above once lymphoma developed. Treatment with tamoxifen resulted in efficient ablation of ATF4 mRNA (Supplementary Fig. 6d) and protein as well as reduction in ATF3, an ATF4 target gene (Fig. 5d) and resulted in reduced lymph node weight (Fig. 5e). Consistent with our in vitro data (Fig. 4b and Supplementary Fig. 4c), 4E-BP1 levels were significantly reduced in lymphoma cells following ATF4 ablation (Fig. 5f). However, ATF4 expression returned to levels comparable to those of vehicle-treated groups by the endpoint of the experiment, suggesting that lymphoma cells that escaped ATF4 excision eventually formed lymphomas (Supplementary Fig. 6e).
Similar to the lymphoma model, knockdown of ATF4 in DLD-1:MycER cells expressing a doxycycline inducible short-hairpin RNA against ATF4, significantly delayed tumor growth (Fig. 5g). Collectively, the in vivo data support our hypothesis that ATF4 is critical for tumor growth driven by hyperactive MYC and indicate that targeting ATF4 in the context of activated MYC elicits potent anti-tumor effects.
We next assessed the levels of ISR signaling in B cells isolated from WT, pre-lymphoma and lymphoma bearing Eμ-myc mice. We noted higher expression of ATF4 in lymphoma samples compared to WT and pre-lymphoma samples (Fig. 6a). Interestingly, levels of p-eIF2α, p-4E-BP1, 4E-BP1 and p-p70S6K were higher in both pre-lymphoma and lymphoma samples (Fig. 6a). Human Burkitt’s lymphoma cell lines also displayed robust increases in ISR signaling and 4E-BP1 levels (Fig. 6b). Similar results were seen in human breast and colon cancer cell lines compared to a human immortalized breast epithelial cell line and normal human colonocytes respectively (Fig. 6c and Fig. 6d).
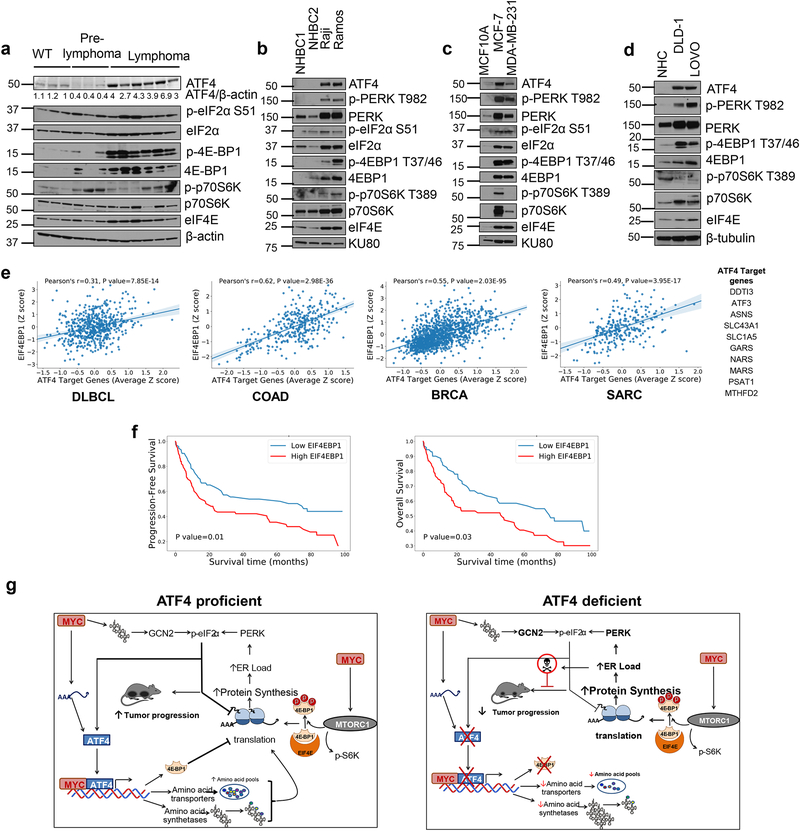
Figure 6. EIF4EBP1 positively correlates with ATF4 target gene expression and associates with poor prognosis.
a. Immunoblot of B cells isolated from WT (n=3), pre-lymphoma Eμ-myc (n=3) or lymphoma bearing Eμ-myc mice (n=6), lysates from different mice per group were used. b. Immunoblot from two normal human B cells (NHBC) and Burkitt’s lymphoma cell lines. c. Immunoblot from normal human colonocytes (NHC) or colon cancer cell lines. d. Immunoblot from MCF10 (non-tumorigenic breast epithelial cell line) or breast cancer cell lines. e. Pearson correlation between EIF4EBP1 and ATF4 target gene expression in Diffused Large B cell Lymphoma (DLBCL, n=562), Colorectal Adenocarcinoma (COAD, n=329), Breast Cancer (BRCA, n=1218) and Sarcoma (SARC, n=265). The center lines depict linear regression lines and shaded regions are 95% confidence intervals for regression lines. Datasets analyzed are listed in the methods section. Previously known ATF4 target gene list used in this analysis is shown in table. f. Kaplan-Meier plots of progression free survival (left) and overall survival (right) of DLBCL patients with high or low EIF4EBP1 expression. The survival analysis using Kaplan-Meier and two-sided log-rank test between high and low EIF4EBP1 expression groups was performed on all the patients with records of progression-free survival (left, n= 173) and overall survival (right, n=171), respectively. g. Proposed model of ATF4 and MYC co-operation in tumor progression. All immunoblots are from n=1 and unprocessed scans of blots are shown in Supplementary Fig 7.
To investigate the relevance of our findings in human malignancy, we analyzed the expression of 4E-BP1 and tested its correlation with ATF4 activity in different cohorts of cancer patients by mining the TCGA database. Since ATF4 is primarily regulated at the translational level, we used a group of 10 well-characterized ATF4 targets (including ASNS, MTHFD2, CHOP) as a surrogate for ATF4 activation4, 25. We found that 4E-BP1 levels displayed a significant positive correlation with ATF4 target genes in DLBCL, colorectal cancer, breast cancer and sarcoma (Fig. 6e). Notably, high expression of 4E-BP1 also correlated with poor prognosis in DLBCL patients (Fig. 6f). These data further support the notion that ATF4-dependent modulation of MYC-driven translational alterations may play a role in progression of MYC-driven lymphomas.
Discussion
MYC activation is invariably linked with enhanced protein synthesis critical for enhanced proliferation and manifestation of MYC’s tumorigenic properties40. However, increased translation places heightened demands on the ER, the site for glycosylation and folding of client proteins41. While PERK-mediated phosphorylation of eIF2α can provide initial relief from increased translation rates11, this mechanism may be insufficient for long-term survival due to a negative feedback loop that promotes dephosphorylation of eIF2α42. This work demonstrates that ATF4 is critical for MYC-dependent tumor progression by not only supporting protein synthesis via expression of amino acid transporters and tRNA synthetases but also by preventing “runaway” mRNA translation and subsequent proteotoxicity via prolonged upregulation of 4E-BP1 levels (Fig. 6g).
Since ablating ATF4 expression did not increase global translation levels following MYC dysregulation, it is likely that ATF4 negatively regulates the expression of a specific set of proteins that traffic through the ER. This notion is supported by the following findings: (a) Absence of ATF4 in a background of MYC activation results in robust PERK phosphorylation, a sensitive sensor of ER stress; (b) Rapamycin or CHX treatment rescues MYC-driven, ATF4-deficient cells from apoptosis; and (c) A chemical ER chaperone rescues ATF4 deficient cells. In the future, it will be important to identify the specific mRNAs that are selectively translated under the control of the ATF4–4E-BP1 axis.
4E-BP1 has been shown to inhibit cap dependent translation and to promote survival under conditions of nutrient and oxidative stress43. Interestingly, higher expression of 4E-BP1 has been noted in multiple cancers44–46 and its high expression is associated with poor prognosis45–47. While ATF4 has been shown to regulate 4E-BP1 in the context of extrinsic physiologic stresses48, 49, its role in regulating 4E-BP1 in the context of MYC-deregulated tumor progression had not been previously reported. Higher expression of 4E-BP1 seems paradoxical, as a hyperactive form of 4E-BP1 blocks MYC-induced tumorigenesis50. Indeed, our findings provide a novel context as to why 4E-BP1 expression is upregulated in tumors: rather than downregulating its levels, tumors maintain high levels of 4E-BP1 whose activity can be readily modulated by rapid phosphorylation-dephosphorylation depending on favorable/unfavorable growth and proliferation conditions.
Our study also highlights the importance of using combined inhibition of both PERK and GCN2 in MYC driven cancers to avoid compensation of ISR kinases in regulating p-eIF2α levels. Indeed, pharmacologic targeting of eIF2α phosphorylation in patient-derived prostate xenograft models is effective in initiating cytotoxic response inhibiting tumor growth38. However, in the lymphoma model tested here, ATF4 levels remained high despite the near complete inhibition of upstream canonical pathways (potentially via translational regulation by mTOR)51 and acute ATF4 ablation significantly decreased tumor growth and prolonged survival. This strongly implies that for certain malignancies, it might be essential to directly inhibit ATF4 activity for optimal antitumor outcome. Taken together, the ISR transcription factor ATF4 is necessary for supporting and censoring protein translation thus promoting progression of MYC driven tumorigenesis.
Methods
Cell culture and reagents.
Human colon adenocarcinoma cell line DLD-1, Burkitt’s lymphoma cell lines Raji and Ramos as well as human breast cancer cell lines, MCF10A, MCF-7 and MDA-MB-231 were purchased from American Type Culture Collection (ATCC). DLD-1 cells were maintained in DMEM (Invitrogen #21063–029) supplemented with 10% FBS (Sigma) and 1% penicillin-streptomycin (Invitrogen). P493–6 cells were provided by Dirk Eick, German Res. center for Environmental Health, Helmholtz center, Munich. P493–6, Raji and Ramos were maintained in RPMI-1640 supplemented with 10% FBS and 1% Penicillin-Streptomycin. MYC expression was turned off in P4936 cells by adding 0.1μg/ml Tetracycline. Wild-type and Gcn2-knockout, Sv40 immortalized, mouse embryonic fibroblasts (MEFs) were purchased from ATCC. Atf4-knockout Sv40 immortalized MEFs were a gift from David Ron (Cambridge Institute for Medical Research (CIMR). Wild-type, Atf4-knockout, Gcn2-knockout, Sv40 immortalized, MEFs were maintained in DMEM supplemented with 10% FBS, 1% Penicillin-Streptomycin, 1x non-essential amino acids (Invitrogen, #11140050) and 55 μM β-mercaptoethanol (Millipore, #ES-007-E). DLD-1 cells and MEFs were infected with retroviruses expressing MycER. MycER was induced with 250nM or 500nM of 4-hydroxytamoxifen (4-OHT, Sigma-H7904) in MEFs or DLD-1 cells, respectively. All cells were determined to be free of mycoplasma and cultured in 5% CO2 at 37°C. Normal human colonocytes were purchased from Cell Applications Inc (732Cn-10a). Normal primary B cells were purchased from the Human Immunology Core at University of Pennsylvania. RNA POL III inhibitor ML60218 (577784-91-9), cyclohexamide (C7698) and rapamycin (R8781) were purchased from Sigma. 4PBA (130380250) was purchased from Fisher Scientific. siRNAs against ATF4 (L-005125–00), PERK (L-004883–00), GCN2 (L-005314–00), p70S6K (L-003616–00), eIF4E (L-003884–00) and non-targeting siRNA (D-001810–10) control were purchased from Dharmacon. LY-4 was provided by Eli Lilly.
Plasmids and retroviruses.
MycER plasmid was provided by Dr. Andrei Thomas-Tikhonenko (The Children’s Hospital of Philadelphia). Retrovirus construct and packaging plasmids, pEco-clontech (for mouse cells), pAmpho-clontech (for human cells) and gag pol were co-transfected into 293T cells by lipofectamine 2000 (Invitrogen), according to manufacturer’s instructions. Retroviral supernatant was collected 48–72hours post transfection and supplied with 8μg/ml polybrene (28728-55-4 Sigma) to infect target cells. Lentiviral TRIPZ inducible shRNA against ATF4 plasmid was purchased from Dharmacon (V3THS_132755). Lentivirus was made in 293T cells by co-transfecting lentiviral vector and packaging plasmids. Lentiviral supernatant was collected 48–72 hours post transfection and used to infect target cells. The dominant negative mutant of 4EBP1 (DN-4EBP1) cDNA has been previously described52. The DN-4EBP1 cDNA was subcloned into pMSCV-hygro to generate a constitutive mammalian expression vector with N-terminal flag tag (4E-BP1 Mutant) used in this study. Retrovirus was made as described earlier to transfect DLD-1, MycER cells. Transduced cells were selected with 400μg/ml Hygromycin to make stable cell lines.
Immunoblotting and antibodies.
Cells were harvested in ice cold PBS and nuclear and cytoplasmic fractions were isolated by BioVision Nuclear Cytoplasm fractionation kit according to manufacturer’s instructions (BioVision K266). RIPA buffer supplied with protease inhibitors (Roche 11836153001) and phosphatase inhibitors (p5726, p0044, Sigma) was used for whole lysate isolation. Protein concentrations were determined by DC protein assay (BioRad). Equal protein was loaded and resolved on to sodium dodecyl sulfate polyacrylamide gels and transferred to polyvinylidene fluoride membranes. All antibodies were incubated overnight at 40C overnight in 5% TBS (20mM Tris-Base and 150mM NaCl), 0.1% Tween-20. Membranes were washed and incubated with secondary antibodies for 1 hour. Membranes were exposed to autoradiography films after washing with TBS, 0.1% tween. The following antibodies were used for detecting proteins: ATF4 (cat# sc-200x, c-20), MYC (cat# sc-764, N-262), ATF3 (cat# sc-188, C-19) were purchased from Santa Cruz. Rabbit β-tubulin (2146), mouse cl-PARP (mouse specific, #9548, 7C9), rabbit cl-PARP (human specific, cat#9541), rabbit cl-caspase3 (cat# 9661), rabbit GCN2 (cat# 3302), rabbit PERK (cat# 3192, C33E10), rabbit p-PERK T980 (cat# 3179,16F8), rabbit p-4EBP1 T37/46 (cat# 2855, 236B4), rabbit 4EBP1 (cat#9452), rabbit eIF4e (cat#9742), rabbit p-p70S6K T389 (cat# 9205), rabbit p70S6K (cat#2708, 49D7), rabbit p-eIF2α S51 (cat# 3597, 119A11) and rabbit eIF2α (cat# 9722) were all purchased from Cell Signaling Technology. Rabbit p-PERK T982 was provided by Eli Lilly. Rabbit p-GCN2 T899 (cat# ab 75836) was purchased from Abcam. Mouse β-actin (cat#A5441, AC-15) was purchased from Sigma. Mouse RNA POL II (cat# 39097) was purchased from Active Motif. Horseradish peroxidase- conjugated secondary antibodies, goat anti mouse (cat#31430) and goat anti rabbit (cat#31460) were purchased from Thermo Scientific.
Clonogenic survival assay.
Cells were grown in complete media in 60mm dishes or in 6-well plates. The following day, cells were treated with 4-OHT. Media was changed after 24 hours. Colonies that formed after a week were fixed with a solution of 10% methanol and 10% acetic acid and then stained with 0.4% crystal violet in 20% ethanol.
35S Methionine and Cysteine labeling.
Cells were plated in 6 well dish. The following day, cells were treated with 4-OHT for indicated times and cells were labeled with 50μCi Met/Cys Express Mix (PerkinElmer, NEG772014MC) for 1hr in Methionine and cysteine free DMEM (21013024, Invitrogen). Cells were washed with ice cold PBS and cell lysates were harvested for protein. Equal protein was loaded and resolved on to sodium dodecyl sulfate polyacrylamide gels and transferred on to polyvinylidene fluoride membranes. Membranes were exposed to autoradiography films. 35S incorporation was quantified by Image J software. β-actin was used as a loading control.
Magnetic isolation of mouse B cells.
Spleens and lymph nodes were excised from euthanized mice and immediately passed through 70μm cell strainer. Normal splenic B cells as well as B cells in lymphoma were isolated by using mouse B cell isolation kit (130-090-862, Miltenyi Biotec) according to manufacturer’s instructions.
Quantitative Real Time PCR.
RNA was harvested for qPCR analysis by using Qiagen RNeasy Mini Kit (74104) according to manufacturer’s instructions. RNA was reverse transcribed by using AMV reverse transcriptase (Promega, M5108) in the presence of RNase inhibitor (Promega, N2111). qPCR was performed with Power SYBR green PCR master mix (Applied Biosystems, 4367659). QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems) was used for data analysis. Primers used for qPCR are listed in Supplementary Table 1.
Apoptosis analysis.
Apoptosis was detected using the annexin V apoptosis detection kit (Invitrogen, # A23204) according to the manufacturer’s instructions. Briefly, Cells were collected and 1×105 cells were resuspended in 100μl annexin-binding buffer. 5μl of the annexin V conjugate was added to each 100μl of cell suspension and incubated at room temperature for 15 minutes. An additional 400μl of annexin-binding buffer was added to cells. Cells were immediately analyzed on BD FACS Canto flow cytometer.
tRNA charging microarray.
About 8 million DLD-1, MycER cells were treated with 500nM 4-OHT for 2h, 4h, 8h or EtOH was used as vehicle. To determine the fraction of charged tRNAs, total RNA was isolated in mild acidic conditions using acidic phenol (pH 4.5) whose low pH preserves the aminoacyl-moiety. Each sample was split into two aliquots and one was oxidized with periodate which oxidizes the free non-aminoacyl groups leaving the changed tRNAs intact. Following subsequent deacylation the amino acid-protected tRNAs hybridize to Cy3-labeled RNA/DNA stem-loop oligonucleotide. The second aliquot was deacylated in 100 mM Tris (pH 9.0) at 37°C for 45 min and hybridized to Atto647-labeled RNA/DNA stem-loop oligonucleotide and designated as total tRNA. Both aliquots were loaded on tRNA microarrays with tRNA probes covering the full-length sequence of cytoplasmic tRNA species as described previously53. For tRNA abundance, total RNA was isolated with TriReagent (Sigma Aldrich) which alkaline pH simultaneously deacylates all tRNAs. The tRNAs were subsequently labeled with by ligating Cy3-labeled RNA/DNA stem-loop oligonucleotide. The tRNAs isolated from the vehicle treated sample were labeled with Att647-labeled RNA/DNA stem-loop oligonucleotide, loaded on all arrays and all other samples were compared to it. The arrays were normalized to spike-in standards which were present in equimolar rations in both Cy3- and Atto647-labeled aliquots. The array processing and quantification was performed with in-house python and R scripts. The data was submitted to the Gene Expression Omnibus (GEO) database and can be accessed using accession number GSE116812.
Chromatin immunoprecipitation.
ChIP was performed as previously described54. Chromatin fragments were prepared from DLD-1: MycER cells following 8hr of MYC activation (EtOH or 4-OHT treatment). DNA was sonicated using Covaris 200 instrument at settings of Temp 5–9, PP200, DF 10, CB 200, 720sec. 50μg DNA was used for each ChIP. The following antibodies were used to perform ChIP at a concentration of 5μg: ATF4 (cat# sc-200x, lot# G0115, c-20), MYC (cat# sc-764, lo#D0413, N-262). Rabbit IgG (cat# sc-2027x, lot# G2516) was used as a negative control. All antibodies were purchased from Santa Cruz Biotechnology. Protein G Dynabeads (10003D) were purchased from Invitrogen. Primers used for qPCR are listed in Supplementary Table 1.
Library preparation, sequencing and analysis.
ChIP DNA from two independent experiments was submitted to Wistar genomics core, (Wistar institute, Philadelphia) for library production using NEBNext Ultra II DNA Library Prep Kit for Illumina #E7645S and NEBNext Multiplex Oligos for Illumina #E7335S according to manufacturer’s instructions. The library fragments were assessed on an Agilent Technologies 2100 Bioanalyzer and yielded 150–350bp products. Illumina NextSeq 500 instrument was used for sequencing. Select targets were validated by qPCR and primers used for qPCR are listed.
CHIP-seq data was aligned using bowtie55 against hg38 version of the human genome and HOMER software56 was used call significant peaks against IgG control or between corresponding replicate pairs of samples using–style factor option and only uniquely aligned reads with duplicates removed. Significant peaks that passed FDR<5% threshold and at least 4 fold over IgG control were used to identify unique binding sites. Only results significant in comparison for both replicates were considered. De-novo motif analysis was performed using HOMER software among the list of sites with significant ATF4 binding in at least one condition (both replicates). Overlap of binding sites with genes was done using Ensemble 84 transcriptome database. Genes with a binding site within 5kb from TSS were considered. Significance of overlap was tested using hypergeometric test using 23,869 Ensemble genes with Entrez ID as a population size. Gene set enrichment analysis of gene sets was done using QIAGEN’s Ingenuity® Pathway Analysis software (IPA®, QIAGEN Redwood City,www.qiagen.com/ingenuity) using “Canonical Pathways” and “Upstream Analysis” options. Pathways with at least 2 member genes that passed FDR<20%, enrichment at least 5 fold threshold and upstream regulators (transcription factors only) that passed p<0.05 and had at least 5 target genes were considered. Functional and pathway enrichment analysis was done use DAVID software57 and FDR<20% categories enriched at least 5 fold were considered. The data was submitted to the Gene Expression Omnibus (GEO) database and can be accessed using accession number: GSE117240
Animal Studies.
All animal experiments were approved by Institutional Animal Care and Use Committee at University of Pennsylvania and comply with all regulations for ethical conduct of research. The Eμ-myc/+ (stock no. 002728), GCN2−/− (stock no. 008240) and Rosa26-CreERT2/+ (stock no. 008463) transgenic mice were purchased from The Jackson Laboratory. Athymic nude mice were purchased from Charles River Laboratory (stock no. 490). ATF4fl/fl mice under C57BL/6 background were generated by Cyagen Biosciences Inc. Briefly, LoxP sites were inserted flanking exon 2 and 3 of Atf4 locus to create a conditional knockout when crossed with Rosa26-CreERT2/+. Eμ-myc/+; GCN2+/+ mice were crossed with GCN2−/− to generate Eμ-myc/+; GCN2+/+, Eμ-myc/+; GCN2+/− and Eμ-myc/+; GCN2−/− mice. ATF4fl/fl mice were crossed with Rosa26-CreERT2/+ to generate ATF4fl/fl; Rosa26-CreERT2/+ which were then crossed with Eμ-myc/+, ATF4 +/+ mice. The following mice were obtained and used for transplantable lymphoma experiments- Eμ-myc/+; Rosa26-CreERT2/+; ATF4wt/wt and Eμ-myc/+; Rosa26-CreERT2/+; ATF4fl/fl. Primers used for genotyping are listed in Supplementary Table 1.
For transplantable lymphoma experiments, mice harboring lymphoma were euthanized according to IACUC guidelines. Lymph nodes were collected immediately on ice and minced and passed through 70μm cell strainer in 50% Isclove’s media and 50% DMEM supplemented with 10% FBS, 1% Penicillin-Streptomycin and 4mM glutamine. Dead cells were removed by spinning cells in Ficoll-Paque PLUS (GE healthcare, # 17-1440-02), 800g for 10min. Lymphoma cells were washed in PBS and 2 million cells were injected into 9 weeks old male C57BL/6J mice. Mice were monitored for lymphoma development by palpation every other day.
For PERK inhibitor (LY-4) experiments, 9 weeks old male mice were injected with 2 milliion Eμ-myc/+; GCN2+/+ or Eμ-myc/+; GCN2−/− lymphoma cells. After 3 days mice were randomized to receive vehicle (Captisol, CYDEX) or LY-4 100mg/kg twice a day by oral gavage three days following lymphoma injection for the duration of the experiment. Moribund mice were euthanized as per IACUC guidelines and lymph nodes and spleen were harvested and assessed for protein levels by immunoblot.
For ATF4 excision experiment, 2 million Eμ-myc/+; Rosa26-CreERT2/+; ATF4wt/wt or Eμ-myc/+; Rosa26-CreERT2/+; ATF4fl/fl lymphoma cells were injected via tail vein into 9-weeks old male C57BL/6J mice. Three days following lymphoma injection, mice were randomized to receive vehicle (peanut oil, Sigma) or 4mg/20g Tamoxifen (T5648, Sigma) by oral gavage for 5 consecutive days. Mice were monitored every other day for lymphoma development by palpation.
3 million DLD-1, MycER, ishATF4 cells were injected in the flanks of 11-weeks old male nude mice and MYC was activated by treating mice with 1mg/mouse tamoxifen 3 days following tumor injection. Once tumors reached 100 cm3, 2mg/mouse doxycycline treatment was started for every other day until duration of experiment. Tumor volume was measured every 2–3 days and calculated as V = (L × W × H)/2, where L is tumor length, W is tumor width and H is height.
Statistics and Reproducibility.
All cell culture experiments were performed three times unless otherwise noted in the legend. GraphPad Prism 7 and Excel 2010 were used for statistical analysis. Error bars indicate mean ± S.D. or mean ± SEM (as indicated in Figure legends) and statistical significance was determined by unpaired, two tailed student’s t-test. One-way ANOVA analysis was used to determine statistical differences in the tRNA microarray data. A p value less than 0.05 was considered statistically significant. For mouse survival analysis, Kaplan-Meier curves and log-rank test were generated in GraphPad Prism 7 software. For xenograft experiment, two-way ANOVA was used.
Patient data analysis.
The gene expression using RNA-seq and survival information of DLBCL dataset58 were obtained from The NCI Center for Cancer Genomics (CCG) website, and gene expression information of 3 TCGA datasets was from UCSC Xena59 (Goldman et al. 2015). For each dataset, the normalized gene expression of EIF4EBP1 gene and other 10 ATF4 targeted genes (DDTI3, ATF3, ASNS, SLC43A1, SLC1A5, GARS, NARS, MARS, PSAT1, MTHFD2) was standardized to Z-score then the Pearson correlation between EIF4EBP1 and average ATF4 target genes was estimated. The visualization of linear relationships on Fig.6e was performed using seaborn software (https://zenodo.org/record/883859#.Ww2av0gvzAQ). In addition, patients in DLBCL dataset were divided into two groups according to ATF4/EIF4EBP1 gene expression: low ATF4/EIF4EBP1 expression (</= median) and high ATF4/EIF4EBP1 expression (> median). The survival analysis using Kaplan-Meier and log-rank test between high and low ATF4/EIF4EBP1 expression groups were performed using lifelines software (https://zenodo.org/record/1252342#.Ww2WRUgvzAQ), as shown on Fig. 6f.
Data Availability
tRNA microarray and ChIP-seq data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession codes GSE116812 and GSE117240 respectively.
The human COAD, BRCA and SARC datasets were derived from the TCGA Data Hub on UCSC Xena platform (http://xena.ucsc.edu/).
The dataset derived from this resource that supports the findings of this study is available in links below:
COAD:
BRCA:
SARC:
The human DLBCL data was derived from NCI Center for Cancer Genomics (CCG) website: https://gdc.cancer.gov. The data-set derived from this resource that supports the findings of this study is available in https://gdc.cancer.gov/about-data/publications/DLBCL-2018.
Statistics Source Data giving rise to graphical representations and statistical analysis in Figures 1–6 and Supplemental Figures 1–6 have been provided as Supplementary Table 3. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Supplementary Material
Acknowledgements
We thank David M. Feldser for critically reading the manuscript. We thank the Koumenis and Maity lab members for helpful discussions. This work was supported by National Institutes of Health grants P01CA165997 (D.R., S.Y.F., J.A.D. and C.K.) and 5R01CA198015–04 to R.K.A. F.T. was supported by NIH F31CA183569.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Pakos-Zebrucka K et al. The integrated stress response. EMBO Rep 17, 1374–1395 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wek RC, Jiang HY & Anthony TG Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans 34, 7–11 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Dong J, Qiu H, Garcia-Barrio M, Anderson J & Hinnebusch AG Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol Cell 6, 269–279 (2000). [DOI] [PubMed] [Google Scholar]
- 4.Harding HP et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell 11, 619–633 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Dey S et al. ATF4-dependent induction of heme oxygenase 1 prevents anoikis and promotes metastasis. J Clin Invest 125, 2592–2608 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gardner BM, Pincus D, Gotthardt K, Gallagher CM & Walter P Endoplasmic reticulum stress sensing in the unfolded protein response. Cold Spring Harb Perspect Biol 5, a013169 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ye J et al. The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. EMBO J 29, 2082–2096 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bi M et al. ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. EMBO J 24, 3470–3481 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fels DR & Koumenis C The PERK/eIF2alpha/ATF4 module of the UPR in hypoxia resistance and tumor growth. Cancer Biol Ther 5, 723–728 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Ozcan U et al. Loss of the tuberous sclerosis complex tumor suppressors triggers the unfolded protein response to regulate insulin signaling and apoptosis. Mol Cell 29, 541–551 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hart LS et al. ER stress-mediated autophagy promotes Myc-dependent transformation and tumor growth. J Clin Invest 122, 4621–4634 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tameire F, Verginadis II & Koumenis C Cell intrinsic and extrinsic activators of the unfolded protein response in cancer: Mechanisms and targets for therapy. Semin Cancer Biol (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beroukhim R et al. The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iritani BM & Eisenman RN c-Myc enhances protein synthesis and cell size during B lymphocyte development. Proc Natl Acad Sci U S A 96, 13180–13185 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stine ZE, Walton ZE, Altman BJ, Hsieh AL & Dang CV MYC, Metabolism, and Cancer. Cancer Discov 5, 1024–1039 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barna M et al. Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature 456, 971–975 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin CJ et al. Targeting synthetic lethal interactions between Myc and the eIF4F complex impedes tumorigenesis. Cell Rep 1, 325–333 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harding HP, Zhang Y, Bertolotti A, Zeng H & Ron D Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell 5, 897–904 (2000). [DOI] [PubMed] [Google Scholar]
- 19.Nagy P, Varga A, Pircs K, Hegedus K & Juhasz G Myc-driven overgrowth requires unfolded protein response-mediated induction of autophagy and antioxidant responses in Drosophila melanogaster. PLoS Genet 9, e1003664 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y et al. Amino acid deprivation promotes tumor angiogenesis through the GCN2/ATF4 pathway. Neoplasia 15, 989–997 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sood R, Porter AC, Olsen DA, Cavener DR & Wek RC A mammalian homologue of GCN2 protein kinase important for translational control by phosphorylation of eukaryotic initiation factor-2alpha. Genetics 154, 787–801 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gomez-Roman N, Grandori C, Eisenman RN & White RJ Direct activation of RNA polymerase III transcription by c-Myc. Nature 421, 290–294 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Wu L et al. Novel small-molecule inhibitors of RNA polymerase III. Eukaryot Cell 2, 256–264 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Avcilar-Kucukgoze I et al. Discharging tRNAs: a tug of war between translation and detoxification in Escherichia coli. Nucleic Acids Res 44, 8324–8334 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han J et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat Cell Biol 15, 481–490 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen H et al. ATF4 regulates SREBP1c expression to control fatty acids synthesis in 3T3-L1 adipocytes differentiation. Biochim Biophys Acta 1859, 1459–1469 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Saxton RA & Sabatini DM mTOR Signaling in Growth, Metabolism, and Disease. Cell 168, 960–976 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hay N & Sonenberg N Upstream and downstream of mTOR. Genes Dev 18, 1926–1945 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Pause A et al. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5’-cap function. Nature 371, 762–767 (1994). [DOI] [PubMed] [Google Scholar]
- 30.Ozcan U et al. Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science 313, 1137–1140 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsieh AC et al. Genetic dissection of the oncogenic mTOR pathway reveals druggable addiction to translational control via 4EBP-eIF4E. Cancer Cell 17, 249–261 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adams JM et al. The C-Myc Oncogene Driven by Immunoglobulin Enhancers Induces Lymphoid Malignancy in Transgenic Mice. Nature 318, 533–538 (1985). [DOI] [PubMed] [Google Scholar]
- 33.Anthony TG et al. Preservation of liver protein synthesis during dietary leucine deprivation occurs at the expense of skeletal muscle mass in mice deleted for eIF2 kinase GCN2. J Biol Chem 279, 36553–36561 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Zhang P et al. The GCN2 eIF2alpha kinase is required for adaptation to amino acid deprivation in mice. Mol Cell Biol 22, 6681–6688 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masuoka HC & Townes TM Targeted disruption of the activating transcription factor 4 gene results in severe fetal anemia in mice. Blood 99, 736–745 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Pytel D et al. PERK Is a Haploinsufficient Tumor Suppressor: Gene Dose Determines Tumor-Suppressive Versus Tumor Promoting Properties of PERK in Melanoma. PLoS Genet 12, e1006518 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao Y et al. PERK is required in the adult pancreas and is essential for maintenance of glucose homeostasis. Mol Cell Biol 32, 5129–5139 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nguyen HG et al. Development of a stress response therapy targeting aggressive prostate cancer. Sci Transl Med 10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ventura A et al. Restoration of p53 function leads to tumour regression in vivo. Nature 445, 661–665 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Ruggero D The role of Myc-induced protein synthesis in cancer. Cancer Res 69, 8839–8843 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu C & Ng DT Glycosylation-directed quality control of protein folding. Nat Rev Mol Cell Biol 16, 742–752 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Novoa I, Zeng H, Harding HP & Ron D Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J Cell Biol 153, 1011–1022 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tettweiler G, Miron M, Jenkins M, Sonenberg N & Lasko PF Starvation and oxidative stress resistance in Drosophila are mediated through the eIF4E-binding protein, d4E-BP. Genes Dev 19, 1840–1843 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kremer CL et al. Expression of mTOR signaling pathway markers in prostate cancer progression. Prostate 66, 1203–1212 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Karlsson E et al. The mTOR effectors 4EBP1 and S6K2 are frequently coexpressed, and associated with a poor prognosis and endocrine resistance in breast cancer: a retrospective study including patients from the randomised Stockholm tamoxifen trials. Breast Cancer Res 15, R96 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cha YL et al. EIF4EBP1 overexpression is associated with poor survival and disease progression in patients with hepatocellular carcinoma. PLoS One 10, e0117493 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miao Y et al. Increased phosphorylation of 4Ebinding protein 1 predicts poor prognosis for patients with colorectal cancer. Mol Med Rep 15, 3099–3104 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Kang MJ et al. 4E-BP is a target of the GCN2-ATF4 pathway during Drosophila development and aging. J Cell Biol 216, 115–129 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamaguchi S et al. ATF4-mediated induction of 4E-BP1 contributes to pancreatic beta cell survival under endoplasmic reticulum stress. Cell Metab 7, 269–276 (2008). [DOI] [PubMed] [Google Scholar]
- 50.Pourdehnad M et al. Myc and mTOR converge on a common node in protein synthesis control that confers synthetic lethality in Myc-driven cancers. Proc Natl Acad Sci U S A 110, 11988–11993 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ben-Sahra I, Hoxhaj G, Ricoult SJH, Asara JM & Manning BD mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science 351, 728–733 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cunningham JT, Moreno MV, Lodi A, Ronen SM & Ruggero D Protein and nucleotide biosynthesis are coupled by a single rate-limiting enzyme, PRPS2, to drive cancer. Cell 157, 1088–1103 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kirchner S et al. Alteration of protein function by a silent polymorphism linked to tRNA abundance. PLoS Biol 15, e2000779 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Soufi A, Donahue G & Zaret KS Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell 151, 994–1004 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Langmead B, Trapnell C, Pop M & Salzberg SL Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heinz S et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang da W, Sherman BT & Lempicki RA Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 58.Schmitz R et al. Genetics and Pathogenesis of Diffuse Large B-Cell Lymphoma. N Engl J Med 378, 1396–1407 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goldman M et al. The UCSC Cancer Genomics Browser: update 2015. Nucleic Acids Res 43, D812–817 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
tRNA microarray and ChIP-seq data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession codes GSE116812 and GSE117240 respectively.
The human COAD, BRCA and SARC datasets were derived from the TCGA Data Hub on UCSC Xena platform (http://xena.ucsc.edu/).
The dataset derived from this resource that supports the findings of this study is available in links below:
COAD:
BRCA:
SARC:
The human DLBCL data was derived from NCI Center for Cancer Genomics (CCG) website: https://gdc.cancer.gov. The data-set derived from this resource that supports the findings of this study is available in https://gdc.cancer.gov/about-data/publications/DLBCL-2018.
Statistics Source Data giving rise to graphical representations and statistical analysis in Figures 1–6 and Supplemental Figures 1–6 have been provided as Supplementary Table 3. All other data supporting the findings of this study are available from the corresponding author on reasonable request.