Abstract
Enterohepatic circulation is responsible for the capture of bile acids and other steroids produced or metabolized in the liver and secreted to the intestine, for reabsorption back into the circulation and transport back to the liver. Bile acids are secreted from the liver in the form of mixed micelles that also contain phosphatidylcholines and cholesterol that facilitate the uptake of fats and vitamins from the diet due to the surfactant properties of bile acids and lipids. Bile acids are synthesized in the liver from cholesterol by a cascade of enzymes that carry out oxidation and conjugation reactions, and transported to the bile duct and gall bladder where they are stored before being released into the intestine. Bile flow from the gall bladder to the small intestine is triggered by food intake in accordance with its role in lipid and vitamin absorption from the diet. Bile acids are further metabolized by gut bacteria and are transported back to the circulation. Metabolites produced in the liver are termed primary bile acids or primary conjugated bile salts, while the metabolites generated by bacterial are called secondary bile acids. About 95% of bile acids are reabsorbed in the proximal and distal ileum into the hepatic portal vein and then into the liver sinusoids, where they are efficiently transported into the liver with little remaining in circulation. Each bile acid is reabsorbed about 20 times on average before being eliminated. Enterohepatic circulation is under tight regulation by nuclear receptor signaling, notably by the farnesoid X receptor (FXR).
Enterohepatic Circulation of Bile Acids
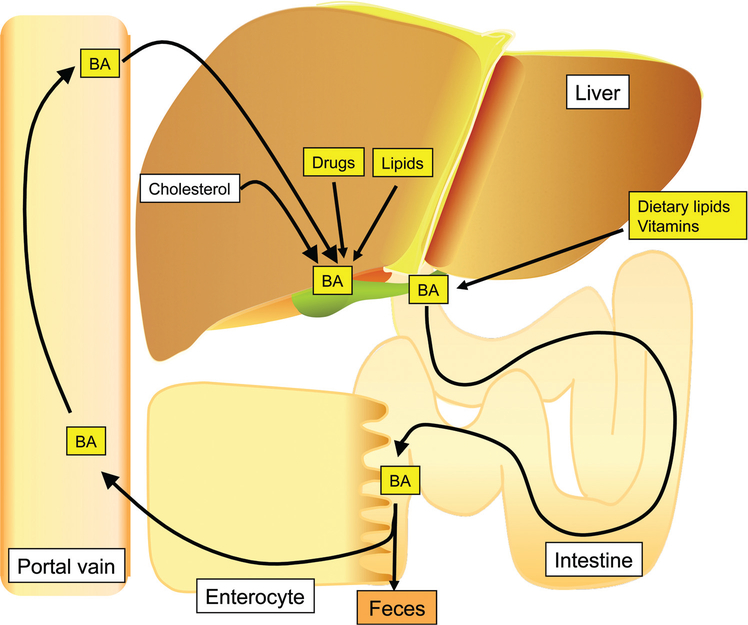
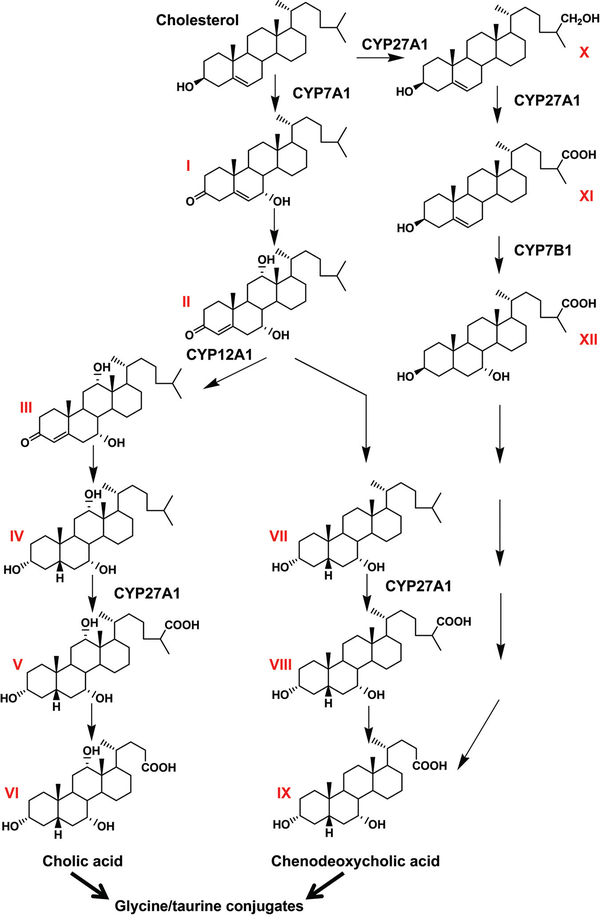
Enterohepatic circulation is responsible for the capture of bile acids and other steroids produced or metabolized in the liver and secreted to the intestine, for reabsorption back into the circulation and transport back to the liver (Fig. 1). Bile acids are synthesized in the liver from cholesterol by a series of reactions that include oxidation by cytochromes P450s (CYP) CYP7A1, CYP6B1, and CYP27A1, followed by conjugation with taurine or glycine by bile acid-CoA ligase (BAL) and bile acid-CoA:amino acid N-acyltransferase (BAT) (126). There are two pathways for bile acid synthesis in the liver, the neutral pathway that proceeds through CYP7A1 metabolism of cholesterol leading to cholic acid (CA) and chenodeoxycholic acid (CDCA), and an acidic pathway initiated by CYP27A1 oxidation of cholesterol resulting in CDCA (Fig. 2). Both pathways ultimately terminate in amino acid-conjugated metabolites destined for secretion into the bile. In mice, BAT uses taurine as a cofactor for conjugation while human BAT uses taurine or glycine as a substrate. The resultant metabolites are amphipathic with one hydrophilic face and one hydrophobic face, thus facilitating their incorporation into mixed micelles containing bile acids, phosphatidylcholines, and cholesterol (62). The bile acids produced in liver cannot diffuse across membranes into circulation and thus can only be eliminated by transport out of the hepatocytes to the bile by transporter proteins. In humans, about 0.5 g/d of bile acids produced from cholesterol in the liver is excreted in the feces as a route for elimination of cholesterol. However, most bile acids are reabsorbed in the intestine and transported back to the liver. Approximately 95% of bile acids are reabsorbed in the ileum with about 3 g being recycled several times a day. In the intestine, bile acids are transported or diffuse across the enterocytes lining the intestine to the basolateral membrane and excreted into portal blood for circulation back to the hepatocytes sinusoids. This pathway was exploited as one of the first methods to lower cholesterol in humans by administration of positively charged resins or sequestrants such as cholestyramine (Questran®) that bind to negatively charged bile acids so that they are not transported back to the body but are eliminated in the feces. As more bile acids are removed, less are reabsorbed, thus lowering hepatic bile acid levels. Cholesterol is then converted to bile acids as a mechanism to normalize bile acid levels lost by use of the sequestrants, thus resulting in lower plasma cholesterol concentrations. Bile acid production not only serves to eliminate cholesterol but also facilitates the excretion of lipids and steroids and aids in the absorption of lipids and vitamins from the diet (19, 62). Bile acid micelles markedly increase the absorption of fatty acids and monoglycerides, as well as fat-soluble vitamins into the enterocyte and ultimately the circulation for distribution to the tissues.
Figure 1.
Enterohepatic circulation of bile acids. Bile acids and other steroids produced or metabolized in the liver and secreted to the intestine, for reabsorption back into the circulation and transport back to the liver. Coincident with bile acid recirculation is the removal and reuptake of drugs, usually conjugated high Mr drugs. Cholesterol is the substrate for bile acid synthesis, one of the routes for mammalian disposal of cholesterol. The author thanks Tsutomu Matsubara for help in preparing this figure.
Figure 2.
Pathway for the synthesis of bile acids. Neutral pathway: cholesterol is converted to 7α-hydroxycholesterol (I) by CYP7A1, the rate-limiting enzyme in cholesterol degradation in the major neutral pathway of bile acid synthesis. 7α-Hydroxycholesterol is reduced to 7α-hydroxy-4-cholesten-3-one (II), which is metabolized to 7α,12,-dihydroxy-4-cholesten-3-one (III) by CYP12A1 or reduced to 5β-cholestane-3α,7α-diol (VII). 7α,12,-Dihydroxy-4-cholesten-3-one is oxidized to 5β-cholestane-3α,7α,12,-triol (IV), and 5β-cholestane-3α,7α-diol is oxidized by CYP27A1 to 3α,7α-dihydroxy-5β-cholestanoic acid (VIII), which is then metabolized to the terminal metabolite chenodeoxycholic acid (IX) through a demethylation reaction. 5β-Cholestane-3α,7α,12,-triol is also oxidized to 3α,7α,12,-trihydroxy-5β-cholestanoic acid (V) by CYP27A1, which is converted to the terminal metabolite cholic acid (VI) by a demethylation reaction. Acidic pathway: cholesterol is also converted to 27-hydroxycholesterol (X) and 3β-hydroxy-5-cholestenoic acid (XI) by CYP27A1 in the minor acidic pathway. 3β-Hydroxy-5-cholestenoic acid is metabolized to 3β,7α-hydroxy-5-cholestenoic acid (XII) by CYP7B1. A series of reactions converts 3β,7α-hydroxy-5-cholestenoic acid to chenodeoxycholic acid (IX). The author thanks Fei Li for making this figure.
Bile acid metabolism not only occurs in the liver, but also in the gut by bacteria in the small intestine and colon, thus resulting in a very complex array of metabolites. Among the reactions carried out by anaerobic bacteria such as Staphylococcus and Lactobacillus species, is dehydroxylation and deconjugation of the taurine and glycine conjugates of bile acids produced in the liver by various hydrolases leading to the production of secondary bile acids such as DCA, lithocholic acid (LCA), and ursodeoxycholic acid (124).
The Nuclear Receptor Superfamily
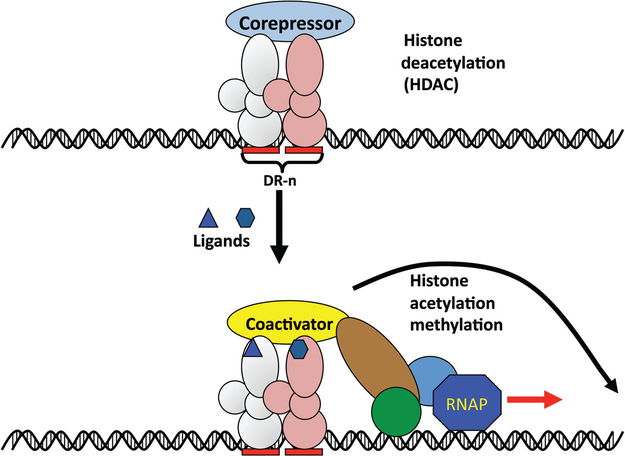
Nuclear receptors are a superfamily of transcription factors that control many aspects of mammalian development and physiology. They consist of 48 members in humans (49 in mice), and an early classification system divided them into four groups, Class I–Class IV, based on their ligand-binding capacity, dimerization properties, and cellular localization(99). Class I receptors that include the steroid hormones such as estrogen receptor (ER; NR3A1), androgen receptor (AR; NR3C4), and progesterone receptor (PR; NR3C3) bind to DNA as homodimers, while Class II receptors, which encompass the retinoic acid receptor γ (RARγ; NR1B3), vitamin D receptor (VDR; NR1I1) and others, bind to DNA as heterodimers, usually with the retinoid X receptor α (RXRα; NR2B1), although there are receptors in this group that form homodimers (110). Class III receptors bind primarily to direct repeats (DRs) as homodimers, while Class IV receptors typically bind to extended core sites as monomers. Nuclear receptors that have no identified ligand have been termed orphan receptors and usually fall into Class III and Class IV. In some cases, ligands have been discovered for orphan receptors while some receptors may activate gene expression in the absence of ligand binding. A more functional classification of nuclear receptors has been generated to distinguish those involved in control of (i) endocrine functions,(ii) metabolism, and (iii) mammalian development, although there is some overlap, particularly between those receptors involved in metabolism and mammalian development (Table 1) (101). Most nuclear receptors share a domain structure consisting of a variable N-terminal domain (AF1), a two zinc finger-containing DNA-binding domain (DBD) that is the most conserved in the nuclear receptor superfamily, a hinge region, ligand-binding domain (LBD), and a highly variable C-terminal domain (AF2) (Fig. 3). The N-terminal and C-terminal AF1 and AF2 domains, respectively, interact with other transcriptional accessory proteins including coactivators and corepressors, and other components of the transcriptional machinery. The general mechanism of transcriptional activation by the nuclear receptors is displayed in Figure 4, showing the metabolic sensor group of receptors that form heterodimers with RXR, that in the absence of a ligand, either reside in the nucleus bound to DNA and are transcriptionally inactive as a result of being associated with corepressors, hi-stone deacetylases (HDAC) and inactive chromatin, or found in the cytoplasm and translocated to the nucleus upon ligand binding. The nonproductive (transcriptionally inactive) DNA binding could be to specific regulatory regions of target genes for the receptors, or nonspecific binding to DNA. For transcriptional activation of target genes, a nuclear receptor binds to specific DNA recognition elements. In the case of those receptors that form heterodimers, such as constitutive androstane receptor (CAR; NR1I3), pregnane X receptor (PXR; NR1I2), FXR (NR1H4), and peroxisome proliferator-activated receptor α (PPARα; NR1C1) (Table 1), these are usually DR elements where one repeat binds the functionally relevant partner while the other repeat binds RXRα. The DRs are separated by one or more nucleotides. For example, a direct repeat-1 (DR-1) is a DR separated by one nucleotide while a DR-2 is a DR separated by two nucleotides. This binding initiates a cascade of events that involve reorganization of chromatin through recruitment of coactivators, histone acetylation and histone methylation and other events (Fig. 4). Among the coactivators for nuclear receptors in liver are the steroid receptor coactivators 1 (SRC-1), transcription intermediary factor 2/glucocorticoid receptor interacting protein or TIF/GRIP (SRC-2), cAMP-response element-binding protein-binding protein (CREB) and peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α), among others (161).
Table 1.
The Nuclear Receptor Superfamily
| Endocrine | Metabolic | Developmental | |||
|---|---|---|---|---|---|
| Receptor | Ligand | Receptor | Ligand | Receptor | Ligand |
| AR | Androgens | CAR | Xenobiotics | ERR | Xenobiotics |
| ER | Estrogens | FXR | Bile acids | COUP-TFα, β, γ | |
| GR | Glucocorticoids | HNF4α, γ | Fatty acids (embedded) | DAX-1 | |
| MR | Mineral corticoids | LXRα, β | Oxysterols | GCNF | |
| PR | Progesterone | PPARα, β, γ | Xenobiotics, fatty acids | LRH-1 | Phospholipids |
| RARα, β, γ | Retinoic acid | PXR | Xenobiotics | NGF-lα, β, γ | |
| TR | Thyroid hormones | RXRα, β, γ | Retinoids | PNR | |
| VDR | Vitamin D, bile acids | Rev-erbα, β | Heme | ||
| RORα, β, γ | Fatty acids, sterols | ||||
| SHP | |||||
| SF-1 | Phospholipids | ||||
| TLX | |||||
| TR2,4 | |||||
Figure 3.
Domain structure of nuclear receptors. The nuclear receptor superfamily proteins have several functional domains; the activation function 1 (AF1) domain at the N terminus, the DNA-binding domain (DBD), a hinge region, a ligand-binding domain (LBD) and another activation function 2 (AF2) domain at the C terminus. The small heterodimer protein (SHP) that has a major role in the regulation of genes involved in bile acid synthesis and transport, lacks the AF1 and DBD domains. Derived in part, with permission, from Science Slides (scienceslides.com).
Figure 4.
Nuclear receptor control of gene expression. Nuclear receptors (NR) bind to cis-acting elements usually located upstream of target genes. In the case of those receptors involved in metabolic control that bind as heterodimers with the retinoid X receptor, the binding site is composed of a direct repeat element (DR) separated by 1 to 4 nucleotides. In the absence of ligand, the NR is either unbound or bound to DNA. If bound to DNA in the absence of ligand, the receptor is complexed to a corepressor and histone deacetylases (HDAC) or demethylases, enzymes that remove acetyl or methyl groups from histones, which serve to keep the chromatin compact. In the presence of ligands, the corepressor is released and coactivators bind along with histone acetyltransferases and histone methyltransferases and components of the RNA polymerase (RNAP) complex resulting in gene transcription. Derived in part, with permission, from Science Slides (scienceslides.com).
Farnesoid X receptor
Bile acid synthesis and enterohepatic circulation is tightly controlled by nuclear receptors, notably the FXR. FXR was first cloned from mouse based on its interaction with the RXRα and was designated as an orphan receptor due to the lack of an identified ligand (128). However, others who cloned the rat homolog found that it was activated by farnesol and related metabolites, thus establishing the original name of this receptor (41). Later studies revealed that bile acids were actually the bona fide high-affinity endogenous ligands for FXR (98, 112, 148). Subsequently, a number of compounds were found to activate FXR with varying degrees of affinity including not only endogenous bile acids, but also synthetic ligands, steroids, aromatics, terpenoids, alkaloids, and fatty acids, thus revealing that this receptor, like other receptors involved in the control of metabolism, has a rather broad ligand-binding specificity, although it is not as promiscuous as the drug receptor PXR (67). Bile acids, including the primary bile acids CA and CDCA and the secondary bile acids DCA and LCA, are the most important physiologically relevant ligands for FXR with the potency to activate FXR CDCA>DCA>LCA>CA (112). The chemical structure of bile acids consists of a concave hydrophilic face and a convex hydrophobic face; bile acids bind with the hydrophobic pocket of the FXR LBD mainly through the hydrophobic face (104). The hydroxyl groups in the hydrophilic portion of the compounds affect the affinity of bile acids with FXR. Semisynthetic bile acid derivatives were developed as FXR ligands, notably 6α-ethylchenodeoxycholic acid (6-ECDCA), showing an even higher affinity for FXR than do bile acids (115). Other FXR agonists and antagonists, such as forskolin (64), guggulsterone (141), fexaramine (33), androsterone (146), and stigmasterol (11), have also been uncovered. The mechanism of control of gene activation by FXR and the role of numerous transcription cofactors, posttranscriptional control, and posttranscriptional protein modifications, has been reviewed (76).
Fxr-Null and Conditional-Null Mouse Models
The role of FXR in modulating bile acid synthesis, transport, and enterohepatic circulation in an intact animal was firmly established by the development and characterization of the Fxr-null mouse (132). Surprisingly, mice lacking expression of FXR develop normally and are fertile, thus indicating that this receptor has no major role in mammalian development, similar to that noted for mice lacking expression of the PXR(82), PPARα (89), and liver X receptor α (LXRα; NR1H3) (114), but unlike some of the other members of the metabolic sensor class of nuclear receptors such as PPARγ (NR1H4)(7) and hepatocyte nuclear factor 4α (HNF4α; NR2A1) (16) in which the knockout mice were embryonic lethal. However, Fxr-null mice were found to have elevated serum bile acids, cholesterol, and triglycerides, and increased hepatic cholesterol and triglycerides that confirms a critical role for FXR in the control of bile acid production and transport (132). These mice also exhibited a proatherogenic serum lipoprotein profile. In confirmation of the data showing that bile acids could activate FXR, the Fxr-null mice had reduced bile acid pools and fecal bile acid excretion as a result of decreased expression of genes encoding proteins involved in bile acid synthesis and transport at the hepatic canalicular membrane, thus establishing in vivo, the major regulatory role for FXR in the control of bile acid and lipid homeostasis, enterohepatic circulation of bile acids, and confirming in vivo that this receptor is an intracellular bile acid sensor. In addition to the Fxr-null mice that lack expression of the receptor in all tissues, conditional knockout mice were generated using the Cre recombinase-LoxP system (54) that lack expression of FXR in liver or intestine (78). These mice provided evidence that repression of bile acid synthesis requires FXR expression in both liver and intestine for feedback repression of CYP7A1 and CYP8B1, enzymes that are required to control hepatic bile acid production and that are suppressed through enterohepatic circulation and increased hepatic bile acids levels (see below). Thus, the Fxr-null mouse line (132) and conditional knockout mice lacking expression of FXR in liver and intestine (78) have been of great value in determining the function of this receptor in the control of bile acid synthesis and transport and in establishing that the physiological changes that occur as a result of altered hepatic and intestinal bile acid levels is target specific to the FXR. A large number of studies of hepatic disease and the role of FXR activators in modulation of metabolism, diabetes, cancer, and liver disease have used the Fxr-null mice.
Hepatic Bile Acid Synthesis
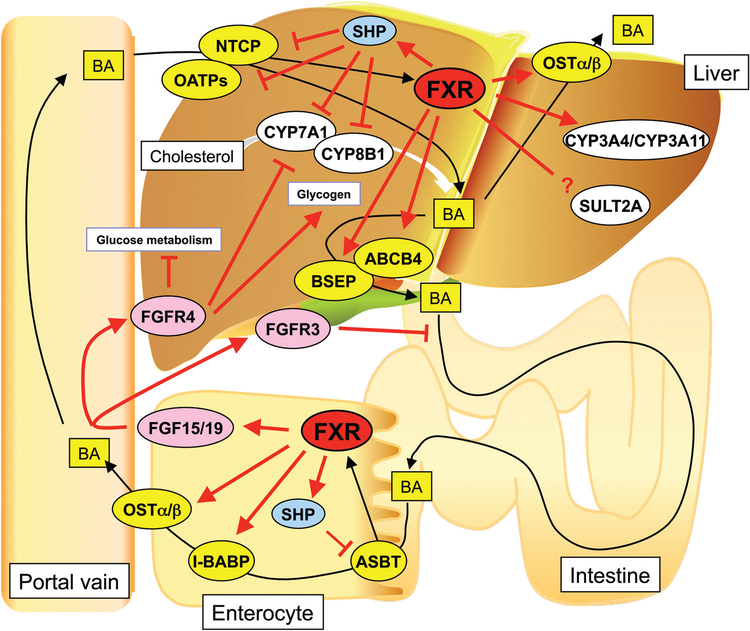
Hepatic bile acid levels are maintained by control of hepatic bile acid uptake, synthesis from cholesterol, and export to the bile duct/gall bladder and reuptake from the intestine to the circulation. CYP7A1 and CYP8B1 are two enzymes that catalyze formation of the primary bile acids CA and CDCA from cholesterol (19) (Fig. 2). The gene encoding CYP7A1 is under complex control by a number of transcription factors, notably albumin D region-binding protein (DBP) (90), LXR (91, 114), HNF4α (71, 139), liver receptor homolog-1 (LRH-1; NR5A2) and Rev-erbα, which indirectly controls circadian expression of the Cyp7a1 gene (36, 86). There is evidence that CYP7A1 is negatively regulated via the hepatic FXR through its induction of another nuclear receptor small heterodimer partner (SHP; NR0B2) and by the intestinal FXR-fibroblast growth factor (FGF)15/19 pathway (see below). SHP has a putative LBD based on sequence comparison with other nuclear receptor superfamily members, but it does not have a corresponding DBD (Fig. 3) (129). The lack of a DBD revealed that SHP likely exerts its regulatory activities through protein-protein interactions with other nuclear receptors, with most evidence to date indicating that it renders positively activating nuclear receptors transcriptionally inactive (166). FXR, LXRα, LRH-1, HNF4α and several other nuclear receptors control expression of the Shp gene (164). In the absence of FXR expression in the Fxr-null mouse model, Shp is expressed at a very low level in liver (132), thus suggesting that FXR is one of the major regulators of Shp and is implicated in the control of bile acid synthesis and transport. However, the phenotype of the Fxr-null mice is not solely driven by the lack of Shp induction, as revealed by the pheno-type of the Fxr/Shp-double null mouse that differs from the Fxr-null mouse phenotype (4,83,164).
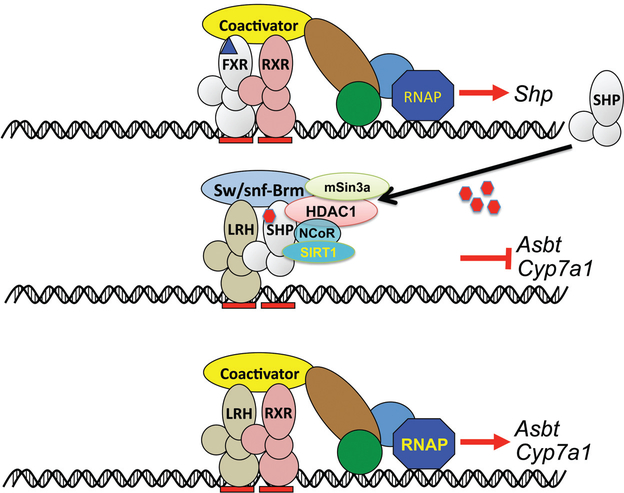
FXR regulation of Shp has been confirmed at the molecular level. There are two enhancer regions in the 5’ and 3’ end of the Shp gene that bind FXR resulting in head to tail chromatin looping that potentiates transcription (92). When SHP accumulates in the liver, it could inhibit the Cyp7a1 gene by interaction with the orphan nuclear receptor LRH-1, a positive regulator of Cyp7a1 expression, resulting in a nonproductive transcription factor (51) (Fig. 5). However, the Cyp7a1 gene has an element in its promoter that binds HNF4α as well as LRH-1 (26,30) and there are binding sites for other transcription factors. Thus, it remains to be determined which transcription factor predominates in the constitutive expression of Cyp7a1 in the intact liver. An LRH-1-responsive element is also located in the regulatory region of the Shp gene; overexpression of LRH-1 can activate the Shp promoter in mice and tissue culture cells (15, 95). Silencing of CYP7A1 is due in part to the recruitment of the sirtuin silent mating type information regulation 2 homolog 1 (SIRT1) and histone deacetylase protein by SHP to mediate inhibition of LRH-1-dependent suppression of CYP7A1 transcription, as revealed largely using cultured human HepG2 cells(12). LRH-1 activation of CYP7A1 and SHP gene transcription was significantly repressed by both SHP and SIRT1 while the inhibition of SIRT1 activity by inhibitors or a dominant negative SIRT1 or knockdown of SHP released the inhibitory effect. Other accessory factors, such as the Brahma chromatin remodeling protein Sw/snf-Brm, Sin3a scaffold corepressor, and histone deacetylase-1 (HDAC1), are also recruited to the promoter by SHP binding and increase the occupancy of SHP at the CYP7A1 promoter in HepG2 cells (105). In addition, ligand binding can also influence SHP; a retinoid-like compound, 4-[3-(1-adamantyl)-4-hydroxyphenyl]-3-chlorocinnamic acid (3Cl-AHPC), was found to bind to SHP and increase its interaction with LRH-1 (105). These studies on SIRT1, Brahma, and 3Cl-AHPC still require validation in intact liver that has a more complex array of transcription machinery than cultured hepatoma cells that may influence Cyp7a1 transcription.
Figure 5.
Repression of the Cyp7a1 gene by small heterodimer protein (SHP). Farnesoid X receptor (FXR) activates expression of SHP in the presence of bile acids agonist. SHP then binds to the positive regulator liver receptor homolog 1 (LRH-1) and inhibits its transactivation of Cyp7a1 and other target genes such as Asbt. Derived in part, with permission, from Science Slides (scienceslides.com) and reference (76).
Bile acids have also been reported to suppress the human CYP7A1 expression through blocking HNF4α recruitment of coactivators PGC-1α and CREB to the CYP7A1 promoter(27). However, it should be noted that these studies were largely done using cultured hepatoma cells and reporter gene transfections and the conclusions are complicated further by the fact that the Cyp7a1 gene is under complex positive con trol by multiple transcription factors and circadian regulation in liver as noted above. Data derived using a panel of genetically modified mice lacking FXR and SHP suggest that the FXR-SHP pathway has only a minor role in suppression of hepatic Cyp7a1 while the FXR-FGF15 mechanism described below predominates in the bile acid suppression of this gene (4,83,164). The controversial role of the FXR-SHP pathway in suppression of Cyp7a1 expression by bile acids is also suggested by the finding that the phenotype of the Fxr-null mice differ from the Fxr/Shp-null mice (4, 83, 164). The precise mechanism for this FXR repression of CYP7A1 requires further studies in intact liver and primary hepatocyte cultures. In addition, there exists a major species difference in the control of CYP7A1 expression, as noted below, that needs to be taken into account when attempting to develop models for suppression of this enzyme by bile acids.
In addition to its regulation by LRH-1 and SHP, the mouse Cyp7a1 is positively regulated by the LXRα, a nuclear receptor that is activated by oxysterols, oxidized metabolites of cholesterol (8, 114). LXRα is preferentially activated by a cholesterol-rich diet and facilitates cholesterol degradation and elimination. However, a major species difference exists; human CYP7A1 is not upregulated by LXRα (52). In contrast to mice and rats, CYP7A1 is downregulated by LXRα through induction of the SHP, by a similar mechanism as FXR suppression of Cyp7a1 (Fig. 5). SHP is regulated directly by LXRα through a DNA response element that overlaps with the bile acid response element that binds to FXR. These data suggest that different species employ distinct mechanisms for regulating cholesterol levels and bile acid synthesis. As noted by others (52), rats and mice lower cholesterol levels by conversion to bile acids, while humans modulate cholesterol levels by reducing absorption in the intestine, largely through decreasing bile acid production. Species differences between rodents and humans in the regulation of CYP7A1 may reflect the amount of cholesterol ingested in the diet, with primates having evolved on a lower cholesterol diet. This species difference in CYP7A1 regulation could account for a greater susceptibility of humans to diet-induced hyper-cholesterolemia as compared to mice. LXRα also induces expression of the ATP-binding cassette transporters ABCG5 and ABCG8 in liver that are involved in cholesterol transport to the bile through the canalicular membrane (123). The combined induction of CYP7A1, ABCG5, and ABCG8 facilitate cholesterol elimination.
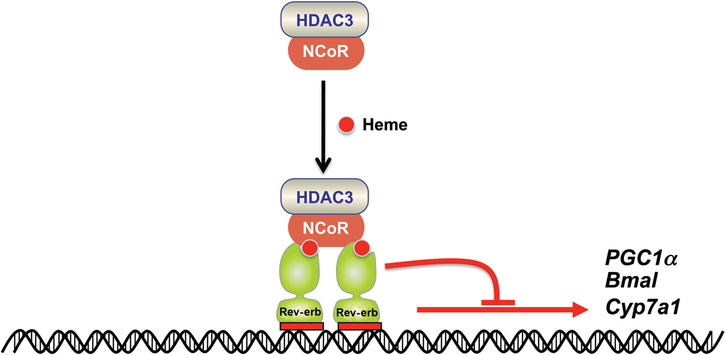
CYP7A1 is under diurnal regulation in rats and mice, with higher expression at night and lower expression during the day, corresponding to the low and high demands for bile acid synthesis in rodents that feed at night. CYP7A1 is also under diurnal regulation in humans (34). The expression, of CYP7A1 was correlated with circulating levels of corticosteroid hormone (48). Earlier studies revealed that this was due in part to regulation of the mouse Cyp7a1 promoter by DBP(90). Indeed, DBP expression is controlled by glucocorticoids (156). However, levels of DBD are not highly correlated with CYP7A1 expression, thus suggesting that the Cyp7a1 gene is under more complex regulation as revealed by numerous subsequent studies as described above. More recent work has revealed that the nuclear receptor Rev-erbα has a major role in regulating the diurnal expression of genes involved in lipid metabolism and adipogenesis (159,160). Rev-erbα is an atypical orphan nuclear receptor that binds heme and represses gene transcription by recruitment of the nuclear hormone receptor corepressor histone deactylase 3 (NCoR-HDAC3) that has histone deacetylase activity and promotes chromatin condensation (35,57). Rev-erbα is under circadian rhythm and is a critical negative limb of the core circadian clock by directly repressing expression of the positive clock component, brain and muscle aryl hydrocarbon receptor nuclear translocator-like (Bmal1) that controls circadian rhythm (56,113,160). Evidence that Rev-erbα is involved in the regulation of bile acid metabolism was obtained from mice deficient in Rev-erbα that display a lower bile acid synthesis rate and an impaired excretion of bile acids into the bile and feces. Mice lacking expression of Rev-erbα have decreased levels of CYP7A1, while forced expression of Rev-erbα using adenovirus restored hepatic expression of this enzyme (36). However, bile acids still suppress hepatic CYP7A1 expression in Rev-erbα-deficient mice; the authors suggest that this is the result of increased expression of Shp, a putative Rev-erbα-target gene and nuclear factor, interleukin 3 regulated (E4BP4/NFIL3). Both SHP and E4BP4 are negative regulators of Cyp7a1. The circadian rhythms of liver CYP7A1 were markedly altered in Rev-erbα-deficient mice, thus indicating a role for Rev-erbα in the regulating CYP7A1 and bile acid synthesis. However, a second study revealed that SHP and E4BP4/NFIL3 expression were not significantly altered in another line of Rev-erbα and proposed that CYP7A1 circadian rhythm was controlled indirectly through direct inhibition of insulin-induced gene 2 (Insig2) by Rev-erbα (86). They proposed that Insig2 inhibits the proteolytic release and activation of sterol regulatory element-binding proteins (SREBP) leading to altered production cholesterol and oxysterols, the latter of which can activate LXRα, a positive regulator of Cyp7a1 expression. A number of other genes involved in diurnal regulation have also been implicated in the regulation of CYP7A1, including adenovirus E4 promoter-binding protein (E4BP4), and PPARα (109).
The other major enzymes involved in bile acid synthesis are CYP8B1 and CYP27A1 (Fig. 2). The human CYP8B1 promoter also contains a negative bile acid response element harboring overlapping binding sites for HNF4α (71, 163) and LRH-1 (29), and induction of SHP by FXR inhibits the Cyp8b1 by negative interference of transcriptional activity of HNF4α and LRH-1 similar to the inhibitory effects that SHP has on Cyp7a1. However, in vivo studies with Fxr-null mice suggested that feedback inhibition of CYP8B1 can also be FXR-independent (132), although recent evidence in genetically altered mouse models support a major role for the FXR-CHP pathway in suppression of Cyp8b1 expression by bile acids (83). Thus, there may be species differences between humans and mice in the regulation of this enzyme by bile acids. In addition, CYP27A1, an enzyme catalyzing the acidic pathway of bile acid biosynthesis, is suppressed by bile acids in a human cell line (14). The CYP27A1 gene, unlike those genes encoding CYP7A1 and CYP8B1, is not specifically expressed in liver, in part due to the presence of binding sites for the ubiquitous transcription factor specificity protein 1 (Sp1) (44). The CYP27A1 promoter also binds to and is activated by the positive regulator HNF4α (14, 44). Definitive in vivo results demonstrating the mechanism of negative regulation of CYP27A1 by bile acids are lacking.
FXR in the intestine has a major role in control of bile acid synthesis in the liver through a mechanism involving FGF19 in humans, and its homolog FGF15 in mice (sometimes referred to as FGF15/19). FGF15/19 is produced in the small intestine and secreted from the intestine to the bloodstream where it circulates to the liver and suppresses bile acid synthesis through binding and activation of the FGF receptor 4 (FGFR4) complex with β-Klotho located on the plasma membrane surface of hepatocytes and other epithelial cells (66,72,81,157). Binding of FGF15/19 to the FGFR4/β-Klotho complex stimulates the c-Jun N-terminal kinase pathway to suppress transcription of the Cyp7a1 gene (63, 68). Thus, the bile acid-activated intestinal FXR downregulates CYP7A1 expression indirectly through direct the activation of intestinal FGF15/19 synthesis and secretion. FGF15/19 also functions to facilitate gall bladder filling via binding to FGFR3 expressed in the gall bladder (22). FGF15/19 signaling in the liver increases hepatic glycogen synthesis through elevated glycogen synthase kinase 3 activity (80, 81). Coincident with increased glycogen synthesis, FGF15/19 inhibits hepatic gluconeogenesis (119) through modulation of the CREB-peroxisome PPARγ coactivator protein-1α (PGC-1α) pathway. Extracellular signal-regulated protein (ERK1) is activated by the FGF15/19-stimulated FGFR4/β-Klotho complex, is stimulated by hepatic glycogen synthesis, and is inhibited by and glucose metabolism independent of insulin signaling. These studies revealed that bile acids have an important role in maintaining hepatic glucose homeostasis through FGF15/19, independent of hepatic FXR, and that FGF15/19 is a postprandial regulator of hepatic carbohydrate homeostasis (119).
HNF4α is also involved in the control of hepatic bile acid synthesis. HNF4α is a nuclear receptor that controls liver development in the embryo, and the expression of liver-specific genes in the adult (150). HNF4α liver-specific null mice had low expression of the mRNAs encoding CYP7A1, CYP8B1, and sterol carrier protein 2, thus indicating that HNF4α controls constitutive expression of these genes in liver. An HNF4α-binding site was found in the mouse Cyp8b1 promoter that was able to direct HNF4α-dependent transcription. Transfection studies revealed an HNF4α response element in the human CYP7A1 gene (139). In HNF4α liver-specific null mice, Cyp7a1 expression was only diminished in the dark cycle (71). Thus, HNF4α has a role in the control of BA homeostasis through the regulation of genes involved in BA biosynthesis, including hydroxylation and side chain β-oxidation of cholesterol in vivo. Mice lacking hepatic HNF4α also exhibited markedly decreased expression of the very long chain acyl-CoA synthase-related gene (VLACSR), the mouse homolog for BAL, and BAT (70). This was associated with elevated levels of unconjugated and glycineconjugated bile acids in gallbladder of HNF4α liver-specific null mice. HNF4α also bound to the promoters of the genes encoding VLACSR and BAT and directed transcription in reporter gene transfection assays. Thus, any perturbation of HNF4α could result in alterations in bile acid metabolism, possibly resulting in disease. However, it is still unclear whether HNF4α is involved in the metabolic control of levels of hepatic lipids and bile acids since there is no clear evidence that it is regulated by ligands. HNF4α is constitutively active during mammalian development and in adult liver, thus suggesting that it was not subject to control by ligands, similar to other nuclear receptors. However, fatty acyl-CoA thioesters were found to be involved in modulating the expression of HNF4α-controlled genes, presumably through interaction with the LBD (58, 59, 116). Structural studies on bacterially expressed protein revealed that it has a mixture of embedded fatty acids at its ligand-binding site that are not easily released (32,153). These fatty acids are likely incorporated into the bacterially-expressed protein during translation and folding. Direct chemical analysis of HNF4α expressed in mammalian cells and liver revealed that it is largely bound with linoleic acid and that the binding is possibly reversible, but not in the classic way that other members of the metabolic sensor family of receptors interact with ligands to modulate gene expression (162). Thus, alteration in activity through ligand binding may require chronic exposure to cellular metabolites and the ligand is incorporated into the LBD while HNF4α is being translated and folded.
All bile acids are conjugated prior to transport from the liver. FXR regulates two major enzymes involved in bile acid conjugation with taurine and glycine, the bile acid-CoA synthetase (BACS, also called SLC27A5) and bile acid-CoA:aminoacid N-acetyltransferase (BAT). BACS catalyzes the formation of the CoA thioester intermediate while BAT produces the amino acid conjugate. The FXR activators CDCA and GW4064 induce Bacs and Bat expression in hepatocytes and Fisher rats and FXR response elements were found in the promoter of BACS and in an intron of the BAT gene (118). Further, mutational analysis confirmed that the IR-1 element of the human BACS and BAT genes allows the binding of FXR-RXR heterodimers.
Hepatic Transport of Bile Acids to the Intestine
Bile salt-exporting pump (BSEP, ABCB11) is a major efflux transporter of bile acids from liver to gallbladder (Fig. 7). ABCB11 deficiencies are associated with progressive familial intrahepatic cholestasis type 2, benign recurrent intrahepatic cholestasis type 2 (BRIC2), and several acquired forms of cholestasis (137). Fxr-null mice fed a CA-supplement diet showed intrahepatic cholestasis, similar to that observed with ABCB11 deficiency in humans. Evidence that BSEP is an FXR target gene was revealed by mutation of the FXR regulatory element in the BSEP promoter that repressed FXR-dependent induction of BSEP expression; this regulatory element binds FXR/RXRα heterodimers (5). BSEP expression was significantly reduced and the FXR agonist GW4064 does not induce the expression BSEP gene in mice lacking FXR expression, thus confirming that FXR controls BSEP gene expression (108,127,132). Expression of another ABC transporter ABCB4, also called MDR3 in humans and MDR2 in rodents, is significantly reduced in Fxr-null mice, where GW4064-induced Abcb4 expression is not observed (25). ABCB4 transports phospholipids across canalicular membranes of hepatocytes (142). Phospholipids are transported to gallbladder from liver via ABCB4 and facilitate the formation of mixed micelles containing cholesterol, bile acids, and phospholipids to effectively increase their solubility and reduce their toxicity to the bile duct (62). Similar to BSEP, ABCB4 deficiency in humans can cause progressive familial intrahepatic cholestasis 3 (PFIC3) (28,138). By a mechanism similar to the mouse Abcb4 gene, FXR regulates the human ABCB4 gene through binding an element in the distal promoter (65). Thus, FXR is a critical factor regulating bile acid flow by inducing expression of hepatic ABCB11/BSEP and ABCB4.
Figure 7.
Mechanism for the enterohepatic circulation of bile acids. Bile acids and other steroids produced or metabolized in the liver and secreted to the intestine, for reabsorption back into the circulation and transport back to the liver. Coincident with bile acid recirculation is the removal and reuptake of drugs, usually conjugated high Mr drugs. Cholesterol is the substrate for bile acid synthesis, one of the routes for mammalian disposal of cholesterol. The author thanks Tsutomu Matsubara for help in preparing this figure.
Intestinal Bile Acid Uptake
FXR controls the intestinal absorption of bile acids through the regulation of expression of four important transporters, apical sodium-dependent transporter (ASBT; SLC10A2), fatty acid-binding protein subclass 6 (FABP6), also known as intestinal bile acid-binding protein, organic solute transporter alpha (OSTα), and OSTβ, that facilitate the transport of bile acids from the intestine to the portal system (Fig. 7). ASBT, the major bile acid transport system in ileal enterocytes, transports bile acids across the ileal enterocyte brush border (apical) membrane (131). In accordance with this function, ASBT deficiency causes significant bile acid malabsorption disease (60,111). Intestinal ASBT expression is inhibited when mice are administered the natural bile acids and FXR ligands CA and taurocholic acid (TCA) (13). Bile acids exert a negative feedback on ASBT expression by FXR induction of SHP that binds to and represses the transcriptional activation properties of LRH-1 as described above for the Cyp7a1 gene. While the negative regulation of ASBT expression is found in mice, it was not observed in rats due to the absence of an LRH-1 responsive element within the rat ASBT promoter (13). After transport inside ileal enterocytes by ASBT, bile acids are reversibly bound to FABP6 expressed in the ileum (84, 155). FABP6 has an important role in enterohepatic circulation through the regulation of bile acid trafficking where it shuttles bile acids from the apical to basolateral membrane in the enterocyte (24). Bile acid-activated FXR can increase FABP6 gene expression through binding to a regulatory element in the promoter (53). Finally, OSTα and OSTβ move bile salts to blood vessels, in accordance with its location at the basolateral membrane (25). OSTα and OSTβ are expressed not only in the ileum, but also in the liver and kidney (25). Ileum expression of both the genes is induced in wild-type mice after exposure to the FXR activator CA or the synthetic ligand GW4064 (168). FXR regulatory elements are found in the promoters of the genes encoding OSTα and OSTβ in both humans and mice (88). Thus, FXR mainly controls the entire transport of bile acids from the intestinal lumen to the enterocyte, within the enterocyte and ultimately to the blood vessel for transport to the liver (Fig. 7).
Hepatic Bile Acid Uptake
Na+-taurocholate cotransporting polypeptide (NTCP; SLC10A1) and organic anion-transporting peptides (OATPs) are the major transporters for hepatocellular basolateral membrane uptake of bile acids and organic solutes from portal vein to liver (55,136). NTCP is responsible for the uptake of conjugated bile acids, while the OATPs control the uptake of unconjugated bile acids (Fig. 7). The gene encoding NTCP is under complex regulation by multiple transcription factors(47). In the mouse cholestasis model of liver disease, expression of both NTCP and OATPs are significantly reduced as compensation for a decrease in the accumulation of bile acids (167). NTCP expression is also negatively regulated by FXR indirectly by induction of SHP that in turn inhibits transcription by a similar mechanism as that described for Cyp7a1(31). The Ntcp gene is positively regulated by retinoids through the RARα/RXRα. After induction of SHP by FXR, SHP inhibits activation of the Ntcp gene by retinoids, possibly through interference with RARα/RXRα, although the exact mechanism of this interference has not been established (31). In humans, the glucocorticoid receptor (GR; NR3C1) is a transcriptional activator of the NTCP gene (37). Ligand-activated GR is suppressed by CDCA or GW4046 indirectly through FXR induction of SHP. Other studies revealed that HNF4α constitutively regulates the mouse Ntcp (46) and rat(74) NTCP promoters, while the CCAAT/enhancer binding protein β (C/EBPβ) controls the mouse Ntcp and human NTCP genes (74). Other studies described a potential role for the homeodomain-containing transcription factor hepatocyte nuclear factor 1α (HNF1α) in regulation of the mouse Ntcp gene (45). Since HNF4α is influenced by SHP, bile acids could also downregulate Ntcp expression by interfering with HNF4α expression, although the specificity of this response would have to be determined since HNF4α controls the expression of many genes in the liver (20, 150). It should be emphasized that many studies reporting on control of gene expression are done in cultured cell lines, usually tumor cells, and by use of transfection and DNA binding for promoter analysis; the results obtained may not reflect the corresponding regulation of these genes in intact tissues. The most reliable results are those involving a combination of in vitro and in vivo studies using gene knockout mice and chromatin immunoprecipitation (ChIP) studies to assess the roles of individual transcription factors in control of specific genes. However, in vivo studies are not always possible when studying the regulation of human gene expression.
The OATP family has 11 members that collectively facilitate the Na+-independent transmembrane transport of endogenous and exogenous compounds including bile acids, bilirubin, steroid hormone conjugates, thyroid hormones, prostaglandins, clinically used drugs, and toxins (140). Some of the genes encoding these transporters are repressed while others are induced. FXR regulates hepatic expression of OATP1 (SLCO1A1) and OATP2 (SLCO1A4) in mice (97). In mice, FXR downregulates hepatic expression of Oatp1 and induces Oatp2, although the mechanisms for these alterations in gene expression by FXR have not been elucidated(97). In human liver, bile acids also repress expression of the OATP1B1 gene, but similar to the mouse Oatp1, the mechanism of repression and the role of FXR has not been established (73). Expression of OATP1B3 was not altered in these studies. Other nuclear receptors including LXR, CAR, PXR, PPARα, and HNF4α were also reported to regulate various OATP genes in liver and other tissues, such as kidney (140). In addition, the aryl hydrocarbon receptor was shown to influence OATP gene expression (18), thus indicating the complex metabolic control of this superfamily of transporters.
Expression of the major bile acid uptake system transporter OATP4 (SLC21A6) is dependent on HNF1α, a homeodomain-containing transcription factor that is not a member of the nuclear receptor superfamily. CDCA administration to cultured hepatoma cells results in decreased expression of HNF1α (75). However, the influence of bile acids on HNF1α is indirect. HNF1α is regulated by HNF4α through a binding site in the Hnf1α promoter (85). In the presence of bile acids, SHP is induced by FXR and decreases the transactivation of HNF4α by direct interaction, similar to the mechanism by which SHP inhibits LRH-1 transactivation (Fig. 5). However, this mechanism was developed in cultured hepatoma cells using promoter cotransfection assays and thus it remains to be determined whether it is the same mechanism in intact liver. In addition, the specificity of this response needs to be investigated since HNF4α controls a large number of genes in the liver unrelated to bile acid synthesis and transport as noted above.
Role of CYP3A4, UGT2B4, and SULT2A1 in Bile Acid Metabolism
An enzyme responsible for the metabolism of many clinically used drugs, CYP3A4 (CYP3A11 in mice), is also involved in bile acid metabolism by catalyzing hydroxylation of CDCA, LCA, and DCA (6, 10). The nuclear receptor PXR is expressed in liver and intestine, and controls transcription of the genes encoding CYP3A4 and CYP3A11, and other genes involved in the metabolism and transport of drugs and various xenobiotics (17). CYP3A11 is also under control of FXR; a significant induction of CYP3A11 expression was observed in mice treated with the FXR ligand GW4064 (49). Evidence also exists in cultured human hepatoma cells that FXR regulates CYP3A4 expression and that CDCA can induce the expression of CYP3A4 in human liver (77). Thus, FXR may be involved in bile acid-induced CYP3A4/CYP3A11 expression. CYP3A4 can metabolize LCA, a bile acid that when present at high levels, causes liver toxicity (61). LCA, produced from CDCA by gut bacterial 7-dehydroxylation, is normally present at low levels but is markedly increased in patients with liver disease (61). In particular, LCA is reabsorbed from the gut to the liver and can cause intrahepatic cholestasis in animal models. In human intestinal cells, LCA can induce expression of FGF19 (154). LCA also induces disruption of phospholipid/sphingolipid homeostasis through TGFβ signaling (103). Through PXR, LCA induces CYP3A11, which offers a protective response against LCA-induced hepatotoxicity since CYP3A11 can metabolize LCA (103,135). PXR can also influence enterohepatic circulation. Tauro-3α,6,7α,12α-tetrol was found to be a marker for efficient hydroxylation of toxic bile acids possibly through induction of CYP3A11 as revealed in cholestatic Fxr-null mice on a CA diet (21). Thus, cholestasis induced by LCA in Fxr-null mice revealed that enhanced expression of CYP3A11 is the major defense mechanism to detoxify cholestatic bile acids.
In addition to inducing bile acid oxidizing enzymes, FXR upregulates expression of the conjugating enzyme uridine 5’-diphosphate-glucuronosyltransferase 2B4 (UGT2B4) (9) that catalyzes production of glucuronidated 6α-hydroxylated bile acids such as hyodeoxycholic acid (117). FXR also induces expression of sulfotransferase 2A1 (SULT2A1) (134) that carries out sulfate conjugation of many hydroxysteroid substrates, including bile acids, pregnenolone, and estrogens (151). Treatment of hepatocytes and HepG2 cells with the FXR agonists CDCA and GW4064 increases UGT2B4 expression and activity (9). Endogenous SULT2A1 expression was decreased in HepG2 cells treated with the FXR agonist CDCA or GW4064 (106). Thus, expression of UGT2B4 and SULT2A1 are modulated by FXR and facilitate bile acid metabolism along with CYP3A.
FXR in the Metabolism and Transport of Drugs
In addition to bile acid homeostasis, FXR can contribute to the metabolism and elimination of drugs and toxicants through the regulation of phase I and II metabolizing enzymes and transporters as noted above. FXR also can regulate the several important phase II metabolism enzymes, such as UGTs and SULTs, converting the hydrophobic compounds to more hydrophilic and less toxic conjugated derivatives that can more easily be eliminated from the body. In accordance with its role in regulating xenobiotic metabolism, activation of FXR protects mice from acetaminophen (APAP)-induced heptotoxicity (87). Under normal therapeutic dosing, APAP is metabolized in the liver mainly through direct conjugation by UGTs and SULTs. However, excessive APAP will saturate both glucuronidation and sulfation pathways, leading to production and accumulation of the toxic N-acetyl-p-benzoquinone imine (NAPQI) metabolite produced by CYPs, notably CYP2E1(50). NAPQI is also subject to conjugation by glutathione S-transferase (GST) but under conditions of high doses of APAP, the amount of NAPQI exceeds the conjugation capacity and the cofactor for GST, glutathione, is depleted. The liver toxicity induced by high dose APAP could be attenuated by upregulation of several phase II enzymes through FXR. To identify which drug metabolizing enzymes might be regulated by FXR, three models were employed, a constitutively active form of FXR (FXR-VP16) where FXR is fused to the coactivator VP-16, native FXR, and by treating wild-type and Fxr-null mice with an FXR agonist. The expression levels of several GSTs (GSTα3, GSTα4, GSTμ1, and GSTμ3), SULTS (SULT1A1 and SULT1A2), and UGTs (UGT1A1), were induced by FXR activation (87). FXR response elements were identified in some of these gene promoters by use of ChIP genome-wide response element analysis.
Enterohepatic circulation is not restricted to bile acids but also involves certain drugs and hormones including thyroid hormone and steroid hormones (125,133,144,145,152). The chemical structure, polarity, and molecular size, and the hep atic and gut microbe metabolism influences whether a drug is transported through cannicular membranes and reabsorbed in the intestine to complete the enterohepatic cycle. Drug bioavailability is affected by the extent of intestinal absorption, hepatic uptake, and metabolism. In general, enterohepatic recirculation may prolong the pharmacological effect of certain drugs and drug metabolites as the compound will remain in the body for longer periods of time than a drug that is primarily subjected to urinary excretion. The amplifying effect of enterohepatic variability can influence bioavailability, volume of distribution, and clearance of a given compound. Genetic abnormalities, disease states, orally administered adsorbents such as charcoal, ion-exchange resins (Amberlite) and aluminum phyllosilicate (Bentonite), and certain coad-ministered drugs can all affect enterohepatic recycling and thus the bioavailability of a drug (125). Any abnormality in bile acid synthesis and bile flow has the potential to markedly effect the clearance of drugs subjected to excretion through the bile and enterohepatic circulation. Serum levels of hormones that are subjected to enterohepatic circulation can also be influenced by alterations in bile flow. Hormone levels can also be altered by metabolism carried out by gut bacteria. For example, steroid hormones conjugated in the liver can be deconjugated by obligate anaerobes that account for 99.99% of the fecal flora; deconjugated metabolites can then be reabsorbed in the intestine. The most common transformation is hydrolysis of conjugated steroids by glucuronidases synthesized by Escherichia coli and Bacteroides species.
Role of FXR in Hepatic Toxicity and Cancer
FXR has a protective role in liver toxicity and cancer as revealed by the finding that upon exposure to a 1% CA diet, Fxr-null mice develop cholestasis, while wild-type mice have slightly decreased body weight but no significant differences in liver toxicity (107). The mechanism of protection was examined by analyzing the metabolic profile of Fxr-null mice fed a CA-diet by use of metabolomics, using a liquid chromatography-mass spectrometry based platform (21). These studies revealed the activation of adaptive metabolic pathways upon bile acid challenge. Levels of p-cresol, corticosterone, and cholic acid were elevated in Fxr-null mice on a CA diet. In addition, taurine-conjugated tetrahydroxy bile acids were increased in Fxr-null mice, likely as a result of induction of CYP3A11, not typically associated with bile acid metabolism. In LCA-induced cholestasis, the excretion of the similar taurine-conjugated tetrahydroxy bile acid was also greatly increased in urine (21). The excreted tetrahydroxy bile acids in LCA-treated Fxr-null mice that are resistant to LCA-induced intrahepatic cholestasis was greater than in LCA-treated wild-type mice, thus suggesting that hydroxylation of bile acids contributes to the detoxification of cholestatic bile acids in Fxr-null mice. These results also demonstrate that tetrahydroxyl bile acids are potential biomarkers for hepatotoxicity and cholestasis. Enhanced serum corticosterone that is elevated in cholestatic animal models and humans, was also observed in CA-treated Fxr-null mice. Abnormal corticosterone metabolism in CA-treated Fxr-null mice was revealed by the increased excretion of corticosterone metabolites 11β-hydroxy-3,20-dioxopregn-4-en-21-oic acid (HDOPA), 11β,20α-dihydroxy-3-oxo-pregn-4-en-21-oic acid (DHOPA), and their hydroxylation metabolites (21,147). Future studies will be required to further determine the molecular mechanism linking liver injury (cholestasis, hepatitis) with hepatic and adrenal steroid metabolism.
FXR deficiency leads to the development of liver and intestine cancer. A high incidence of hepatocellular adenoma, carcinoma, and hepatocholangiocellular carcinoma was detected in 12-month-old male and female Fxr-null mice (79,158). Lowering of bile acids by cholestyramine treatment reduced the spontaneous tumors in Fxr-null mice, and liver cancer produced in wild-type mice by N-nitrosodiethylamine was increased, thus implicating high hepatic bile acids in the carcinogenesis process (158). The increased cell proliferation and liver cancer was associated with elevated expression of proinflammatory cytokines, hepatocyte proliferation, and hepatocyte regeneration. Of particular note was the finding of increased IL-1β in younger Fxr-null mice noted before tumor development (79). This is in agreement with studies showing a role for inflammation in liver cancer (96). Another study also indicated that FXR deficiency is associated with increased colon carcinogenesis in mice treated with the colon carcinogen azoxymethane (100). The increased colon cell proliferation was correlated with a hyperproliferative state of the colon epithelium including increased expression of the cell cycle control protein cyclin D1, and inflammation as revealed by elevated IL-6 expression. These results suggest that activation of FXR by its ligands may protect against liver and intestinal carcinogenesis.
FXR Ligand as Therapeutic Agents
Since FXR is a modulator of the metabolism and transport of bile acids in liver and intestine, and indirectly modulates glucose metabolism and inflammation, clinical activation or inhibition of FXR could potentially be used for therapy against intra- and extracholestasis (38), inflammatory bowel disease(42), and obesity-associated type 2 diabetes (120, 165). Extracts of the resin of the guggul tree (Commiphora mukul) are known to lower low-density lipoprotein cholesterol levels in humans. Guggul extracts are found in many nutritional supplements. One of the earliest indications that targeting FXR could be used for treatment of disease was uncovered by study of the plant sterol guggulsterone [4,17(20)-pregnadiene-3,16-dione], the active agent in this extract, in wild-type and Fxr-null mice (141). Guggulsterone was found to be an efficacious antagonist of FXR. Treatment of high-cholesterol fed wild-type mice diet with this compound decreases hepatic cholesterol; it is not effective in Fxr-null mice, thus re vealing that inhibition of FXR activation is the basis for the cholesterol-lowering activity of guggulsterone and indicating that inhibition of FXR may be a suitable clinical target for lowering cholesterol.
Fxr-null mice are protected against liver injury caused by bile acid overload in rodent models of cholestasis. Thus, antagonism of FXR could also protect mice from cholestasis. An antagonist theonellasterol, a 4-methylene-24-ethylsteroid, was isolated from the marine sponge Theonella swinhoei and found to be highly specific toward FXR (122). The bile duct ligation (BDL) model of obstructive cholestasis was used to test the therapeutic efficacy of theonellasterol. Liver injury caused by BDL was attenuated by treatment of mice with theonellasterol. The mechanism of protection was likely due to an increase in expression of the basolateral transporter MRP4 that is negatively regulated by FXR. In this study, administration of BDL mice with an FXR agonist did not rescue mice from liver injury.
In animal models, GW4064, an aromatic ligand and agonist for FXR, shows efficacy toward several types of diseases through its target-specific effect on FXR. GW4064 protects against cholestatic liver damage in a rat model of intrahepatic and extrahepatic cholestasis (94). The efficacy of FXR activators in the treatment of disease has been studied in preclinical models such as the naphthylisothiocyanate (ANIT) and BDL rat and mouse models for intrahepatic and extrahepatic cholestasis, respectively. Treatment of both cholestasis models with GW4064 results in significant reduction in liver damage (94). GW4064-treated cholestatic rats have decreased expression of the bile acid biosynthetic enzymes CYP7A1 and CYP8B1, and increased expression of genes encoding the bile acid transporters BSEP and MRP2. This suggests that GW4064 or other FXR activators with favorable pharmacokinetic properties could have potential in the prevention of cholesterol gallstone disease. GW4064 treatment can reduce cholesterol precipitation and gallstone formation through the induction of Abcb11 and Abcb4 and the resultant increased biliary concentrations of bile salts and phospholipids in the gallstone-susceptible C57L/J mouse gallstone model (108). In CYP7A1-overexpressing mice with high biliary and fecal cholesterol, GW4064 treatment induces hepatic ABCG5/G8 expression through FXR activation, suggesting that GW4064 could reduce gallstone formation by increasing the transport of cholesterol in the liver (93). Finally, GW4064 may prevent epithelial deterioration and bacterial translocation in patients with impaired bile flow. BDL can increase the number of aerobic and anaerobic bacteria in the ileum and cecum. Treatment of BDL mice with GW4064 inhibits bacterial overgrowth in the lumen of ileum and cecum (69). Thus, either activation or inhibition of FXR could be used to treat cholestatic liver disease as revealed in rodent models, although studies in primates and humans need to be carried out.
GW4064 also may be another route for the treatment of certain types of diabetes mellitus. Administration of GW4064 for 4 and 11 days significantly decreased levels of plasma glucose, triglycerides, and cholesterol in wild-type and genetically obese, diabetic, leptin-deficient db/db mice, and these effects were dependent on activation of FXR (165). Other studies revealed that GLUT4, the main insulin-responsive glucose transporter, which has a role in maintaining systemic glucose homeostasis, is induced by FXR, thus further implicating this receptor as a potential target for type 2 diabetes (130). FXR could also have a role in treatment of diabetic nephropathy associated with type 1 diabetes (149).
6-ECDCA has been used as an FXR agonist for testing the benefit of FXR activation in the treatment of various diseases involving bile acid dysfunction, such as liver fibrosis(38) and inflammatory bowel disease (42). The Food and Drug Administration and the European Medicines Agency have granted this agent, also known as INT-747, orphan drug status for the treatment of primary biliary cirrhosis (PBC) (2, 3). 6-ECDCA was evaluated in phase I clinical trials in healthy volunteers, and phase II clinical trials in patients with type 2 diabetes mellitus, nonalcoholic fatty liver disease (NAFLD) and PBC. 6-ECDCA showed antifibrotic activity in three liver fibrosis models through activation FXR. In the porcine serum-induced rat liver fibrosis model, 6-ECDCA can reduce expression of α1(I) collagen, transforming growth factor-β1 (TGF-β1), and α-smooth muscle actin in liver (38). In the rat BDL model, 6-ECDCA can reduce liver fibrosis and α1(I) collagen, TGF-β1, α-SMA, as well as tissue metalloproteinase inhibitor (TIMP)-1 and 2 mRNA by 70% to 80% (38). In the CCl4 liver toxicity model, 6-ECDCA administration results in induction of SHP, prevents upregulation of tissue inhibitor of metalloproteinase 1 (TIMP-1) mRNA, and accelerates collagen elimination (121). 6-ECDCA also shows anticholeretic activity on two cholestasis models (40). In LCA-induced cholestasis, 6-ECDCA treatment can fully reverse the reduced bile flow and transiently protect against the liver injury (115). In estrone-induced cholestasis, administration of 6-ECDCA reduces serum ALP activity and improves the cholestatic changes caused by estrogen. 6-ECDCA partially abrogates the reduction of bile acid output through increased β-muricholic acid (β-MCA) and taurochenodeoxycholic acid (TCDCA) secretion (39). Additionally, 6-ECDCA treatment can decrease the level of glucose, free fatty acid and HDL in plasma, and the triglyceride, free fatty acid, cholesterol, and glycogen content in the liver via FXR activation(23), thus suggesting that 6-ECDCA treatment is a potential therapy for nonalcoholic steatohepatitis/NAFLD. In clinical trials, 6-ECDCA can significantly decrease the serum levels of ALT, ALP, and gamma glutamyl transpeptidase in primary PBC patients (1). In patients with type 2 diabetes mellitus and NAFLD, 6-ECDCA can increase glucose disposal rates and reduce body weight (102). Thus, compounds that activate or inhibit FXR may be of great value for the treatment of various hepatic and metabolic diseases.
Conclusions
Ingested endogenous dietary chemicals, toxicants, and xeno-biotic compounds are absorbed in the small intestine and transported to the liver and other tissues. In intestine and liver exposed to high concentrations of bile acids, FXR plays an important role in endogenous chemical homeostasis and protection from potential toxicity (Fig. 3). Recent discoveries suggest that alteration of hepatic and intestinal FXR signaling is involved in multiple diseases. Further understanding of the role of FXR in the enterohepatic system can contribute to development of new clinical agents and therapeutic strategies. In addition to being a major regulator of homeostasis, FXR plays an important role in intestinal defense against inflammation, interacting with nuclear factor-kappaB (NF-κB) signaling (143). Exposure of LPS-activated macrophages to an FXR ligand leads to a reciprocal regulation of NF-κB-dependent genes such as TNFα and IL-1α. Intestinal FXR activation in response to certain bile acid metabolites, can possibly control bacterial growth and maintain mucosal integrity, by regulating expression of a variety of genes involved in defense against inflammation and mucosa protection. Thus, FXR is a potentially crucial factor that regulates intestinal innate immunity and homeostasis. More studies are warranted to determine the involvement of bile acids in diseases, in particular, the potential interplay between bile acids and the gut microbiota that are associated with numerous diseases of the GI tract, and metabolic disorders such as type 2 diabetes and obesity (43).
Figure 6.
Mechanism for gene suppression by Rev-erbα. Rev-erbα binds heme and represses gene transcription by recruitment of NCoR-HDAC3 corepressor complex and promotes chromatin condensation. Rev-erbα suppresses expression of Cyp7a1 and the positive clock component, Bmal1 that controls circadian rhythm. Derived in part, with permission, from Science Slides (scienceslides.com) and reference (160).
References
- 1.Intercept Pharmaceuticals Inc press release. 2010, July 28.
- 2.Intercept Pharmaceuticals Inc Company World Wide Web Site. 2010, June 18.
- 3.Akwabi-Ameyaw A, Bass JY, Caldwell RD, Caravella JA, Chen LH, Creech KL, Deaton DN, Madauss KP, Marr HB, McFadyen RB, Miller AB, Navas F, Parks DJ, Spearing PK, Todd D, Williams SP, Wisely GB. FXR agonist activity of conformationally constrained analogs of GW 4064. Bioorg Med Chem Lett 19: 4733–4739, 2009. [DOI] [PubMed] [Google Scholar]
- 4.Anakk S, Watanabe M, Ochsner SA, McKenna NJ, Finegold MJ, Moore DD. Combined deletion of Fxr and Shp in mice induces Cyp17a1 and results in juvenile onset cholestasis. J Clin Invest 121: 86–95, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ananthanarayanan M, Balasubramanian N, Makishima M, Mangelsdorf DJ, Suchy FJ. Human bile salt export pump promoter is transactivated by the farnesoid X receptor/bile acid receptor. J Biol Chem 276: 28857–28865, 2001. [DOI] [PubMed] [Google Scholar]
- 6.Araya Z, Wikvall K. 6alpha-hydroxylation of taurochenodeoxycholic acid and lithocholic acid by CYP3A4 in human liver microsomes. Biochim Biophys Acta 1438: 47–54, 1999. [DOI] [PubMed] [Google Scholar]
- 7.Barak Y, Nelson MC, Ong ES, Jones YZ, Ruiz-Lozano P, Chien KR, Koder A, Evans RM. PPAR gamma is required for placental, cardiac, and adipose tissue development. Molecular Cell 4: 585–595, 1999. [DOI] [PubMed] [Google Scholar]
- 8.Baranowski M Biological role of liver X receptors. J Physiol Pharmacol 59(Suppl 7): 31–55, 2008. [PubMed] [Google Scholar]
- 9.Barbier O, Torra IP, Sirvent A, Claudel T, Blanquart C, Duran-Sandoval D, Kuipers F, Kosykh V, Fruchart JC, Staels B. FXR induces the UGT2B4 enzyme in hepatocytes: A potential mechanism of negative feedback control of FXR activity. Gastroenterology 124: 1926–1940, 2003. [DOI] [PubMed] [Google Scholar]
- 10.Bodin K, Lindbom U, Diczfalusy U. Novel pathways of bile acid metabolism involving CYP3A4. Biochim Biophys Acta 1687: 84–93, 2005. [DOI] [PubMed] [Google Scholar]
- 11.Carter BA, Taylor OA, Prendergast DR, Zimmerman TL, Von Furstenberg R, Moore DD, Karpen SJ. Stigmasterol, a soy lipid-derived phytosterol, is an antagonist of the bile acid nuclear receptor FXR. Pediatr Res 62: 301–306, 2007. [DOI] [PubMed] [Google Scholar]
- 12.Chanda D, Xie YB, Choi HS. Transcriptional corepressor SHP recruits SIRT1 histone deacetylase to inhibit LRH-1 transactivation. Nucleic Acids Res 38: 4607–4619, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen F, Ma L, Dawson PA, Sinal CJ, Sehayek E, Gonzalez FJ, Breslow J, Ananthanarayanan M, Shneider BL. Liver receptor homologue-1 mediates species- and cell line-specific bile acid-dependent negative feedback regulation of the apical sodium-dependent bile acid transporter. J Biol Chem 278: 19909–19916, 2003. [DOI] [PubMed] [Google Scholar]
- 14.Chen W, Chiang JY. Regulation of human sterol 27-hydroxylase gene (CYP27A1) by bile acids and hepatocyte nuclear factor 4alpha (HNF4alpha). Gene 313: 71–82, 2003. [DOI] [PubMed] [Google Scholar]
- 15.Chen W, Owsley E, Yang Y, Stroup D, Chiang JY. Nuclear receptor-mediated repression of human cholesterol 7alpha-hydroxylase gene transcription by bile acids. J Lipid Res 42: 1402–1412, 2001. [PubMed] [Google Scholar]
- 16.Chen WS, Manova K, Weinstein DC, Duncan SA, Plump AS, Prezioso VR, Bachvarova RF, Darnell JE. Disruption of the Hnf-4 gene, expressed in visceral endoderm, leads to cell-death in embryonic ectoderm and impaired gastrulation of mouse embryos. Gene Dev 8: 2466–2477, 1994. [DOI] [PubMed] [Google Scholar]
- 17.Cheng J, Ma X, Gonzalez FJ. Pregnane X receptor- and CYP3A4-humanized mouse models and their applications. Br J Pharmacol 163: 461–468, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng X, Maher J, Dieter MZ, Klaassen CD. Regulation of mouse organic anion-transporting polypeptides (Oatps) in liver by prototypical microsomal enzyme inducers that activate distinct transcription factor pathways. Drug Metab Dispos 33: 1276–1282, 2005. [DOI] [PubMed] [Google Scholar]
- 19.Chiang JY. Bile acids: Regulation of synthesis. J Lipid Res 50: 1955–1966, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiang JY. Hepatocyte nuclear factor 4alpha regulation of bile acid and drug metabolism. Expert Opin Drug Metab Toxicol 5: 137–147, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho JY, Matsubara T, Kang DW, Ahn SH, Krausz KW, Idle JR, Luecke H, Gonzalez FJ. Urinary metabolomics in Fxr-null mice reveals activated adaptive metabolic pathways upon bile acid challenge. J Lipid Res 51: 1063–1074, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi M, Moschetta A, Bookout AL, Peng L, Umetani M, Holmstrom SR, Suino-Powell K, Xu HE, Richardson JA, Gerard RD, Mangelsdorf DJ, Kliewer SA. Identification of a hormonal basis for gallbladder filling. Nat Med 12: 1253–1255, 2006. [DOI] [PubMed] [Google Scholar]
- 23.Cipriani S, Mencarelli A, Palladino G, Fiorucci S. FXR activation reverses insulin resistance and lipid abnormalities and protects against liver steatosis in Zucker (fa/fa) obese rats. J Lipid Res 51: 771–784, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coppola CP, Gosche JR, Arrese M, Ancowitz B, Madsen J, Vanderhoof J, Shneider BL. Molecular analysis of the adaptive response of intestinal bile acid transport after ileal resection in the rat. Gastroenterology 115: 1172–1178, 1998. [DOI] [PubMed] [Google Scholar]
- 25.Dawson PA, Hubbert M, Haywood J, Craddock AL, Zerangue N, Christian WV, Ballatori N. The heteromeric organic solute transporter alpha-beta, Ostalpha-Ostbeta, is an ileal basolateral bile acid transporter. J Biol Chem 280: 6960–6968, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Fabiani E, Mitro N, Anzulovich AC, Pinelli A, Galli G, Crestani M. The negative effects of bile acids and tumor necrosis factor-alpha on the transcription of cholesterol 7alpha-hydroxylase gene (CYP7A1) converge to hepatic nuclear factor-4: A novel mechanism of feedback regulation of bile acid synthesis mediated by nuclear receptors. J Biol Chem 276: 30708–30716, 2001. [DOI] [PubMed] [Google Scholar]
- 27.De Fabiani E, Mitro N, Gilardi F, Caruso D, Galli G, Crestani M. Coordinated control of cholesterol catabolism to bile acids and of gluconeogenesis via a novel mechanism of transcription regulation linked to the fasted-to-fed cycle. J Biol Chem 278: 39124–39132, 2003. [DOI] [PubMed] [Google Scholar]
- 28.De Vree JM, Jacquemin E, Sturm E, Cresteil D, Bosma PJ, Aten J, Deleuze JF, Desrochers M, Burdelski M, Bernard O, Oude Elferink RP, Hadchouel M. Mutations in the MDR3 gene cause progressive familial intrahepatic cholestasis. Proc Natl Acad Sci U S A 95: 282–287, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Del Castillo-Olivares A, Gil G. Alpha 1-fetoprotein transcription factor is required for the expression of sterol 12alpha-hydroxylase, the specific enzyme for cholic acid synthesis. Potential role in the bile acid-mediated regulation of gene transcription. J Biol Chem 275: 17793–17799, 2000. [DOI] [PubMed] [Google Scholar]
- 30.Del Castillo-Olivares A, Gil G. Role of FXR and FTF in bile acid-mediated suppression of cholesterol 7alpha-hydroxylase transcription. Nucleic Acids Res 28: 3587–3593, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Denson LA, Sturm E, Echevarria W, Zimmerman TL, Makishima M, Mangelsdorf DJ, Karpen SJ. The orphan nuclear receptor, shp, mediates bile acid-induced inhibition of the rat bile acid transporter, ntcp. Gastroenterology 121: 140–147, 2001. [DOI] [PubMed] [Google Scholar]
- 32.Dhe-Paganon S, Duda K, Iwamoto M, Chi YI, Shoelson SE. Crystal structure of the HNF4 alpha ligand binding domain in complex with endogenous fatty acid ligand. J Biol Chem 277: 37973–37976, 2002. [DOI] [PubMed] [Google Scholar]
- 33.Downes M, Verdecia MA, Roecker AJ, Hughes R, Hogenesch JB, Kast-Woelbern HR, Bowman ME, Ferrer JL, Anisfeld AM, Edwards PA, Rosenfeld JM, Alvarez JG, Noel JP, Nicolaou KC, Evans RM. A chemical, genetic, and structural analysis of the nuclear bile acid receptor FXR. Mol Cell 11: 1079–1092, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duane WC, Levitt DG, Mueller SM, Behrens JC. Regulation of bile-acid synthesis in man - presence of a diurnal rhythm. J Clin Invest 72: 1930–1936, 1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duez H, Staels B. Rev-erb-alpha: An integrator of circadian rhythms and metabolism. J Appl Physiol 107: 1972–1980, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duez H, Van Der Veen JN, Duhem C, Pourcet B, Touvier T, Fontaine C, Derudas B, Bauge E, Havinga R, Bloks VW, Wolters H, Van Der Sluijs FH, Vennstrom B, Kuipers F, Staels B. Regulation of bile acid synthesis by the nuclear receptor Rev-erb alpha. Gastroenterology 135: 689–698, 2008. [DOI] [PubMed] [Google Scholar]
- 37.Eloranta JJ, Jung D, Kullak-Ublick GA. The human Na+-taurocholate cotransporting polypeptide gene is activated by glucocorticoid receptor and peroxisome proliferator-activated receptor-gamma coactivator-1alpha, and suppressed by bile acids via a small heterodimer partner-dependent mechanism. Mol Endocrinol 20: 65–79, 2006. [DOI] [PubMed] [Google Scholar]
- 38.Fiorucci S, Antonelli E, Rizzo G, Renga B, Mencarelli A, Riccardi L, Orlandi S, Pellicciari R, Morelli A. The nuclear receptor SHP mediates inhibition of hepatic stellate cells by FXR and protects against liver fibrosis. Gastroenterology 127: 1497–1512, 2004. [DOI] [PubMed] [Google Scholar]
- 39.Fiorucci S, Clerici C, Antonelli E, Orlandi S, Goodwin B, Sadeghpour BM, Sabatino G, Russo G, Castellani D, Willson TM, Pruzanski M, Pellicciari R, Morelli A. Protective effects of 6-ethyl chenodeoxycholic acid, a farnesoid X receptor ligand, in estrogen-induced cholestasis. J Pharmacol Exp Ther 313: 604–612, 2005. [DOI] [PubMed] [Google Scholar]
- 40.Fiorucci S, Rizzo G, Antonelli E, Renga B, Mencarelli A, Riccardi L, Orlandi S, Pruzanski M, Morelli A, Pellicciari R. A farnesoid x receptor-small heterodimer partner regulatory cascade modulates tissue metalloproteinase inhibitor-1 and matrix metalloprotease expression in hepatic stellate cells and promotes resolution of liver fibrosis. J Pharmacol Exp Ther 314: 584–595, 2005. [DOI] [PubMed] [Google Scholar]
- 41.Forman BM, Goode E, Chen J, Oro AE, Bradley DJ, Perlmann T, Noonan DJ, Burka LT, McMorris T, Lamph WW, Evans RM, Weinberger C. Identification of a nuclear receptor that is activated by farnesol metabolites. Cell 81: 687–693, 1995. [DOI] [PubMed] [Google Scholar]
- 42.Gadaleta RM, van Erpecum KJ, Oldenburg B, Willemsen EC, Renooij W, Murzilli S, Klomp LW, Siersema PD, Schipper ME, Danese S, Penna G, Laverny G, Adorini L, Moschetta A, van Mil SW. Farnesoid X receptor activation inhibits inflammation and preserves the intestinal barrier in inflammatory bowel disease. Gut 60: 463–472, 2011. [DOI] [PubMed] [Google Scholar]
- 43.Garrett WS, Gordon JI, Glimcher LH. Homeostasis and inflammation in the intestine. Cell 140: 859–870, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garuti R, Croce MA, Piccinini L, Tiozzo R, Bertolini S, Calandra S. Functional analysis of the promoter of human sterol 27-hydroxylase gene in HepG2 cells. Gene 283: 133–143, 2002. [DOI] [PubMed] [Google Scholar]
- 45.Geier A, Dietrich CG, Voigt S, Kim SK, Gerloff T, Kullak-Ublick GA, Lorenzen J, Matern S, Gartung C. Effects of proinflammatory cytokines on rat organic anion transporters during toxic liver injury and cholestasis. Hepatology 38: 345–354, 2003. [DOI] [PubMed] [Google Scholar]
- 46.Geier A, Martin IV, Dietrich CG, Balasubramaniyan N, Strauch S, Suchy FJ, Gartung C, Trautwein C, Ananthanarayanan M. Hepatocyte nuclear factor-4 alpha is a central transactivator of the mouse Ntcp gene. Am J Physiol-Gastrointest Liver Physiol 295: G226–G233, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Geier A, Wagner M, Dietrich CG, Trauner M. Principles of hepatic organic anion transporter regulation during cholestasis, inflammation and liver regeneration. Bba-Mol Cell Res 1773: 283–308, 2007. [DOI] [PubMed] [Google Scholar]
- 48.Gielen J, Vancantfort J, Robaye B, Renson J. Rat-Liver Cholesterol 7alpha-hydroxylase.3. New results about its circadian-rhythm. Eur J Biochem 55: 41–48, 1975. [DOI] [PubMed] [Google Scholar]
- 49.Gnerre C, Blattler S, Kaufmann MR, Looser R, Meyer UA. Regulation of CYP3A4 by the bile acid receptor FXR: Evidence for functional binding sites in the CYP3A4 gene. Pharmacogenetics 14: 635–645, 2004. [DOI] [PubMed] [Google Scholar]
- 50.Gonzalez FJ. The 2006 Bernard B. Brodie Award Lecture. Cyp2e1. Drug Metab Dispos 35: 1–8, 2007. [DOI] [PubMed] [Google Scholar]
- 51.Goodwin B, Jones SA, Price RR, Watson MA, McKee DD, Moore LB, Galardi C, Wilson JG, Lewis MC, Roth ME, Maloney PR, Willson TM, Kliewer SA. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol Cell 6: 517–526, 2000. [DOI] [PubMed] [Google Scholar]
- 52.Goodwin B, Watson MA, Kim H, Miao J, Kemper JK, Kliewer SA. Differential regulation of rat and human CYP7A1 by the nuclear oxysterol receptor liver X receptor-alpha. Mol Endocrinol 17: 386–394, 2003. [DOI] [PubMed] [Google Scholar]
- 53.Grober J, Zaghini I, Fujii H, Jones SA, Kliewer SA, Willson TM, Ono T, Besnard P. Identification of a bile acid-responsive element in the human ileal bile acid-binding protein gene. Involvement of the farnesoid X receptor/9-cis-retinoic acid receptor heterodimer. J Biol Chem 274: 29749–29754, 1999. [DOI] [PubMed] [Google Scholar]
- 54.Hadjantonakis AK, Pirity M, Nagy A. Cre recombinase mediated alterations of the mouse genome using embryonic stem cells. Methods Mol Biol 461: 111–132, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hagenbuch B, Meier PJ. Molecular cloning, chromosomal localization, and functional characterization of a human liver Na+/bile acid cotransporter. J Clin Invest 93: 1326–1331, 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hardin PE, Yu W. Circadian transcription: Passing the HAT to CLOCK. Cell 125: 424–426, 2006. [DOI] [PubMed] [Google Scholar]
- 57.Harding HP, Lazar MA. The orphan receptor Rev-ErbA alpha activates transcription via a novel response element. Mol Cell Biol 13: 3113–3121, 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hertz R, Ben-Haim N, Petrescu AD, Kalderon B, Berman I, Eldad N, Schroeder F, Bar-Tana J. Rescue of MODY-1 by agonist ligands of hepatocyte nuclear factor-4 alpha. J Biol Chem 278: 22578–22585, 2003. [DOI] [PubMed] [Google Scholar]
- 59.Hertz R, Magenheim J, Berman I, Bar-Tana J. Fatty acyl-CoA thioesters are ligands of hepatic nuclear factor-4 alpha. Nature 392: 512–516, 1998. [DOI] [PubMed] [Google Scholar]
- 60.Heubi JE, Balistreri WF, Fondacaro JD, Partin JC, Schubert WK. Primary bile acid malabsorption: Defective in vitro ileal active bile acid transport. Gastroenterology 83: 804–811, 1982. [PubMed] [Google Scholar]
- 61.Hofmann AF. Detoxification of lithocholic acid, a toxic bile acid: Relevance to drug hepatotoxicity. Drug Metab Rev 36: 703–722, 2004. [DOI] [PubMed] [Google Scholar]
- 62.Hofmann AF. The enterohepatic circulation of bile acids in mammals: Form and functions. Front Biosci 14: 2584–2598, 2009. [DOI] [PubMed] [Google Scholar]
- 63.Holt JA, Luo G, Billin AN, Bisi J, McNeill YY, Kozarsky KF, Donahee M, Wang DY, Mansfield TA, Kliewer SA, Goodwin B, Jones SA. Definition of a novel growth factor-dependent signal cascade for the suppression of bile acid biosynthesis. Genes Dev 17: 1581–1591, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Howard WR, Pospisil JA, Njolito E, Noonan DJ. Catabolites of cholesterol synthesis pathways and forskolin as activators of the farnesoid X-activated nuclear receptor. Toxicol Appl Pharmacol 163: 195–202, 2000. [DOI] [PubMed] [Google Scholar]
- 65.Huang L, Zhao A, Lew JL, Zhang T, Hrywna Y, Thompson JR, de Pedro N, Royo I, Blevins RA, Pelaez F, Wright SD, Cui J. Farnesoid X receptor activates transcription of the phospholipid pump MDR3. J Biol Chem 278: 51085–51090, 2003. [DOI] [PubMed] [Google Scholar]
- 66.Hughes SE. Differential expression of the fibroblast growth factor receptor (FGFR) multigene family in normal human adult tissues. J Histochem Cytochem 45: 1005–1019, 1997. [DOI] [PubMed] [Google Scholar]
- 67.Ihunnah CA, Jiang M, Xie W. Nuclear receptor PXR, transcriptional circuits and metabolic relevance. Biochim Biophys Acta 1812: 956–963, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Inagaki T, Choi M, Moschetta A, Peng L, Cummins CL, McDonald JG, Luo G, Jones SA, Goodwin B, Richardson JA, Gerard RD, Repa JJ, Mangelsdorf DJ, Kliewer SA. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab 2: 217–225, 2005. [DOI] [PubMed] [Google Scholar]
- 69.Inagaki T, Moschetta A, Lee YK, Peng L, Zhao G, Downes M, Yu RT, Shelton JM, Richardson JA, Repa JJ, Mangelsdorf DJ, Kliewer SA. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci U S A 103: 3920–3925, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Inoue Y, Yu AM, Inoue J, Gonzalez FJ. Hepatocyte nuclear factor 4alpha is a central regulator of bile acid conjugation. J Biol Chem 279: 2480–2489, 2004. [DOI] [PubMed] [Google Scholar]
- 71.Inoue Y, Yu AM, Yim SH, Ma X, Krausz KW, Inoue J, Xiang CC, Brownstein MJ, Eggertsen G, Bjorkhem I, Gonzalez FJ. Regulation of bile acid biosynthesis by hepatocyte nuclear factor 4alpha. J Lipid Res 47: 215–227, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jones S Mini-review: Endocrine actions of fibroblast growth factor 19. Mol Pharm 5: 42–48, 2008. [DOI] [PubMed] [Google Scholar]
- 73.Jung D, Elferink MGL, Stellaard F, Groothuis GMM. Analysis of bile acid-induced regulation of FXR target genes in human liver slices. Liver Int 27: 137–144, 2007. [DOI] [PubMed] [Google Scholar]
- 74.Jung D, Hagenbuch B, Fried M, Meier PJ, Kullak-Ublick GA. Role of liver-enriched transcription factors and nuclear receptors in regulating the human, mouse, and rat NTCP gene. Am J Physiol Gastrointest Liver Physiol 286: G752–G761, 2004. [DOI] [PubMed] [Google Scholar]
- 75.Jung D, Kullak-Ublick GA. Hepatocyte nuclear factor 1 alpha: A key mediator of the effect of bile acids on gene expression. Hepatology 37: 622–631, 2003. [DOI] [PubMed] [Google Scholar]
- 76.Kemper JK. Regulation of FXR transcriptional activity in health and disease: Emerging roles of FXR cofactors and post-translational modifications. Biochim Biophys Acta 1812: 842–850, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Khan AA, Chow EC, van Loenen-Weemaes AM, Porte RJ, Pang KS, Groothuis GM. Comparison of effects of VDR versus PXR, FXR and GR ligands on the regulation of CYP3A isozymes in rat and human intestine and liver. Eur J Pharm Sci 37: 115–125, 2009. [DOI] [PubMed] [Google Scholar]
- 78.Kim I, Ahn SH, Inagaki T, Choi M, Ito S, Guo GL, Kliewer SA, Gonzalez FJ. Differential regulation of bile acid homeostasis by the farnesoid X receptor in liver and intestine. J Lipid Res 48: 2664–2672, 2007. [DOI] [PubMed] [Google Scholar]
- 79.Kim I, Morimura K, Shah Y, Yang Q, Ward JM, Gonzalez FJ. Spontaneous hepatocarcinogenesis in farnesoid X receptor-null mice. Carcinogenesis 28: 940–946, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kir S, Beddow SA, Samuel VT, Miller P, Previs SF, Suino-Powell K, Xu HE, Shulman GI, Kliewer SA, Mangelsdorf DJ. FGF19 as a post-prandial, insulin-independent activator of hepatic protein and glycogen Synthesis. Science 331: 1621–1624, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kir S, Kliewer SA, Mangelsdorf DJ. Roles of FGF19 in Liver Metabolism. Cold Spring Harb Symp Quant Biol 76: 139–144, 2011. [DOI] [PubMed] [Google Scholar]
- 82.Kliewer SA, Moore JT, Wade L, Staudinger JL, Watson MA, Jones SA, McKee DD, Oliver BB, Willson TM, Zetterstrom RH, Perlmann T, Lehmann JM. An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell 92: 73–82, 1998. [DOI] [PubMed] [Google Scholar]
- 83.Kong B, Wang L, Chiang JY, Zhang Y, Klaassen CD, Guo GL. Mechanism of tissue-specific farnesoid X receptor in suppressing the expression of genes in bile-acid synthesis in mice. Hepatology 2012. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kramer W, Girbig F, Gutjahr U, Kowalewski S, Jouvenal K, Muller G, Tripier D, Wess G. Intestinal bile acid absorption. Na(+)-dependent bile acid transport activity in rabbit small intestine correlates with the coexpression of an integral 93-kDa and a peripheral 14-kDa bile acid-binding membrane protein along the duodenum-ileum axis. J Biol Chem 268: 18035–18046, 1993. [PubMed] [Google Scholar]
- 85.Kuo CJ, Conley PB, Chen L, Sladek FM, Darnell JE Jr., Crabtree GR. A transcriptional hierarchy involved in mammalian cell-type specification. Nature 355: 457–461, 1992. [DOI] [PubMed] [Google Scholar]
- 86.Le Martelot G, Claudel T, Gatfield D, Schaad O, Kornmann B, Sasso GL, Moschetta A, Schibler U. REV-ERBalpha participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol 7: e1000181, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee FY, de Aguiar Vallim TQ, Chong HK, Zhang Y, Liu Y, Jones SA, Osborne TF, Edwards PA. Activation of the farnesoid X receptor provides protection against acetaminophen-induced hepatic toxicity. Mol Endocrinol 24: 1626–1636, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lee H, Zhang Y, Lee FY, Nelson SF, Gonzalez FJ, Edwards PA. FXR regulates organic solute transporters alpha and beta in the adrenal gland, kidney, and intestine. J Lipid Res 47: 201–214, 2006. [DOI] [PubMed] [Google Scholar]
- 89.Lee SS, Pineau T, Drago J, Lee EJ, Owens JW, Kroetz DL, Fernandez-Salguero PM, Westphal H, Gonzalez FJ. Targeted disruption of the alpha isoform of the peroxisome proliferator-activated receptor gene in mice results in abolishment of the pleiotropic effects of peroxisome proliferators. Mol Cell Biol 15: 3012–3022, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lee YH, Alberta JA, Gonzalez FJ, Waxman DJ. Multiple, functional Dbp sites on the promoter of the cholesterol 7-alpha-hydroxylase P450 gene, cyp7 - proposed role in diurnal regulation of liver gene-expression. J Biol Chem 269: 14681–14689, 1994. [PubMed] [Google Scholar]
- 91.Lehmann JM, Kliewer SA, Moore LB, Smith-Oliver TA, Oliver BB, Su JL, Sundseth SS, Winegar DA, Blanchard DE, Spencer TA, Willson TM. Activation of the nuclear receptor LXR by oxysterols defines a new hormone response pathway. J Biol Chem 272: 3137–3140, 1997. [DOI] [PubMed] [Google Scholar]
- 92.Li GD, Thomas AM, Hart SN, Zhong XB, Wu DQ, Guo GL. Farnesoid X receptor activation mediates head-to-tail chromatin looping in the Nr0b2 gene encoding small heterodimer partner. Mol Endocrinol 24: 1404–1412, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li T, Matozel M, Boehme S, Kong B, Nilsson LM, Guo G, Ellis E, Chiang JY. Overexpression of cholesterol 7alpha-hydroxylase promotes hepatic bile acid synthesis and secretion and maintains cholesterol homeostasis. Hepatology 53: 996–1006, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu Y, Binz J, Numerick MJ, Dennis S, Luo G, Desai B, MacKenzie KI, Mansfield TA, Kliewer SA, Goodwin B, Jones SA. Hepatoprotection by the farnesoid X receptor agonist GW4064 in rat models of intra- and extrahepatic cholestasis. J Clin Invest 112: 1678–1687, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lu TT, Makishima M, Repa JJ, Schoonjans K, Kerr TA, Auwerx J, Mangelsdorf DJ. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol Cell 6: 507–515, 2000. [DOI] [PubMed] [Google Scholar]
- 96.Maeda S, Kamata H, Luo JL, Leffert H, Karin M. IKK beta couples hepatocyte death to cytokine-driven compensatory proliferation that promotes chemical hepatocarcinogenesis. Cell 121: 977–990, 2005. [DOI] [PubMed] [Google Scholar]
- 97.Maeda T, Miyata M, Yotsumoto T, Kobayashi D, Nozawa T, Toyama K, Gonzalez FJ, Yamazoe Y, Tamai I. Regulation of drug transporters by the farnesoid X receptor in mice. Mol Pharm 1: 281–289, 2004. [DOI] [PubMed] [Google Scholar]
- 98.Makishima M, Okamoto AY, Repa JJ, Tu H, Learned RM, Luk A, Hull MV, Lustig KD, Mangelsdorf DJ, Shan B. Identification of a nuclear receptor for bile acids. Science 284: 1362–1365, 1999. [DOI] [PubMed] [Google Scholar]
- 99.Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans RM. The nuclear receptor superfamily: The second decade. Cell 83: 835–839, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Maran RR, Thomas A, Roth M, Sheng Z, Esterly N, Pinson D, Gao X, Zhang Y, Ganapathy V, Gonzalez FJ, Guo GL. Farnesoid X receptor deficiency in mice leads to increased intestinal epithelial cell proliferation and tumor development. J Pharmacol Exp Ther 328: 469–477, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Margolis RN, Christakos S. The nuclear receptor superfamily of steroid hormones and vitamin D gene regulation An update. Ann Ny Acad Sci 1192: 208–214, 2010. [DOI] [PubMed] [Google Scholar]
- 102.Mason A, Luketic V, Lindor K, Hirschfield G, Gordon S, Mayo M, Kowdley K, Parés A, Trauner M, Castelloe E, Sciacca C, Jones TB, Böhm O, Shapiro D. Farnesoid-X receptor agonists: A new class of drugs for the treatment of PBC? An international study evaluating the addition of INT-747 to ursodeoxycholicacid. J Hepatol 52: S1–S2, 2010. [Google Scholar]
- 103.Matsubara T, Tanaka N, Patterson AD, Cho JY, Krausz KW, Gonzalez FJ. Lithocholic acid disrupts phospholipid and sphingolipid homeostasis leading to cholestasis in mice. Hepatology 53: 1282–1293, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mi LZ, Devarakonda S, Harp JM, Han C, Pellicciari R, Willson TM, Khorasanizadeh S, Rastinejad F. Structural basis for bile acid binding and activation of the nuclear receptor FXR. Molecular Cell 11: 1093–1100, 2003. [DOI] [PubMed] [Google Scholar]
- 105.Miao J, Choi SE, Seok SM, Yang L, Zuercher WJ, Xu Y, Willson TM,Xu HE, Kemper JK. Ligand-Dependent Regulation of the Activity of the Orphan Nuclear Receptor, Small Heterodimer Partner (SHP), in the Repression of Bile Acid Biosynthetic CYP7A1 and CYP8B1 Genes. Mol Endocrinol 25: 1159–1169, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Miyata M, Matsuda Y, Tsuchiya H, Kitada H, Akase T, Shimada M, Nagata K, Gonzalez FJ, Yamazoe Y. Chenodeoxycholic acid-mediated activation of the farnesoid X receptor negatively regulates hydroxysteroid sulfotransferase. Drug Metab Pharmacokinet 21: 315–323, 2006. [DOI] [PubMed] [Google Scholar]
- 107.Miyata M, Tozawa A, Otsuka H, Nakamura T, Nagata K, Gonzalez FJ, Yamazoe Y. Role of farnesoid X receptor in the enhancement of canalicular bile acid output and excretion of unconjugated bile acids: A mechanism for protection against cholic acid-induced liver toxicity. J Pharmacol Exp Ther 312: 759–766, 2005. [DOI] [PubMed] [Google Scholar]
- 108.Moschetta A, Bookout AL, Mangelsdorf DJ. Prevention of cholesterol gallstone disease by FXR agonists in a mouse model. Nat Med 10: 1352–1358, 2004. [DOI] [PubMed] [Google Scholar]
- 109.Noshiro M, Usui E, Kawamoto T, Kubo H, Fujimoto K, Furukawa M, Honma S, Makishima M, Honma K, Kato Y. Multiple mechanisms regulate circadian expression of the gene, for cholesterol 7 alpha-hydroxylase (Cyp7a), a key enzyme in hepatic bile acid biosynthesis. J Biol Rhythm 22: 299–311, 2007. [DOI] [PubMed] [Google Scholar]
- 110.Novac N, Heinzel T. Nuclear receptors: Overview and classification.Curr Drug Targets Inflamm Allergy 3: 335–346, 2004. [DOI] [PubMed] [Google Scholar]
- 111.Oelkers P, Kirby LC, Heubi JE, Dawson PA. Primary bile acid malabsorption caused by mutations in the ileal sodium-dependent bile acid transporter gene (SLC10A2). J Clin Invest 99: 1880–1887, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Parks DJ, Blanchard SG, Bledsoe RK, Chandra G, Consler TG, Kliewer SA, Stimmel JB, Willson TM, Zavacki AM, Moore DD, Lehmann JM. Bile acids: Natural ligands for an orphan nuclear receptor. Science 284: 1365–1368, 1999. [DOI] [PubMed] [Google Scholar]
- 113.Paschos GK, Baggs JE, Hogenesch JB, FitzGerald GA. The role of clock genes in pharmacology. Annu Rev Pharmacol 50: 187–214, 2010. [DOI] [PubMed] [Google Scholar]
- 114.Peet DJ, Turley SD, Ma WZ, Janowski BA, Lobaccaro JMA, Hammer RE, Mangelsdorf DJ. Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXR alpha. Cell 93: 693–704, 1998. [DOI] [PubMed] [Google Scholar]
- 115.Pellicciari R, Fiorucci S, Camaioni E, Clerici C, Costantino G, Maloney PR, Morelli A, Parks DJ, Willson TM. 6alpha-ethyl-chenodeoxycholic acid (6-ECDCA), a potent and selective FXR agonist endowed with anticholestatic activity. J Med Chem 45: 3569–3572, 2002. [DOI] [PubMed] [Google Scholar]
- 116.Petrescu AD, Payne HR, Boedecker A, Chao H, Hertz R, Bar-Tana J, Schroeder F, Kier AB. Physical and functional interaction of acyl-CoA-binding protein with hepatocyte nuclear factor-4 alpha. J Biol Chem 278: 51813–1824, 2003. [DOI] [PubMed] [Google Scholar]
- 117.Pillot T, Ouzzine M, Fournel-Gigleux S, Lafaurie C, Radominska A, Burchell B, Siest G, Magdalou J. Glucuronidation of hyodeoxycholic acid in human liver. Evidence for a selective role of UDP-glucuronosyltransferase 2B4. J Biol Chem 268: 25636–25642, 1993. [PubMed] [Google Scholar]
- 118.Pircher PC, Kitto JL, Petrowski ML, Tangirala RK, Bischoff ED, Schulman IG, Westin SK. Farnesoid X receptor regulates bile acid-amino acid conjugation. J Biol Chem 278: 27703–27711, 2003. [DOI] [PubMed] [Google Scholar]
- 119.Potthoff MJ, Boney-Montoya J, Choi M, He T, Sunny NE, Satapati S, Suino-Powell K, Xu HE, Gerard RD, Finck BN, Burgess SC, Mangelsdorf DJ, Kliewer SA. FGF15/19 regulates hepatic glucose metabolism by inhibiting the CREB-PGC-1alpha pathway. Cell Metab 13: 729–738, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Prawitt J, Abdelkarim M, Stroeve JHM, Popescu I, Duez H, Velagapudi VR, Dumont J, Bouchaert E, van Dijk TH, Lucas A, Dorchies E, Daoudi M, Lestavel S, Gonzalez FJ, Oresic M, Cariou B, Kuipers F, Caron S, Staels B. Farnesoid X receptor deficiency improves glucose homeostasis in mouse models of obesity. Diabetes 60: 1861–1871, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Renga B, Mencarelli A, Cipriani S, D’Amore C, Zampella A, Monti MC, Distrutti E, Fiorucci S. The nuclear receptor FXR regulates hepatic transport and metabolism of glutamine and glutamate. Bba-Mol Basis Dis 1812: 1522–1531, 2011. [DOI] [PubMed] [Google Scholar]
- 122.Renga B, Mencarelli A, D’Amore C, Cipriani S, D’Auria MV, Sepe V, Chini MG, Monti MC, Bifulco G, Zampella A, Fiorucci S. Discovery that theonellasterol a marine sponge sterol Is a highly selective FXR antagonist that protects against liver injury in cholestasis. Plos One 7: e30443, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Repa JJ, Berge KE, Pomajzl C, Richardson JA, Hobbs H, Mangelsdorf DJ. Regulation of ATP-binding cassette sterol transporters ABCG5 and ABCG8 by the liver X receptors alpha and beta. J Biol Chem 277: 18793–18800, 2002. [DOI] [PubMed] [Google Scholar]
- 124.Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res 47: 241–259, 2006. [DOI] [PubMed] [Google Scholar]
- 125.Roberts MS, Magnusson BM, Burczynski FJ, Weiss M. Enterohepatic circulation: Physiological, pharmacokinetic and clinical implications. Clin Pharmacokinet 41: 751–790, 2002. [DOI] [PubMed] [Google Scholar]
- 126.Russell DW. Fifty years of advances in bile acid synthesis and metabolism. J Lipid Res 50: S120–S125, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Schuetz EG, Strom S, Yasuda K, Lecureur V, Assem M, Brimer C, Lamba J, Kim RB, Ramachandran V, Komoroski BJ, Venkataramanan R, Cai HB, Sinal CJ, Gonzalez FJ, Schuetz JD. Disrupted bile acid homeostasis reveals an unexpected interaction among nuclear hormone receptors, transporters, and cytochrome p450. J Biol Chem 276: 39411–39418, 2001. [DOI] [PubMed] [Google Scholar]
- 128.Seol W, Choi HS, Moore DD. Isolation of proteins that interact specifically with the retinoid X receptor: Two novel orphan receptors. Mol Endocrinol 9: 72–85, 1995. [DOI] [PubMed] [Google Scholar]
- 129.Seol W, Choi HS, Moore DD. An orphan nuclear hormone receptor that lacks a DNA binding domain and heterodimerizes with other receptors. Science 272: 1336–1339, 1996. [DOI] [PubMed] [Google Scholar]
- 130.Shen H, Zhang Y, Ding H, Wang X, Chen LL, Jiang HL, Shen X. Farnesoid X receptor induces GLUT4 expression through FXR response element in the GLUT4 promoter. Cell Physiol Biochem 22: 1–14, 2008. [DOI] [PubMed] [Google Scholar]
- 131.Shneider BL. Intestinal bile acid transport: Biology, physiology, and pathophysiology. J Pediatr Gastroenterol Nutr 32: 407–417, 2001. [DOI] [PubMed] [Google Scholar]
- 132.Sinal CJ, Tohkin M, Miyata M, Ward JM, Lambert G, Gonzalez FJ. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell 102: 731–744, 2000. [DOI] [PubMed] [Google Scholar]
- 133.Smith RL. The biliary excretion and enterohepatic circulation of drugs and other organic compounds. Fortschr Arzneimittelforsch 9: 299–360, 1966. [DOI] [PubMed] [Google Scholar]
- 134.Song CS, Echchgadda I, Baek BS, Ahn SC, Oh T, Roy AK, Chatterjee B. Dehydroepiandrosterone sulfotransferase gene induction by bile acid activated farnesoid X receptor. J Biol Chem 276: 42549–42556, 2001. [DOI] [PubMed] [Google Scholar]
- 135.Staudinger JL, Goodwin B, Jones SA, Hawkins-Brown D, MacKenzie KI, LaTour A, Liu Y, Klaassen CD, Brown KK, Reinhard J, Willson TM, Koller BH, Kliewer SA. The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc Natl Acad Sci U S A 98: 3369–3374, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Stieger B, Hagenbuch B, Landmann L, Hochli M, Schroeder A, Meier PJ. In situ localization of the hepatocytic Na+/Taurocholate cotransporting polypeptide in rat liver. Gastroenterology 107: 1781–1787, 1994. [DOI] [PubMed] [Google Scholar]
- 137.Stieger B, Meier Y, Meier PJ. The bile salt export pump. Pflugers Arch 453: 611–620, 2007. [DOI] [PubMed] [Google Scholar]
- 138.Strautnieks SS, Bull LN, Knisely AS, Kocoshis SA, Dahl N, Arnell H, Sokal E, Dahan K, Childs S, Ling V, Tanner MS, Kagalwalla AF, Nemeth A, Pawlowska J, Baker A, Mieli-Vergani G, Freimer NB, Gar-diner RM, Thompson RJ. A gene encoding a liver-specific ABC transporter is mutated in progressive familial intrahepatic cholestasis. Nat Genet 20: 233–238, 1998. [DOI] [PubMed] [Google Scholar]
- 139.Stroup D, Chiang JY. HNF4 and COUP-TFII interact to modulate transcription of the cholesterol 7alpha-hydroxylase gene (CYP7A1). J Lipid Res 41: 1–11, 2000. [PubMed] [Google Scholar]
- 140.Svoboda M, Riha J, Wlcek K, Jaeger W, Thalhammer T. Organic anion transporting polypeptides (OATPs): Regulation of expression and function. Curr Drug Metab 12: 139–153, 2011. [DOI] [PubMed] [Google Scholar]
- 141.Urizar NL, Liverman AB, Dodds DT, Silva FV, Ordentlich P, Yan Y, Gonzalez FJ, Heyman RA, Mangelsdorf DJ, Moore DD. A natural product that lowers cholesterol as an antagonist ligand for FXR. Science 296: 1703–1706, 2002. [DOI] [PubMed] [Google Scholar]
- 142.Van Helvoort A, Smith AJ, Sprong H, Fritzsche I, Schinkel AH, Borst P, van Meer G. MDR1 P-glycoprotein is a lipid translocase of broad specificity, while MDR3 P-glycoprotein specifically translocates phosphatidylcholine. Cell 87: 507–517, 1996. [DOI] [PubMed] [Google Scholar]
- 143.Vavassori P, Mencarelli A, Renga B, Distrutti E, Fiorucci S. The bile acid receptor FXR is a modulator of intestinal innate immunity. J Immunol 183: 6251–6261, 2009. [DOI] [PubMed] [Google Scholar]
- 144.Visser TJ, Rutgers M, de Herder WW, Rooda SJ, Hazenberg MP. Hepatic metabolism, biliary clearance and enterohepatic circulation of thyroid hormone. Acta Med Austriaca 15(Suppl 1): 37–39, 1988. [PubMed] [Google Scholar]
- 145.Vree TB, Timmer CJ. Enterohepatic cycling and pharmacokinetics of oestradiol in postmenopausal women. J Pharm Pharmacol 50: 857–864, 1998. [DOI] [PubMed] [Google Scholar]
- 146.Wang S, Lai K, Moy FJ, Bhat A, Hartman HB, Evans MJ. The nuclear hormone receptor farnesoid X receptor (FXR) is activated by androsterone. Endocrinology 147: 4025–4033, 2006. [DOI] [PubMed] [Google Scholar]
- 147.Wang T, Shah YM, Matsubara T, Zhen YY, Tanabe T, Nagano T, Fotso S, Krausz KW, Zabriskie TM, Idle JR, Gonzalez FJ. Control of steroid 21-oic acid synthesis by peroxisome proliferator-activated receptor alpha and role of the hypothalamic-pituitary-adrenal axis. J Biol Chem 285: 7670–7685, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wang XX, Jiang T, Shen Y, Adorini L, Pruzanski M, Gonzalez FJ, Scherzer P, Lewis L, Miyazaki-Anzai S, Levi M. The farnesoid X receptor modulates renal lipid metabolism and diet-induced renal inflammation, fibrosis, and proteinuria. Am J Physiol-Renal 297: F1587–F1596, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang XXX, Jiang T, Shen Y, Caldas Y, Miyazaki-Anzai S, Santamaria H, Urbanek C, Solis N, Scherzer P, Lewis L, Gonzalez FJ, Adorini L, Pruzanski M, Kopp JB, Verlander JW, Levi M. Diabetic nephropathy is accelerated by farnesoid X receptor deficiency and inhibited by farnesoid X receptor activation in a type 1 diabetes model. Diabetes 59: 2916–2927, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Watt AJ, Garrison WD, Duncan SA. HNF4: A central regulator of hepatocyte differentiation and function. Hepatology 37: 1249–1253, 2003. [DOI] [PubMed] [Google Scholar]
- 151.Weinshilboum RM, Otterness DM, Aksoy IA, Wood TC, Her C, Raftogianis RB. Sulfation and sulfotransferases 1: Sulfotransferase molecular biology: cDNAs and genes. FASEB J 11: 3–14, 1997. [PubMed] [Google Scholar]
- 152.Winter J, Bokkenheuser VD. Bacterial metabolism of natural and synthetic sex-hormones undergoing enterohepatic circulation. J Steroid Biochem 27: 1145–1149, 1987. [DOI] [PubMed] [Google Scholar]
- 153.Wisely GB, Miller AB, Davis RG, Thornquest AD, Johnson R, Spitzer T, Sefler A, Shearer B, Moore JT, Miller AB, Willson TM, Williams SP. Hepatocyte nuclear factor 4 is a transcription factor that constitutively binds fatty acids. Structure 10: 1225–1234, 2002. [DOI] [PubMed] [Google Scholar]
- 154.Wistuba W, Gnewuch C, Liebisch G, Schmitz G, Langmann T. Lithocholic acid induction of the FGF19 promoter in intestinal cells is mediated by PXR. World J Gastroenterol 13: 4230–4235, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wong MH, Oelkers P, Craddock AL, Dawson PA. Expression cloning and characterization of the hamster ileal sodium-dependent bile acid transporter. J Biol Chem 269: 1340–1347, 1994. [PubMed] [Google Scholar]
- 156.Wuarin J, Falvey E, Lavery D, Talbot D, Schmidt E, Ossipow V, Fonjallaz P, Schibler U. The role of the transcriptional activator protein Dbp in circadian liver gene-expression. J Cell Sci 16: 123–127, 1992. [DOI] [PubMed] [Google Scholar]
- 157.Xie MH, Holcomb I, Deuel B, Dowd P, Huang A, Vagts A, Foster J, Liang J, Brush J, Gu Q, Hillan K, Goddard A, Gurney AL. FGF-19, a novel fibroblast growth factor with unique specificity for FGFR4. Cytokine 11: 729–735, 1999. [DOI] [PubMed] [Google Scholar]
- 158.Yang F, Huang XF, Yi TS, Yen Y, Moore DD, Huang WD. Spontaneous development of liver tumors in the absence of the bile acid receptor farnesoid X receptor. Cancer Res 67: 863–867, 2007. [DOI] [PubMed] [Google Scholar]
- 159.Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA, Waitt GM, Parks DJ, Pearce KH, Wisely GB, Lazar MA. Rev-erb alpha, a heme sensor that coordinates metabolic and circadian pathways. Science 318: 1786–1789, 2007. [DOI] [PubMed] [Google Scholar]
- 160.Yin L, Wu N, Lazar MA. Nuclear receptor Rev-erbalpha: A heme receptor that coordinates circadian rhythm and metabolism. Nucl Recept Signal 16: e001, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.York B, O’Malley BW. Steroid receptor coactivator (SRC) family: Masters of systems biology. J Biol Chem 285: 38743–38750, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Yuan XH, Ta TC, Lin M, Evans JR, Dong YC, Bolotin E, Sherman MA, Forman BM, Sladek FM. Identification of an endogenous ligand bound to a “native orphan nuclear receptor. Plos One 4: e5609, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zhang M, Chiang JY. Transcriptional regulation of the human sterol 12alpha-hydroxylase gene (CYP8B1): roles of heaptocyte nuclear factor 4alpha in mediating bile acid repression. J Biol Chem 276: 41690–41699, 2001. [DOI] [PubMed] [Google Scholar]
- 164.Zhang Y, Hagedorn CH, Wang L. Role of nuclear receptor SHP in metabolism and cancer. Biochim Biophys Acta 1812: 893–908, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zhang Y, Lee FY, Barrera G, Lee H, Vales C, Gonzalez FJ, Willson TM, Edwards PA. Activation of the nuclear receptor FXR improves hyperglycemia and hyperlipidemia in diabetic mice. Proc Natl Acad Sci U S A 103: 1006–1011, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zhang Y, Wang L. Nuclear receptor small heterodimer partner in apoptosis signaling and liver cancer. Cancers 3: 198–212, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zollner G, Fickert P, Zenz R, Fuchsbichler A, Stumptner C, Kenner L, Ferenci P, Stauber RE, Krejs GJ, Denk H, Zatloukal K, Trauner M. Hepatobiliary transporter expression in percutaneous liver biopsies of patients with cholestatic liver diseases. Hepatology 33: 633–646, 2001. [DOI] [PubMed] [Google Scholar]
- 168.Zollner G, Wagner M, Moustafa T, Fickert P, Silbert D, Gumhold J, Fuchsbichler A, Halilbasic E, Denk H, Marschall HU, Trauner M. Coordinated induction of bile acid detoxification and alternative elimination in mice: Role of FXR-regulated organic solute transporter-alpha/beta in the adaptive response to bile acids. Am J Physiol Gastrointest Liver Physiol 290: G923–932, 2006. [DOI] [PubMed] [Google Scholar]