Abstract
Genome mutagenesis can be achieved in a variety of ways, though a select few are suitable for therapeutic settings. Among them, the harnessing of intracellular homologous recombination affords the safety and efficacy profile suitable for such settings. Recombinagenic donor DNA and mutagenic triplex-forming molecules co-opt this natural recombination phenomenon to enable the specific, heritable editing and targeting of the genome. Editing the genome is achieved by designing the sequence-specific recombinagenic donor DNA to have base mismatches, insertions, and deletions that will be incorporated into the genome when it is used as a template for recombination. Targeting the genome is similarly achieved by designing the sequence-specific mutagenic triplex-forming molecules to further recruit the recombination machinery thereby upregulating its activity with the recombinagenic donor DNA. This combination of extracellularly introduced, designed synthetic molecules and intercellularly ubiquitous, evolved natural machinery enables the mutagenesis of chromosomes and engineering of whole genomes with great fidelity while limiting nonspecific interactions.
Herein, we demonstrate the harnessing of recombinagenic donor DNA and mutagenic triplex-forming molecular technology for potential therapeutic applications. These demonstrations involve, among others, utilizing this technology to correct genes so that they become physiologically functional, to induce dormant yet functional genes in place of non-functional counterparts, to place induced genes under regulatory elements, and to disrupt genes to abrogate a cellular vulnerability. Ancillary demonstrations of the design and synthesis of this recombinagenic and mutagenic molecular technology as well as their delivery and assayed interaction with duplex DNA reveal a potent technological platform for engineering specific changes into the living genome.
Keywords: Triplex-forming oligonucleotide (TFO), Triplex-forming peptide nucleic acid (PNA), Donor DNA, Recombination, Molecular delivery, Genome targeting, Genome editing, Genome mutagenesis, Genome engineering
1. Introduction
For genome mutagenesis, significant sequence-specificity metrics should be satisfied. For therapeutic genome mutagenesis these standards are raised even higher so that significant on-target and off-target safety metrics are met as well. Among the means of chromosome and whole genome mutagenesis methods that meet these raised standards are those that utilize the intracellular phenomena of homologous recombination using extracellularly introduced recombinagenic donor DNA and mutagenic triplex-forming molecular technology.
Mutagenesis by homologous recombination involves the artificial and deliberate exchange and integration of disparate pieces of DNA that share some sequence similarity, but with tolerance for dissimilarity. Naturally occurring homologous recombination transpires to create genetic variation and diversification on a microscopic level in order to tolerate and adapt during the course of evolution at a macroscopic level. Artificial insertion of DNA sequences into human genomic loci, such as for the chromosomal beta-globin locus, has been achieved by homologous recombination [1]. The technological applications for homologous recombination include intracellular gene targeting and editing [2] and genome and protein engineering [3]. Specifically, the nucleotide excision repair, or NER, pathway has been implicated to be involved in triplex-formation induced recombination [4] and this induced recombination has even been triggered by intracellular generation of single-stranded DNA for chromosomal triplex formation [5]. The endogenous repair machinery recruited for this NER-based recombination by the triplex-forming molecules utilizes includes xeroderma pigmentosum, complementation group A (XPA) protein [6, 7], xeroderma pigmentosum, complementation group C (XPC) [8, 9], replication protein A (RPA) [6], and Cockayne’s syndrome, group B (CSB) protein [10], among others, and complexes thereof to reduce nonspecific recombination. In doing so, targeted chromosome and genome mutagenesis is feasible in mammalian cells in a safe and effective manner [11, 12].
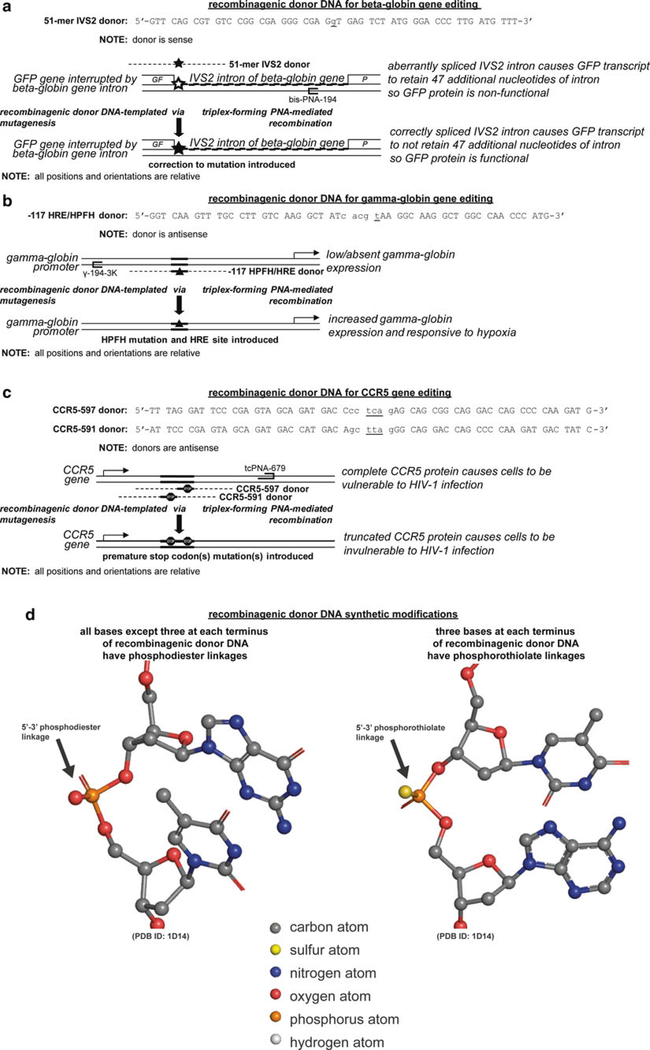
Cells proficient in this repair machinery can be treated with recombinagenic donor DNA molecules as templates for genome editing at a recombination-potentiated editing site. These recombinagenic donor DNA molecules are single-stranded and nearly homologous to the genome editing site and neighboring bases on one strand of an allele of the chromosome, except for the mismatched nucleobases, as genome mutation, insertion, or deletion edits. This recombinagenic donor DNA preferentially localizes directly to the homologous genome editing site upon transfection. Given that there are a number of possible recombinagenic donor DNA, that vary in sequence length, sequence off-centering from the genome editing site, and sequence homology and mismatches to the strand to be edited, or the complementary strand, a variety of molecules can be designed and empirically evaluated for a genomic locus for recombination efficiency. Further efficiency, along with specificity, can be gained by coupling the activities of this recombinagenic donor DNA with the mutagenic capacity of triplex-forming molecules. In doing so for gene editing, a 51-mer recombinagenic donor DNA has designed to correct the beta-globin gene from mis-splicing (Fig. 1a), a 54-mer recombinagenic donor DNA has been designed to induce the gamma-globin gene from postnatal dormancy and place it under exquisite control of the in vivo hypoxia microenvironment (Fig. 1b), and two 60-mers recombinagenic donor DNA have been designed to disrupt the CCR5 gene with premature stop codons (Fig. 1c).
Fig. 1.
(continued) Synthetic recombinagenic donor DNA molecules and their genomic editing sites. Recombinagenic donor DNA molecules have been designed to edit genomic sites for (a) the correction of a splice site mutation in the beta-globin gene to reestablish functional beta-globin expression, (b) the induction of expression, and regulation by hypoxia, of a silenced, but functional, fetal gamma-globin gene to supplant non-functional beta-globin expression in adults, (c) and the disruption of the CCR5 gene to eliminate a receptor-mediated entry of HIV-1. (d) For these exogenously introduced recombinagenic donor DNA molecules, positive design using Watson-Crick oriented base pairing with genomic DNA is implemented, as well as negative design using synthetic modifications to the linkages at each terminus to prevent endogenous nuclease-mediated degradation. Linkages for recombinagenic donor DNA molecular modeled from [97]
Since recombinagenic donor DNA are foreign matter, they encounter the cellular defense machinery as well. In order to combat this, recombinagenic donor DNA can be synthetically modified to resist these defenses. For example, three nucleotides at each terminus of the recombinagenic donor DNA are modified to phosphorothioate linkages, so that phosphodiester linkage recognizing nucleophilic enzymes are unable to attack the molecule (Fig. 1d). While this modification resists nucleases from digesting the recombinagenic donor DNA, this internucleotide linkage modification does not prevent the recombination machinery proteins from using it as a template for recombination.
While modest levels of recombination and, thus, modification frequency occur when recombinagenic donor DNA is utilized by the recombination machinery, significantly greater levels are achieved through the use of triplex-forming molecules that heighten the recruitment and activity of this machinery in a dose-dependent manner. It has only been a little over six decades since the discovery of the duplex structure of deoxyribose nucleic acid (DNA) was made by Watson and Crick [13]. Four brief years after this discovery, the discovery of the triplex structure of DNA, whereby a third strand of polyuracil bound a duplex of polyadenine and polyuracil, was made by Felsenfeld, Davies, and Rich [14]. In the mid-1980s, further elucidation not just of their structure, but of some of their mechanism of action, such as sequence-specific distortion and cleavage was made by Dervan and coworkers as well as by Helene and coworkers [15, 16].
These discoveries have led to greater understanding and application of a diverse array of triplex-forming molecules that are designed for a variety of purposes. Triplex-forming molecules that are composed of oligonucleotides, such as those used in the first triplexes discovered, are termed triplex-forming oligonucleotides (TFOs). These oligonucleotides are made of nucleobases and linked by phosphodiester linkages, except for some notable synthetic modifications that are suited to their genome binding and nuclease resistance roles. The TFO molecule also has an orientation, just as other oligonucleotides, whereby it is parallel or antiparallel in polarity to the polypurine strand of genomic DNA. Furthermore, the major groove binding to duplex DNA necessitates Hoogsteen or reverse Hoogsteen base pairing, as the natural DNA Watson-Crick donor-acceptor base pairing atoms are already occupied. Thus, Hoogsteen base pairing occurs so that adenine, or A, guanine, or G, and thymine or T, base pair with adenine, guanine, and adenine, respectively, of the Watson-Crick-base-paired DNA duplex to form the triplex. A special case occurs with cytosine, or C, on this third strand as its N3 atom lacks the hydrogen atom at physiological pH to be a Hoogsteen base pairing hydrogen bond donor. Thus, while TFOs in which this N3 atom is protonated in acidic pHs are able to Hoogsteen base pair with duplex DNA to form triplex structures, an unnatural nucleobase such as pseudoisocytidine or J, a, C-nucleoside analog that mimics the N3 protonation of cytosine in triplex-forming peptide nucleic acids (PNAs), can Hoogsteen base pair with the guanine of a Watson-Crick base pair without this pH dependency.
The major groove of duplex genomic DNA provides the steric and conformational clearance for TFOs and other triplex-forming molecules to bind and form this triplex molecule. In the triplex code dictated by Hoogsteen base pairing, the third strand triplex-forming molecule can bind duplex genomic DNA in a sequence-specific manner in either a parallel or antiparallel motif. The antiparallel purine motif involves a polypurine TFO that binds to the purine strand of the duplex genomic DNA through reverse Hoogsteen base pairing and in an orientation that has antiparallel polarity [17]; whereas the parallel pyrimidine motif involves a polypyrimidine TFO that binds to the purine strand of the duplex genomic DNA through regular, not reverse, Hoogsteen base pairing and in an orientation that has parallel polarity [18, 19].
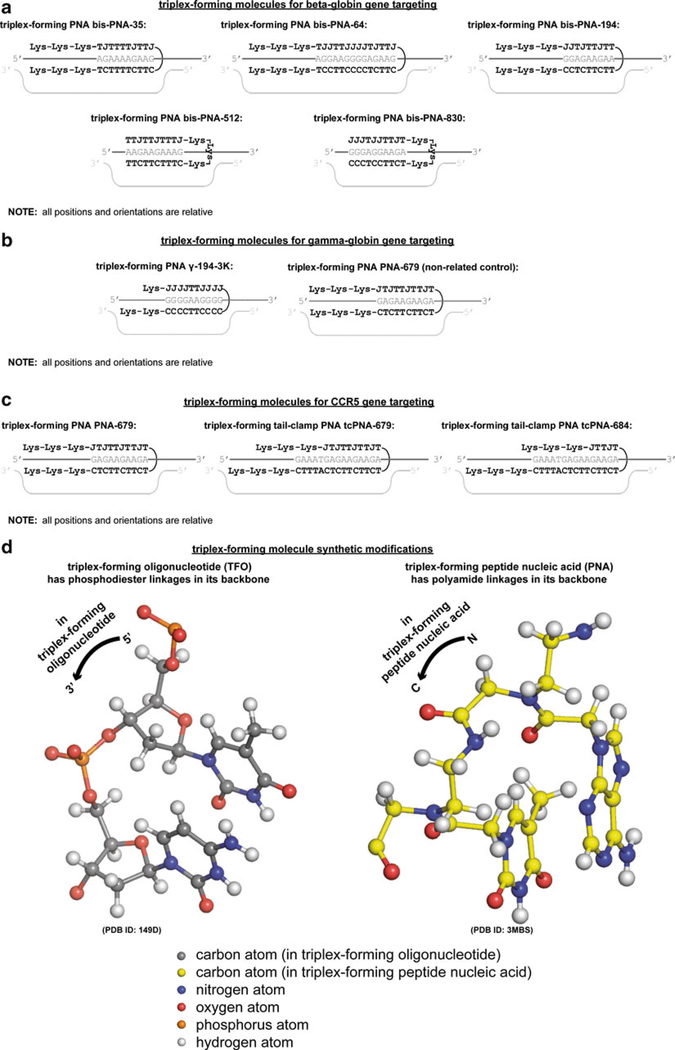
The chemistry of triplex-forming molecules can be further modified from TFOs in triplex-forming PNAs. These triplex-forming PNAs have been designed to upregulate the mutagenic potential of a recombination machinery interacting with recombinagenic donor DNA for the beta-globin gene (Fig. 2a), the gamma-globin gene (Fig. 2b), and the CCR5 gene. In these triplex-forming PNAs, not only are the termini internucleoside linkages, but rather the all of the internucleoside linkages are modified so that the negatively charged phosphodiester linkages are replaced by uncharged polyamide linkages [20] (Fig. 2d). This neither wholly nucleic nor amino acid biopolymer is not recognized by cellular nucleases and proteases and, in addition, is better electrostatically complemented by the negatively charged genomic DNA phosphodiester linkages [21]. Furthermore, triplex-forming PNAs are resistant to polymerase recognition and thus will not contribute to the crosstalk noise by itself providing a template for replication or transcription [22].
Fig. 2.
(continued) Synthetic mutagenic triplex-forming molecules and their genomic targeting sites. Triplex-forming molecules have been designed to target genomic sites, thereby upregulating homologous recombination with the recombinagenic donor DNA molecules, or as non-related controls, to examine targeting specificity, for (a) the beta-globin gene, where triplex-forming bis-PNAs are used, (b) the gamma-globin gene, where triplex-forming PNAs were used, (c) and the CCR5 gene, where triplex-forming PNAs and triplex-forming tail-clamp PNAs were used. (d) For these exogenously introduced triplex-forming molecules, positive design is implemented based on major groove binding Hoogsteen oriented base pairing and strand-invading Watson-Crick oriented base pairing with genomic DNA, as well as negative design based on synthetic modifications to the linkages in the backbone to prevent endogenous nuclease and protease-mediated degradation and enhance electrostatic complementarity to negatively charged genomic DNA. Linkages for triplex-forming oligonucleotide and triplex-forming peptide nucleic acid molecular modeled from [98] and [99], respectively
Antiparallel and parallel triplex-forming DNA stimulates homologous recombination in human cells [19, 23]. TFOs and triplex-forming PNAs retain the ability to bind sites on genomic and episomal DNA in a recombination-like motif with high thermodynamic stability to form triplexes [24]. Unlike the recombinagenic donor DNA molecule, which localizes directly to the genome editing site, the mutagenic triplex-forming molecule can potentiate the recombination machinery upon binding to a genome targeting site that is immediately overlapping the genome editing site, or as distant as almost as a kilobase away. Under acidic conditions, the natural base cytosine in a TFO can be protonated so that it can Hoogsteen bind to guanine bases in the genomic target. Similar to TFOs, triplex-forming PNAs can form triplexes in the major groove of DNA [25] under normal physiological conditions using the unnatural base pseudoisocytidine. By linking a triplex-forming PNA with a strand-invading PNA through a flexible linker, a triplex-forming bis-PNA can simultaneously Hoogsteen and Watson-Crick hydrogen base pair to the same base in duplex genomic DNA in a sequence-specific manner at physiologically high melting temperatures [26]. Further extensions of the strand-invading PNA “tail” Watson-Crick base pairing domain beyond that of the triplex PNA “clamp” Hoogsteen base pairing domain creates a triplex-forming tail-clamp PNA that garners greater affinity as well as specificity for the DNA targeting site [27]. These binding affinities of TFOs and triplex-forming PNAs have been demonstrated to be considerable enough in vitro to achieve a variety of perturbations to cellular processes, including the obstructing DNA polymerization [28], inhibiting transcription initiation and elongation [29], and preventing sequence-specific protein binding [30]. In addition, the conformational capacity of triplex-forming PNAs to potentially form D-loop open DNA conformations have been demonstrated to act as artificial transcriptional promoters [31–35] and diethylene glycol (miniPEG)-based triplex-forming gamma PNAs have been demonstrated to act as an antisense gene targeting agent to silence the CCR5 gene [36]. This triplex-forming molecular activity can further alter genomic conditions by inducing [37] or inhibiting [38] DNA transcription, DNA replication [39–41], and protein-DNA interactions [42–45], and promoting site-specific DNA damage [10, 46–48], mutagenesis [49, 50], or recombination on chromosomal and episomal DNA [7, 51, 52].
There still exist many challenges in efficiently delivering recombinagenic donor DNA and mutagenic triplex-forming technology in vitro and in vivo, though a number of strategies have advanced the state-of-the-art (see Subheading 3). Interactions of triplex-forming PNAs with cellular and nuclear membranes and negatively-charged genomic DNA have been improved through the decoration with positively charged lysine residues [37] and conjugation to cell-penetrating peptides such as Antennapedia and transactivator of transcription (TAT) [53] as well as polymeric nanoparticles [54]. This is significant, since upon arrival into the cytoplasm, the functionality of triplex-forming molecules can be inhibited by cellular conditions, such as concentrations of cytosolic potassium and magnesium. Within the nucleus, triplex-forming molecular interaction may also be limited by the accessibility of its target genomic site as a result of its chromosomal location and interaction with other genomic components, such as chromatin, regulatory proteins, and other molecules competing for access to the same genomic targeting site. A nuclear localization signal when conjugated to TFO and triplex-forming PNA molecules has been shown to increase directed delivery to the nucleus [55]. The aforementioned modifications, among others, have been used to increase binding affinity as well. For example, binding affinity of TFOs in vitro has been increased by modifying the linkages between bases, such as by replacing the phosphodiester linkages with cationic phosphoramidate linkages, N, N-diethylethylenediamine or N, N-dimethyl-aminopropylamine [56]. In G-rich oligonucleotides, the use of N, N-diethylethylene-diamine-modified bases may also mitigate G-quartet formation. The use of pseudoisocytosine, J, rather than cytosine, C, in triplex-forming molecules in order to Hoogsteen pair with guanine, G, in a DNA targeting site at physiological pH has already been mentioned. Thus, in addition to the linkages between bases, the bases themselves can be modified to increase the binding affinity of TFOs in the pyrimidine motif, for example by using 5-methyl-2-deoxyuridine and 5-methyl-2-deoxycytidine, as too can the sugar moieties to which they are covalently linked, for example to a 2-O-(2-aminoethyl)-ribose [57, 58].
Upon delivery, triplex formation at a specific targeting site leads to gene targeting by provoking the cell’s own DNA repair pathways, primarily the NER and recombination machinery, to localize near that site in a similarly safe manner to that which has evolved already to repair naturally occurring triplex adducts [59]. This a versatile mechanism of mutagenesis, as the inherent sequence specificity of a triplex-forming molecule can be coupled to a DNA damage-causing mutagen that is not sequence or structure specific, such as psoralen (pso), to a choromosomal locus of interest [60, 61]. These pso-TFOs have induced mutations to a target site on plasmids in vitro, when transfected into mammalian cells, and on chromosomes [46, 49, 62]. Furthermore, the influence of the cell cycle on genome accessibility and recombination machinery availability has been elucidated, showing that by synchronizing and treating cells in late S-phase this targeted mutagenesis could be increased 5.5-fold over those in G1, and 2.5-fold over cells in early S-phase [63]. By modulating the cell cycle or transcriptional state, it may be possible to increase the efficiency of triplex-forming molecule induced mutagenesis.
The detection and quantification of these mutations induced by triplex-forming molecules, such as TFOs, on plasmids in vitro, has been developed in a blue and white cell reporter system. This reporter system is composed of the supF reporter gene, which encodes an amber suppressor tyrosine tRNA having a TFO targeting site, cloned into an SV40 plasmid vector. When cells carrying this cloned plasmid vector are plated in the presence of 4-chloro-5-bromo-3-indolyl-β-D-galactopyranoside and isopropyl-β-D-1-thiogalactopyranoside, bacteria having an amber mutation in the lacZ gene but a functional copy of the supF gene produce blue colonies. However, a nonfunctional, or mutated, copy of the supF gene due to the mutagenic triplex-forming molecule produces white colonies. The number and ratio of white colonies to blue colonies can be used to calculate the mutagenesis frequency [46, 64]. Using this reporter system, mutagenesis frequencies due to pso-TFOs have been observed and quantified in murine cells containing a chromosomally integrated copy of the supF reporter gene [65]. Whereas psoralen has been shown to induce mutations, formation of a triplex structure alone using either TFOs or triplex-forming PNAs has also been shown to be sufficient to stimulate mutagenesis and treated by the cellular repair and recombination machinery accordingly [25, 65]. Extending these studies even further, in transgenic mice containing the supF reporter gene integrated in the chromosome, intraperitoneal injection of a TFO, AG30, also led to site-specific mutations [49]. Note that when this mutagenesis occurs at a low frequency tolerated by the cell studies have indicated that the cell fate is one of repairing the target site and environs by upregulating the frequency of homologous recombination [66]. To exploit this idea, pso-TFOs have been used to create site-specific damage in order to sensitize a target site for subsequent homologous recombination [67, 68]. It was found that not only could pso-TFO-associated DNA damage increase intrachromosomal recombination [4], but also a TFO without a DNA-damaging agent was sufficient to induce homologous recombination [7, 52, 69]. Triplex formation increased the level of recombination at a targeting site and led to gene correction of a specific mutation [7]. Using a plasmid with two tandem supF genes, each containing different point mutations, increased intramolecular recombination upon binding of a TFO was demonstrated and resulted in gene correction of one copy of the gene [67]. Furthermore, homologous recombination was stimulated by TFOs with or without pso at a frequency of 1 %, which is 2,000-fold over with the following, and with italics on the words thymidine kinase: background in the nuclei of murine cells containing two mutant copies of the herpes simplex virus thymidine kinase gene [52]. Given the potential for a TFO to be mutagenic, and given the potential for the recombination machinery to be recruited by this mutagenic induction, it then followed that linking a TFO to a short donor fragment, would act to upregulate recombination near a co-localized editing site and recombinagenic donor DNA [51]. That recombinagenic DNA donor fragment is almost homologous to the targeting site, except for the edited bases [70]. For certain editing sites, the more effective designed recombinagenic donor DNA have been antisense (homologous to the transcribed strand) [69] but this is not universally the case for other sites and sense counterparts [26]. A designed recombinagenic donor also need not be for an editing site near the targeting site to be effective since recombination has been detected at sites up to three-fourths of a kbp away from the targeting site [71]. Site-specific intermolecular homologous recombination in a singe genomic targeting site up to a frequency of 0.11 % has been achieved in mammalian cells by TFOs designed employing either motifs [69].
Recombinagenic donor DNA and mutagenic triplex forming molecules, when used for gene editing and targeting of chromosomes and genomes, has numerous potential therapeutic applications. In particular, the editing and targeting of a gene or genes in living cells, and the subsequent propagation of the results of those targetings and editings in progeny provides a means of permanently transforming cells from disease-prone to disease-free. In order to implement recombinagenic donor DNA and mutagenic triplex-forming molecular technology, a series of methods are recommended, such as for identifying genomic editing and targeting sites, designing the synthetic molecules to interact with these sites, and then assessing the binding affinities and modification frequencies (see Subheading 3).
There are a variety of therapeutic gene targeting and editing capabilities of triplex-forming oligonucleotides and recombinagenic donor DNA molecules. Among these capabilities are gene correction, induction with regulation, and disruption, as discussed below.
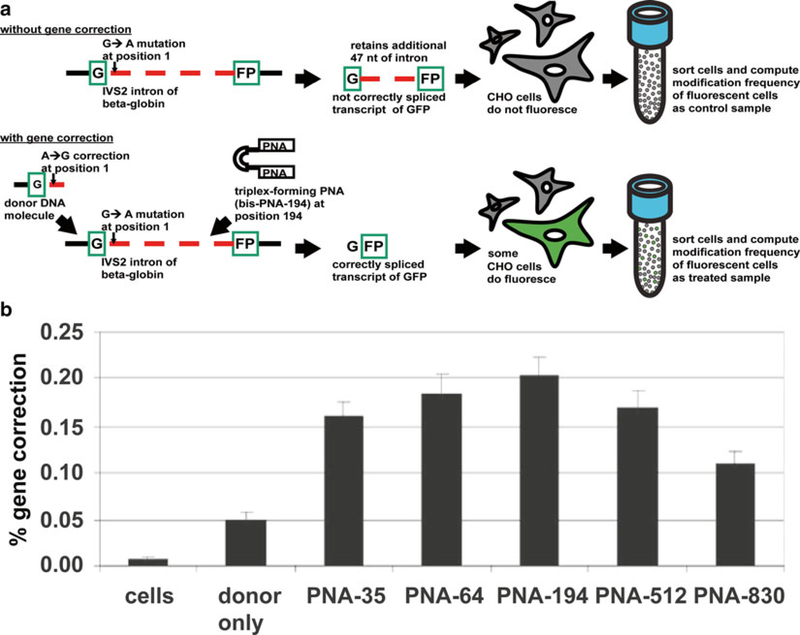
For gene correction, donor DNA carry one or more corrected bases that will be utilized by a homologous recombination machinery, which is upregulated due to a triplex-forming molecule, to correct a particular disease-causing gene. In the case of a hematologic disorder, beta-thalassemia a common mutation, that of a guanine to an adenine mutation (G → A), on the second intron at position 1 (IVS2–1) of the beta-globin gene residing on the short arm of human chromosome 11 has been successfully modified to a healthy state using recombinagenic donor DNA and mutagenic triplex-forming technology in living cells [26, 72]. In a dualpronged approach, using Chinese hamster ovary (CHO) GFP reporter cells the correction of this particular splicing mutation was demonstrated, as was the creation of this beta-thalassemia mutation in healthy human CD34+ (hCD34+) cells.
To elaborate, the CHO GFP reporter cell system integrated a genomic green fluorescent protein (GFP) gene interrupted by the IVS2 intron of the beta-globin gene with a G → A mutation at its position 1 [26]. When this mutation went uncorrected, then the expressed transcript from this locus was not correctly spliced and retained an 47 additional nucleotides arising from the betaglobin intron causing the GFP protein to be improperly translated and the CHO GFP reporter cell carrying it to not be fluorescent. However, if this mutation is corrected using a triplex-forming bis-PNA at position 194 in conjunction with a recombinagenic donor DNA carrying the A → G correction at position 1, then proper splicing and translation occurs so that the CHO GFP reporter cell carrying it does become fluorescent. Each fluorescent or non-fluorescent cell can be accurately measured by fluorescence activated cell sorting (FACS) of these CHO model system cells and the recombinagenic and mutagenic potential of donor DNA and triplex-forming bis-PNA to cause gene correction can be quantified. In addition to the doses of triplex-forming bis-PNAs and donor DNAs to further increase their availability from cellular sequestration by lysosomes chloroquine can be added to the treatment regimen. The correction percentage can also be modulated by synchronizing the CHO cells at various cell cycle phases just before the treatment regimen as well as increase accessibility of the genomic DNA (Table 1).
Table 1.
Recombinagenic and mutagenic frequencies of therapeutic targeting and editing of genomic loci to ameliorate disease etiologies
| Disease etiology | Genomic locus | Recombinagenic and mutagenic molecules | Cell type | Molecular delivery modality | Recombinagenic and mutagenic frequency (%) |
|---|---|---|---|---|---|
| Beta-thalassemia | Beta-globin | Donor DNA | CHO | electroporation | 0.05 |
| Beta-thalassemia | Beta-globin | Donor DNA + triplex-forming PNA | CHO | electroporation | 0.20 |
| Hemoglobinopathies, such as sickle cell disease | Gamma-globin | Donor DNA | K562 | electroporation | 0.29 |
| Hemoglobinopathies, such as sickle cell disease | Gamma-globin | Donor DNA + triplex-forming PNA | K562 | electroporation | 1.63 |
| HIV-1 | CCR5 | Donor DNA + triplex-forming PNA | hCD34+ | nucleofection | 0.10 |
| HIV-1 | CCR5 | Donor DNA + triplex-forming PNA | hCD34+ | nanoparticles | 0.91 |
| HIV-1 | CCR5 | Donor DNA + triplex-forming PNA | THP-1 | electroporation | 2.46 |
Recombinagenic and mutagenic frequencies on the chromosome or episome are influenced by a number of factors, including the particular endogenous genomic locus, exogenously introduced molecules, cell type, and molecular delivery modality
These modifications can be detected at the phenotypic level, and also at the genotypic level. A phenotypic assay can be such as the aforementioned fluorescent protein reporter system can be used to detect the consequence of a modifiction or at the genomic level, allele-specific polymerase chain reaction (PCR) can be used to detect a sequence modification. Extending this further, allele specific quantitative PCR (qPCR) can detect modification as well as quantify modification frequencies in treated cells relative to cells with genomes having known modification frequencies. Furthermore, at the transcriptional mRNA level, combined quantitative reverse transcriptase real time PCR (qRT-PR) can show the presence and amount of gene expression from the modification of the genome.
For gene induction and regulation disruption, donor DNA and triplex-forming molecule modify the silenced promoters of functional but dormant genes and perturb them by editing in a regulatory element. These induced gene products can mitigate hematologic disease [37]. For example, a cause of thalassemias, sickle cell disease, and other hemoglobinopathies is a non-functional beta-globin gene and subunit in adult hemoglobin. The hereditary persistence of fetal hemoglobin (HPFH) is a benign condition in which a functional, fetal gamma-globin subunit is expressed in adults and can outcompete the polymerization of non-functional beta-globin subunits with alpha-globin subunits, can alleviate hemoglobinopathies, such as thalassemias, arising from the non-fUnctional beta-globin [73, 74]. Chin, Reza, and Glazer demonstrated that the targeted induction of the functional fetal gamma-globin gene also on the short arm of human chromosome 11 in cells carrying the non-functional adult beta-globin gene can mimic HPFH and restore the working state of hemoglobin, thus lessening the severity of these diseases. A triplex-forming bis-PNA targeting the gamma-globin promoter along with a HFPH recombinagenic donor DNA at the −117 position that also edited in a hypoxia responsive element (HRE) to regulate the HPFH state was used in the treatment. An unrelated control triplex-forming bis-PNA was also evaluated to examine whether the triplex-forming PNAs were acting as artificial promoters of gene expression. A triplex-forming PNA and either −117 HRE/HPFH donor DNA or control donor that introduced the HPFH mutation but not HRE were used to treat hCD34+ cells, that were subsequently subjected to a hypoxic environment for two days. The harvested RNA of treated cells, as well as those of control cells from a normal oxygen tension environment, were subjected to qRT-PCR with TaqMan® probes indicated that gamma-globin was being expressed and regulated by hypoxia. In contrast, cell samples treated with a gamma-globin triplex-forming PNA and an unrelated control triplex-forming PNA both showed minimal induction of expression of gamma-globin, thus suggesting that the donor DNA editings were the cause for these changes. Allele-specific qPCR relative amplification values were correlated with modification frequencies to demonstrate that greater recombinagenic and mutagenic frequencies were achieved when using donor DNA and triplex-forming PNA concurrently (Table 1).
For gene disruption, donor DNA and triplex-forming tail-clamp PNA create premature stop codons and, in turn, create nonfunctional gene products. Premature stop codons can thus stop the production of a cellular component that makes the cell vulnerable to disease or infection. A naturally occurring mutation, called CCR5-delta32 to the chemokine receptor gene, prevent proper CCR5 chemokine receptor trafficking and ultimately an inability of human immunodeficiency virus, type 1 (HIV-1) to infect human cells [75]. This suggests various strategies of recapitulating this mutation to prevent HIV-1 propagation, such as by intracellular immunization by a CCR5 blocking single-chain antibody therapy [76], or by non-autologous transplantation of stem cells homozygous for this CCR5-delta32 mutation [77], or by knockdown siRNAs of CCR5 delivered by lentiviral vectors [78], but, given their transient nature and infectious delivery mechanism respectively, their translational potential as therapeutics is uncertain [79]. Disruption of the CCR5 gene by using donor DNA and tail-clamp PNA molecules delivered to hCD34+ hematopoietic stem cells (HSCs) by nanoparticles successfully resulted in permanent, heritable, non-immunogenic, and non-infectious disruption of HIV-1 [27, 80]. the self-renewal and heritable capabilities of (HSCs) are advantageous for gene targeting and editing therapeutic intervention that persists and propagates further. For HIV-1 infections, hCD34+ HSCs are also advantageous since they have been shown to remain unaffected [81].
Co-transfection of a triplex-forming tail-clamp PNA and donor DNA led to the greatest amount of gene disruption, enough to nullify CCR5 expression over single-cell clones and their progeny. Specific on-target effects were 43-fold greater than the negligible off-target effects, as determined by DNA sequencing from single-cell cloning assays. In doing so, the differentiation capacity of THP-1 human acute monocytic leukemia cells into a macrophage-like state was maintained, as was high levels of CCR5 cell surface expression. The triplex-forming PNAs and donor DNAs introduced to create both 591 and 597 gene disruptions double mutant resulted in differentiated parental THP-1 cells with negligible cell surface staining for CCR5.
This disruption of the CCR5 protein now led to resistance to HIV-1 infection. Cloned THP-1 cells treated with these molecules were isolated, expanded, and induced to express CCR5 and then challenged with live HIV-1. The treated cells, as opposed to the parental cells, had significantly lower core protein p24 antigen levels, which indicate the amount of HIV-1 infection in cells.
Molecular delivery of these mutagenic and recombinagenic molecules using electroporation, nucleofection, cell-penetrating peptides, and nanoparticles, have been applied to in vitro and in vivo targeting and site-specific editing [54]. Human HSCs engrafted in NOD-scid IL2rγnull mice were treated with biodegradable nanoparticles carrying recombinagenic triplex-forming PNAs, recombinagenic donor DNA molecules, or both. An in vitro screen indicated that nanoparticles encapsulating both more effectively achieved genome targeting and editing of human hematopoietic cells [82]. Further direct deep sequencing of the CCR5 gene had modification on-target at frequencies of 0.43 % in these stem cells found in the spleen, and at 0.05 % in the bone marrow, and off-target at two order of magnitudes lower off-target to related genes.
Thus, recombinagenic donor DNA and mutagenic triplex-forming molecular technology have the demonstrated potential and performance record to target and edit chromosomes and engineer genomes in a safe and effective heritable fashion. This technology may play a pivotal therapeutic role in transforming and then transferring a patient’s own once disease-causing cells to those that are disease-free by genomes and chromosomes that have had their genes corrected, induced, regulated, or disrupted by recombinagenic donor DNA and mutagenic triplex-forming molecules.
2. Materials
Synthetic molecules, such as the single-stranded recombinagenic donor DNA molecules and the mutagenic triplex-forming molecules, TFO, triplex-forming peptide nucleic acid, mentioned herein, can be procured from commercial vendors worldwide, an institutional oligonucleotides facility, or synthesized in the laboratory. Upon synthesis, it is highly recommended that these oligonucleotides be purified using high-pressure liquid chromatography or gel purification to remove synthesis reagents or impurities prior their use in the treatment of cells. Further best practices are highly recommended for industrial formulation, scale-up, packaging, and quality control.
2.1. Recombinagenic DNA Donors
-
Recombinagenic donor DNA can be synthesized in the laboratory using sequentially coupled solid-phase phosphoramidite chemistry, consisting of synthesis cycles of deblocking (detritylation) of a acid-labile DMT (4,4′-dimethoxytrityl) group on the 5′-hydroxyl group, coupling with a nucleoside phosphoramidite in acetonitrile that is catalyzed by an acidic azole catalyst, capping with a mixture of acetic anhydride and 1-methylimidazole, and lastly oxidation with iodine and water having a weak base (see Notes 1 and 2).
Alternatively, long double-stranded recombinagenic donor DNA can be synthesized by PCR amplification from a plasmid carrying the donor sequence of interest.
Recombinagenic donor DNA can be procured from an institutional oligonucleotide synthesis facility.
Recombinagenic donor DNA can be procured from a commercial supplier, such as The Midland Certified Reagent Company (Midland, TX, USA) or Panagene (South Korea) (see Note 3).
2.2. Triplex-Forming Molecules (TFOs and Triplex-Forming PNAs)
TFOs can be synthesized can be synthesized in the laboratory using, again, sequentially coupled phosphoramidite chemistry.
TFOs can be procured from an institutional oligonucleotide synthesis facility.
TFOs can be procured from a commercial supplier, such as The Midland Certified Reagent Company (see Notes 4 and 5).
Triplex-forming PNAs can be synthesized in the laboratory using an α-amino protecting group based on 9-fluorenylmethyloxycarbonyl, or Fmoc, or based on tert-butyloxycarbonyl, or Boc, solid-phase peptide synthesis chemistry. Fmoc and Boc are protecting groups that permit proper peptide linked moiety synthesis by blocking nonspecific reactions. The chemistry for either consists of an initial activation step, followed by synthesis cycles of coupling and deblocking of the Fmoc or Boc protecting group, and then a final deblocking step (see Note 6).
Triplex-forming PNAs can be procured from an institutional peptide synthesis facility (see Note 7).
Triplex-forming PNAs can be procured from a commercial supplier, such as Bio-Synthesis (Lewisville, TX, USA), PNA BIO (Thousand Oaks, CA, USA), or Panagene (see Notes 8–10).
2.3. Cells and Vectors
Various mammalian cell types have been amenable to treatment with recombinagenic and mutagenic molecules including: Chinese hamster ovary (CHO) cells, human K562 myelogenous leukemia cells, human THP-1 acute monocytic leukemia cells, and human CD34+ primary hematopoietic progenitor cells.
Various plasmid vectors have been suitable for gel shift assays due to their supercoiled conformations reminiscent of genomic DNA (Fig. 3a) and allele-specific qPCR assays due to their relatively smaller size as compared to the human genome (Fig. 5a), such as pBlueScript II-SK (Agilent Technologies, Santa Clara, CA, USA) and FLuc+ from pGL3-Basic Vector (Promega, Madison, WI, USA).
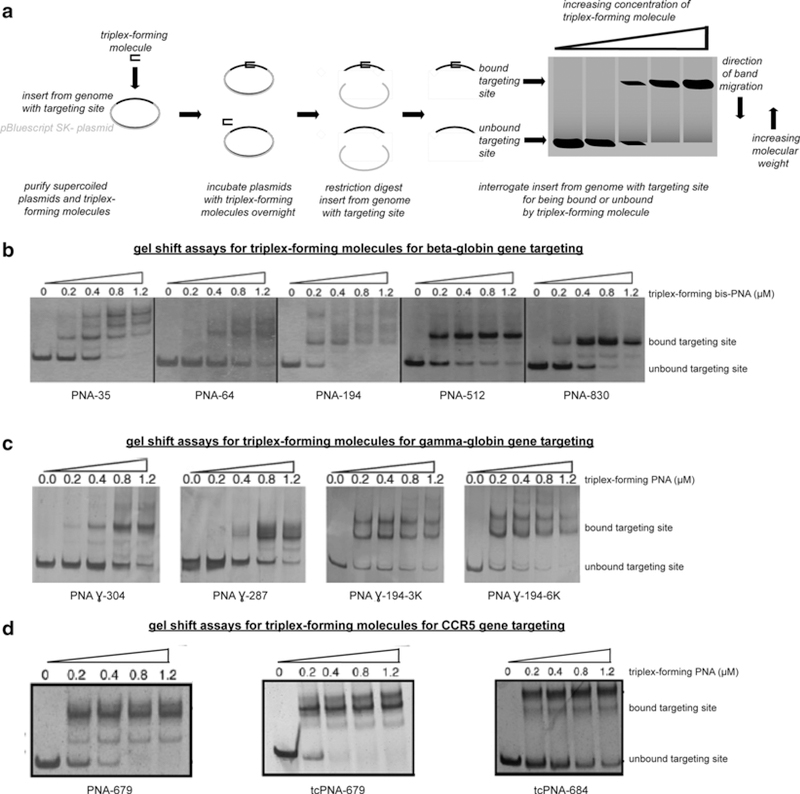
Fig. 3.
Assessing triplex-forming molecule binding to targeting sites with gel shift assays. Gel shift assays are used to (a) assess the binding potential of triplex-forming molecules to genomic DNA, using supercoiled plasmid DNA with an insert from the genome with the targeting site as surrogates, and a shift in position of the visualized band in an electrophoresed gel to indicate that the triplex-forming molecule has bound this targeting site in (b) the beta-globin gene, (c) the gamma-globin gene, (d) and the CCR5 gene
Fig. 5.
Quantifying recombinagenic and mutagenic frequencies of gene induction and regulation with allele-specific qPCR assays of the gamma-globin genomic locus. Allele-specific qPCR assays are used to (a) quantify the recombinagenic and mutagenic frequency of recombinagenic donor DNA or recombinagenic donor DNA co-transfected with mutagenic triplex-forming molecules, in the gene induction and regulation of the gammaglobin genomic locus by correlating the relative qPCR amplification values with modification frequencies, (b) with varying frequencies of gene induction and regulation attributed to the particular molecules used in the treatment
2.3.1. Chinese Hamster Ovary (CHO) Cell Cultures
Chinese hamster ovary cell medium: Ham’s F12 medium, 10 % supplementation of fetal bovine serum, and 2 mM of l-glutamine. Vacuum-filter.
If genomic cassettes are integrated in the genome, such as those in the cells of the beta-globin fluorescent assay (Fig. 4a), then in order to maintain the integrations further supplementation of this aforementioned culture medium with a selection marker, such as hygromycin, that is also filtered, is highly suggested (see Note 11).
Fig. 4.
Quantifying recombinagenic and mutagenic frequencies of gene correction with fluorescent assays of the beta-globin genomic locus. Fluorescent assays are used to (a) quantify the recombinagenic and mutagenic frequency of recombinagenic donor DNA or recombinagenic donor DNA with triplex-forming molecules, in the gene correction of the beta-globin genomic locus to permit correct splicing of transcript of GFP fluorescent reporter, (b) with varying frequencies of gene correction attributed to the particular molecules used in the treatment
2.3.2. Human K562 Myelogenous Leukemia Cell Cultures
K562 myelogenous leukemia cell medium: RPMI medium, 10 % supplementation of fetal bovine serum. Vacuum-filter.
If viability of these cell cultures is compromised due to contamination, antibiotics may be considered, such as penicillin and streptomycin at the requisite concentrations (see Note 12).
2.3.3. Human Primary CD34+ Hematopoietic Progenitor Cell Cultures
The StemSpan Serum-Free Expansion Medium (StemCell Technologies Inc., Vancouver, Canada): bovine serum albumin, recombinant human insulin, human transferrin (iron-saturated), 2-mercaptoethanol, Iscove’s Modified Dulbecco’s Medium, and other supplements.
To permit proper maintenance of human primary CD34+ hematopoietic progenitor cells in this media, a supplement of StemSpan CC110 cytokine mixture (StemCell Technologies Inc.) should also be used. The StemSpan CC110 cytokine mixture: 100 ng/mL rh Flt-3 Ligand, 20 ng/mL rh IL-3, 20 ng/mL rh IL-6, and 100 ng/mL rh Stem Cell Factor (see Note 13).
2.4. Solutions, Buffers, and Other Reagents
The triplex-binding buffer in the TFO binding assay: 10 mM of Tris-HCl (pH 7.6), 0.1 mM of MgCl2, 1 mM of spermine, 10 % of glycerol (with or without 140 mM potassium) (see Note 14).
The silver stain solution in the triplex-forming PNA binding assay: sodium borohydrate in 0.1 % silver nitrate (Sigma Aldrich, St. Louis, MO, USA), e.g., 1.0 g of AgNO3 in 1.0 L of dH2O (see Note 15).
The developer solution in the triplex-forming PNA binding assay: 15.0 g of NaOH, 0.1 g of NaBH4, 5.0 mL of formaldehyde (see Note 16).
3. Methods
3.1. Recombinagenic donor DNA and Their Editing Sites
A recombinagenic donor DNA is an exogenously introduced molecule, which is nearly homologous to the genomic DNA editing site of interest sans the mismatched, inserted, or deleted bases that it is designed to harbor, that the endogenous cellular homologous recombination machinery utilizes as a template to introduce recombinagenic edits to the genome with great specificity.
3.1.1. Design of Recombinagenic donor DNA
The sequence length of single-stranded recombinagenic donor DNA molecules ranges from 30 to 2,000 bases in length, with sequence content that is homologous to the editing site, sans the mismatching base edits to be made flanked by a sufficient number of matching bases, and with sequence binding site proximity to within 750 bp of the triplex-forming molecule’s binding site [69], and sequence orientation either as antisense (binding to the sense strand of the DNA) or sense (binding to the antisense strand of the DNA) (see Note 17). It is highly suggested that recombinagenic donor DNA molecules of both orientations are synthesized and used to treat cells, as overall mutagenic recombination efficiencies with genomic DNA editing site can vary due to a number of features of the DNA donors as well of the genomic DNA editing site by themselves, such as the intramolecular self-assembly or folding potential of either, and their interactions together, such as the intermolecular self-assembly or hybridization potential of both in the cellular milieu. Further considerations include additional cellular and genomic components, such as other potentially hybridizable nucleic acid molecules and chromatin structure, respectively, that can adversely affect the availability of recombinagenic donor DNA and the accessibility of the genomic editing site.
3.1.2. Synthesis of Recombinagenic donor DNA
Recombinagenic donor DNA molecules can be synthesized internally in the laboratory through sequentially coupled solid-phase phosphoramidite chemistry or long double-stranded donors can be synthesized by PCR amplification of a plasmid, or procured externally from an institutional oligonucleotide facility or commercial supplier. To inhibit nuclease degradation, additional synthesis modifications of the first and last three DNA donor bases is highly suggested, so that they are covalently connected to each other and to the rest of the DNA donor bases by phosphorothioate, rather than phosphodiester, linkages.
3.2. Mutagenic Triplex-Forming Molecules and Their Targeting Sites
A triplex-forming molecule is also an exogenously introduced molecule, which binds the major groove of duplex DNA targeting site to form a triplex, that, in turn, upregulates the recruitment of the endogenous cellular recombination machinery for mutagenic purposes upon the genome with the recombinagenic donor DNA with great specificity.
The concurrent treatment of the cellular genome with a highly specific triplex-forming molecule and a highly specific recombinagenic donor DNA ensures conditionally co-localized mutagenesis and recombination at targeting and editing sites in a safe and efficacious manner.
3.2.1. Design of Triplex-Forming Oligonucleotides (TFOs)
TFOs bind with high affinity to the genomic DNA duplex, either in the antiparallel purine motif, by being polypurine in TFO sequence content and binding antiparallel to the polypurine strand of the genomic DNA duplex, or in the parallel pyrimidine motif, by being polypyrimidine in TFO sequence content and binding parallel to the polypurine strand of the genomic DNA duplex [83] in both antiparallel and parallel motifs a genomic targeting site of successive homopurine, or polypurine, bases, at a length of 14–30 bases, should be chosen.
If this chosen genomic targeting site of polypurines is A-rich, then the pyrimidine motif is preferred and requires the TFO sequence content to be C and T or their analogs (C+ will form two Hoogsteen bonds with a G in a G:C base pair, and T will form two Hoogsteen bonds with the A in an A:T base pair) [83].
Alternatively, if this chosen genomic targeting site of polypurines is G-rich, then the purine motif is preferred and requires the TFO sequence content to be A (or T) and G or their analogs (G forms two reverse Hoogsteen bonds with the G in a G:C base pair, and A forms two reverser Hoogsteen bonds with the A in an A:T base pair) [83].
Like recombinagenic donor DNA, additional cellular and genomic components, such as other potentially hybridizable nucleic acid molecules and chromatin structure, respectively, can adversely affect the availability of TFOs and the accessibility of the genomic editing sites, and thus several genome targeting sites should be identified, for which TFOs should be designed and empirically evaluated accordingly.
3.2.2. Synthesis of Triplex-Forming Oligonucleotides (TFOs)
TFOs can also be can be synthesized internally in the laboratory through sequentially coupled solid-phase phosphoramidite chemistry, or procured externally from an institutional oligonucleotide facility or commercial supplier to create a molecule with a high binding affinity to the genomic DNA duplex. To inhibit nuclease degradation, additional synthesis modifications of the bases is highly suggested, so that they are covalently connected to each other by phosphoramidite, N,N-diethylethylenediamine (DEED), or phosphorothioate linkages, rather than phosphodiester, linkages (see Note 18).
3.2.3. Design of Triplex-forming Peptide Nucleic Acids (PNAs)
Triplex-forming PNAs also bind with high affinity to the genomic DNA duplex, either in aforementioned the antiparallel purine motif or parallel pyrimidine motif. Note again that in both antiparallel and parallel motifs a genomic targeting site of successive homopurine, or polypurine, bases, but, as opposed to TFOs, only at a lesser length of 8–10 bases due to PNA’s greater electrostatic complementarity, should be chosen. A preference for the pyrimidine motif for A-rich genomic targeting sites and for the purine motif for the G-rich genomic targeting sites also remains. Furthermore, as opposed to TFOs, triplex-forming PNAs in the pyrimidine motif can forgo the N3 atom protonation requirement of the C+ base in order to form Hoogsteen bonds with a G in a G:C base pair, a requirement that cannot usually be met in a physiological or therapeutic setting, by instead using a cytosine analog, pseudoisocytosine, J, to create two Hoogsteen bonds with the G in a G:C base pair on this triplex-forming strand [84].
In addition to the triplex-forming PNA, a second strand-invading PNA can be designed so that they form a triplex-forming bis-PNA. Usually this triplex-forming bis-PNA contains two pyrimidine PNAs, one that binds as a triplex to the duplex genomic DNA via Hoogsteen bonds and another that displaces one of the two duplex genomic DNA strands and binds the other genomic DNA strand via Watson-Crick bonds. These two pyrimidine PNAs are covalently bonded through a conformationally flexible linker, typically composed of (8-amino-3,6-dioxaoctanoic acid). Furthermore, the orientation of the N-to-C termini polyamide linkages in this bis-PNA peptide aligns in the same direction as the 5′−3′ phosphodiester linkages in the bound genomic DNA strand.
Furthermore, this triplex-forming bis-PNA design can be developed so that the stranding-invading PNA Watson-Crick bonding portion is lengthened into a “tail” portion beyond those genomic DNA bases bound by the triplex-forming Hoogsteen bonding, or “clamp,” portion. This triplex-forming tail-clamp PNA facilitates greater specificity by expanding the genomic DNA targeting site beyond the polypurines, and also lends itself to greater affinity created by the additional Watson-Crick bonding [27].
Additional affinity for the triplex-forming bis-PNA or tail-clamp PNA to the genomic DNA can be implemented by design through the conjugation of lysine residues, which are positively charged at physiological pH. This covalent addition of several lysine residues on the Watson-Crick bonding strand or as part of the linker confers electrostatic complementarity to the negatively charged genomic DNA and thus facilitates strand invasion. As an additional benefit, these positively charged lysine residues promote molecular transport into cells [85]. This transcellular uptake and nuclear affinity phenomena are not limited to lysine residues, but possible through other cell-penetrating peptide conjugations [86], including Antennapedia, which is composed of the third helix of the Drosophila homeodomain transcription factor [87, 88], a transduction domain from the trans-activator of transcription (Tat) protein, which is translated from the HIV genome as YGRKKRRQRRR [89], and a nuclear localization signal also of Tat, which is translated from the HIV genome as GRKKR [90, 91].
3.2.4. Synthesis of Triplex-Forming Peptide Nucleic Acids (PNAs)
Like TFOs, triplex-forming PNAs can be synthesized internally in the laboratory, or procured externally from an institutional facility or commercial supplier, in this case for peptide synthesis, to create a molecule with a high binding affinity to the genomic DNA duplex. Unlike TFOs, which can be synthesized using sequentially coupled solid-phase phosphoramidite chemistry, triplex-forming PNAs can be synthesized using Fmoc or Boc solid-phase peptide synthesis chemistry. Given the hybrid nature of triplex-forming PNAs, i.e., that they have nucleic acid bases but polyamide linkages and, thus, are neither wholly nucleic acids nor peptides, they are naturally resistant to cellular nucleases and proteases. Furthermore, as opposed to the negatively charged phosphodiester linkages in TFOs, the uncharged polyamide linkages in triplex-forming peptide-nucleic acids experience lesser electrostatic repulsion from the negatively charged phosphodiester linkages of the genomic DNA.
Like recombinagenic donor DNA and TFOs, additional cellular and genomic components, such as other potentially hybridizable nucleic acid molecules and chromatin structure, respectively, can adversely affect the availability of triplex-forming PNAs and the accessibility of the genomic editing sites, and thus several genome targeting sites should be identified, for which triplex-forming PNAs should be designed and empirically evaluated accordingly (see Note 19).
3.3. Molecular Delivery of Mutagenic and Recombinagenic Molecules
A number of delivery modalities have been developed to transport mutagenic and recombinagenic molecules into cells, including electroporation, nucleofection, cationic lipids, cell-penetrating peptides, and nanoparticles.
3.3.1. Electroporation
Electroporation utilizes an electric current between two conductive plates to temporarily destabilize the cellular membrane and thus permit the exogenously located recombinagenic and mutagenic molecules to enter the cells. These conductive plates are embedded in sterile cuvettes that hold the cells, the treatment recombinagenic molecules, and the mutagenic molecules, suspended in their native media or phosphate buffered solution (PBS). Upon electroporation, it is not unusual for a fraction of the cells to undergo apoptosis immediately, and thus appearing as cellular debris floating or falling out of suspension, or shortly thereafter when the electroporated sample is rescued onto fresh media plates for expansion. When electroporating a new cell type, it is suggested that, rather than recombinagenic or mutagenic molecules, a fluorescent reporter plasmid such as GFP be used as a surrogate to optimize the conditions for electroporation of the exogenously introduced molecules. After optimizing electroporation conditions, it is suggested that a dose response standard curve be performed to optimize the subsequent cellular utilization conditions for the exogenously introduced molecules.
3.3.2. Nucleofection
Nucleofection as a delivery modality for recombinagenic and mutagenic molecules can be applied to cells that are particularly not amenable to the aforementioned electroporation protocol. The nucleofection cuvettes and solutions are adapted to particular cell types to permit greater molecular uptake. However, as a consequence of its greater disturbance of the cellular membrane, only approximately a tenth of the cells that undergo nucleofection remain viable after a day [54]. Thus, while nucleofection as a delivery modality may be effective when prototyping the recombinagenic and mutagenic molecules in a laboratory setting, other modalities can be considered when translating the fully developed molecules in a therapeutic setting.
3.3.3. Cationic Lipids
The recombinagenic and mutagenic molecules can also be encapsulated in lipids that are cytosed by the cell membrane. However, there are electrostatic considerations to be met in the encapsulation process. Cationic lipids readily encapsulate the anionic recombinagenic donor DNA or TFOs without lysine residue conjugations; however, they are unable to encapsulate the net charge neutral, or if having lysine residues conjugations then net charge positive, triplex-forming PNAs. Thus cationic lipids are often useful when delivering recombinagenic donor DNA to the genomic DNA editing site, but this approach does not benefit from the concurrent delivery of the triplex-forming PNA to the corresponding targeting site that ultimately yields greater modification frequencies.
3.3.4. Cell-Penetrating Peptides
The recombinagenic and mutagenic molecules can also be conjugated with peptides that signal the cell for cellular transmembrane transport. These cell-penetrating peptides can be developed either by-design or empirically, or biologically inspired from nature, such as from the Antennapedia or Tat protein domains of the Drosophila homeodomain transcription factor and HIV Tat proteins, respectively. while these cell-penetrating peptides facilitate uptake without the harshness of electroporation and nucleofection, they may or may not inadvertently affect the interaction of the molecules to which they are conjugated with the genomic DNA. Once again, it is suggested that various designs of these molecules with various conjugations be designed and empirically evaluated.
3.3.5. Nanoparticles
The recombinagenic and mutagenic molecules and also be encapsulated in nanoparticles that are cytosed by the cell membrane. The nanoparticle formulations vary and thus are often more versatile than the cationic lipids to ferry a host of neutral, net positively, or net negatively charged molecules, either alone or encapsulated together across the cell membrane. For example, nanoparticles with the requisite molecular payload can be constructed using double emulsion solvent encapsulation of poly-lactic-co-glycolic acid (PLGA) alone or with poly-ethylene glycol (PEG), both suitable for a therapeutic setting across the world as they are well-characterized and regulatory agency-approved biopolymers [92]. Furthermore, upon molecular uptake, these nanoparticles often disintegrate into metabolites that are not cytotoxic and do not further interfere with the interactions of the recombinagenic and mutagenic molecules with the genomic DNA [93] (see Note 20).
3.4. Assessing Triplex-Forming Molecule Binding to Targeting Sites with Gel Shift Assays
3.4.1. TFO Binding Gel Shift Assay
A gel shift assay can be performed to assess the binding of TFOs. In preparation, a duplex DNA fragment with the TFO targeting site can be created by synthesizing and then annealing complementary oligomers, where one of the oligomers contains the site. To isolate successfully duplexed DNA from the non-duplexed oligomers, 5′ end-labeling with T4 Polynucleotide kinase and (γ−32P) dATP can be done with 10−6 M of duplexed DNA in a total volume of 20 μL. To purify this labeled, duplexed DNA, a 15 % polyacrylamide electrophoretic gel can be prepared and used for electroelution of the duplex DNA. To concentration the purified duplex, a centrifugal column concentrator, such as a Centricon-3 column from Millipore can be applied (see Note 21).
To assess the TFO binding, incubate a series of reactions of fixed concentration of duplex DNA and increasing concentrations of TFO in TFO triplex-binding buffer at 37 °C for 12–24 h. A typical reaction is 20 μL, with 2 μL (10−6 M) of labeled duplex and 2 μL of tenfold dilutions of TFO (10−12-10−7 M). This incubation will permit the steady-state binding of the TFO to its targeting site on the duplex DNA. To resolve the state of binding, the reactions should be electrophoresed on a 12 % native (19:1 acrylamide-bisacrylamide) gel at 60–70 V in gel running buffer to discriminate the bound triplex from the unbound duplex DNA. To image the run gel, a PhosphorImager can be used and each band shift can be assessed. Image processing software can be utilized to estimate relative kinetic dissociation constants and, more generally, ranges of TFO concentrations for subsequent empirical studies.
3.4.2. Triplex-Forming PNA Binding Gel Shift Assay
A gel shift assay can also be performed to assess the binding of triplex-forming PNAs (Fig. 3a).
In preparation, plasmid DNA with the target site, from a genomic insert or mutagenized plasmid segment or annealed duplex DNA insert, flanked by restriction sites should be prepared. This plasmid DNA will afford the supercoiled DNA duplex that is reminiscent of genomic DNA.
To assess the triplex-forming PNA binding, incubate a series of reactions a fixed concentration of this plasmid DNA, such as 2 μg in a final volume of 10 μL TE, with the target site and increasing concentrations of triplex-forming PNAs ranging from 0 to 1 μM, 10 μM of KCl at 37 °C for 24 h. Post-incubation, each reaction should be restriction digested in a final volume of 20 μL at a temperature at which all restriction enzymes remain active for 1–2 h. Then, each digestion should be stopped by heat inactivation and electrophoresed to yield the targeting site without the rest of the plasmid and with the bound triplex-forming PNA. The electrophoresis components include DNA loading dye for each sample and migration on a 10 % polyacrylamide gel or 8 % native gel (19:1 in TBE) until bands are well separated. In other words, a typical electrophoresis involves a 8 % native gel that can be made with 10 mL of 40 % 19:1 bis-acrylamide-acrylamide, 10 mL of 5× TB, 200 μL of 0.5 M EDTA, 29.25 mL of dH2O, 500 μL of 10 % ammonium persulfate, and 50 μL of TEMED, for a final volume of 50 mL. To resolve the resulting gel shift, initially stain with silver stain solution for 10 min, and then rinse the gel with dH2O for 1 min, and then develop with developer solution for 1 min, and again rinse the gel with dH2O for 1 min. With increasing concentrations of triplex-forming PNA, a single band representing the duplex DNA fragment containing the targeting site should transition to multiple bands, that represent the duplex DNA fragment with different triplex-binding modes [94], to shift higher, i.e., migrate less in the gel (see Note 22). To image the run gel, a standard computer scanner and scanning software can be used. In this manner, the triplex-forming PNA binding potentials for various designed synthetic molecules have been assessed for beta-globin targeting sites (Fig. 3b), gamma-globin targeting sites (Fig. 3c), and CCR5 targeting sites (Fig. 3d).
3.5. Quantifying Recombinagenic and Mutagenic Frequencies of Gene Correction with Fluorescent Assays of the Beta-Globin Genomic Locus
Fluorescent assays permit a facile means of sorting, counting, and thus quantifying cells that have undergone treatment with recombinagenic and mutagenic molecules. CHO reporter cells having a genome that harbors a split GFP transcript interrupted by the beta-globin IVS2 intron splicing mutation that causes beta-thalassemia provides a platform for such quantification of gene correction. Treatment of approximately one million of these CHO reporter cells with the recombinagenic donor carrying the A to G correction at position 1 of this IVS2 intron leads to a transcript of GFP that is also correctly spliced, thus yielding cells that fluoresce green (Fig. 4a). Further concurrent treatment of the CHO reporter cells with recombinagenic donor DNA and the triplex-forming PNA yields more cells that fluoresce green, and thus a greater gene correction frequency (Fig. 4b). Sort cells by fluorescent activated cell sorting (FACS) and analyze using flow cytometry software, such as FlowJo (see Note 23).
3.6. Quantifying Recombinagenic and Mutagenic Frequencies of Gene Induction and Regulation with Allele-Specific qPCR Assays of the Gamma-Globin Genomic Locus
Allele-specific PCR is a technique that is sensitive to genomic changes, and one that can combined with a qPCR technique that provides quantification by amplification, to produce an allele-specific qPCR assay. Different applications of this powerful technique can provide both qualitative and quantitative assessments of genome modification frequencies after treatment with recombinagenic donor DNA and triplex-forming molecules.
3.6.1. Genotyping Analysis by Allele-Specific PCR
After treating cells containing the target gene with TFO, triplex-forming PNA, and/or donor DNA, gene modification can be detected by allele-specific PCR [95, 96]. Harvest genomic DNA from the treated cells and dilute it to approximately 50 μg/μL. A forward primer can be designed with its 3′ end containing the desired mutation, and the reverse primer should be similar in length and Tm. Add 50 ng of the genomic DNA to a 25 μL PCR reaction and a gradient run to determine the optimal annealing temperature of the primers. Plasmids containing the wild-type and mutant gene can be used as control templates (see Note 24). The expected results are that a band should be present on the gel in lanes with mutant template and no bands for wild-type DNA (see Note 25). When combined with qPCR, allele-specific qPCR can use relative known frequencies of surrogates to extrapolate the gene induction and regulation frequency of, for example the usually dormant gamma-globin gene (Fig. 5a). After treatment with the recombinagenic and mutagenic molecules, the treated genome can be harvested and its unknown modification frequency can be extrapolated by correlating its allele-specific qPCR amplification values alongside those of known concentrations and thus modification frequencies of plasmids carrying the same uninduced gene sequence, i.e., the endogenous plasmid, and induced gene sequence, i.e., the codon-modified plasmid, in a milieu of genomic DNA, as well as those of a known concentration the untreated genome (see Note 26). In the case of gamma-globin, the cotreatment with the designed recombinagenic donor DNA and mutagenic triplex-forming PNA molecules yield a sequence modification frequency that is more than 560 % greater than that of treatment with the designed recombinagenic donor DNA alone (Fig. 5b).
3.7. Quantifying Recombinagenic and Mutagenic Frequencies of Gene Disruption with Single-Cell Cloning Assays of the CCR5 Genomic Locus
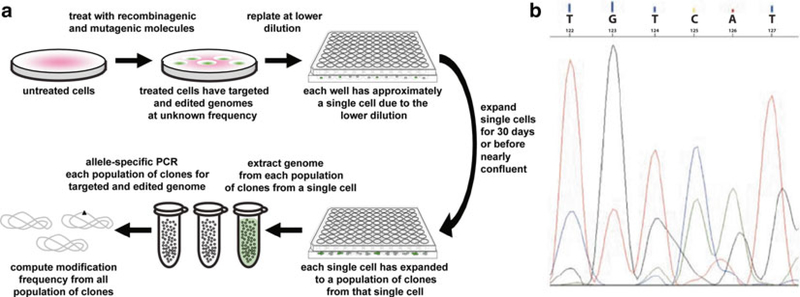
To quantify the amount of gene modification in cells treated with TFO, triplex-forming PNA, and/or donor DNA, the genomes of single-cell clones can be directly expanded and counted or deep sequenced [27, 82].
For counting and frequency quantification, such as those for the CCR5 gene, the treated cells can be replated at lower dilution. Cells can be treated in bulk and then diluted into multi-well dishes so that approximately a single cell on average occupies a single well. Each single-cell clone can proliferate within its dish to a density sufficient to extract the genomic DNA. a population of cell clones, rather than a single cell, is used to provide sufficient identical genomes for allele-specific PCR or direct sequencing (Fig. 6a). Based on the presence or absence or absence of the genome modification in each population, the cell that gave rise to that population can be assigned the same state, and all cells can thus be categorized to compute the modification frequency.
Fig. 6.
Quantifying recombinagenic and mutagenic frequencies of gene disruption with single-cell cloning assays of the CCR5 genomic locus. Single-cell cloning assays are used to (a) quantify the recombinagenic and mutagenic frequency of recombinagenic donor DNA alone or recombinagenic donor DNA co-transfected with mutagenic triplex-forming molecules, in the gene disruption of the CCR5 genomic locus by expanding each single cell to a population of clones sufficient in number to enable allele-specific PCR-based verification and calculation of modification frequency, (b) with sequencing chromatographic mixed peaks indicating a heterozygous clone (wild type sequence being TGTCAT; modified sequence being CTGAGG)
For direct deep sequencing, forward and reverse primers can be designed to insert barcode-tagging and to encapsulate the desired locus with the location of the mutation position centered, and be similar in length and Tm. Determine the optimal anneal temperature of the primers using a gradient run and 50 ng of the genomic DNA to a 25 μL PCR reaction. Gel-electrophorese the PCR products and purify the desired bands using a gel purification kit. Ligate the direct deep sequencing PCR samples to adapters and sequence with 75 base-pair paired-end reads on an Illumina HiSeq platform. The expected results are that a band should be present on the gel representing the amplified locus that was flanked by the primers. If one of the two alleles undergoes modification, a heterozygous clone with be created, as evidenced by mixed peaks in sequencing chromatograms (Fig. 5b). The ratio of the number of clones harboring the targeted modification to the total number of single-cell clones assayed represents the targeting frequency.
Acknowledgement
We gratefully acknowledge members of the Glazer Laboratory for helpful discussions. This work was supported by a National Institutes of Health (NIH) grant R01HL082655 and by a Doris Duke Innovations in Clinical Research Award (to P.M.G.). A National Institute of Diabetes and Digestive and Kidney Diseases Experimental and Human Pathobiology Postdoctoral Fellowship from NIH grant T32DK007556 also provided support (to F.R.). The authors declared no conflict of interest.
Footnotes
To increase resistance to intracellular nucleases, and thus bioavailability, the synthesis of the recombinagenic donor DNA should proceed with phosphorothioate, rather than phosphodiester, termini linkages on the first three bases on 5′ end and last three bases on 3′ end, and then these three subsequences, i.e., the two phosphorothiolated termini and the intervening phosphodiester sequence, can be linked.
If recombinagenic donor DNA is synthesized on a solid support, then a final deblocking followed by cleavage and deprotection step is necessary to free the final product from the support surface.
In order to maintain the integrity of recombinagenic donor DNAs when not in use, they should be aliquoted into smaller portions and stored at −20 °C. Also when in use for experiments, aliquots of recombinagenic donor DNA can be thawed and kept on ice.
To increase resistance to intracellular nucleases, and thus bioavailability, the synthesis of TFOs can progress with phosphoramidate, N, N-diethylethylenediamine (DEED), or phosphorothioate linkages.
The TFOs should be modified with one or more amine groups on the 3′ end to promote resistance to nucleases.
If triplex-forming peptide nucleic acid is synthesized on a solid support, then a final deblocking followed by cleavage and deprotection step is necessary to free the final product from the support surface.
Unnatural bases such as pseudoisocytosine should be procured and supplied to the institutional peptide synthesis facility.
The triplex-forming PNA should be modified with one or more terminal lysine residues to increase electrostatic complementarity, and thus affinity, to negatively charged genomic DNA.
While synthetically modified TFOs and triplex-forming PNAs are less prone to intracellular nuclease and nuclease and protease degradation, respectively, the latter also affords better electrostatic complementarity with the negatively charged genomic DNA. As such, triplex-forming PNAs are recommended over TFOs for use when possible.
Triplex-forming PNAs and TFOs should be aliquoted and keep in the −20 °C freezer for storage. Also during usage for experiments, an aliquot of triplex-forming PNAs can be thawed but kept cold on ice.
An antibiotic like hygromycin is light-sensitive. Thus, exercise caution when supplementing medium with hygromycin by shielding the medium flask with aluminum foil wrapping and/or use amber glass containers.
Antibiotics like penicillin and streptomycin are not light-sensitive. Shielding with aluminum foil wrapping or amber glass containers is not necessary.
Human primary CD34+ hematopoietic progenitor cells cannot be maintained indefinitely in their pluripotent state without differentiating. It is highly suggested that fresh or pre-frozen aliquots of these cells be procured immediately prior to use and then used within a few days.
This buffer has been composed so that it is appropriate for evaluating TFO and triplex-forming PNA binding under physiological conditions.
For ideal gel staining, silver stain solution should be fresh and, if possible, during the time the loaded gel is running through the electrophoresis apparatus.
For ideal gel developing, developer solution should be fresh and, if possible, during the time the loaded gel is being stained.
The optimal donor length and offsets from the recombination center will vary from editing site to site. This often makes it necessary to try various donor candidates and at various concentrations.
To prevent degradation by 3′ exonucleases it is recommended that TFOs have a 3′ end cap of an amine group chemical modification.
Triplex-forming PNAs do not require this chemical modification, since their peptide backbones make them resistant to degradation.
Use transfection methods and parameters that are suitable for the cell type in use. Some of these transfection methods include digitonin permeabilization, electroporation, cationic lipids, or nanoparticles. It is advisable to optimize the transfection method and parameters that provide high delivery efficiency and low cell damage and/or death.
The appropriate size Centricon column should be used based on the molecular weight of the duplex and/or triplex molecule to be filtered.
Rather than a single band, multiple bands may appear during the gel shift. This is not unexpected, since many triplex-forming PNAs have the ability to form multiple complex triplex structures with their target DNA.
Immediately after harvesting CHO reporter cells by trypsining the focal adhesions and washing with PBS, but prior to cell sorting, it is recommended that they be sterile-filtered through a 40 μm filter to further separate clusters of cells into individuals for sorting.
First the wild-type gene can be cloned, to which mutations can be introduced with site-directed mutagenesis via the QuikChange Site-Directed Mutagenesis Kit (Stratagene, LaJolla, CA, USA).
Initially, 40 cycles of PCR can be applied during allele-specific PCR using genomic DNA. In general, this provides sufficient amplification over background. For allele-specific PCR using plasmid DNA, since it is much easier to PCR-amplify from the latter, it is preferable to reduce the concentration rather than modify the allele-specific PCR cycling program, if possible.
Codon modifications, are silent modifications, and thus permit the wild-type protein to be transcribed and translated from a modified gene sequence of usually 6–8 base pairs, necessary for the sensitivity threshold of allele-specific PCR.
References
- 1.Smithies O, Gregg RG, Boggs SS, Koralewski MA, Kucherlapati RS (1985) Insertion of DNA sequences into the human chromosomal betaglobin locus by homologous recombination. Nature 317:230–234 [DOI] [PubMed] [Google Scholar]
- 2.Thomas KR, Capecchi MR (1987) Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell 51:503–512 [DOI] [PubMed] [Google Scholar]
- 3.Reza F, Zuo P, Tian J (2007) Protein interfacial pocket engineering via coupled computational filtering and biological focusing criterion. Ann Biomed Eng 35:1026–1036 [DOI] [PubMed] [Google Scholar]
- 4.Faruqi AF, Datta HJ, Carroll D, Seidman MM, Glazer PM (2000) Triple-helix formation induces recombination in mammalian cells via a nucleotide excision repair-dependent pathway. Mol Cell Biol 20:990–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Datta HJ, Glazer PM (2001) Intracellular generation of single-stranded DNA for chromosomal triplex formation and induced recombination. Nucleic Acids Res 29:5140–5147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vasquez KM, Christensen J, Li L, Finch RA, Glazer PM (2002) Human XPA and RPA DNA repair proteins participate in specific recognition of triplex-induced helical distortions. Proc Natl Acad Sci U S A 99:5848–5853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Datta HJ, Chan PP, Vasquez KM, Gupta RC, Glazer PM (2001) Triplex-induced recombination in human cell-free extracts - dependence on XPA and HsRad51. J Biol Chem 276: 18018–18023 [DOI] [PubMed] [Google Scholar]
- 8.Thoma BS, Vasquez KM (2003) Critical DNA damage recognition functions of XPC-hHR23B and XPA-RPA in nucleotide excision repair. Mol Carcinog 38:1–13 [DOI] [PubMed] [Google Scholar]
- 9.Thoma BS, Wakasugi M, Christensen J, Reddy MC, Vasquez KM (2005) Human XPC-hHR23B interacts with XPA-RPA in the recognition of triplex-directed psoralen DNA interstrand crosslinks. Nucleic Acids Res 33: 2993–3001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang G, Seidman MM, Glazer PM (1996) Mutagenesis in mammalian cells induced by triple helix formation and transcription-coupled repair. Science 271:802–805 [DOI] [PubMed] [Google Scholar]
- 11.Wang G, Levy DD, Seidman MM, Glazer PM (1995) Targeted mutagenesis in mammalian cells mediated by intracellular triple helix formation. Mol Cell Biol 15:1759–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Summers WC, Sarkar SN, Glazer PM (1985) Direct and inducible mutagenesis in mammalian cells. Cancer Surv 4:517–528 [PubMed] [Google Scholar]
- 13.Watson JD, Crick FHC (1953) Molecular structure of nucleic acids - a structure for deoxyribose nucleic acid. Nature 171:737–738 [DOI] [PubMed] [Google Scholar]
- 14.Felsenfeld G, Davies DR, Rich A (1957) Formation of a 3-stranded polynucleotide molecule. J Am Chem Soc 79:2023–2024 [Google Scholar]
- 15.Moser HE, Dervan PB (1987) Sequence-specific cleavage of double helical DNA by triple helix formation. Science 238:645–650 [DOI] [PubMed] [Google Scholar]
- 16.Le Doan T, Perrouault L, Praseuth D, Habhoub N, Decout JL, Thuong NT et al. (1987) Sequence-specific recognition, photocrosslinking and cleavage of the DNA double helix by an oligo-[alpha]-thymidylate covalently linked to an azidoproflavine derivative. Nucleic Acids Res 15:7749–7760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faruqi AF, Krawczyk SH, Matteucci MD, Glazer PM (1997) Potassium-resistant triple helix formation and improved intracellular gene targeting by oligodeoxyribonucleotides containing 7-deazaxanthine. Nucleic Acids Res 25:633–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalish JM, Seidman MM, Weeks DL, Glazer PM (2005) Triplex-induced recombination and repair in the pyrimidine motif. Nucleic Acids Res 33:3492–3502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Camerini-Otero RD, Hsieh P (1993) Parallel DNA triplexes, homologous recombination, and other homology-dependent DNA interactions. Cell 73:217–223 [DOI] [PubMed] [Google Scholar]
- 20.Seidman MM (2004) Oligonucleotide mediated gene targeting in mammalian cells. Curr Pharm Biotechnol 5:421–430 [DOI] [PubMed] [Google Scholar]
- 21.Demidov VV, Potaman VN, Frankkamenetskii MD, Egholm M, Buchard O, Sonnichsen SH et al. (1994) Stability of peptide nucleic-acids in human serum and cellular-extracts. Biochem Pharmacol 48:1310–1313 [DOI] [PubMed] [Google Scholar]
- 22.Ray A, Norden B (2000) Peptide nucleic acid (PNA): its medical and biotechnical applications and promise for the future. FASEB J 14: 1041–1060 [DOI] [PubMed] [Google Scholar]
- 23.Rooney SM, Moore PD (1995) Antiparallel, intramolecular triplex DNA stimulates homologous recombination in human cells. Proc Natl Acad Sci U S A 92:2141–2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walter A, Schutz H, Simon H, Birch-Hirschfeld E (2001) Evidence for a DNA triplex in a recombination-like motif: I. Recognition of Watson-Crick base pairs by natural bases in a high-stability triplex. J Mol Recog 14: 122–139 [DOI] [PubMed] [Google Scholar]
- 25.Faruqi AF, Egholm M, Glazer PM (1998) Peptide nucleic acid-targeted mutagenesis of a chromosomal gene in mouse cells. Proc Natl Acad Sci U S A 95:1398–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chin JY, Kuan JY, Lonkar PS, Krause DS, Seidman MM, Peterson KR et al. (2008) Correction of a splice-site mutation in the betaglobin gene stimulated by triplex-forming peptide nucleic acids. Proc Natl Acad Sci U S A 105:13514–13519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schleifman EB, Bindra R, Leif J, Del Campo J, Rogers FA, Uchil P et al. (2011) Targeted disruption of the CCR5 gene in human hematopoietic stem cells stimulated by peptide nucleic acids. Chem Biol 18:1189–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guieysse AL, Praseuth D, Francois JC, Helene C (1995) Inhibition of replication initiation by triple helix-forming oligonucleotides. Biochem Biophys Res Commun 217:186–194 [DOI] [PubMed] [Google Scholar]
- 29.Jain A, Magistri M, Napoli S, Carbone GM, Catapano CV (2010) Mechanisms of triplex DNA-mediated inhibition of transcription initiation in cells. Biochimie 92:317–320 [DOI] [PubMed] [Google Scholar]
- 30.Ferdous A, Akaike T, Maruyama A (2000) Inhibition of sequence-specific protein-DNA interaction and restriction endonuclease cleavage via triplex stabilization by poly(L-lysine)-graft-dextran copolymer. Biomacromolecules 1:186–193 [DOI] [PubMed] [Google Scholar]
- 31.Hanvey JC, Peffer NJ, Bisi JE, Thomson SA, Cadilla R, Josey JA et al. (1992) Antisense and antigene properties of peptide nucleic-acids. Science 258:1481–1485 [DOI] [PubMed] [Google Scholar]
- 32.Koppelhus U, Zachar V, Nielsen PE, Liu XD, EugenOlsen J, Ebbesen P (1997) Efficient in vitro inhibition of HIV-1 gag reverse transcription by peptide nucleic acid (PNA) at minimal ratios of PNA/RNA. Nucleic Acids Res 25:2167–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Praseuth D, Grigoriev M, Guieysse AL, Pritchard LL, HarelBellan A, Nielsen PE et al. (1996) Peptide nucleic acids directed to the promoter of the alpha-chain of the interleukin-2 receptor. Biochim Biophys Acta 1309:226–238 [DOI] [PubMed] [Google Scholar]
- 34.Nielsen PE, Egholm M, Berg RH, Buchardt O (1993) Sequence specific-inhibition of DNA restriction enzyme cleavage by PNA. Nucleic Acids Res 21:197–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mollegaard NE, Buchardt O, Egholm M, Nielsen PE (1994) Peptide nucleic-acid.DNA strand displacement loops as artificial transcription promoters. Proc Natl Acad Sci U S A 91:3892–3895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bahal R, McNeer NA, Ly DH, Saltzman WM, Glazer PM (2013) Nanoparticle for delivery of antisense gammaPNA oligomers targeting CCR5. Artif DNA PNA XNA 4:49–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chin JY, Reza F, Glazer PM (2013) Triplex-forming peptide nucleic acids induce heritable elevations in gamma-globin expression in hematopoietic progenitor cells. Mol Ther 21(3):580–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Faria M, Wood CD, Perrouault L, Nelson JS, Winter A, White MRH et al. (2000) Targeted inhibition of transcription elongation in cells mediated by triplex-forming oligonucleotides. Proc Natl Acad Sci U S A 97:3862–3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Birg F, Praseuth D, Zerial A, Thuong NT, Asseline U, Ledoan T et al. (1990) Inhibition of simian virus-40 DNA-replication in CV-1 cells by an oligodeoxynucleotide covalently linked to an intercalating agent. Nucleic Acids Res 18:2901–2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Volkmann S, Jendis J, Frauendorf A, Moelling K (1995) Inhibition of HIV-1 reverse transcription by triple-helix forming oligonucleotides with viral RNA. Nucleic Acids Res 23: 1204–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Diviacco S, Rapozzi V, Xodo L, Helene C, Quadrifoglio F, Giovannangeli C (2001) Site-directed inhibition of DNA replication by triple helix formation. FASEB J 15:2660–2668 [DOI] [PubMed] [Google Scholar]
- 42.Maher LJ, Wold B, Dervan PB (1989) Inhibition of DNA-binding proteins by oligonucleotide-directed triple helix formation. Science 245:725–730 [DOI] [PubMed] [Google Scholar]
- 43.Francois JC, Saisonbehmoaras T, Thuong NT, Helene C (1989) Inhibition of restriction endonuclease cleavage via triple helix formation by homopyrimidine oligonucleotides. Biochemistry 28:9617–9619 [DOI] [PubMed] [Google Scholar]
- 44.Hanvey JC, Shimizu M, Wells RD (1990) Site-specific inhibition of EcoRI restriction modification enzymes by a DNA triple helix. Nucleic Acids Res 18:157–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mayfield C, Ebbinghaus S, Gee J, Jones D, Rodu B, Squibb M et al. (1994) Triplex formation by the human Ha-Ras promoter inhibits Sp1 binding and in-vitro transcription. J Biol Chem 269:18232–18238 [PubMed] [Google Scholar]
- 46.Havre PA, Gunther EJ, Gasparro FP, Glazer PM (1993) Targeted mutagenesis of DNA using triple helix-forming oligonucleotides linked to psoralen. Proc Natl Acad Sci U S A 90:7879–7883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takasugi M, Guendouz A, Chassignol M, Decout JL, Lhomme J, Thuong NT et al. (1991) Sequence-specific photoinduced crosslinking of the 2 strands of double-helical DNA by a psoralen covalently linked to a triple helixforming oligonucleotide. Proc Natl Acad Sci U S A 88:5602–5606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vasquez KM, Wensel TG, Hogan ME, Wilson JH (1996) High-efficiency triple-helix-mediated photo-cross-linking at a targeted site within a selectable mammalian gene. Biochemistry 35: 10712–10719 [DOI] [PubMed] [Google Scholar]
- 49.Vasquez KM, Narayanan L, Glazer PM (2000) Specific mutations induced by triplex-forming oligonucleotides in mice. Science 290:530–533 [DOI] [PubMed] [Google Scholar]
- 50.Wang X, Tolstonog G, Shoeman RL, Traub P (1996) Selective binding of specific mouse genomic DNA fragments by mouse vimentin filaments in vitro. DNA Cell Biol 15:209–225 [DOI] [PubMed] [Google Scholar]
- 51.Chan PP, Lin M, Faruqi AF, Powell J, Seidman MM, Glazer PM (1999) Targeted correction of an episomal gene in mammalian cells by a short DNA fragment tethered to a triplex-forming oligonucleotide. J Biol Chem 274:11541–11548 [DOI] [PubMed] [Google Scholar]
- 52.Luo ZJ, Macris MA, Faruqi AF, Glazer PM (2000) High-frequency intrachromosomal gene conversion induced by triplex-forming oligonucleotides microinjected into mouse cells. Proc Natl Acad Sci U S A 97:9003–9008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rogers FA, Manoharan M, Rabinovitch P, Ward DC, Glazer PM (2004) Peptide conjugates for chromosomal gene targeting by triplex-forming oligonucleotides. Nucleic Acids Res 32:6595–6604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McNeer NA, Chin JY, Schleifman EB, Fields RJ, Glazer PM, Saltzman WM (2011) Nanoparticles deliver triplex-forming PNAs for site-specific genomic recombination in CD34+ human hematopoietic progenitors. Mol Ther 19: 172–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Branden LJ, Mohamed AJ, Smith CIE (1999) A peptide nucleic acid-nuclear localization signal fusion that mediates nuclear transport of DNA. Nat Biotechnol 17:784–787 [DOI] [PubMed] [Google Scholar]
- 56.Vasquez KM, Dagle JM, Weeks DL, Glazer PM (2001) Chromosome targeting at short polypurine sites by cationic triplex-forming oligonucleotides. J Biol Chem 276:38536–38541 [DOI] [PubMed] [Google Scholar]
- 57.Lacroix L, Lacoste J, Reddoch JF, Mergny JL, Levy DD, Seidman MM et al. (1999) Triplex formation by oligonucleotides containing 5-(1-propynyl)-2′-deoxyuridine: decreased magnesium dependence and improved intracellular gene targeting. Biochemistry 38:1893–1901 [DOI] [PubMed] [Google Scholar]
- 58.Puri N, Majumdar A, Cuenoud B, Natt F, Martin P, Boyd A et al. (2002) Minimum number of 2′-O-(2-aminoethyl) residues required for gene knockout activity by triple helix forming oligonucleotides. Biochemistry 41:7716–7724 [DOI] [PubMed] [Google Scholar]
- 59.Chin JY, Schleifman EB, Glazer PM (2007) Repair and recombination induced by triple helix DNA. Front Biosci 12:4288–4297 [DOI] [PubMed] [Google Scholar]
- 60.Raha M, Lacroix L, Glazer PM (1998) Mutagenesis mediated by triple helix-forming oligonucleotides conjugated to psoralen: effects of linker arm length and sequence context. Photochem Photobiol 67:289–294 [PubMed] [Google Scholar]
- 61.Raha M, Wang G, Seidman MM, Glazer PM (1996) Mutagenesis by third-strand-directed psoralen adducts in repair-deficient human cells: high frequency and altered spectrum in a xeroderma pigmentosum variant. Proc Natl Acad Sci U S A 93:2941–2946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang G, Glazer PM (1995) Altered repair of targeted psoralen photoadducts in the context of an oligonucleotide-mediated triple-helix. J Biol Chem 270:22595–22601 [DOI] [PubMed] [Google Scholar]
- 63.Majumdar A, Puri N, Cuenoud B, Natt F, Martin P, Khorlin A et al. (2003) Cell cycle modulation of gene targeting by a triple helixforming oligonucleotide. J Biol Chem 278: 11072–11077 [DOI] [PubMed] [Google Scholar]
- 64.Macris MA, Glazer PM (2003) Transcription dependence of chromosomal gene targeting by triplex-forming oligonucleotides. J Biol Chem 278:3357–3362 [DOI] [PubMed] [Google Scholar]
- 65.Vasquez KM, Wang G, Havre PA, Glazer PM (1999) Chromosomal mutations induced by triplex-forming oligonucleotides in mammalian cells. Nucleic Acids Res 27:1176–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sargent RG, Rolig RL, Kilburn AE, Adair GM, Wilson JH, Nairn RS (1997) Recombination-dependent deletion formation in mammalian cells deficient in the nucleotide excision repair gene ERCC1. Proc Natl Acad Sci U S A 94:13122–13127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Faruqi AF, Seidman MM, Segal DJ, Carroll D, Glazer PM (1996) Recombination induced by triple-helix-targeted DNA damage in mammalian cells. Mol Cell Biol 16:6820–6828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sandor Z, Bredberg A (1995) Triple-helix directed psoralen adducts induce a low-frequency of recombination in an Sv40 shuttle vector. Biochim Biophys Acta 1263:235–240 [DOI] [PubMed] [Google Scholar]
- 69.Knauert MP, Kalish JM, Hegan DC, Glazer PM (2006) Triplex-stimulated intermolecular recombination at a single-copy genomic target. Mol Ther 14:392–400 [DOI] [PubMed] [Google Scholar]
- 70.Schleifman EB, Chin JY, Glazer PM (2008) Triplex-mediated gene modification. Methods Mol Biol 435:175–190 [DOI] [PubMed] [Google Scholar]
- 71.Knauert MP, Lloyd JA, Rogers FA, Datta HJ, Bennett ML, Weeks DL et al. (2005) Distance and affinity dependence of triplex-induced recombination. Biochemistry 44:3856–3864 [DOI] [PubMed] [Google Scholar]
- 72.Shahid KA, Majumdar A, Alam R, Liu ST, Kuan JY, Sui XF et al. (2006) Targeted crosslinking of the human beta-globin gene in living cells mediated by a triple helix forming oligonucleotide. Biochemistry 45:1970–1978 [DOI] [PubMed] [Google Scholar]
- 73.Friedman S, Schwartz E (1976) Hereditary persistence of foetal haemoglobin with β-chain synthesis in cis position (Gy-β + -HPFH) in a negro family. Nature 259:138–140 [DOI] [PubMed] [Google Scholar]
- 74.Xu XS, Glazer PM, Wang G (2000) Activation of human gamma-globin gene expression via triplex-forming oligonucleotide (TFO)-directed mutations in the gamma-globin gene 5′ flanking region. Gene 242:219–228 [DOI] [PubMed] [Google Scholar]
- 75.Samson M, Libert F, Doranz BJ, Rucker J, Liesnard C, Farber CM et al. (1996) Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature 382:722–725 [DOI] [PubMed] [Google Scholar]
- 76.Steinberger P, Andris-Widhopf J, Buhler B, Torbett BE, Barbas CF 3rd (2000) Functional deletion of the CCR5 receptor by intracellular immunization produces cells that are refractory to CCR5-dependent HIV-1 infection and cell fusion. Proc Natl Acad Sci U S A 97:805–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hutter G, Nowak D, Mossner M, Ganepola S, Mussig A, Allers K et al. (2009) Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med 360: 692–698 [DOI] [PubMed] [Google Scholar]
- 78.Anderson J, Akkina R (2007) Complete knockdown of CCR5 by lentiviral vector-expressed siRNAs and protection of transgenic macrophages against HIV-1 infection. Gene Ther 14:1287–1297 [DOI] [PubMed] [Google Scholar]
- 79.Gorman M, Glazer PM (2006) Directed gene modification via triple helix formation. Curr Mol Med 1:391–399 [DOI] [PubMed] [Google Scholar]
- 80.Schleifman EB, Glazer PM (2014) Peptide nucleic acid-mediated recombination for targeted genomic repair and modification. Methods Mol Biol 1050:207–222 [DOI] [PubMed] [Google Scholar]
- 81.Shen H, Cheng T, Preffer FI, Dombkowski D, Tomasson MH, Golan DE et al. (1999) Intrinsic human immunodeficiency virus type 1 resistance of hematopoietic stem cells despite coreceptor expression. J Virol 73:728–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McNeer NA, Schleifman EB, Cuthbert A, Brehm M, Jackson A, Cheng C et al. (2013) Systemic delivery of triplex-forming PNA and donor DNA by nanoparticles mediates site-specific genome editing of human hematopoietic cells in vivo. Gene Ther 20:658–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Reza F, Glazer PM (2014) Triplex-mediated genome targeting and editing. Methods Mol Biol 1114:115–142 [DOI] [PubMed] [Google Scholar]
- 84.Egholm M, Christensen L, Dueholm KL, Buchardt O, Coull J, Nielsen PE (1995) Efficient Ph-independent sequence-specific DNA-binding by pseudoisocytosine-containing Bis-Pna. Nucleic Acids Res 23:217–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sazani P, Kang SH, Maier MA, Wei CF, Dillman J, Summerton J et al. (2001) Nuclear antisense effects of neutral, anionic and cationic oligonucleotide analogs. Nucleic Acids Res 29:3965–3974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Koppelhus U, Awasthi SK, Zachar V, Holst HU, Ebbesen P, Nielsen PE (2002) Cell-dependent differential cellular uptake of PNA, peptides, and PNA-peptide conjugates. Antisense Nucleic Acid Drug Dev 12:51–63 [DOI] [PubMed] [Google Scholar]
- 87.Karagiannis ED, Urbanska AM, Sahay G, Pelet JM, Jhunjhunwala S, Langer R et al. (2013) Rational design of a biomimetic cell penetrating peptide library. ACS Nano 7:8616–8626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sagan S, Burlina F, Alves ID, Bechara C, Dupont E, Joliot A (2013) Homeoproteins and homeoprotein-derived peptides: going in and out. Curr Pharm Des 19:2851–2862 [DOI] [PubMed] [Google Scholar]
- 89.Schwarze SR, Hruska KA, Dowdy SF (2000) Protein transduction: unrestricted delivery into all cells? Trends Cell Biol 10:290–295 [DOI] [PubMed] [Google Scholar]
- 90.Hauber J, Malim MH, Cullen BR (1989) Mutational analysis of the conserved basic domain of human immunodeficiency virus tat protein. J Virol 63:1181–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ruben S, Perkins A, Purcell R, Joung K, Sia R, Burghoff R et al. (1989) Structural and functional characterization of human immunodeficiency virus tat protein. J Virol 63:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schleifman EB, McNeer NA, Jackson A, Yamtich J, Brehm MA, Shultz LD et al. (2013) Site-specific genome editing in PBMCs with PLGA nanoparticle-delivered PNAs confers HIV-1 resistance in humanized mice. Mol Ther Nucleic Acids 2:e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.McNeer NA, Schleifman EB, Glazer PM, Saltzman WM (2011) Polymer delivery systems for site-specific genome editing. J Control Rel 155:312–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hansen GI, Bentin T, Larsen HJ, Nielsen PE (2001) Structural isomers of bis-PNA bound to a target in duplex DNA. J Mol Biol 307:67–74 [DOI] [PubMed] [Google Scholar]
- 95.Orou A, Fechner B, Utermann G, Menzel HJ (1995) Allele-specific competitive blocker Pcr - a one-step method with applicability to pool screening. Hum Mutat 6:163–169 [DOI] [PubMed] [Google Scholar]
- 96.Parsons BL, McKinzie PB, Heflich RH (2005) Allele-specific competitive blocker-PCR detection of rare base substitution. Methods Mol Biol 291:235–245 [DOI] [PubMed] [Google Scholar]
- 97.Williams LD, Egli M, Ughetto G, van der Marel GA, van Boom JH, Quigley GJ et al. (1990) Structure of 11-deoxydaunomycin bound to DNA containing a phosphorothioate. J Mol Biol 215:313–320 [DOI] [PubMed] [Google Scholar]
- 98.Radhakrishnan I, Patel DJ (1994) Solution structure of a pyrimidine.purine.pyrimidine DNA triplex containing T.AT, C+.GC and G.TA triples. Structure 2:17–32 [DOI] [PubMed] [Google Scholar]
- 99.Yeh JI, Pohl E, Truan D, He W, Sheldrick GM, Du S et al. (2010) The crystal structure of non-modified and bipyridine-modified PNA duplexes. Chemistry 16:11867–11875 [DOI] [PMC free article] [PubMed] [Google Scholar]