Abstract.
Schistosomiasis is a blood parasitic disease caused by trematode parasites of the genus Schistosoma. Schistosoma mansoni is one of the main contributors of the disease and 90% of the global burden of schistosomiasis is in Africa. Mass drug administration (MDA) has been implemented to reduce the disease burden in endemic areas. Because of MDA, the diagnostic sensitivity and specificity for classical diagnostic tests are reduced. In any disease situation, diagnosis is vital in determining asymptomatic, concurrent, current, new, and reinfection cases to evaluate the efficacy of any control program. We have evaluated the positive infection for S. mansoni from filtered urine samples collected from Zambian school children after MDA using loop-mediated isothermal amplification (LAMP) and compared its sensitivity and specificity with polymerase chain reaction (PCR). One hundred eleven urine samples collected from school children aged between 7 and 15 years from Siavonga district in southern Zambia were evaluated by PCR and LAMP for DNA extracted by two different protocols (filter-based versus crude extraction). The infection prevalence was 77% with PCR and almost 94% with mansoni-LAMP. Also, LAMP detected 16% (Qiagen extraction) and 10% (LAMP-Procedure for Ultra Rapid Extraction) more positive S. mansoni infection than PCR. We have demonstrated the efficacy of LAMP in a laboratory setup after MDA. The possible inclusion of LAMP as a field-based point-of-care test for surveillance can provide reliable prevalence of schistosomiasis after MDA and help in determining the efficacy of a control program.
INTRODUCTION
The London Declaration 2020 aims to implement interventions necessary to control or eliminate several neglected tropical diseases, including schistosomiasis.1 Schistosomiasis in Africa is an ongoing public health problem, which currently infects close to 300 million individuals and more than 700 million people are at risk of getting infected.2 The most prominent human schistosome, Schistosoma mansoni has been endemic in 54 countries,3 mostly in Africa. There is a strong age-specific relationship with the human population. In the age group of 6–15 years, the infection prevalence and intensity peak, resulting into consequences of growth and cognition delays, attention deficit, poor performance in school, and a negative effect on the overall growth and quality of a child’s life.4
Currently, schistosomiasis has been treated routinely with mass drug administration (MDA5). Because of MDA control intervention, the detection limit for remaining or new S. mansoni infection poses a challenge for gold standard (WHO-recommended) tests and is often missed.6 The gold standard diagnostic test, Kato-Katz (KK—parasitological) is low cost and most commonly used, but it lacks sensitivity for low endemic areas and for the posttreatment situation.7 This diagnostic problem is exacerbated as elimination campaigns progress, and the test becomes less effective and will miss out detection of asymptomatic carriers, which could be the likely source of continued transmission. Evaluation of control programs and disease reemergence needs more sensitive, specific, easy-to-use diagnostic tests.8 To address this issue, there is a need to develop a highly sensitive diagnostic method, which will use species-specific DNA detection equally well in high-, medium-, and low-intensity infection settings.
Recently, tests involving nucleic acid amplification, such as polymerase chain reaction (PCR), have shown great sensitivity and specificity for S. mansoni for various types of samples.9–12 We have previously detected an S. mansoni–specific cell-free repeat DNA fragment captured on filter paper through urine filtration via PCR.6,13,14 This approach is devoid of stool testing and reliance on eggs.15 The inherent technological limitations of PCR, such as longer time requirement, requirement of gel electrophoresis for visualization, expertise in molecular biology, and expensive equipment, make it unusable in the field or in resource-poor endemic settings.16,17
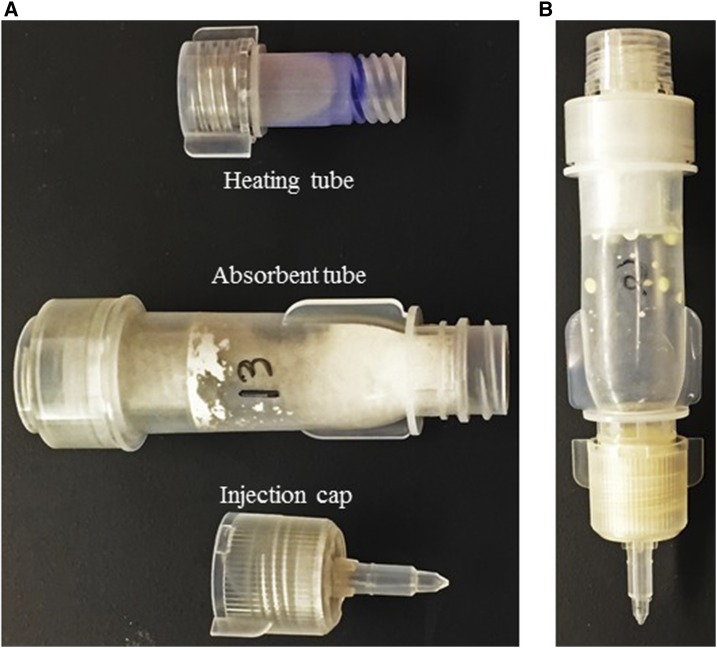
It has, however, been proposed that loop-mediated isothermal amplification (LAMP) can be used as a point-of-care (POC) assay for field diagnostics.18 Loop-mediated isothermal amplification uses four primers which amplify six different regions of the target DNA, which occurs at one constant temperature, and positive reactions will yield changes in color or turbidity.19 It is more resistant to inhibitors than PCR because it uses Bst polymerase. The sensitivity of LAMP has been demonstrated to be greater than that of PCR.20 Loop-mediated isothermal amplification reactions occur quicker than PCR, amplify DNA fragments independent of the standard thermocycler and electrophoresis, and remove the need for special reagents. This saves time and money. To use LAMP as a POC diagnostic test, commercially available Procedure for Ultra Rapid Extraction (PURE; Figure 1) kit (Eiken Chemical Co., Ltd, Tokyo, Japan) has been used to rapidly isolate DNA from clinical samples without potential inhibitors.16,21 This procedure has been used for tuberculosis,16 pneumonia,21 malaria,9,16,20 and other infectious diseases.
Figure 1.
Loop-mediated isothermal amplification-Procedure for Ultra Rapid Extraction DNA extraction kit. (A) Three components of the extraction kit. (B) All attached components for DNA extraction represents a closed environment for rapid DNA extraction. This figure appears in color at www.ajtmh.org.
In this study, we have evaluated the performance of LAMP (as POC diagnosis) for S. mansoni by amplifying the species-specific cell-free repeat DNA fragment (121-bp Sm1-7 repeat fragment, GenBank: M61098.1) from filtered urine samples collected from Zambian school children after MDA and compared its sensitivity and specificity against PCR. In addition, two different DNA extraction kits (Qiagen and PURE kits) were used for isolation of DNA, which have been compared and evaluated to see the impact of extracted DNA on the diagnostic performances of LAMP and PCR.
MATERIALS AND METHODS
Study design and sample population.
This was a cross-sectional study conducted in July 2016 in Siavonga District in the Southern Province of Zambia. The sample population involved school children aged 7–15 years, who provided consent to participate in the study from their parents either verbally or in written. Urine and stool samples were collected 1 month after the administration of praziquantel (given at 40 mg/kg). Ethical clearance for conducting this study was obtained from the Institutional Review Board (IRB) of ERES Coverage of Zambia (IRB # 2016-Apr-002) and from Marquette University, Milwaukee, WI (IRB # HR-3116).
Sample collection.
A total of 111 urine samples were collected from school children (50 males and 60 females). The same reference number was used for two different sample types, and two plastic cups were provided in two consecutive days to the participants for collection of urine on the first day and stool on the second day. About 30–40 mL of urine was filtered through Whatman #3 filter paper (Sigma-Aldrich, St. Louis, MO), which was marked with the same reference number before filtering the urine. Filter papers were then left to dry under a fly-proof net and on drying, packed in individual Ziploc bags with desiccant and shipped to Marquette University, Milwaukee, WI, where they were stored in a refrigerator at 4°C for further testing. For every participant, age, gender, locality, and result of the parasitological test were recorded.
Parasitological examination of stool samples for S. mansoni eggs.
The Kato-Katz kit, a WHO-recommended kit (WHO, Geneva, Switzerland) was used to detect the presence of S. mansoni eggs.22 Two consecutive stool smears were evaluated for the presence of eggs. Briefly, feces were pressed through a mesh screen to remove large particles. A portion of the sieved stool sample was then transferred to the hole of a template placed on a slide. After filling the hole on the template, the template was removed, and the remaining sample was covered with a piece of cellophane previously soaked in glycerol–malachite green. The slide was then examined under a microscope for the presence of helminth eggs.
DNA extraction and quantification.
One hundred eleven filter papers were used for DNA extraction by the QIAmp DNeasy® Blood and Tissue Kit (Qiagen, Hilden, Germany) and by the LAMP-PURE extraction kit (Eiken Chemical Co., Ltd). For Qiagen extraction, each filter paper disc was divided into four quadrants, and 12 punches (∼1 mm in diameter) were removed from one quadrant using a regular paper punch. The scissors and paper punch were washed with 10% bleach and water between each sample to prevent any contamination. The filter paper punches were mixed with 800 µL of DNA–RNA-free water (Sigma-Aldrich) and heat-shocked at 95°C for 10 minutes. The samples were then left on a shaker overnight to finish the extraction with the Qiagen kit the next day by following the manufacturer’s protocol. All extracted DNA samples were quantified via NanoDrop (Thermo Fisher Scientific, Waltham, MA) and aliquoted into two tubes of 50 µL each. The stock and aliquot DNA were stored at −20°C for amplification.
The LAMP-PURE extraction kit composed of three different units, namely, heating tube, adsorbent tube, and injection cap (Figure 1). For extraction, 800 µL of the sample was added to the heating tube and attached with the adsorbent tube, as the heating was already performed. The powder in the adsorbent tube removed any possible inhibitor. Then, the injection cap was attached. The adsorbent tube was squeezed over a 1.5-mL Eppendorf tube, and approximately 600 µL of the extracted DNA was collected per sample. All samples were quantified. Loop-mediated isothermal amplification-Procedure for Ultra Rapid Extraction extraction was a crude process, which resulted in very high DNA concentrations; all samples were subjected to dilution using the formula: C1V1 = C2V2 to concentrations of approximately 5 ng/µL of 100 µL each for two aliquots. One aliquot for each sample was then re-quantified to confirm the concentration. The stock and aliquot DNA were then stored in a −20°C freezer for PCR and LAMP amplification.
Amplification of the S. mansoni repeat DNA via PCR and LAMP.
All 111 samples extracted by the Qiagen kit and the LAMP–PURE kit were amplified by PCR and LAMP. Polymerase chain reaction amplification was carried out in 10 µL volume with the S. mansoni genomic DNA (BEI Resources, Manassas, VA) as the positive control, the Schistosoma haematobium genomic DNA (BEI Resources) as the negative control, template DNA controls were collected from people in the United States, who have never been exposed to schistosomiasis, and nuclease-free water as the water control (Sigma-Aldrich). For PCR amplification, the reaction volume consisted of 5 μL of the PCR mastermix (New England Biolabs Inc., Ipswich, MA), 0.5 μL (10 μM concentration) each of forward and reverse primers, 0.5 μL of 25 mM MgCl2, 2 μL of DNA (concentration: 4–6 ng/µL), and rest nuclease-free water. The amplification profile included initial denaturation at 95°C for 10 minutes; then 35 cycles at 95°C for 30 seconds, 57°C for 90 seconds, and 72°C for 45 seconds; and a final extension at 72°C for 10 minutes. The same PCR amplification protocol and arrangement was followed for both Qiagen- and LAMP–PURE-extracted DNA. To confirm amplification and correct amplicon size, PCR products were visualized on a 2% agarose gel stained with SYBR Green (Thermo Scientific) with a 50-bp reference DNA marker (New England BioLabs Inc.).
Loop-mediated isothermal amplification was performed using the LAMP ready-to-use buffer mix for two separate DNA templates. The 2× ready-to-use buffer mix was composed of 10× LAMP buffer (Eiken Chemical Co., Ltd), 5 M betaine (Sigma-Aldrich), and 10 mM dNTPs (Promega, Madison, WI). The reaction volume for all LAMP amplification was 10 μL, which was composed of 4 μL of ready mix buffer, 0.5 μL each of LAMP primers (5 pmoles of F3 and B3 and 40 pmoles of forward inner primer [FIP] and backward inner primer [BIP]), 1 μL of Bst DNA polymerase (New England BioLabs Inc.), 2–3 μL of extracted DNA (4–5 ng/µL in concentration), and 1 μL of nuclease-free water. The amplification was carried out for 2 hours at 63°C with inactivation for 5 minutes at 80°C. Loop-mediated isothermal amplification products were detected by adding 1 μL of SYBR Green (1:20 dilution), and a picture was taken using a cell phone. To confirm the correct amplified product, all LAMP products were visualized on 2% agarose gel stained with SYBR Green and run with a 50-bp reference ladder. Gel pictures were captured using the Azure c200 system (Azure Biosystems, Dublin, CA). The primers used for PCR and LAMP were reported in earlier publications.6,13
Statistical analysis.
We performed quantitative assessment to evaluate the sensitivity, specificity, disease prevalence, positive predictive value (PPV), negative predictive value (NPV), and accuracy of PCR and LAMP amplification for S. mansoni by comparing the two different extraction methods. The abovementioned statistical analysis was performed by comparing positive and negative infection detection by KK and PCR and LAMP for two different DNA extractions. Disease prevalence was determined based on the number of positive cases by each diagnostic test against the total number of samples evaluated. Accuracy was determined based on the probability of a test of correctly diagnosing as a positive case. For quantitative analysis, MedCalc 12.4.0 (MedCalc Software, Ostend, Belgium) was used. Data were processed through JMP 12 (JMP® v12, SAS Institute Inc., Cary, NC) and converted to numerical values (1 = positive and 0 = negative) for statistical analysis. JMP was also used to calculate the agreement statistics by measuring the kappa value and Bowker’s symmetry. The kappa coefficient determined the agreement between two tests, where −1 = negative association, 0 = random, and +1 = full agreement. The Bowker’s symmetry was a disagreement statistic, which was trying to determine the symmetry between two tests based on the χ2 approximation of the distribution of the test statistic.23
RESULTS
Detection of positive and negative infection by PCR and LAMP for two different DNA extraction methods.
When evaluated for PCR amplification, both extraction methods (Qiagen and LAMP-PURE) yielded the same number of positives and negatives. The positive infection rate was 77.5% (86/111) with PCR for both DNA extractions, whereas it was almost 94% (104/111) for Qiagen extraction and 87% (97/111) for LAMP-PURE with LAMP amplification (Table 1). Loop-mediated isothermal amplification detected more positive infections than PCR, 18 for Qiagen (16% more) and 11 for LAMP-PURE (10% more; Table 1). The findings were consistent with previous study findings about LAMP being more sensitive than PCR.24
Table 1.
Detection of positive and negative infection by KK, PCR, and LAMP for Schistosoma mansoni for both Qiagen- and LAMP-PURE–extracted DNA
| A. Comparison of positive and negative infection detection by PCR and LAMP | ||||
|---|---|---|---|---|
| N: 111 samples | PCR positive | PCR negative | LAMP positive | LAMP negative |
| Qiagen | 86 (77.5%) | 25 | 104 (93.7%) | 7 |
| LAMP-PURE | 86 (77.5%) | 25 | 97 (87.4%) | 14 |
| B. Comparison of positive and negative infection detection by KK and PCR | ||||
|---|---|---|---|---|
| N: 111 samples | PCR Qiagen positive | PCR Qiagen negative | PCR LAMP-PURE positive | PCR LAMP-PURE negative |
| KK positive | 9 (8.1%) | 25 | 7 (6.3%) | 2 |
| KK negative | 77 (69.3%) | 0 | 79 (71.8%) | 23 |
| C. Comparison of positive and negative infection detection by KK and LAMP | ||||
|---|---|---|---|---|
| N: 111 samples | LAMP Qiagen positive | LAMP Qiagen negative | LAMP LAMP-PURE positive | LAMP LAMP-PURE negative |
| KK positive | 9 (8.1%) | 0 | 8 (7.2%) | 1 |
| KK negative | 95 (85.6%) | 7 | 89 (80.1%) | 13 |
KK = Kato-Katz; LAMP-PURE = loop-mediated isothermal amplification-Procedure for Ultra Rapid Extraction; PCR = polymerase chain reaction.
Detection of positive and negative infection by KK and comparison against PCR and LAMP.
The amplification by PCR and LAMP for Qiagen extraction identified all of the samples positively identified by KK, but yielded significantly more positives by PCR (69.3%) and LAMP (85.6%), which were eventually KK negative (Table 1). The amplification for LAMP-PURE samples by PCR and LAMP also identified significantly more KK-negative samples, but few KK positives (two for PCR and one for LAMP) came out as negative (Table 1).
Comparative analysis of efficacy for amplification methods for different DNA extraction methods.
Loop-mediated isothermal amplification for Qiagen-extracted samples yielded 18 more positive infections, which were negative by PCR (total 86) for the same extraction. Similarly, LAMP-PURE yielded 17 more positive infections than PCR (total 80), although failed to detect six PCR-positive infections (Table 2). When PCR for LAMP-PURE–extracted samples were compared against PCR LAMP-PURE, only 74 were matched for both, with overlap of negative amplification by the one came out as positive by another amplification. Loop-mediated isothermal amplification for both extracted DNA improved the outcome with 91 positives for both extractions, with the 13 remaining undetected for LAMP-PURE extraction (came out positive for Qiagen). Loop-mediated isothermal amplification-Procedure for Ultra Rapid Extraction–extracted DNA resulted in some discrepancies for both PCR and LAMP amplification (Table 2).
Table 2.
Diagnostic comparison of amplification efficacy by PCR and LAMP for Qiagen- and LAMP-PURE–extracted DNA for Schistosoma mansoni
| LAMP-Qiagen | PCR-Qiagen | LAMP LAMP-PURE | PCR LAMP-PURE | ||
|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | ||
| Positive | 86 | 18 | Positive | 74 | 23 |
| Negative | 0 | 7 | Negative | 12 | 2 |
| LAMP_LAMP_PURE | LAMP-Qiagen | LAMP_LAMP_PURE | PCR-Qiagen | ||
|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | ||
| Positive | 91 | 6 | Positive | 80 | 17 |
| Negative | 13 | 1 | Negative | 6 | 8 |
LAMP-PURE = loop-mediated isothermal amplification-Procedure for Ultra Rapid Extraction; PCR = polymerase chain reaction.
Disease prevalence estimation, comparison statistics, and agreement statistics analysis.
The diagnostic parameter calculation showed 100% sensitivity (95% CI: 96–100) and 100% specificity (95% CI: 59–100) with 94% disease prevalence for LAMP-Qiagen (Table 3). The predictive value estimation and accuracy were also 100% for LAMP-Qiagen. Polymerase chain reaction-Qiagen was the second best test with 83% sensitivity (95% CI: 74–89) and 100% specificity (95% CI: 59–100) and similar disease prevalence (94%). The PPV was 100% for PCR-Qiagen, but the NPV (28%) and accuracy (84%) were lower than those of LAMP-Qiagen (Table 3). Loop-mediated isothermal amplification for LAMP-PURE showed higher sensitivity (88%; 95% CI: 80–93) and higher disease prevalence (94%), although it showed lower specificity (only 14%; 95% CI: 0.4–58). These findings were comparatively lower for PCR for LAMP-PURE with 79% sensitivity (95% CI: 69–86), 31% specificity (95% CI: 9–61), 88% disease prevalence, and lowest accuracy value (73%) of them all (Table 3). The PPV values for all four combinations were close and ranged from 90% to 100%. However, NPVs were variable, as it was 100% for LAMP-Qiagen and significantly lower for LAMP_LAMP-PURE (7%) and PCR_LAMP-PURE (16%; Table 3).
Table 3.
Comparison statistics measured for PCR and LAMP amplification for Schistosoma mansoni based on two different extraction techniques
| PCR-Qiagen (95% CI) | PCR_LAMP-PURE (95% CI) | LAMP-Qiagen (95% CI) | LAMP_LAMP_PURE (95% CI) | |
|---|---|---|---|---|
| Sensitivity | 83% (74–89%) | 79% (69–86%) | 100% (96–100%) | 88% (80–93%) |
| Specificity | 100% (59–100%) | 31% (9–61%) | 100% (59–100%) | 14% (0.4–58%) |
| Prevalence | 94% | 88% | 94% | 94% |
| PPV | 100% | 90% | 100% | 94% |
| NPV | 28% | 16% | 100% | 7% |
| Accuracy | 84% | 73% | 100% | 83% |
LAMP-PURE = loop-mediated isothermal amplification-Procedure for Ultra Rapid Extraction; NPV = negative predictive value; PCR = polymerase chain reaction; PPV = positive predictive value. The infection prevalence identified by each extraction technique is highlighted in bold to emphasize the difference.
Test positives for PCR-Qiagen were highly likely to be the same with LAMP-Qiagen, although LAMP was more sensitive for Qiagen-extracted samples (Bowker = 18, P = 0.001). In addition, moderate agreement (kappa = 0.38) was seen for both PCR and LAMP amplification for Qiagen (Table 4). Similarly, PCR for Qiagen and LAMP for LAMP-PURE showed moderate agreement (kappa = 0.30), but the test positives may not be similar as evidenced from the Bowker’s symmetry (5.26, P = 0.0218). On the other hand, 1) PCR-Qiagen and PCR_LAMP-PURE and 2) LAMP-Qiagen and LAMP_LAMP-PURE were highly unlikely to yield always the same result (Table 4).
Table 4.
Agreement statistics estimation (kappa coefficient and Bowker symmetry test) comparing species-specific DNA amplification by PCR and LAMP for both Qiagen- and LAMP-PURE–extracted DNA for Schistosoma mansoni
| Comparison of diagnostic tests | Kappa coefficient | Bowker’s symmetry test* | ||
|---|---|---|---|---|
| Degree of agreement | 95% CI | Symmetry of disagreement | P-value† | |
| PCR-Qiagen vs. LAMP-Qiagen | 0.38 | 0.17–0.58 | 18 | 0.0001‡ |
| PCR-Qiagen vs. PCR_LAMP-PURE | −0.08 | −0.25–0.09 | 0 | 1.0000 |
| PCR-Qiagen vs. LAMP_LAMP-PURE | 0.30 | 0.08–0.51 | 5.26 | 0.0218‡ |
| LAMP-Qiagen vs. LAMP_LAMP-PURE | 0.01 | −0.17–0.19 | 2.58 | 0.1083 |
LAMP-PURE = loop-mediated isothermal amplification-Procedure for Ultra Rapid Extraction; PCR = polymerase chain reaction.
* Bowker’s symmetry test = this test checks for symmetry in two-way tables and the test decision is based on a χ2 approximation of the distribution of the test statistic.
† α level was set at 0.05.
‡ Significant. P-values are highlighted in bold.
DISCUSSION
We found that LAMP is more sensitive in detecting low-level infections, especially after MDA. Using well-defined positive and negative controls and 111 filtered urine samples collected from school children after MDA, the LAMP assay for the Qiagen-extracted DNA achieved 100% sensitivity, specificity, and accuracy with 94% disease prevalence (Table 3). Moreover, LAMP-Qiagen detected 16% more positive infection than PCR amplification for the Qiagen-extracted DNA (Table 1). Polymerase chain reaction also performed well with 100% specificity and 83% sensitivity for Qiagen extraction. The sensitivity of LAMP is being considered as 10 times higher than PCR by previous study,24 and our findings are consistent with such findings. The findings also signify the need for such sensitive assays for low infection intensities, especially after MDA, as it is evident from the performance of KK. The sensitivity of PCR and LAMP (Qiagen extraction only) is eight and 11 times higher than that of KK for the samples evaluated for this particular study (Table 1). In addition, the findings in our study for PCR and LAMP amplification for PURE extraction is consistent with findings reported elsewhere. The sensitivity is high, ranging from 79% (PCR) to 88% (LAMP), but specificity is the issue (31% for PCR and only 14% for LAMP). As PCR and LAMP both are sensitive molecular methods, higher sensitivity is expected. The lower specificity could probably be associated with the presence of inhibitors in the extracted DNA.
The Qiagen kit is very reliable and produced quality DNA in our previous studies. Also, it is a membrane-based DNA extraction kit, which usually takes care of the inhibitors present in extracted DNA along with some protein and other cellular debris. On the other hand, PURE is an enclosed system devoid of membrane, so there is less chance of contamination, but higher chances of having impurities in the extracted DNA. The possible presence of impurities in the extracted DNA may have led to the misdiagnosis of positive samples and regarded them as true negatives. The presence of inhibitors could have hindered the amplification of the species-specific repeat fragment by PCR and LAMP. Considering the fact that PURE has been designed for quick DNA extraction, which is suitable for field, it can, thus, be an integral part of LAMP diagnosis. Other studies have been planned to address this issue specially to evaluate the efficacy of LAMP amplification based on PURE extraction for asymptomatic individuals.
This study, however, may not provide a conclusive outcome about LAMP’s superiority over PCR because of the limitation of the sample size. To substantiate the assertion that LAMP is superior over PCR, there would be a need to conduct studies in various settings considering different prevalence rates of schistosomiasis and with varying disease burdens among the infected population. Another limitation of the study is the absence of baseline data that probably would have reflected the efficacy of the MDA for this particular population.
To determine the species-specific amplification, we have sequenced random samples to verify the amplified fragments by both PCR and LAMP. All the sequences are matched against NCBI GenBank, and they matched with the 121-bp Sm1-7 repeat fragment (GenBank: M61098.1) for S. mansoni with 100% identity. This indicates the specificity of the LAMP primers and the uniqueness of the cell-free repeat fragment for S. mansoni.
Misdiagnosis or underdiagnosis occurs often when the infection level is low, and this is true for the MDA program, where traditional tests usually fail to detect the remaining infection or reinfection.25 This study shows the usefulness of the LAMP assay in the diagnosis of these low-level infections after MDA, which is evident from the poor performance of KK. This is highly important in determining the applicability of LAMP as a field diagnostic technique. Although LAMP may not be able to quantify the disease burden, but increased sensitivity and specificity will aid into identification of the remaining infection that has escaped the MDA. This will help in determining the actual infection prevalence for the particular population going through treatment and will ultimately determine the efficacy of the control program. Moreover, LAMP can be implemented as an integrated diagnostic approach (POC test) for surveillance and to determine the transmission foci by extending the testing for infected snails. Using LAMP as a POC diagnosis to determine the disease prevalence, especially after MDA, will help stakeholders to make informed decision for control intervention.
Given the advantages and demonstrated performance of LAMP, we anticipate that it will be suitable to use not only for resource-poor environments but also for well-equipped health facilities. Procedure for Ultra Rapid Extraction kit can also be part of this process, and overall, LAMP-PURE can play an important role in molecular xenomonitoring for S. mansoni.
Acknowledgments:
We would like to acknowledge the participating schools and their pupils in the study districts in Zambia; our field technicians, H. Sinsungwe and B. Malunda, for their technical assistance in sample processing and microscopic examination; and the participants in the United States, who provided urine samples, which are used as negative control. Schistosoma mansoni genomic DNA is acquired from the Schistosomiasis Resource Center for distribution by BEI Resources, NIAID, NIH: Genomic DNA from Adult Male and Female S. mansoni, Strain NMRI, NR-28910.
REFERENCES
- 1.Hotez PJ, Damania A, Barua A, Stanaway J, 2017. The first “London Declaration”: the Commonwealth and its neglected tropical diseases. PLoS Negl Trop Dis 11: e0005321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herricks JR, et al. 2017. The global burden of disease study 2013: what does it mean for the NTDs? PLoS Negl Trop Dis 11: e0005424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colley DG, Bustinduy AL, Secor WE, King CH, 2014. Human schistosomiasis. Lancet 383: 2253–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.King CH, Dangerfield-Cha M, 2008. The unacknowledged impact of chronic schistosomiasis. Chronic Ill 4: 65–79. [DOI] [PubMed] [Google Scholar]
- 5.Hotez PJ, Fenwick A, 2009. Schistosomiasis in Africa: an emerging tragedy in our new global health decade. PLoS Negl Trop Dis 3: e485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lodh N, Mwansa JC, Mutengo MM, Shiff CJ, 2013. Diagnosis of Schistosoma mansoni without the stool: comparison of three diagnostic tests to detect Schistosoma mansoni infection from filtered urine in Zambia. Am J Trop Med Hyg 89: 46–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu J, Guan Z-X, Zhao B, Wang Y-Y, Cao Y, Zhang H-Q, Zhu X-Q, He Y-K, Xia C-M, 2015. DNA detection of Schistosoma japonicum: diagnostic validity of a LAMP assay for low-intensity infection and effects of chemotherapy in humans. PLoS Negl Trop Dis 9: e0003668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hotez PJ, Pecoul B, Rijal S, Boehme C, Aksoy S, Malecela M, Tapia-Conyer R, Reeder JC, 2016. Eliminating the neglected tropical diseases: translational science and new technologies. PLoS Negl Trop Dis 10: e0003895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enk MJ, Oliveira e Silva G, Rodrigues NB, 2012. Diagnostic accuracy and applicability of a PCR system for the detection of Schistosoma mansoni DNA in human urine samples from an endemic area. PLoS One 7: e38947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamburger J, He N, Abbasi I, Ramzy RM, Jourdane J, Ruppel A, 2001. Polymerase chain reaction assay based on a highly repeated sequence of Schistosoma haematobium: a potential tool for monitoring schistosome-infested water. Am J Trop Med Hyg 65: 907–911. [DOI] [PubMed] [Google Scholar]
- 11.Pontes LA, Dias-Neto E, Rabello A, 2002. Detection by polymerase chain reaction of Schistosoma mansoni DNA in human serum and feces. Am J Trop Med Hyg 66: 157–162. [DOI] [PubMed] [Google Scholar]
- 12.Sandoval N, Siles-Lucas M, Perez-Arellano JL, Carranza C, Puente S, Lopez-Aban J, Muro A, 2006. A new PCR-based approach for the specific amplification of DNA from different Schistosoma species applicable to human urine samples. Parasitology 133: 581–587. [DOI] [PubMed] [Google Scholar]
- 13.Lodh N, Mikita K, Bosompem KM, Anyan WK, Quartey JK, Otchere J, Shiff CJ, 2017. Point of care diagnosis of multiple schistosome parasites: species-specific DNA detection in urine by loop-mediated isothermal amplification (LAMP). Acta Trop 173: 125–129. [DOI] [PubMed] [Google Scholar]
- 14.Lodh N, Naples JM, Bosompem KM, Quartey J, Shiff CJ, 2014. Detection of parasite-specific DNA in urine sediment obtained by filtration differentiates between single and mixed infections of Schistosoma mansoni and S. haematobium from endemic areas in Ghana. PLoS One 9: e91144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ibironke O, Koukounari A, Asaolu S, Moustaki I, Shiff C, 2012. Validation of a new test for Schistosoma haematobium based on detection of Dra1 DNA fragments in urine: evaluation through latent class analysis. PLoS Negl Trop Dis 6: e1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kouzaki Y, et al. 2015. PURE-LAMP procedure for the diagnosis of extrapulmonary tuberculosis: a case series. Intern Med 54: 1447–1450. [DOI] [PubMed] [Google Scholar]
- 17.Verweij JJ, Brienen EA, Ziem J, Yelifari L, Polderman AM, Van Lieshout L, 2007. Simultaneous detection and quantification of Ancylostoma duodenale, Necator americanus, and Oesophagostomum bifurcum in fecal samples using multiplex real-time PCR. Am J Trop Med Hyg 77: 685–690. [PubMed] [Google Scholar]
- 18.Abbasi I, King CH, Muchiri EM, Hamburger J, 2010. Detection of Schistosoma mansoni and Schistosoma haematobium DNA by loop-mediated isothermal amplification: identification of infected snails from early prepatency. Am J Trop Med Hyg 83: 427–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T, 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28: E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gandasegui J, Fernández-Soto P, Muro A, Simões Barbosa C, Lopes de Melo F, Loyo R, de Souza Gomes EC, 2018. A field survey using LAMP assay for detection of Schistosoma mansoni in a low-transmission area of schistosomiasis in Umbuzeiro, Brazil: assessment in human and snail samples. PLoS Negl Trop Dis 12: e0006314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawano S, Maeda T, Suzuki T, Abe T, Mikita K, Hamakawa Y, Ono T, Sonehara W, Miyahira Y, Kawana A, 2015. Loop-mediated isothermal amplification with the procedure for ultra rapid extraction kit for the diagnosis of pneumocystis pneumonia. J Infect Chemother 21: 224–226. [DOI] [PubMed] [Google Scholar]
- 22.Lamberton PHL, Kabatereine NB, Oguttu DW, Fenwick A, Webster JP, 2014. Sensitivity and specificity of multiple Kato-Katz thick smears and a circulating cathodic antigen test for Schistosoma mansoni diagnosis pre- and post-repeated-praziquantel treatment. PLoS Negl Trop Dis 8: e3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anne K, Sonja K, 2007. Bowker’s test for symmetry and modifications within the algebraic framework. Comput Stat Data Anal 51: 4124–4142. [Google Scholar]
- 24.Hamburger J, Abbasi I, Kariuki C, Wanjala A, Mzungu E, Mungai P, Muchiri E, King CH, 2013. Evaluation of loop-mediated isothermal amplification suitable for molecular monitoring of schistosome-infected snails in field laboratories. Am J Trop Med Hyg 88: 344–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Basuni M, Muhi J, Othman N, Verweij JJ, Ahmad M, Miswan N, Rahumatullah A, Aziz FA, Zainudin NS, Noordin R, 2011. A pentaplex real-time polymerase chain reaction assay for detection of four species of soil-transmitted helminths. Am J Trop Med Hyg 84: 338–343. [DOI] [PMC free article] [PubMed] [Google Scholar]