Abstract.
Cutaneous leishmaniasis (CL) remains one of the world’s most prevalent neglected diseases, particularly in developing countries. Identification of the involved Leishmania species is an important step in the diagnosis and case management process. In this study, we tested simple, rapid, and highly sensitive loop-mediated isothermal amplification (LAMP) assays for Leishmania DNA species-specific detection from cutaneous lesions. Two LAMP assays, targeting cysteine protease B (cpb) gene, were developed to detect and identify Leishmania major and Leishmania tropica species. Loop-mediated isothermal amplification specificity was examined using DNA samples from other Leishmania species and Trypanosoma species. No cross-reactions were detected. The developed LAMP assays exhibited sensitivity with a detection limit of 20 fg and 200 fg for L. major and L. tropica, respectively. Both tests were applied on clinical samples of CL suspected patients living in endemic Tunisian regions and compared with kinetoplast DNA quantitative PCR (qPCR), microscopic, and conventional cpb-based polymerase chain reaction (PCR) assays. Our LAMP tests were able to discriminate between L. major and L. tropica species and showed a sensitivity of 84% and a specificity of 100%. However, when compared with the performance of the diagnostic tests with latent class analysis (LCA), our LAMP assays show a sensitivity of 100%. These assays can be used as a first-line molecular test for early diagnosis and prompt management of CL cases in public health programs.
INTRODUCTION
Cutaneous leishmaniasis (CL) is a disease caused by flagellated protozoan parasites of the genus Leishmania, which belong to the order of Kinetoplastida. It causes single or multiple, sometimes persisting, often ulcerated skin lesions on the uncovered parts of the body. Cutaneous leishmaniasis is endemic in around 98 countries of the New and Old World. Between 0.7 and 1.2 million new cases are reported each year,1 and 350 million people are at risk of contracting the disease. Three clinico-epidemiological CL forms occur in Tunisia: zoonotic CL (ZCL), chronic CL (CCL), and sporadic CL, caused by Leishmania (L.) major, Leishmania tropica, and Leishmania infantum, respectively.2,3 Recently, L. major has largely spread to the central and southern parts of the country, whereas L. tropica distribution is no longer restricted to southeastern Tunisia.4 This distribution has favored the development of CL mixed foci in which both L. tropica and L. major are transmitted especially in the Tataouine governorate, southeast Tunisia, where around 400 cases are reported yearly.5
Rapid and accurate diagnosis of leishmaniasis is crucial to initiate early management of infected cases and to control transmission according to the identified species. Laboratory diagnosis is based on parasitological methods (microscopic examination and culture), serological assays, and molecular tools (conventional PCR and quantitative PCR [qPCR]).6–10 Microscopic examination is widely used and exhibits relatively low sensitivity, underestimating the infection rate and missing many cases. PCR techniques, especially qPCR technology, exhibit higher sensitivity but require costly equipment which are not available in rural areas, where most cases are observed. Because of the absence of a gold standard for CL diagnosis,11–14 many in-house PCR protocols have been developed and used for Leishmania detection.15
Correct CL diagnosis and species identification would require a highly sensitive, specific, and easy-to-use diagnostic tool (or a combination of tools).16 DNA loop-mediated isothermal amplification (LAMP) could meet such criteria.17 The process has the potential to combine the high sensitivity and specificity of a molecular diagnostic test with the ability to operate the test under the limited resources of field conditions. Indeed, the LAMP has been used as a powerful diagnostic tool and successfully used for the diagnosis of several protozoan parasitic diseases, such as malaria,18,19 trypanosomiasis,20–22 and leishmaniasis.23–25
In the presented study, we developed two species-specific LAMP assays for the diagnosis of CL due to L. major and L. tropica, targeting the cysteine protease B (cpb) multi-copy gene. The multi-copy nature of the cpb genes and their polymorphism allow major structural change between Leishmania species and strains,26 and make it an excellent target gene for the development of both species-sensitive and specific LAMP assays.27 Whereas L. tropica contains an unknown number of cpb copy genes, L. major contains eight tandemly arranged cpb genes present in two variants: variant 1 (1,047 bp), present in five copies, and variant 2 (1,332 bp), present in three copies.28 Leishmania chagasi, Leishmania donovani/L. infantum, and Leishmania mexicana present five, six, and 19 tandemly arranged cpb genes, respectively.29,30
To assess our developed LAMP assay, we locally collected skin samples from CL suspected patients living in endemic regions of Tunisia. Sensitivity and specificity of our LAMP assay were compared with those of qPCR, cpb PCR, and microscopic techniques.
MATERIALS AND METHODS
Study sites.
The study was carried out between September 2015 and July 2016 at three endemic Tunisian governorates: two in central (Sidi Bouzid and Kairouan) and one in southern (Tataouine) Tunisia.
Study participants.
Samples were collected from patients with suggestive CL symptoms, referred to public health centers by their physicians as part of the CL diagnostic procedure. In total, 81 patients were recruited.
Ethical concerns.
Informed consent was requested from all adults recruited for the study. Consent was obtained from parents or guardians for the inclusion of young children. Study procedures were approved by the Committee of Bio-Medical Ethics of Institut Pasteur de Tunis (Ref 2015/06/I/LR11IPT06/V2).
Sample collection.
The tissue material was aspirated after actively scraping the skin lesions’ peripheral edge. Smears were prepared and stained on the site for microscopic examination. Part of the collected serosities was placed in an Eppendorf tube containing phosphate-buffered saline, coded and enclosed in a self-sealing bag, and stored at 4°C for further molecular testing.
Microscopic confirmation.
A positive test was based on the microscopic demonstration of Leishmania amastigotes on smear slides stained by May-Grunwald Giemsa stain and screened at ×100 objective.
Culture.
A total of 26 cultures were carried out on Novy, McNeal, Nicolle (NNN) media, prepared according to Nicolle and Berrebi recommendations.31,32 Inoculation of dermal scraping was immediately performed after sampling. The NNN tubes were incubated between 22°C and 26°C.31 Microscopic examination and subculturing were performed weekly for 5 weeks. Positive cultures were conserved for isoenzyme typing.
DNA extraction.
DNA was extracted from tissue materials, aspirated from skin lesions using the Wizard Genomic DNA Purification kit (Promega, Madison, WI) according to the manufacturer’s instructions. Each DNA sample was investigated with different molecular tools.
Cysteine protease B conventional PCR amplification.
Different cpb gene copies were amplified using three different species-specific PCR assays for the L. major, L. tropica, and L. infantum complex. Amplification sizes were 1,176 bp for L. major, 600 bp for L. tropica, and 325 bp for L. infantum.33
Polymerase chain reactions targeting the cpb gene were performed following the modified protocol described by Chaouch et al.23,33 using a 25-µL final volume, containing 5 µL of the sample, 1.5 mM of MgCl2, 0.2 µM of each deoxynucleotide, 50 pmol of each primer, and 1.25 U of Taq DNA polymerase. Reactions were conducted in a TECHNE TC512 machine following the cycling conditions: initial denaturation step at 94°C for 5 minutes, followed by 35 cycles at 94°C for 1 minute, 62°C for 1 minute, and 72°C for 1 minute, with a final elongation step at 72°C for 10 minutes. The PCR products were electrophoresed on 1% agarose gels, stained with ethidium bromide, and then visualized under UV light. A 1-kb ladder (Fermentas, Germany) was used to size amplicons.
Quantitative real-time PCR assay.
A qPCR was conducted according Mary et al.34 and as described by Chouihi et al.35 Each sample was tested in duplicate. A distilled water sample and DNA extract from L. infantum promastigotes were included as negative and positive controls, respectively. Amplification was conducted in an ABI PRISM 7700 sequence detector (Applied Biosystems, Foster City, CA).
Loop-mediated isothermal amplification.
Primer design.
The LAMP primer sets were designed using the Primer Explorer ver.4 software (Eiken Chemical, Japan [http://primerexplorer.jp/lamp4.0.0/index.html]), using L. major and L. tropica consensus sequences of the cpb multi-copy gene, accessible on the GenBank database.33 For each species, a set of four LAMP primers, recognizing six specific sections of L. major and L. tropica cpb genes, were designed (Table 1). Two additional loop primers, loop forward (LF) and loop backward (LoR),15 were manually designed. The loop primers were added to increase the number of loops in the reaction, thereby increasing the reaction speed. Final specificity verification was performed using the Basic Local Alignment Search Tool (BLAST) (http://blast.ncbi.nlm.nih.gov/Blast.cgi) analysis against human DNA and any other organisms included in leishmaniasis differential diagnosis.
Table 1.
Species-specific LAMP primer sequences used in the study
| Species | Oligo name | Sequence (5´–3´) |
|---|---|---|
| Leishmania tropica | Tropica-F3 | GCAGGCGTTCGAGTGG |
| Tropica-B3 | CAATCGAGATGGGGCCATT | |
| Tropica-LF | GGACACGTAGGGGTAGCTGTCC | |
| Tropica-LB | GACGATCGAAAGCAGCGAAA | |
| Tropica-FIP | CGGGCACATAACCGCTGCTGAACGGGACCATGTTCACG | |
| Tropica-BIP | AGTGCTCGAACAGCAGTCAACTTAACCGTTTCGCTGCTTTCG | |
| Leishmania major | Major-F3 | GTGCATGTGTCGCAGAGC |
| Major-B3 | TCAGCAACTTGTGCAGACTG | |
| Major-FIP | TCGTGCAGCACATGTCGCTTGCTAGCACGGAGAGCGAAGA | |
| Major-BIP | TTCTCACCGCCAACAAGTGTGCCACTTGGTCCTAGAGGAGCC | |
| Major-LF | TCTCCTCCATCGTCGCCCG | |
| Major-LB | TGCTACGAGAACCGGGGGC | |
| Leishmania infantum33 | Infantum-F3 | GCGATGACAAAGACAATGGC |
| Infantum-B3 | TCATCACGTAGCCGTCGA | |
| Infantum-FIP | TCTCCGTGAACACGATCCCGTCTGATGCTGCAGGCGTTC | |
| Infantum-BIP | AGAGCTACCCCTACACGTCCGTTTGCGCGCCGGGAAC | |
| Infantum-LF | ACATGTGTCGCAGCAGCCAC | |
| Infantum-LB | GGTGATGTGGCCGAGTGCTT |
LF = loop forward.
Loop-mediated isothermal amplification reactions.
Leishmania major and L. tropica specific LAMP reactions were standardized for optimal temperature after gradient analysis (64–68°C) and time (45–50 minutes) using L. major and L. tropica reference strains. Fifty-one different parasite strains (including L. infantum, L. donovani, L. major, L. tropica, Leishmania turanica, Leishmania gerbilii, Leishmania tarentolea, and Trypanosoma cruzi) were used as references to test the LAMP assay specificity. The analytical sensitivity of the LAMP assays was evaluated after serial dilutions (10-fold serial dilutions from 0.00002 ng to 20 ng) of reference DNA samples. Briefly, the LAMP assay was achieved in a 25-µL reaction mixture containing in-house buffer (2.5 mM), forward inner primer (FIP) and backward inner primer (BIP) (1.6 mM), loop-F and loop-B (0.8 µM), F3 and B3 primers (0.2 mM), Bst DNA polymerase (8 U, GeneON, Germany), betaine (10 µM), deoxynucleotide triphosphates (2.5 mM), MgSO4 (10 mM), ddH2O, and DNA template (2.5 µL). The LAMP test was performed for 50 minutes at 67°C and completed by increasing the temperature to 80°C for 5 minutes in a heat block. To confirm positivity, amplicons were electrophoresed in 1% agarose gels stained with ethidium bromide and visualized under UV light.
Statistical analysis.
Data were captured anonymously. The sensitivity and specificity of the techniques used were estimated with 2 × 2 contingency tables using qPCR as the gold standard. Sensitivity and specificity were also estimated by using the latent class analysis (LCA),36 without considering a gold standard. In this LCA, unobserved or latent Leishmania infection is used to explain dependencies between the observed diagnostics. In basic latent class models, the observed variables are assumed to be independent conditions of latent class; that is, there are no associations between the observed variables within each category of the latent variable. The latent variable is the true status of the disease, and the hypothesis is that there are two latent classes (presence or absence of disease). Molecular tests and microscopic examination are assumed to be conditionally independent, given the true unobserved infection status. Latent class analysis was performed using the LEM package “(Vermunt, unpublished data)”, including the latent variable “Cutaneous Leishmaniasis disease” (X) and four observed diagnostic test variables: q-PCR, LAMP, cpb PCR, and microcopy.
RESULTS
Detection threshold and analytical specificity of species-specific LAMPs.
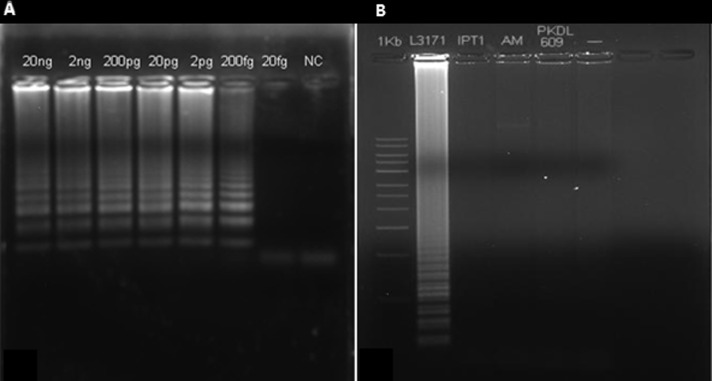
A set of specific oligonucleotide primers were designed to amplify the sequences of the target cpb genes from L. major and L. tropica genomic DNA. Serially diluted samples (20 ng–20 fg) of L. major and L. tropica promastigote DNA were used as references. Loop-mediated isothermal amplification was able to detect 20 fg and 200 fg of L. major and L. tropica DNA, respectively (Figure 1A). Loop-mediated isothermal amplification reaction specificity was next evaluated using DNA samples from other Trypanosoma genera. No amplicon was detected using up to 20 ng of DNA and no cross-reactions were observed, particularly between endemic Leishmania species in Tunisia. Our results showed that LAMP cpb is species specific for L. major and L. tropica detection (Figure 1B).
Figure 1.
(A) Sensitivity of LAMP assays for the detection of Leishmania DNA using serial dilutions of Leishmania tropica. After incubation at 67°C for 50 minutes, LAMP reactions were inspected by agarose gel electrophoresis of LAMP products. Lanes 1–6 show the typical ladder-shaped pattern of a positive reaction. (B) Specificity of Leishmania major LAMP assay tested on different species: lane 1: 1-kb ladder, lane 2: L. major (L3171), lane 3: Leishmania infantum (MHOM/TN/1980/IPT1), lane 4: L. tropica (AM), lane 5: Leishmania donovani (PKDL306), and lane 6: negative control.
Diagnostics evaluation.
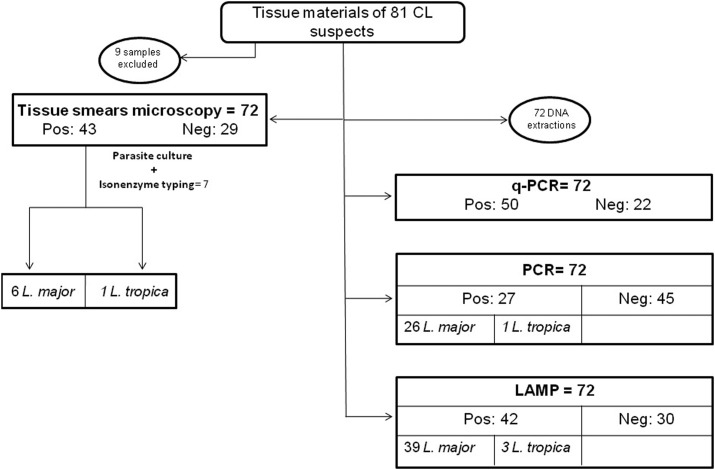
A total of 81 suspected CL patients were sampled for this study. Figure 2 shows the workflow applied for the tested samples. Seventy-two of the 81 samples were tested using either microscopy or molecular tools, whereas nine samples were excluded because of the absence of slides (four samples) or poor DNA quality (five samples).
Figure 2.
Diagram describing the workflow of samples from cutaneous leishmaniasis suspects in the study.
Microscopy and culture.
Leishmania amastigotes were detected by microscopy in 43 cases (59.7%) (29 slides were considered as negative). Among the microscopy-positive samples, seven showed positive cultures and were identified by isoenzyme typing as L. major (six isolates) and L. tropica (one isolate).
Molecular tools.
Fifty of the 72 (69.4%) samples were tested positive using the qPCR assays targeting the kinetoplast Leishmania genome, and were thus considered as confirmed CL cases. Among those, 42 were also found to be positive using the LAMP assay, but only 27 were found to be positive by cpb PCR. Thus, 15 samples were positive by LAMP and negative by cpb PCR, and eight samples were positive by qPCR and negative using LAMP. This low sensitivity of PCR can be explained by the large size of the amplified fragment.
Among the 27 samples (37.5%) found to be positive by species-specific applied cpb PCRs, 26 corresponded to L. major, one amplification corresponded to L. tropica, and none corresponded to L. infantum PCR. No mixed amplifications were observed using species-specific PCR.
The species-specific LAMPs showed 42 positive reactions (58.3%), with 39 L. major and three L. tropica. Isoenzyme typing results were concordant with those of LAMP and cpb PCR identifications, except for one sample which was identified as L. major by isoenzyme typing but amplified with an L. tropica LAMP.
Statistics.
Performance of the techniques was assessed using a statistical 2 × 2 contingency table, assuming qPCR as the gold standard. Microscopy, LAMP, and cpb PCR showed sensitivities of 82%, 84%, and 54%, respectively, and specificities of 90.9%, 100%, and 100%, respectively (Table 2). The lowest negative likelihood ratio (LR−) was achieved by the LAMP with 0.16 (a LR− below 0.1 virtually rules out the chance that a patient has the disease) (Table 2). Moreover, we compared the results of the 2 × 2 contingency tables with those of the LCA. Loop-mediated isothermal amplification sensitivity and specificity estimated from the 2 × 2 contingency tables were broadly corroborated by the LCA (LAMP: Se 100%–Sp 100%) (Table 3).
Table 2.
Performance of the different assays for the diagnosis of cutaneous leishmaniasis by classical validation (2 × 2) (95% CI) (qPCR as gold standard)
| Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | LR+ | LR− | Kappa (κ) | |
|---|---|---|---|---|---|---|---|
| Microscopy | 82 (70–90) | 90.9 (70–98) | 95.3 (89–100) | 69 (52.1–85.8) | 9 (2.3–43) | 0.19 (0.1–0.3) | 0.66 |
| LAMP | 84 (71.1–91.8) | 100 (82–100) | 100 (100–100) | 73.3 (57.5–89.2) | – | 0.16 (0.08–0.30) | 0.76 |
| PCR | 54 (40.4–67) | 100 (82.1–100) | 100 (100–100) | 48.9 (34.3–63.5) | – | 0.46 (0.3–0.6) | 0.41 |
LAMP = loop-mediated isothermal amplification; LR = likelihood ratio; NPV = negative predictive value; PPV = positive predictive value.
Table 3.
Performance of the different assays for the diagnosis of CL by classical validation (2 × 2) and through LCA (95% CI)
| Sensitivity (%) | Specificity (%) | |||
|---|---|---|---|---|
| 2 × 2 analysis | LCA | 2 × 2 analysis | LCA | |
| qPCR | 100* | 100 (95.2–99.9) | 100* | 73.3 (65.3–81.3) |
| Microscopy | 82 (70–90) | 88.1 (83.1–93.1) | 90.9 (70–98) | 80 (68.9–84.4) |
| LAMP | 84 (71.1–91.8) | 100 (98–100) | 100 (82–100) | 100 (96–100) |
| PCR | 54 (40.4–67) | 64.3 (57.0–71.6) | 100 (82.1–100) | 100 (89–100) |
LAMP = loop-mediated isothermal amplification; LCA = latent class analysis; qPCR = quantitative PCR.
* By definition (qPCR reference test).
DISCUSSION
With an increased number of cases and geographical distribution, CL is becoming an important health concern in endemic regions, especially in Maghreb countries.37 Moreover, conventional diagnostic methods (microscopy and PCRs) require expertise and advanced facilities, and are time-consuming. Therefore, simplifying and improving CL diagnostic modalities is becoming critical, especially in low-resource areas that are most affected by the disease. A reliable, rapid, and cost-effective diagnostic tool is required for effective CL control and efficient case management, especially when more than one Leishmania species is present in one region. This is indeed the case in different countries, including Tunisia, where the spreading of the parasite leads to the coexistence in the same region, for example, of L. major and L. tropica in the southeast37; therefore, species characterization is crucial for case management and epidemiological purposes. Accordingly, LAMP assays targeting the cpb multi-copy gene were developed to complement the CL diagnosis process and reinforce epidemiological studies. This was tested on skin samples collected from 72 patients with suspected CL. The results were compared with those obtained by microscopic, qPCR (targeting kDNA), and conventional cpb PCR analyses. The developed LAMP systems exhibited great sensitivity by detecting very low Leishmania DNA concentrations (20 fg and 200 fg for L. major and L. tropica, respectively). The threshold variations were likely due to the difference in the cpb gene copy number present in the studied species’ genomes.38 Adams et al.39 stated that the parasite load of skin lesion swab samples taken from CL patients should be high enough to achieve direct gene amplification, whereas van der Meide et al.40 reported that a 2-mm-diameter skin biopsy sample from a CL patient contains 6,100–94,100 parasites.
When compared with qPCR, the LAMP assay exhibits high sensitivity (84%) and specificity (100%), with an interesting positive predictive value (PPV = 100) and acceptable negative predictive value (NPV = 73.3). The sensitivity and specificity of the LAMP were greatly improved after applying the LCA statistical test (Table 3). The positive likelihood ratio (LR+) confirms that the patients diagnosed as positive using the LAMP are potentially true positives. The negative likelihood ratio (LR−) is greater than 0.1 (0.16) and lower than 1, supporting the possibility of low false-negative results. Using Kappa, the LAMP exhibited a higher agreement degree (0.76) than microscopy (0.66) and conventional cpb PCR (0.41), although it is simpler and less expensive as reported in the literature.41 By comparing the positive results of all these techniques, we noted good concordance with a reduced number of false positives and false negatives.
Concerning species identification (n = 42), the majority of our strains corresponded to L. major (92.9%), confirming the large predominance of this species throughout the Tunisian territory. In fact, ZCL due to L. major is responsible for more than 90% of CL cases in Tunisia, with an annual incidence exceeding 5,000 cases during epidemic years, especially in the center and the south.37 Our specific LAMPs identified more strains than conventional cpb PCR for either L. major (39 versus 26) or L. tropica (3 versus 1). No L. infantum strains, which are endemic predominantly in the northern parts of the country, were detected.
The three L. tropica strains were observed in the historical focus of Tataouine, southeast Tunisia, where CCL is most prevalent.37
Only one discrepancy over seven identified strains was observed between the LAMP and isoenzyme typing: The DNA extracted from the concerned sample was amplified and identified as L. tropica with the LAMP, whereas isoenzyme electrophoresis pointed it as L. major. This discrepancy could be due to a possible contamination during the culture process needed for the isoenzyme typing.
Compared with other molecular techniques, the LAMP assay has many advantages. The amplification is fast and implementable with basic equipment and post-amplification handling.42,43 In fact, the average time taken for the LAMP assay was 50 minutes, regardless of DNA extraction that could be avoided, whereas the qPCR and conventional PCR take approximately 3 hours. Moreover, the LAMP reaction is unaffected by PCR inhibitors found in biological components.39,44,45 The use of agarose gel migration can also be avoided and the result assessed by staining with different stains such as malachite green,25 FDR,46 and SYBR green.47 Furthermore, the stability of the LAMP reagents facilitates the field deployment of the technique.48
LAMP tests have been developed and applied to different cases of leishmaniasis, including Leishmania infection screening in dogs and detection of human asymptomatic subjects.23,47,49 Loop-mediated isothermal amplification tests targeting the kDNA minicircles (104 copies) showed improved performance compared with microscopy for CL diagnosis in Sri Lanka and Colombia.50,51 Similar results were obtained by Verma et al. who used the LAMP test targeting the 18S rRNA gene (104.8 copies) for CL and post-kala-azar dermal leishmaniasis (PKDL) diagnosis in India.49,52 Despite targeting the cpb gene that is present in reduced number of copies compared with the kDNA and the 18S rRNA genes, our LAMP assay showed promising performance. Otherwise, our methodology permitted, for the first time, to our knowledge, the identification of Leishmania species using a noninvasive technique and a fairly representative sampling (Table 4).
Table 4.
Details and summary of the diagnostic performance of different studies using LAMP for CL diagnosis compared with our study
| Kothalawala et al.50 | Adams et al.51 | Verma et al.49 | Verma et al.52 | This study: 3 different species-specific LAMP | |
|---|---|---|---|---|---|
| Country | Suriname | Colombia | India | India | Tunisia |
| LAMP target | 18S rRNA | 18S rRNA | kDNA | kDNA | cpb |
| Primer specificity | L. donovani | Leishmania genus | L. donovani, L. major, L. tropica | L. donovani | L. major, L. tropica, L. infantum |
| Study population | 17 CL | 105 CL | 10 CL | 62 PKDL | 82 CL suspects |
| Clinical specimen tested | Lesion aspirate | Lesion swab | Skin biopsy | Skin biopsy | Lesion aspirate |
| DNA purification method | Boom method | Qiagen DNAeasy blood and tissue kit | QIAamp Mini Kit (QIAGEN, Hilden, Germany) | QIAamp Mini Kit (QIAGEN) | Wizard® Genomic DNA Purification kit (Promega) |
| Reference test | Microscopy | Microscopy and/or culture | rK39 RDT-positive qPCR | rK39 RDT-positive qPCR | qPCR |
| Sensitivity (%) | 82.6 | 95 | 80 | 97 | 84 |
| Specificity (%) | 100 | 86 | 100 | 100 | 100 |
CL = cutaneous leishmaniasis; cpb = cysteine protease B; L. = Leishmania; LAMP = loop-mediated isothermal amplification; qPCR = quantitative PCR.
CONCLUSION
Our developed LAMP assay targeting the multi-copy cpb gene clearly allows the discrimination between different Leishmania parasite species causing CL in north Africa. It is more convenient and less expensive than qPCR41 or PCR53 for the detection and identification of Leishmania species.
Our results demonstrate the high performance of this technique, and the relevance of its implementation, especially in endemic CL countries, such as Tunisia, where more than one species are transmitted. It could help achieve early diagnosis and prompt case management and adapt control programs.
Acknowledgments:
We would like to thank Lamia Guizani-Tabbane, Imen Ben Abda (Institut Pasteur de Tunis, Tunisia), and Lyndon Zass (Centre for Proteomic & Genomic Research, Cape Town, South Africa) for critical reading of the manuscript. We also thank Afif ben Salah for the procurement of biological sampling (Institut Pasteur de Tunis, Tunisia), and Razika Benikhlef and Zoubir Harrat from Institut Pasteur of Algeria for isoenzyme typing of Leishmania isolates. We would like to acknowledge the H3Africa Bioinformatics Network (H3ABioNet) and the Ministry of High Education and Scientific Research (Tunisia) in the frame of the research laboratory LR 11-IPT-06. The American Society of Tropical Medicine and Hygiene (ASTMH) assisted with publication expenses.
REFERENCES
- 1.Alvar J, Velez ID, Bern C, Herrero M, Desjeux P, Cano J, Jannin J, den Boer M; WHO Leishmaniasis Control Team , 2012. Leishmaniasis worldwide and global estimates of its incidence. PLoS One 7: e35671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ben-Ismail R, Khaled S, Makni S, Ben Rachid MS, 1989. Anti-leishmanial antibodies during natural infection of Psammomys obesus and Meriones shawi (Rodentia, Gerbillinae) by Leishmania major. Ann Soc Belg Med Trop 69: 35–40. [PubMed] [Google Scholar]
- 3.Aoun K, Bouratbine A, 2014. Cutaneous leishmaniasis in north Africa: a review. Parasite 21: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haouas N, et al. 2012. Geographical distribution updating of Tunisian leishmaniasis foci: about the isoenzymatic analysis of 694 strains. Acta Tropica 124: 221–228. [DOI] [PubMed] [Google Scholar]
- 5.Tabbabi A, Ghrab J, Aoun K, Ready PD, Bouratbine A, 2011. Habitats of the sandfly vectors of Leishmania tropica and L. major in a mixed focus of cutaneous leishmaniasis in southeast Tunisia. Acta Tropica 119: 131–137. [DOI] [PubMed] [Google Scholar]
- 6.Schallig HD, Oskam L, 2002. Molecular biological applications in the diagnosis and control of leishmaniasis and parasite identification. Trop Med Int Health 7: 641–651. [DOI] [PubMed] [Google Scholar]
- 7.Chargui N, et al. 2005. Usefulness of PCR in the diagnosis of cutaneous leishmaniasis in Tunisia. Trans R Soc Trop Med Hyg 99: 762–768. [DOI] [PubMed] [Google Scholar]
- 8.Chargui N, et al. 2009. Population structure of Tunisian Leishmania infantum and evidence for the existence of hybrids and gene flow between genetically different populations. Int J Parasitol 39: 801–811. [DOI] [PubMed] [Google Scholar]
- 9.Aoun K, et al. 2015. Investigation and analysis of an outbreak of cutaneous leishmaniasis in Ksar Ouled Dabbab, Tataouine (Tunisia), 2012–2013. Med Sante Trop 25: 300–305. [DOI] [PubMed] [Google Scholar]
- 10.Remadi L, Haouas N, Chaara D, Slama D, Chargui N, Dabghi R, Jbeniani H, Mezhoud H, Babba H, 2016. Clinical presentation of cutaneous leishmaniasis caused by Leishmania major. Dermatology 232: 752–759. [DOI] [PubMed] [Google Scholar]
- 11.da Silva ES, van der Meide WF, Schoone GJ, Gontijo CM, Schallig HD, Brazil RP, 2006. Diagnosis of canine leishmaniasis in the endemic area of Belo Horizonte, Minas Gerais, Brazil by parasite, antibody and DNA detection assays. Vet Res Commun 30: 637–643. [DOI] [PubMed] [Google Scholar]
- 12.Reithinger R, Dujardin JC, 2007. Molecular diagnosis of leishmaniasis: current status and future applications. J Clin Microbiol 45: 21–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Lima AC, Zampieri RA, Tomokane TY, Laurenti MD, Silveira FT, Corbett CE, Floeter-Winter LM, Gomes CM, 2011. Leishmania sp. identification by PCR associated with sequencing of target SSU rDNA in paraffin-embedded skin samples stored for more than 30 years. Parasitol Res 108: 1525–1531. [DOI] [PubMed] [Google Scholar]
- 14.Costa MM, Penido M, dos Santos MS, Doro D, de Freitas E, Michalick MS, Grimaldi G, Gazzinelli RT, Fernandes AP, 2012. Improved canine and human visceral leishmaniasis immunodiagnosis using combinations of synthetic peptides in enzyme-linked immunosorbent assay. PLoS Negl Trop Dis 6: e1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodriguez-Cortes A, Ojeda A, Francino O, Lopez-Fuertes L, Timon M, Alberola J, 2010. Leishmania infection: laboratory diagnosing in the absence of a “gold standard”. Am J Trop Med Hyg 82: 251–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reithinger R, Davies CR, 2002. American cutaneous leishmaniasis in domestic dogs: an example of the use of the polymerase chain reaction for mass screening in epidemiological studies. Trans R Soc Trop Med Hyg 96 (Suppl 1): S123–S126. [DOI] [PubMed] [Google Scholar]
- 17.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T, 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28: E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poon LL, et al. 2006. Sensitive and inexpensive molecular test for falciparum malaria: detecting Plasmodium falciparum DNA directly from heat-treated blood by loop-mediated isothermal amplification. Clin Chem 52: 303–306. [DOI] [PubMed] [Google Scholar]
- 19.Han ET, Watanabe R, Sattabongkot J, Khuntirat B, Sirichaisinthop J, Iriko H, Jin L, Takeo S, Tsuboi T, 2007. Detection of four Plasmodium species by genus- and species-specific loop-mediated isothermal amplification for clinical diagnosis. J Clin Microbiol 45: 2521–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Njiru ZK, Mikosza AS, Armstrong T, Enyaru JC, Ndung’u JM, Thompson AR, 2008. Loop-mediated isothermal amplification (LAMP) method for rapid detection of Trypanosoma brucei rhodesiense. PLoS Negl Trop Dis 2: e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Njiru ZK, Mikosza AS, Matovu E, Enyaru JC, Ouma JO, Kibona SN, Thompson RC, Ndung’u JM, 2008. African trypanosomiasis: sensitive and rapid detection of the sub-genus Trypanozoon by loop-mediated isothermal amplification (LAMP) of parasite DNA. Int J Parasitol 38: 589–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Besuschio SA, et al. 2017. Analytical sensitivity and specificity of a loop-mediated isothermal amplification (LAMP) kit prototype for detection of Trypanosoma cruzi DNA in human blood samples. PLoS Negl Trop Dis 11: e0005779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaouch M, Mhadhbi M, Adams ER, Schoone GJ, Limam S, Gharbi Z, Darghouth MA, Guizani I, BenAbderrazak S, 2013. Development and evaluation of a loop-mediated isothermal amplification assay for rapid detection of Leishmania infantum in canine leishmaniasis based on cysteine protease B genes. Vet Parasitol 198: 78–84. [DOI] [PubMed] [Google Scholar]
- 24.Sriworarat C, Phumee A, Mungthin M, Leelayoova S, Siriyasatien P, 2015. Development of loop-mediated isothermal amplification (LAMP) for simple detection of Leishmania infection. Parasit Vectors 8: 591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nzelu CO, et al. 2016. A rapid molecular diagnosis of cutaneous leishmaniasis by colorimetric malachite green-loop-mediated isothermal amplification (LAMP) combined with an FTA card as a direct sampling tool. Acta Tropica 153: 116–119. [DOI] [PubMed] [Google Scholar]
- 26.Rogers MB, et al. 2011. Chromosome and gene copy number variation allow major structural change between species and strains of Leishmania. Genome Res 21: 2129–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuru T, Janusz N, Gadisa E, Gedamu L, Aseffa A, 2011. Leishmania aethiopica: development of specific and sensitive PCR diagnostic test. Exp Parasitol 128: 391–395. [DOI] [PubMed] [Google Scholar]
- 28.Rafati S, Salmanian AH, Hashemi K, Schaff C, Belli S, Fasel N, 2001. Identification of Leishmania major cysteine proteinases as targets of the immune response in humans. Mol Biochem Parasitol 113: 35–43. [DOI] [PubMed] [Google Scholar]
- 29.Mundodi V, Somanna A, Farrell PJ, Gedamu L, 2002. Genomic organization and functional expression of differentially regulated cysteine protease genes of Leishmania donovani complex. Gene 282: 257–265. [DOI] [PubMed] [Google Scholar]
- 30.Mottram JC, Frame MJ, Brooks DR, Tetley L, Hutchison JE, Souza AE, Coombs GH, 1997. The multiple cpb cysteine proteinase genes of Leishmania mexicana encode isoenzymes that differ in their stage regulation and substrate preferences. J Biol Chem 272: 14285–14293. [DOI] [PubMed] [Google Scholar]
- 31.Nicolle C, 1908. Isolement et entretien des corps de Leishman. Arch Inst Past Tunis 2: 55–56. [Google Scholar]
- 32.Berrebi J, 1936. La culture des leishmanies. Arch Inst Past Tunis 25: 89–141. [Google Scholar]
- 33.Chaouch M, Fathallah-Mili A, Driss M, Lahmadi R, Ayari C, Guizani I, Ben Said M, Benabderrazak S, 2013. Identification of Tunisian Leishmania spp. by PCR amplification of cysteine proteinase B (cpb) genes and phylogenetic analysis. Acta Tropica 125: 357–365. [DOI] [PubMed] [Google Scholar]
- 34.Mary C, Faraut F, Lascombe L, Dumon H, 2004. Quantification of Leishmania infantum DNA by a real-time PCR assay with high sensitivity. J Clin Microbiol 42: 5249–5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chouihi E, Amri F, Bouslimi N, Siala E, Selmi K, Zallagua N, Ben Abdallah R, Bouratbine A, Aoun K, 2009. Les cultures sur milieu NNN dans le diagnostic biologique des leishmanioses [in French]. Pathol Biol 57: 219–224. [DOI] [PubMed] [Google Scholar]
- 36.Garrett ES, Zeger SL, 2000. Latent class model diagnosis. Biometrics 56: 1055–1067. [DOI] [PubMed] [Google Scholar]
- 37.Bousslimi N, Aoun K, Ben-Abda I, Ben-Alaya-Bouafif N, Raouane M, Bouratbine A, 2010. Epidemiologic and clinical features of cutaneous leishmaniasis in southeastern Tunisia. Am J Trop Med Hyg 83: 1034–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hide M, Banuls AL, 2008. Polymorphisms of cpb multicopy genes in the Leishmania (Leishmania) donovani complex. Trans R Soc Trop Med Hyg 102: 105–106. [DOI] [PubMed] [Google Scholar]
- 39.Adams ER, Schoone GJ, Ageed AF, Safi SE, Schallig HD, 2010. Development of a reverse transcriptase loop-mediated isothermal amplification (LAMP) assay for the sensitive detection of Leishmania parasites in clinical samples. Am J Trop Med Hyg 82: 591–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van der Meide WF, Schoone GJ, Faber WR, Zeegelaar JE, de Vries HJ, Ozbel Y, Lai AFRF, Coelho LI, Kassi M, Schallig HD, 2005. Quantitative nucleic acid sequence-based assay as a new molecular tool for detection and quantification of Leishmania parasites in skin biopsy samples. J Clin Microbiol 43: 5560–5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mohammadiha A, Mohebali M, Haghighi A, Mandian R, Abadi AR, Zarei Z, Yeganeh F, Kazemi B, Taghipour N, Akhoundi B, 2013. Comparison of real-time PCR and conventional PCR with two DNA targets for detection of Leishmania (Leishmania) infantum infection in human and dog blood samples. Exp Parasitol 133: 89–94. [DOI] [PubMed] [Google Scholar]
- 42.Ndao M, 2009. Diagnosis of parasitic diseases: old and new approaches. Interdiscip Perspect Infect Dis 2009: 278246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amirabadi AR, Shahhosseiny MH, Yousefi JV, Ghahri M, 2012. LOOP mediated isothermal AMPlification (LAMP) in diagnosis of neorocryptococcosis. Afr J Biotechnol 11: 3986–3992. [Google Scholar]
- 44.Al-Soud WA, Radstrom P, 2001. Purification and characterization of PCR-inhibitory components in blood cells. J Clin Microbiol 39: 485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuboki N, Inoue N, Sakurai T, Di Cello F, Grab DJ, Suzuki H, Sugimoto C, Igarashi I, 2003. Loop-mediated isothermal amplification for detection of African trypanosomes. J Clin Microbiol 41: 5517–5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mikita K, Maeda T, Yoshikawa S, Ono T, Miyahira Y, Kawana A, 2014. The Direct Boil-LAMP method: a simple and rapid diagnostic method for cutaneous leishmaniasis. Parasitol Int 63: 785–789. [DOI] [PubMed] [Google Scholar]
- 47.Abbasi I, Kirstein OD, Hailu A, Warburg A, 2016. Optimization of loop-mediated isothermal amplification (LAMP) assays for the detection of Leishmania DNA in human blood samples. Acta Tropica 162: 20–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thekisoe OM, Bazie RS, Coronel-Servian AM, Sugimoto C, Kawazu S, Inoue N, 2009. Stability of loop-mediated isothermal amplification (LAMP) reagents and its amplification efficiency on crude trypanosome DNA templates. J Vet Med Sci 71: 471–475. [DOI] [PubMed] [Google Scholar]
- 49.Verma S, Singh R, Sharma V, Bumb RA, Negi NS, Ramesh V, Salotra P, 2017. Development of a rapid loop-mediated isothermal amplification assay for diagnosis and assessment of cure of Leishmania infection. BMC Infect Dis 17: 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kothalawala HS, Karunaweera ND, 2016. Loop-mediated isothermal amplification assay as a sensitive diagnostic tool for Leishmania donovani infections in Sri Lanka. Ceylon Med J 61: 68–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Adams ER, et al. 2018. Development and evaluation of a novel LAMP assay for the diagnosis of cutaneous and visceral leishmaniasis. J Clin Microbiol 56: e00386-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Verma S, Avishek K, Sharma V, Negi NS, Ramesh V, Salotra P, 2013. Application of loop-mediated isothermal amplification assay for the sensitive and rapid diagnosis of visceral leishmaniasis and post-kala-azar dermal leishmaniasis. Diagn Microbiol Infect Dis 75: 390–395. [DOI] [PubMed] [Google Scholar]
- 53.Takagi H, Itoh M, Islam MZ, Razzaque A, Ekram AR, Hashighuchi Y, Noiri E, Kimura E, 2009. Sensitive, specific, and rapid detection of Leishmania donovani DNA by loop-mediated isothermal amplification. Am J Trop Med Hyg 81: 578–582. [DOI] [PubMed] [Google Scholar]